Abstract
A range of environmental conditions can lead to oxidative stress; thus, a prompt and effective response to oxidative stress is crucial for the survival of plants. Microarray and northern-blot analyses were performed toward the identification of the factors and signaling pathways that enable plants to limit oxidative damage caused by exposure to high light (HL). Arabidopsis plants grown under moderate light (100 μmol m−2 s−1) were exposed to HL (1,000 μmol m−2 s−1) for 1 h. The microarray analyses revealed that exposure of Arabidopsis to HL caused an increase in known antioxidant genes, as well as several unknown genes. Some of these unknown genes had homologies to possible regulatory genes and metabolic enzymes. Furthermore, it was found that a range of chaperones were up-regulated in the HL treatment and that this induction was specifically due to the HL stress. The temporal expression under HL and different oxidative stress conditions of a subset of HL-responsive genes was confirmed via northern-blot analysis. Results from the arrays were also compared with publicly available microarray data sets from a range of different stress conditions at the Arabidopsis Functional Genomics Consortium. This cross comparison enabled the identification of genes that may be induced by changes in redox poise. Finally, to determine if the genes that were differentially expressed by HL stress were under similar transcriptional control, we analyzed the promoter sequences for the presence of common motifs.
Although light is essential for photosynthesis and, thus, crucial for the survival of plants, it can also cause oxidative stress. Exposure of a plant to light exceeding what is utilized in photochemistry leads to inactivation of photosynthetic functions and the production of reactive oxygen species (ROS) such as hydrogen peroxide (H2O2), superoxide (O2−), hydroxyl radicals, and singlet oxygen (1O2; Niyogi, 1999). The ROS produced by exposure to excessive light originates from three sites in the photosynthetic apparatus, the light-harvesting complex associated with PSII, the PSII reaction center, and the PSI acceptor site (Niyogi, 1999). The effects of these ROS can be the oxidation of lipids, proteins, and enzymes necessary for the proper functioning of the chloroplast and the cell as a whole (Foyer et al., 1994). Besides excess light, a range of abiotic environmental conditions such as O3, salt, toxic metals, and temperature can induce increased production of ROS by limiting the ability of a plant to utilize light energy through photosynthesis (Conklin and Last, 1995; Richards et al., 1998; Shinozaki and Yamaguchi-Shinozaki, 2000).
Under non-stressed conditions, plants have evolved several mechanisms to provide protection against the adverse effect of ROS formed during cellular metabolism (Asada, 1999). This protection consists of an antioxidant defense system that provides adequate protection against ROS produced during normal cellular metabolic activity and photosynthesis. This defense mechanism consists of enzymes such as superoxide dismutase (SOD), ascorbate peroxidase (APX), and glutathione-S-transferase (GST) that can dismutate O2− radicals and scavenge H2O2 (for review, see Niyogi, 1999). Because H2O2 is a strong oxidant that rapidly targets thiol groups, its formation by exposure to excessive light needs to be counteracted by the plant for photosynthesis to function because this is dependent on thiol-regulated enzymes (Noctor and Foyer, 1998). In addition, antioxidants such as carotenoids and tocopherols play a role in the dissipation of excess light, preventing lipid oxidation and in the scavenging of ROS (Niyogi, 1999). However, should the plant become affected by oxidative stress due to a disparity in its capacity to generate sufficient antioxidant potential, this will result in reduced productivity and ultimately death.
In an attempt to decrease the production of ROS caused by exposure to excess light, the plant can adjust its light-harvesting antennae size and thermally dissipate excess absorbed light by a process called non-photochemical quenching (Gilmore et al., 1994). Furthermore, an increase in the xanthophyll zeaxanthin, by the de-epoxidation of violaxanthin by violaxanthin de-epoxidase (VDE), is thought to be involved in the thermal dissipation of excess light energy and the protection of photosynthetic membranes against lipid peroxidation. This process is known as the xanthophyll cycle (Yamamoto et al., 1962).
Additional high-light (HL)-specific responses consist of the expression of the early light-induced proteins (ELIPs), which may bind chlorophyll a and lutein (Adamska, 1997). More recently, heat shock proteins (HSPs) have been shown to be induced in Arabidopsis plants treated with H2O2 and in cyanobacteria treated with HL, implicating them in a protective role against HL and its effects (Desikan et al., 2001; Hihara et al., 2001). This becomes all the more likely as an increasing number of studies show the existence of cross tolerance in plants. When a plant is exposed to moderate stress conditions, this often induces resistance to other stresses (Sabehat et al., 1998).
Because diverse abiotic stresses can result in oxidative damage and induce multiple antioxidant mechanisms, it is likely that several stress-sensing pathways converge. The mechanisms and pathways that regulate and coordinate the plant response are complex and poorly understood. However, it is becoming clear that a redox-controlled mechanism might be involved. It has been shown that a light-driven change in the redox potential of plastoquinone (PQ) regulates the expression of two cytosolic peroxidases during HL stress (Karpinski et al., 1999). Furthermore, the redox state of PQ has been shown to be involved in the expression of chloroplast-encoded genes (Pfannschmidt et al., 1999).
Investigation of plant gene expression has been greatly facilitated by microarray analysis (Richmond and Somerville, 2000). Although in the past it was only possible to look at a few genes at the time, it is now possible to measure a large number of gene expression patterns simultaneously and understand global changes in gene expression under a given condition. Here, we report the changes in mRNA level of Arabidopsis genes after exposure to HL conditions. We have identified an increase in expression of known antioxidant genes such as APX1 and dehydroascorbate reductase (DHAR), as well as unknown genes with homologies to regulatory genes and metabolic enzymes. Furthermore, it was found that several HSP genes were up-regulated, implicating them in the antioxidant response in addition to their chaperone function. To further investigate the regulatory circuit(s) of the HL-responsive genes, we carried out promoter analyses by searching for overrepresented motifs found in promoter sequences of these genes using MotifSampler (Thijs et al., 2001).
RESULTS
Light Stress
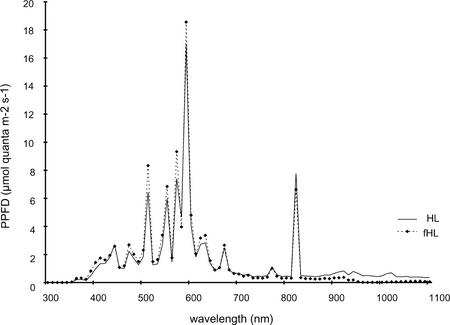
One hour of HL stress is sufficient to impair photosynthetic capacity and induce different and complementary photoprotective mechanisms (Russell et al., 1995). An increase in visible light (photosynthetically active radiation [PAR]) is commonly associated with an increase in the infrared spectrum. To delineate effects specifically due to PAR and those induced by shifts in temperature, we undertook parallel experiments in which infrared was filtered out. Spectral measurements showed that the filter removed 50% of the infrared spectrum compared with unfiltered HL, but had no effect on the low level of UV or the visible spectrum (Fig. 1). The 1-h treatment regimes were: moderate light (ML), 100 μmol m−2 s−1 at 22°C; HL, 1,000 μmol m−2 s−1 at 29°C; fHL, 1,000 μmol m−2 s−1 at 24°C; and warm ML (wML), 100 μmol m−2 s−1 at 27.5°C. Unless otherwise stated, all temperatures are air temperatures. Plants transferred to the HL, fHL, and wML treatments had similar handling and notably no touch-induced genes were observed to be up-regulated. Each of the HL conditions was compared with the ML conditions. One hour of HL and fHL induced the photoprotective xanthophyll cycle, resulting in decreased violaxanthin and a concomitant increase in zeaxanthin and antheraxanthin. Expressed as a ratio, the de-epoxidation state of the xanthophyll cycle pigments increased from 0.01 ± 0.03 at ML to 0.57 ± 0.1 for HL and 0.56 ± 0.13 for fHL. There was a marginal increase in the de-epoxidation state in one of the four plants analyzed, resulting in a ratio of 0.04 ± 0.08 at wML.
Figure 1.
Spectral measurements of HL and infrared-filtered HL (fHL). Spectra for HL (—) and fHL (-♦-) at 1,000 μmol m−2 s−1 of light were measured with a portable spectroradiometer (see “Materials and Methods”).
Expression of Genes by HL
We applied a high degree of stringency to all aspects of the data collection and statistical analysis. First, RNA was extracted and pooled from over 50 plants for each of the biological replicates to ensure consistency of the biological material. Second, the microarrays were undertaken three times for each treatment, including a biological replicate and a technical replicate, which involved a fluorescent dye swap. Third, gene expression data were normalized using Perl scripts with the overall background for each experiment being calculated using a set of 188 control spots on each slide (Schenk et al., 2000). For a gene to be considered as induced or repressed, the gene expression ratio had to exceed a 2 times change for each of the replicates. In addition, its signal intensity had to be 2 times higher than the average background, plus 2 times sds for each of the replicates, as faint hybridization signals are more variable. The average value for genes that met these criteria are presented in Table I and the supplemental data. Finally, a number of genes were represented by more than one spot and the values for these are presented as averages of all spots.
Table I.
Expression data of a subset of genes under the different HL regimes
| Function | Locus | HL Ratio | fHL Ratio |
|---|---|---|---|
| Stress response | |||
| APX1 | At1g07890 | 2.7 | 2.9 |
| APX2 | At3g09640 | 3.1 | 1.8 |
| GST1 | At1g02930 | 1.5 | 2.4 |
| GST6 | At2g47730 | 3.6 | 2.6 |
| Monodehydroascorbate reductase | At3g09940 | 1.0 | 3.3 |
| DHAR | At1g19570 | 2.4 | 3.7 |
| ELIP | At3g22840 | 5.0 | 2.9 |
| Photosynthesis and photoprotection | |||
| VDE precursor | At1g08550 | −2.5 | −1.3 |
| 4-Hydroxyphenylpyruvate dioxygenase (HPPD) | At1g06570 | 2.3 | 1.7 |
| β-Carotene hydroxylase II (BCH II) | At5g52570 | 6.3 | 2.0 |
| Heat shock/chaperone protein | |||
| HSP101 | At1g74310 | 11.1 | 5.7 |
| HSP70-3 | At3g09440 | 5.6 | 4.2 |
| Putative small heat shock protein (sHSP) | At2g29500 | 10.4 | 2.5 |
| BiP luminal-binding protein | At5g42020 | 3.2 | 2.6 |
| HSP90 | At5g56010 | 7.9 | 6.6 |
| HSP81-2 | At5g56030 | 4.8 | 5.5 |
| HSP81-4 | At5g56000 | 5.4 | 5.8 |
| DNAJ protein homolog ATJ | At5g22060 | 2.5 | 2.5 |
| Flowering | |||
| CCA1 | At2g46830 | 5.1 | 1.2 |
| flowering locus F | At5g65070 | 4.6 | 1.4 |
| BEL1 | At5g41410 | −2.5 | −1.3 |
| FHA/CRY2 | At1g04400 | −2.5 | −1.7 |
| Defense | |||
| PAL1 | At2g37040 | 2.4 | 1.4 |
| Chalcone synthase (CHS) | At5g13930 | 8.6 | 1.5 |
| lipoxygenase AtLOX2 | At3g45140 | 1.0 | 2.2 |
| Signaling | |||
| HY5 | At5g11260 | 2.2 | 2.4 |
| Lignin | |||
| Cinnamyl alcohol dehydrogenase | At4g34230 | 1.5 | 3.6 |
| Peroxidase ATP23a | At1g68850 | 3.6 | 2.9 |
| Other | |||
| Blue copper protein | At5g20230 | 1.1 | 3.4 |
| Isocitrate lyase | At3g21720 | 1.5 | 4.3 |
| Beta-amylase | At4g15210 | 6.3 | 5.5 |
| Vegetative storage protein (Vsp2) | At5g24770 | 2.8 | 3.0 |
| Lysosomal pro-X carboxypeptidase | At5g65760 | 5.8 | 2.0 |
| Unknown and hypothetical | |||
| Hypothetical protein | At3g17800 | 3.0 | 3.0 |
| Unknown | At5g61820 | 2.6 | 2.6 |
| Unknown | At1g19180 | −1.3 | 3.8 |
Example of expression of genes induced or repressed by Arabidopsis after exposure to HL and fHL treatments for 1 h (see Fig. 1). Data are an average of three experiments and genes represented by multiple spots were averaged. Ratios < 2× are considered unchanged. The complete list is provided in the supplemental data.
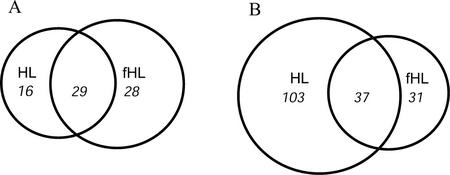
Of the >6,000 genes analyzed in the HL (1,000 μmol m−2 s−1 + infrared) experiment, a total of 185 genes were differentially expressed (defined as ≥2-fold expression) compared with the control ML. Of these, 45 genes were induced and 140 were repressed (Fig. 2). A significant number of induced genes represented chaperones and HSPs (20%; Table I). Other genes induced under HL alone belonged to several different functional classes (Table I). Among these, CHS, which is also induced by UV (Jenkins, 1997) and involved in the anthocyanin pathway, and PAL1, a gene involved in wounding and cold response (Abarca et al., 2001), were up-regulated. Furthermore, the β-carotene hydroxylase II (BCH II) gene, involved in the xanthophyll biosynthesis (zeaxanthin), and the flowering genes CCA1 and a MADS box transcription factor like were induced. APX1, APX2, and DHAR were the only genes of the antioxidant ascorbate-glutathione cycle to be induced to a significant level under this light regime. Among the genes that were repressed, it was interesting to find VDE, an enzyme whose activity is increased by HL. Other repressed genes included the flowering genes BEL1 and FHA and the chlorophyll synthesis enzyme protochlorophyllide oxidoreductase.
Figure 2.
Diagrams depicting total numbers of overlapping and nonoverlapping induced (A) and repressed (B) genes after exposure to two HL conditions. HL, Arabidopsis plants exposed to HL for 1 h. fHL, Arabidopsis plants exposed to fHL for 1 h. Ratios are the average of three individual experiments and genes with equal or greater than 2-fold difference in their ratio were considered to be differentially expressed.
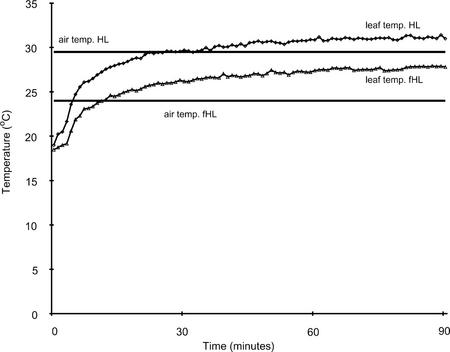
The induction of the heat shock genes was intriguing. As a consequence, we monitored the leaf temperature of plants transferred to wML, HL, and fHL (Fig. 3). Leaf temperatures for HL (30.5°C), fHL (27.5°C), and wML (26°C) were within a few degrees of the respective air temperatures. The rate of increase in temperature was similar for HL, fHL (Fig. 3), and wML after transfer from the ML chamber. It is well established that plant HSPs are not strongly induced until air temperatures reach 37°C and the minimal air temperature for a detectable increase is 34°C (Altschuler and Mascarenhas, 1985; Kimpel and Key, 1985; Wu et al., 1988; Conner et al., 1990). For Arabidopsis leaves, air temperatures ranging from 22°C to 31°C had no effect on levels of a range of HSPs; some started to increase at 34°C and the strongest increase was observed at 37°C (Wu et al., 1988; Conner et al., 1990). Neither air nor leaf temperatures reached this level in any of our experiments (Fig. 3).
Figure 3.
Arabidopsis leaf and growth cabinet air temperature after exposure to HL (♦) and fHL (▴) for 1 h. Leaf temperature of 24-d-old Arabidopsis plants was measured with copper-constantan thermocouples. Four replicates per treatment were averaged and plotted in Excel (Office 97, Microsoft, Redmond, WA). The air temperature was measured in the shade.
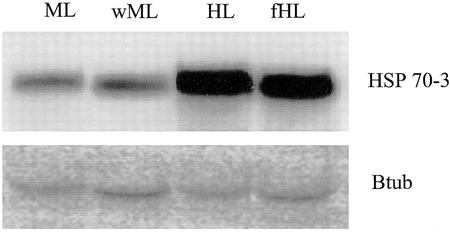
To determine whether the induction of these HSPs was due to HL exposure and not as a result of an increase in leaf surface temperature concomitant with HL, northern-blot analyses were carried out under HL, fHL, and different temperatures with ML. Tissue samples from both HL treatments and ML treatments were used for RNA isolation. The northern blot revealed an increase in expression levels of the HSP70-3 transcript under both HL conditions at 24°C and 29°C but not under ML at 22°C or 27.5°C (air temperatures; Fig. 4), indicating that HL and not the leaf temperature per se caused the increased expression of the HSPs.
Figure 4.
RNA gel-blot analyses of Arabidopsis HSP 70-3. Total RNA was isolated from 24-d-old leaves after exposure for 1 h to: 22°C and 100 μmol m−2 s−1 of light (ML), 27.5°C and 100 μmol m−2 s−1 of light (wML), 29°C and 1,000 μmol m−2 s−1 of light (HL), or 24°C and 1,000 μmol m−2 s−1 fHL. Twenty micrograms of total RNA was probed with HSP70-3 or β-tubulin (Btub).
Expression of Genes by Filtered HL
In the fHL (1,000 μmol m−2 s−1) experiment, 125 genes were differentially expressed compared with the ML-grown plants. Of these, 57 were induced and 68 were repressed (Fig. 2; Table I). Of the genes involved in the antioxidant-scavenging ascorbate-glutathione cycle APX1, monodehydroascorbate reductase and DHAR were found to be significantly induced. Moreover, genes encoding enzymes of cell wall synthesis (isocitrate lyase, peroxidase ATP23a, and cinnamyl alcohol dehydrogenase) were also induced, as well as a transcription factor, HY5, involved in light response signaling. Furthermore, under fHL, there was a marked increase in the induction of the vegetative storage protein (Vsp2), and lipoxygenase AtLOX, which have been found to be wound inducible (Bell and Mullet, 1993; Utsugi et al., 1998), and GST1, which is involved in detoxification of toxic compounds (Droog, 1997). Genes that were repressed under filtered HL included the light-harvesting complex genes (Lhcb2.1, Lhcb2.2, Lhcb2.4, and Lhcb4.2).
Expression of Genes by Both HL Conditions
Despite the minor temperature difference between HL and fHL, only 29 genes were induced by both HL treatments, with 16 induced solely by HL and 28 induced by fHL (Fig. 2). Reduced gene expression was observed for 103 genes by HL alone, 31 genes solely by fHL, and 37 genes were repressed under both conditions. Genes with increased transcript abundance under both regimes included GST6, ELIP, the luminal-binding protein BiP, and β-amylase (Table I). The majority of the genes repressed under both HL conditions encoded for either hypothetical proteins or for genes of unknown function. The gene for VDE was also inhibited under both HL conditions, but to a lesser degree under fHL.
Given that filtering of the heat by water and glass may alter the spectrum and, thus, affect the induction of phytochrome and UV-induced genes, we measured the spectrum of HL and fHL (Fig. 1). The spectra were very consistent for the UV and visible wavelengths, including red and far red. Differences were only observed in the photo flux density of wavelengths above 850 nm, which is in the infrared spectrum. Filtering of HL by glass and water reduced the percentage of infrared of the total visible spectrum by more than 50%.
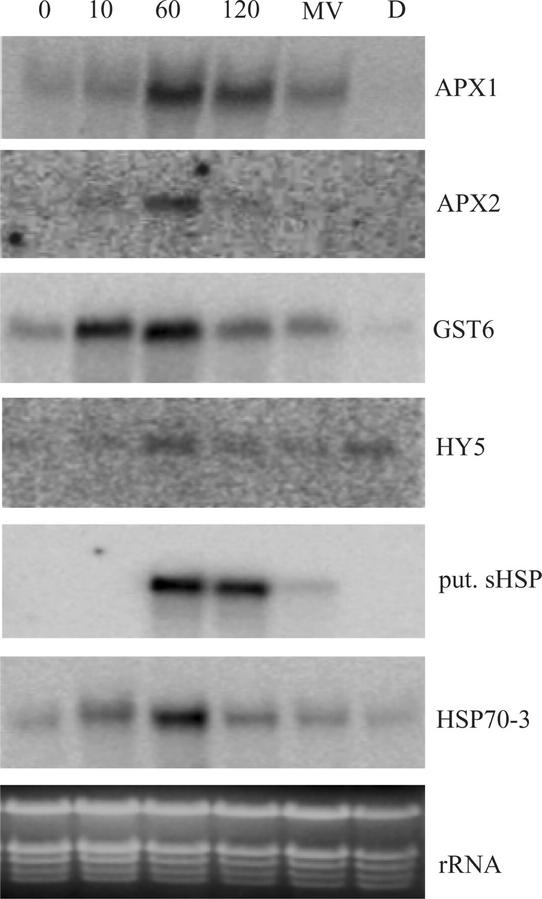
To gain a better understanding of how gene expression is controlled temporally by HL and to confirm array data, we focused on the expression of six representative genes derived from functional groups present in the HL-responsive genes (Table I). All genes probed by northern blots exhibited similar induction levels to those obtained from the microarray analysis after 1 h of HL (Fig. 5). APX1, GST6, and HSP70-3 followed a similar expression pattern, an increase within 10 min of light stress, and decreasing by 2 h. This decline after 2 h may suggest that a degree of acclimatization to HL exposure led to a feedback into APX and GST6 gene expression.
Figure 5.
RNA gel-blot time course analyses of selected HL-induced genes. Twenty micrograms of total RNA isolated from Arabidopsis leaf exposed to 0, 10, 60, and 120 min of HL (1,000 μmol photons m−2 s−1), methyl viologen (MV), or 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU; D) was probed with: APX1, APX2, GST6, long hypocotyl 5 (HY5), putative small HSP (put. sHSP), and HSP70-3. rRNA, Ethidium bromide-stained ribosomal RNA.
To ascertain whether the regulation of these six representative genes was in response to HL per se or in response to side effects of HL, namely a shift in redox poise or ROS levels, plants were treated with herbicides that alter the redox state of the PQ pool and ROS content. Then transcript levels were measured. To test the effect of ROS on gene expression, plants were treated with MV, a chemical that generates O2− and subsequently H2O2 due to the action of SOD. The elevated levels of H2O2 increased expression of APX1, GST6, HSP70-3, and a chloroplast-targeted putative sHSP, whereas the expression of APX2 and HY5 was unaffected (Fig. 5). Photosynthetic electron transfer under HL will reduce the PQ pool, whereas application of the herbicide DCMU will oxidize the PQ pool due to blocking electron transfer at the PSII acceptor site. All of the genes strongly induced by HL were repressed by DCMU in ML, except HY5, for which DCMU had an inducing effect. Thus, both the production of H2O2 and the redox state of the PQ contribute in varying degrees to the expression of different genes.
Expression Profiles by Different Light Regimes and Environmental Stresses
The microarray data and the northern-blot analyses provided a list of genes involved in the response to HL in Arabidopsis and suggested what stimuli they may respond to. However, to further our understanding of the nature of these genes and to determine whether they are specific to HL or are generic stress-related genes, we compared the expression of these genes induced by HL to their expression profiles in 13 different microarray experiments available at the Stanford Microarray Database (SMD; http://afgc.stanford.edu/; Table II).
Table II.
Expression profile of several HL-induced genes under different environmental conditions
| Experiment | Gene
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Putative sHSP | HSC70-G7 | APX1 | ELIP | GST1 | GST6 | CHS | HY5 | Blue Cu2+ protein | Hypothetical protein | Unknown | Unknown | |
| HL plants (1,000 μmol m−2 s−1) | 10.4 | 5.6 | 2.7 | 5.0 | – | 3.6 | 8.6 | 2.2 | – | 3.0 | 2.6 | – |
| IR-filtered HL plants (1,000 μmol m−2 s−1) | 2.5 | 4.2 | 2.9 | 2.9 | 2.4 | 2.6 | – | 2.4 | 3.4 | 3.0 | 2.6 | 3.8 |
| SMD experiment and identification no. | ||||||||||||
| Cells + H2O2 (9,371 and 7,525) | 11.7 | – | – | 3.6 | – | 2.4 | – | 2.5 | 3.4 | – | – | – |
| Antimycin A treatment (5,198 and 5,201) | 3.5 | – | – | – | – | – | −2 | – | 4.1 | – | – | – |
| Chlorotic (cch1) leaves (11,604 and 11,605) | – | – | 2.3 | – | – | 2.4 | 3.1 | 2.1 | – | – | 2.2 | – |
| Circadian rhythm (2,368 and 10,186) | – | – | – | – | – | – | −10 | – | 8.3 | – | – | – |
| Aluminum stress 1 (7,304 and 7,305) | – | – | – | – | – | – | – | – | 2.6 | – | – | – |
| Shade avoidance 3 (8,266 and 8,130) | – | – | – | −5 | – | – | −5 | −3 | – | – | – | – |
| Continuous light (4,188) | – | – | – | – | – | – | – | – | – | – | – | – |
| Pathogen response (9,754 and 9,753) | – | – | – | – | – | – | – | – | 2.0 | – | – | – |
| Iron deficiency (9,549 and 7,114) | – | – | – | – | – | – | 4.6 | – | – | – | – | – |
| Vernalization 2 (5,247 and 5,248) | – | – | – | – | – | – | 2.6 | – | – | – | – | – |
| ABA-insensitive mutant (11,757 and 11,895) | – | – | 3.5 | – | – | 2.1 | 9.1 | 8.6 | – | – | 7.2 | −2 |
| Leaves-etiolated seedlings (3,610) | – | −2.5 | −5 | – | – | – | – | −3 | – | – | – | −2.5 |
| Chilling tolerance (14,448 and 14,449) | −2.5 | 3.6 | 2.8 | – | – | 3.2 | – | – | – | – | 3.2 | – |
Expression ratio of HL-induced genes representing functional groups compared with their expression level under a range of different environmental or genetic experiments from SMD (http://genome_www4.stanford.edu/MicroArray/SMD/). The SMD identification no. is indicated in parentheses. Data are the average of two experiments where possible and values less than 2-fold are considered as unchanged (–).
The small chloroplast-targeted HSP induced by both HL conditions and MV was also strongly induced in H2O2-treated cells (SMD nos. 9,371 and 7,525). This induction was not seen for the cytosolic HSC70-G7, nor was it induced by antimycin A treatment, an inhibitor of the mitochondrial electron transport chain (identification nos. 5,198 and 5,201). CHS was highly induced by HL and was also induced in the abscisic acid (ABA)-insensitive mutant (SMD nos. 11,757 and 11,895) abi1-1, in iron-deficient-grown roots (SMD nos. 9,849 and 7,114) and in the chlorophyll mutant cch1 (gun5; identification nos. 11,604 and 11,605). In contrast, fHL resulted in unchanged levels of CHS and far-red light led (SMD nos. 8,266 and 8,130) to CHS repression, as was the case in the circadian rhythm experiment (SMD nos. 2,368 and 10,186).
The HL-inducible gene, ELIP, was moderately induced by H2O2 treatment and repressed by far-red light. GST1 was only slightly induced by fHL, whereas GST6 was induced by both HL conditions, H2O2, in cch1, in abi1-1, and in the chilling-sensitive cold-treated cls8 mutant (SMD nos. 14,448 and 14,449). These mutants also exhibited increased APX1 gene expression under those conditions. HY5 was induced in cch1 and abi1-1 mutants and repressed by far-red light. The blue copper protein found to be induced by fHL was also induced by several other conditions. Its highest expression was in the circadian rhythm experiment. Other conditions that elicited a transcriptional response of the blue copper protein were H2O2, antimycin A, aluminum (SMD nos. 7,304 and 7,305), and in fungal spore-inoculated leaves in the pathogen response experiment (SMD nos. 9,754 and 9,753).
Of the unknown genes, the gene encoding for the hypothetical protein At3g17800 was only induced to a significant level by both HL conditions. The unknown genes, At1g31460 and At1g19180, were induced by HL and fHL, respectively. Furthermore, an increased transcriptional response of At1g31460 was found in the cch1, abi1-1, and cls8 mutants, whereas abi1-1 and light-grown leaves versus etiolated leaves repressed the transcription of At1g19180.
Promoter Motif Analyses
We have carried out promoter motif analyses of all the genes differentially expressed by the HL treatments for which promoter sequences were available to elucidate known and novel light regulatory motifs for the known and unknown genes. Our data were subdivided into sets of coregulated genes, induced or repressed in HL, fHL, or in the overlap of both HL experiments (Fig. 2). The notion was that similarly expressed genes might be coordinately regulated. Also, the presence of known regulatory motifs may shed light on pathways or cellular functions of the HL-responsive genes. We analyzed 500 bp of sequence immediately upstream of the ATG of each differentially expressed gene for common 6- to 10-bp motifs within each set of coregulated promoters that were present in significantly higher numbers than would be expected by chance (Thijs et al., 2001).
Various motifs were found and all were analyzed with PlantCARE (Rombauts et al., 1999) to determine if these motifs had already been assigned a function. The motifs in common for all sets of coregulated induced genes under all conditions were different motifs involved in light responsiveness (Table III). The different LREs found might suggest that it is the combination of these elements that regulates the genes. Some observations were paradoxical, such as the ABREs being identified in genes repressed by HL and in other genes induced by fHL. This would indicate that factors besides ABA could regulate these genes. Heat stress response elements were found in the subset of promoter sequences of genes induced by both HL experiments. As expected, most of the promoters containing these motifs were from the HSP family. However, a HSE was also found in the promoter of APX1, one of the unknown genes, and in two putative genes. Finally, previously unidentified motifs were identified in both HL conditions.
Table III.
Promoter motifs from genes differentially regulated by both HL conditions
| Clusters and Motifs | Genes | Motif Name | Motif Function |
|---|---|---|---|
| HL-induced genes | |||
| CTrACCGTCC | 7 /15 (0.98) | A box/MBS | Cis-acting regulatory element, MYB-binding site |
| nAmGTTGGyr | 9 /15 (0.85) | CAAT box | Common cis-acting promoter element |
| CTATCT | 7 /15 (0.8) | Sbp-CMA1 | Module involved in light response |
| ATCCGACGTC | 3 /15 (0.99) | – | Unknown |
| AGTAACyA | 8 /15 (0.8) | – | Unknown |
| HL-repressed genes | |||
| AAGAAGAAGA | 23 /77 (0.8) | TCA element | Cis-acting element involved in SA responsiveness |
| CGTnTC | 18 /77 (0.8) | ABA response element (ABRE) | Cis-acting element involved in ABA responsiveness |
| CwTCTTCwTC | 27 /77 (0.8) | – | Unknown |
| CAAAGCAA | 21 /77 (0.8) | – | Unknown |
| fHL-induced genes | |||
| CTTCCn | 7 /28 (0.8) | G box | Cis-acting element involved in light responsiveness |
| nACGTGTmmC | 13 /28 (0.9) | ABRE | Cis-acting element involved in ABA responsiveness |
| CAmmCACmAm | 13 /28 (0.8) | – | Unknown |
| WCTyCmGATT | 13 /28 (0.8) | – | Unknown |
| fHL-repressed genes | |||
| ATAAATAAAA | 13 /28 (0.8) | TATA box | Core promoter element −30 of transcription start |
| GTCTCn | 10 /28 (0.8) | AuxRE | Part of auxin response element |
| CACTTwCC | 17 /28 (0.8) | – | Unknown |
| HL- plus fHL-induced genes | |||
| WTCTCCTTCs | 7 /14 (0.8) | TCCC motif | Part of light-responsive element (LRE) |
| GAAACT | 7 /14 (0.8) | Heat shock element (HSE) AE box | Heat stress response, part of LRE |
| CTTCTC | 9 /14 (0.8) | HSE | Heat stress response |
| TCGGATTCGT | 3 /14 (0.99) | – | Unknown |
| HL- plus fHL-repressed genes | |||
| TAAATAAAnm | 11 /34 (0.8) | 1 box | Part of LRE core promoter element |
| TCTTCTTC | 11 /34 (0.95) | sbpCMA1a | Part of LRE |
| ACCATA | 12 /34 (0.8) | ACA-motif | Part of LRE |
Promoter sequences (500 bp) for genes coregulated by two HL experiments were obtained from The Institute for Genome Research (ftp://www.tigr.org) and searched for overrepresented 6- to 10-motifs by a motif sampler (Thijs et al., 2001). Nos. represent the amount of promoters out of the total amount of promoters analyzed per cluster. Nos. in parentheses represent the probability score of that motif; the closer this score is to 1 the closer the motif resembles the motif model; >0.8 is considered significant. The letters other than A, C, G, and T are degenerate symbols. They represent a possible combination of two letters: A-T = W, A-C = M, A-G = R, C-G = S, C-T = Y, G-T = K, and n can be A, C, G, or T. The returned overrepresented motifs were compared to known plant motifs available from the PlantCARE database to determine function. HL, 1,000 μmol m−2 s−1; fHL, 1,000 μmol m−2 s−1 infrated filtered light. A list of genes containing each promoter motif is provided in the supplemental data.
DISCUSSION
Photoprotection, Antioxidants, and Chaperones
In this study, we have employed DNA microarray technology to analyze transcriptional responses of the Arabidopsis genome to HL. This technology has identified known and unknown genes that were differentially expressed after 1 h of HL exposure. Furthermore, the analyses have shed light on the regulation of some of the protective measures utilized against oxidative stress.
Measurements of leaf and air temperature in combination with HPLC analyses provided evidence that the plants were stressed more by HL treatments than by the associated increase in temperature. This could be concluded from the induction of the photoprotective xanthophyll cycle and that HSP70-3 was not induced by the wML treatment in the absence of light stress (Fig. 4).
Several genes involved in antioxidant biosynthesis were up-regulated by both HL conditions. The genes for BCH II and HPPD (Table I) exhibited an increase in transcript abundance. HPPD is an enzyme involved in the synthesis of PQ and tocopherol and PQ is an essential component of carotenoid biosynthesis by acting as an intermediate electron acceptor between carotenoid desaturases and the photosynthetic electron transport chain (Norris et al., 1995). Tocopherols themselves also have antioxidant functions because they physically quench and scavenge 1O2, O2−, and OH radicals (Niyogi, 1999).
BCH II is one of two genes encoding the BCH that catalyzes the synthesis of zeaxanthin and lutein (Sun et al., 1996). Changes in the expression of carotenoid biosynthetic genes gave examples of the utility of microarrays and a cautionary note on their interpretation with respect to transcriptional and posttranscriptional regulation of biological processes. With respect to their utility, the total size of the xanthophyll cycle pool size increased and although this has been thought to reflect an increase in “free” β-carotene after D1 turnover (Depka et al., 1998), that it coincides with an increase in BCH II mRNA suggests there may be regulation of the level of gene expression as well. On the cautionary side, we observed a decrease in transcript abundance for the xanthophyll cycle enzyme, VDE, as have others for its protein and activity in spinach (Spinacia oleracea) under HL (Eskling and Akerlund, 1998). However, the former may just reflect VDE's very low expression level, which can lead to more variability in microarray datasets (Wang et al., 2001). Regardless, it should be noted that induction of the cycle is a pH-catalyzed posttranslational activation of VDE (Gilmore, 2001), making shifts in transcription unnecessary.
The ELIPs are members of the LHC protein super family (Adamska, 1997) and their induction under both HL conditions is further confirmation of their induction during light stress (Lindahl et al., 1997). The ELIPs were also expressed in Arabidopsis cells treated with H2O2 (Table II). Lower expression level of the ELIP under fHL might be ascribed to a lower H2O2 concentration in these plants if the slightly higher temperature of HL has the same effect as a short burst of hot air on ROS production (Vallelian-Bindschedler et al., 1998). However, measurements to determine if this is the case remain to be done. The reduction in light-harvesting chlorophyll a/b-binding protein expression under HL is in agreement with many studies and reflects a strategy to reduce antenna size under sustained HL (Escoubas et al., 1995).
APX and its substrate ascorbate largely accomplish the removal of H2O2. Our results show that the two cytosolic forms, APX1 and APX2, were induced by both HL conditions, although APX2 transcript induction under fHL was marginal. When the APX1 expression was analyzed in other microarray experiments at SMD, it was found to exhibit increased expression in the chlorophyll mutant cch1, the cold-treated mutant cls8, and the ABA-insensitive mutant abi1-1. The light intensity of 230 μmol m−2 s−1 in the cch1 mutant experiment would be perceived as HL due to the chlorotic state of this mutant, explaining the increase in APX1 expression. Chilling has been found to increase expression of APX and this induction could be attributed to oxidative stress caused by reduced efficiency of PSI and a consequent increase of ROS (Terashima et al., 1998). abi1-1 is unable to close its stomata and, thus, is water stressed, and that will induce APX1 expression in pea (Pisum sativum; Mittler and Zilinskas, 1994).
Under fHL, the expression levels of nearly one-half of the up-regulated genes, including APX2 and GST6, were lower than under HL (Table I). H2O2 induces both cytosolic APX (Conklin and Last, 1995) and GST6 expression (Chen et al., 1996) as also observed in Figure 5, implying the observed decrease in GST6 expression in the fHL may reflect decreased H2O2 and other ROS. A possible explanation could be that the H2O2 concentration in fHL leaves is lower than in HL. In fact, 60-s bursts of 50°C air have been shown to increase O2− and H2O2 in barley (Hordeum vulgare; Vallelian-Bindschedler et al., 1998). However, in our experiments, air temperature was only a few degrees higher and it may be that other processes also contribute to the difference in expression profiles. An alternative explanation is that the PQ pool was not as reduced in fHL as compared with HL because DCMU, which oxidizes the PQ pool, led to reduced APX2 and GST6 expression (Karpinski et al., 1999). However, the mechanism for this is difficult to envisage because the PAR is the same in fHL and HL, implying a higher temperature must be having a synergistic or distinct effect on APX2 and GST6 induction.
Among the genes induced by both HL conditions, the chaperone family of HSPs were highly induced and well represented, forming more than 30% of genes induced by both HL conditions. Why would HSPs be induced by HL? HSP induction is apparently not because of the moderately elevated temperature; rather, it is because of the oxidizing environment of HL (Fig. 4). The functions of different classes of chaperones range from protein transport, folding, assembly, preventing aggregation, and de-aggregation. Because representatives from several classes of chaperones were induced, it implies that all these functions were required. HSP101, exhibiting the highest ratio of transcript increase in the HL and second highest in the fHL experiments, is crucial in the acquisition of thermotolerance in Arabidopsis and it functions by reactivating proteins that have aggregated (Queitsch et al., 2000), as is one of the functions of HSP70. Recent evidence has shown that certain sHSPs provide a protective function against oxidative conditions and that oxidative stress induces expression of HSPs and chaperones. In rice (Oryza sativa) and tomato (Lycopersicon esculentum), the sHSPs OsHSP26 and HSP22 are induced by H2O2 (Banzet et al., 1998; Lee et al., 2000). In cyanobacteria and Arabidopsis, HL and H2O2, respectively, induced chaperones, HSPs, and the heat shock transcription factor (Desikan et al., 2001; Hihara et al., 2001). Furthermore, in Arabidopsis, the small chloroplast HSP undergoes H2O2-dependent conformational changes and may act as a scavenger in addition to its presumed chaperone role (Harndahl et al., 1999). This would seem in accordance with the expression levels of the putative sHSP in the HL and H2O2 microarray experiments, implicating H2O2 in the transcriptional activation of this gene (Tables I and II). Thus, increased expression of chaperones under both HL conditions may possibly be an adaptive response to limit oxidation-mediated disulfide bridge-induced protein aggregation.
The microarray analyses have identified genes previously not thought to be involved in HL-induced oxidative stress; for example, the vegetative storage protein Vsp2. Recently, this gene has been implicated in conferring salt tolerance because it was induced in wild-type Arabidopsis and decreased in the mutant sos3 (salt overly sensitive; Gong et al., 2001). Likewise, the circadian clock-associated gene CCA1 was induced in HL but not fHL. Given that brief illumination of dark-grown plants is sufficient to transiently stimulate CCA1 gene expression (Wang and Tobin, 1998), it is not clear how the difference in the IR spectrum between fHL and HL could be responsible for CCA1 induction.
Regulation of Gene Expression
It is perhaps surprising that the fHL and HL datasets show only a 30% overlap. The light-driven expression of numerous genes is controlled by several photoreceptors that absorb light in the visible and UV range. However, the filter did not reduce the percentage of UV or any component of the visible spectrum, such as red and far red, ruling out phytochrome-, cryptochrome-, and UV-mediated responses. Thus, the strong differential expression of CHS between the HL experiments would not reflect reduced UV levels in fHL, which is known to alter CHS expression in Arabidopsis (Long and Jenkins, 1998). It is worth noting that HY5 is a positive regulator of photomorphogenic development and light activation of the CHS promoter (Oyama et al., 1997; Ang et al., 1998) and CHS induction was mirrored by the HY5 induction pattern under a range of conditions (Table II). However, HY5 and CHS expression levels do not correlate well for fHL and HL, indicating involvement of an unidentified factor in CHS induction.
The question remains as to why there is a difference in HL and fHL gene expression profiles. One possible explanation for some differences is that the stringent threshold for a gene to be considered induced in both sets would require it to be >2 times induced in six replicates. For example, APX2 only just failed to meet this criterion (Table I). More likely, the distinct profiles reflects biological responses, possibly due to synergistic effects of heat and light exacerbating oxidative stress because there is a considerable overlap between processes induced by heat and oxidative stress (Storozhenko et al., 1998). In the alga Chenopodium rubrum, heat and light act synergistically to co-effect HSP expression and the APX1 and APX2 promoters contain functional HSEs (Knack and Kloppstech, 1992; Storozhenko et al., 1998; Panchuk et al., 2002; Table III). As noted above, heat can induce ROS (Vallelian-Bindschedler et al., 1998). Thus, the small increase in temperature may act in concert with HL via heat shock and ROS response elements to induce a suite of genes not induced by fHL and significantly enhance the expression of nearly one-half of the fHL up-regulated genes listed in Table I. The suite of genes uniquely induced by fHL may reflect alternative, moderate stress response pathways that are suppressed by pathways induced by HL. In this context, it may be interesting that At1g19180, an unknown, went from 3.8 in fHL to −1.3 in HL.
The cross comparison between our experiments and those at SMD demonstrates that abiotic stresses do have differing effects on induction of different stress-associated genes. In general, experiments that affected chloroplast function or directly induced oxidative pressure, such as H2O2 and the chlorophyll biosynthetic mutant, cch1 (gun5) (Mochizuki et al., 2001), had the most similar expression profiles to HL treatments. However, other abiotic stresses, such as aluminum toxicity or iron deficiency, induced only one of the 12 genes analyzed. Even for H2O2, only about 50% of the genes were similarly induced. This, in part, reflects the different nature of the experiments (intact leaves + HL verses cell cultures + H2O2) and also reflects the reduction of the PQ pool by HL. Although directly comparing MV, DCMU, and HL treatments requires caution, Figure 5 shows that both oxidative pressure and the REDOX state of the PQ pool contribute to the expression of APX1, GST6, and sHSP. That is, MV induced oxidative damage, but did not fully up-regulate those genes (compare 60-min HL and MV in Fig. 5) and, likewise, oxidation of the PQ pool did not fully down-regulate those genes (compare 0-min HL and DCMU). However, it appears that oxidative stress may be a major determinant for HY5 and HSP70.
To gain further insight from the large quantity of data generated by microarray analyses, differentially expressed genes were clustered according to their expression patterns and subjected to promoter motif analyses (Thijs et al., 2001). Table III lists motifs that were statistically over represented in a given cluster. LREs were found in the induced clusters for HL, fHL, and HL plus fHL (Table III). These findings are not surprising because no single LRE common to all light-regulated genes has been found (Terzaghi and Cashmore, 1995). Explanations for multiple LREs are that a combinatorial interaction of distinct LREs is required for proper light responsiveness (Degenhardt and Tobin, 1996; Chattopadhyay et al., 1998). Furthermore, a combination of cis-elements allows responses to multiple stimuli, whereas a single promoter cis-element is adequate for responding to a particular stimulus (Hill and Treisman, 1995). Intriguingly, only in the fHL plus HL cluster did we find motifs involved in heat stress response. One of these HSE elements was found in the APX promoter, which had previously been found to be heat shock inducible (Sato et al., 2001). In addition, elements involved in ABA responsiveness were identified in clusters of genes induced by fHL and repressed by HL, seeming to indicate the involvement of another as yet unknown factor.
Motifs of unknown function were found in each group of differentially expressed genes and one might speculate that they could play a role in the fine-tuning of a response. Further analyses of these motifs will be required to ascertain function and determine if the unknown motifs are involved in a light stress-specific response. These analyses have also shed light on possible functions of unknown or hypothetical genes because they contained promoter motifs that were also present in known genes such as APX, and several HSPs.
Microarray technology has provided the means to analyze the expression profile of over 6,000 genes in response to two different HL regimes. In addition to the structural genes involved in detoxification, photo-acclimation, protein folding, and de-aggregation, a substantial subset of genes that were differentially expressed are also involved in other stress-sensing responses. Thus, we have commenced the functional analyses of the unknown and hypothetical genes identified by this study. This may identify common regulators for abiotic stress responses in general and the redox state and H2O2 signaling pathways in particular that are implicated in the regulation of genes such as GST6, APX1, and APX2. Finally, systematic and integrated analysis of multiple datasets as outlined in this study will further assist in the elucidation of stress signaling networks.
MATERIALS AND METHODS
Plant Material
Arabidopsis ecotype Colombia plants were grown at low density in 16-h-light/8-h-dark cycles at a temperature of 22°C under 100 to 150 μmol m−2 s−1 of light. Plants were fertilized twice weekly with 0.5× Hoagland medium. Two HL conditions were applied to investigate the plant response. The 1-h treatment regimes were: ML, 100 μmol m−2 s−1 at 22°C; HL, 1,000 μmol m−2 s−1 at 29°C; fHL, 1,000 μmol m−2 s−1 at 24°C; and wML, 100 μmol m−2 s−1 at 27.5°C. For fHL, the light was filtered to minimize any heat effect of HL and was achieved by filtering light through a glass tray with a frosted bottom containing 5 cm of cold water placed over the plants. The wML plants were placed in a growth chamber at 27.5°C and leaves were harvested after 1 h.
Leaf material was harvested from 24-d-old plants after conclusion of the treatment and immediately frozen in liquid nitrogen. To minimize plant-to-plant variation of gene expression, leaf material from more than 50 plants was pooled for RNA extraction. Plants were sprayed with MV (2.5 μm) or DCMU (10 μm) three times over 12 h. The leaves were harvested 12 h after the last application. The effectiveness of DCMU was tested via chlorophyll measurements (data not shown).
Leaf Temperature, Spectral Measurements, and HPLC Analyses
Leaf temperature of 24-d-old Arabidopsis plants was measured with copper-constantan thermocouples (64-μm diameter) referenced against a PT-100 platinum resistance thermometer. Four thermocouples were attached to the underside of four leaves per treatment. The data were averaged and plotted in Excel. The air temperature was measured in the shade.
Pigments were extracted and analyzed by HPLC (Pogson et al., 1998) from 24-d-old leaves taken from plants treated with both HL conditions, from plants grown at 27.5°C for 1 h and from plants under normal growing conditions.
Spectral measurements were carried out using a portable spectroradiometer (model LI-1800, LI-COR, Lincoln, NE) under the HL and fHL conditions used for the microarray analyses and data were plotted in Excel.
Microarray Analyses
Approximately 6,000 clones from the Mendel library were spotted onto glass slides (Helliwell et al., 2001). An additional 220 clones not represented in the library were added. These clones represented genes in pigment biosynthesis, antioxidant biosynthesis, and genes encoding PS-associated proteins. The additional clones were either selected as expressed sequence tags from the Arabidopsis Biological Research Council or amplified via reverse transcriptase (RT)-PCR and cloned into T-easy (Access RT-PCR system, Promega, Madison, WI). Gene-specific primers were designed for each member of the APX, GST, and SOD multigene families.
RNA was isolated with minor modifications to the described protocol (Logemann et al., 1987). Isolated RNA was DNAse treated and further purified with the RNAeasy kit (Qiagen USA, Valencia, CA). One hundred micrograms of total RNA was used in the first strand cDNA synthesis using a modified protocol of two-step probe labeling method (Schenk et al., 2000). cDNA was RNase A treated and purified using the Qiaprep mini prep kit (Qiagen USA) and resuspended in 8 μL of Tris-EDTA (10 mm Tris and 1 mm EDTA, pH 8.2).
Two microliters of resuspended cDNA was used in the labeling reaction. The labeling reaction was purified using the Qiaprep mini prep kit (Qiagen USA). The experimental and control tissues were labeled either with Cy3 or Cy5 fluorescent dye (Amersham-Pharmacia Biotech, Uppsala) and hybridized to a microarray slide as described by Schenk et al. (2000). The slides were scanned with a GenePix 4000 scanner (Axon Instruments, Foster City, CA) and image analysis was performed using GenePix 3.0 software (Axon Instruments). Spot intensities from scanned slides were analyzed and normalized using custom Perl scripts (Schenk et al., 2000; http://cellwall.stanford.edu/scripts/index.html). Genes with equal to or greater than 2-fold difference in their ratio were considered to be differentially expressed. Each microarray experiment was undertaken three times: one replication was technical, using RNA from the same 50+ plants but reversing the dye, and the other was biological, using RNA from another 50+ plants grown under the same conditions. The average value of the three experiments is given in “Results.”
Promoter Analyses
Sequences 500 bp upstream of the ATG site of differentially regulated genes found in the microarray experiments were retrieved from The Institute for Genomic Research ftp site (ftp://www.tigr.org). These sequences were subdivided according to which condition they were differentially regulated. The subgroups were analyzed for overrepresented motifs with MotifSampler (Thijs et al., 2001). We then compared the detected motifs with known motifs available in the PlantCARE database (Rombauts et al., 1999).
RNA Gel-Blot Analyses
Total RNA was extracted as described above. RNA was fractionated on a 1% (w/v) denaturing agarose gel, blotted according to the manufacturer's protocol (Amersham-Pharmacia Biotech), and probed with [α-32P]dCTP-labeled PCR fragments. Probe detection was carried out using a phosphor imager and data analyzed with ImageQuant software (Molecular Dynamics, Sunnyvale, CA). PCR fragments were isolated either from the corresponding expressed sequence tag using M13 forward and reverse primers or via RT-PCR with gene-specific primers.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jun Yang (Commonwealth Scientific and Industrial Research Organisation [CSIRO] Plant Industry, Canberra, Australia) for her technical work in the production of the microarray slides, Dr. Erik Jan Klok (CSIRO Plant Industry) for his help with the promoter motif analyses, and Gavin Kennedy (CSIRO Plant Industry) for his help with the Perl script analyses.
Footnotes
This work was supported by The Australian National University's International Postgraduate Research Award to J.B.R. and by the Australian Research Council (grant no. F00077 to B.J.P.).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005595.
LITERATURE CITED
- Abarca D, Martin M, Sabater B. Differential leaf stress responses in young and senescent plants. Physiol Plant. 2001;113:409–415. doi: 10.1034/j.1399-3054.2001.1130315.x. [DOI] [PubMed] [Google Scholar]
- Adamska I. ELIPs: light-induced stress proteins. Physiol Plant. 1997;100:794–805. [Google Scholar]
- Altschuler M, Mascarenhas JP. Transcription and translation of heat shock proteins in seedlings and developing seeds of soybean exposed to a gradual temperature increase. Plant Mol Biol. 1985;5:291–297. doi: 10.1007/BF00020626. [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Asada K. The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- Banzet N, Richaud C, Deveaux Y, Kazmaier M, Gagnon J, Triantaphylides C. Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. Plant J. 1998;13:519–527. doi: 10.1046/j.1365-313x.1998.00056.x. [DOI] [PubMed] [Google Scholar]
- Bell E, Mullet JE. Characterization of an Arabidopsis-lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol. 1993;103:1133–1137. doi: 10.1104/pp.103.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Puente P, Deng XW, Wei N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998;15:69–77. doi: 10.1046/j.1365-313x.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- Chen WQ, Chao G, Singh KB. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 1996;10:955–966. doi: 10.1046/j.1365-313x.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- Conklin PL, Last RL. Differential accumulation of antioxidant messenger-RNAs in Arabidopsis thaliana exposed to ozone. Plant Physiol. 1995;109:203–212. doi: 10.1104/pp.109.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner TW, Lafayette PR, Nagao RT, Key JL. Sequence and expression of a Hsp83 from Arabidopsis thaliana. Plant Physiol. 1990;94:1689–1695. doi: 10.1104/pp.94.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt J, Tobin EM. A DNA binding activity for one of two closely defined phytochrome regulatory elements in an Lhcb promoter is more abundant in etiolated than in green plants. Plant Cell. 1996;8:31–41. doi: 10.1105/tpc.8.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depka B, Jahns P, Trebst A. Beta-carotene to zeaxanthin conversion in the rapid turnover of the D1 protein of photosystem II. FEBS Lett. 1998;424:267–270. doi: 10.1016/s0014-5793(98)00188-4. [DOI] [PubMed] [Google Scholar]
- Desikan R, Mackerness SAH, Hancock JT, Neill SJ. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001;127:159–172. doi: 10.1104/pp.127.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droog F. Plant glutathione S-transferases, a tale of theta and tau. J Plant Growth Regul. 1997;16:95–107. [Google Scholar]
- Escoubas JM, Lomas M, Laroche J, Falkowski PG. Light-intensity regulation of Cab gene-transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskling M, Akerlund HE. Changes in the quantities of violaxanthin de-epoxidase, xanthophylls and ascorbate in spinach upon shift from low to high light. Photosynth Res. 1998;57:41–50. [Google Scholar]
- Foyer CH, Descourvieres P, Kunert KJ. Protection against oxygen radicals: an important defense mechanism studied in transgenic plants. Plant Cell Environ. 1994;17:507–523. [Google Scholar]
- Gilmore AM. Xanthophyll cycle-dependent nonphotochemical quenching in photosystem II: mechanistic insights gained from Arabidopsis thaliana L. mutants that lack violaxanthin deepoxidase activity and/or lutein. Photosynth Res. 2001;67:89–101. doi: 10.1023/A:1010657000548. [DOI] [PubMed] [Google Scholar]
- Gilmore AM, Mohanty N, Yamamoto HY. Epoxidation of zeaxanthin and antheraxanthin reverses nonphotochemical quenching of photo-system-II chlorophyll-a fluorescence in the presence of trans-thylakoid delta-pH. FEBS Lett. 1994;350:271–274. doi: 10.1016/0014-5793(94)00784-5. [DOI] [PubMed] [Google Scholar]
- Gong ZZ, Koiwa H, Cushman MA, Ray A, Bufford D, Kore-eda S, Matsumoto TK, Zhu JH, Cushman JC, Bressan RA et al. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001;126:363–375. doi: 10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harndahl U, Hall RB, Osteryoung KW, Vierling E, Bornman JF, Sundby C. The chloroplast small heat shock protein undergoes oxidation-dependent conformational changes and may protect plants from oxidative stress. Cell Stress Chaperones. 1999;4:129–138. doi: 10.1379/1466-1268(1999)004<0129:tcshsp>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Chin-Atkins AN, Wilson IW, Chapple R, Dennis ES, Chaudhury A. The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell. 2001;13:2115–2125. doi: 10.1105/TPC.010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell. 2001;13:793–806. doi: 10.1105/tpc.13.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CS, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- Jenkins GI. UV and blue light signal transduction in Arabidopsis. Plant Cell Environ. 1997;20:773–778. doi: 10.1046/j.1365-3040.1997.d01-105.x. [DOI] [PubMed] [Google Scholar]
- Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science. 1999;284:654–657. doi: 10.1126/science.284.5414.654. [DOI] [PubMed] [Google Scholar]
- Kimpel JA, Key JL. Presence of heat shock mRNAs in field grown soybeans. Plant Physiol. 1985;79:672–678. doi: 10.1104/pp.79.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knack G, Kloppstech K. The heat-shock response in a photoautotrophic cell-culture of Chenopodium rubrum: the effects of temperature and light. J Plant Physiol. 1992;140:489–493. [Google Scholar]
- Lee BH, Won SH, Lee HS, Miyao M, Chung WI, Kim IJ, Jo J. Expression of the chloroplast-localized small heat shock protein by oxidative stress in rice. Gene. 2000;245:283–290. doi: 10.1016/s0378-1119(00)00043-3. [DOI] [PubMed] [Google Scholar]
- Lindahl M, Funk C, Webster J, Bingsmark S, Adamska I, Andersson B. Expression of ELIPs and PS II-S protein in spinach during acclimative reduction of the photosystem II antenna in response to increased tight intensities. Photosynth Res. 1997;54:227–236. [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Long JC, Jenkins GI. Involvement of plasma membrane redox activity and calcium homeostasis in the UV-B and UV-A/blue light induction of gene expression in Arabidopsis. Plant Cell. 1998;10:2077–2086. doi: 10.1105/tpc.10.12.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Zilinskas BA. Regulation of pea cytosolic ascorbate peroxidase and other antioxidant enzymes during the progression of drought stress and following recovery from drought. Plant J. 1994;5:397–405. doi: 10.1111/j.1365-313x.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK. Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D. Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell. 1995;7:2139–2149. doi: 10.1105/tpc.7.12.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schoffl F. Heat stress and heat shock transcription factor dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 2002;129:838–853. doi: 10.1104/pp.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397:625–628. [Google Scholar]
- Pogson BJ, Niyogi KK, Bjorkman O, DellaPenna D. Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc Natl Acad Sci USA. 1998;95:13324–13329. doi: 10.1073/pnas.95.22.13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC. Aluminum induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol. 1998;116:409–418. doi: 10.1104/pp.116.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T, Somerville S. Chasing the dream: plant EST microarrays. Curr Opin Plant Biol. 2000;3:108–116. doi: 10.1016/s1369-5266(99)00049-7. [DOI] [PubMed] [Google Scholar]
- Rombauts S, Dehais P, Van Montagu M, Rouze P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999;27:295–296. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AW, Critchley C, Robinson SA, Franklin LA, Seaton GGR, Chow WS, Anderson JM, Osmond CB. Photosystem II regulation and dynamics of the chloroplast D1 protein in Arabidopsis leaves during photosynthesis and photoinhibition. Plant Physiol. 1995;107:943–952. doi: 10.1104/pp.107.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabehat A, Weiss D, Lurie S. Heat-shock proteins and cross-tolerance in plants. Physiol Plant. 1998;103:437–441. [Google Scholar]
- Sato Y, Murakami T, Funatsuki H, Matsuba S, Saruyama H, Tanida M. Heat shock-mediated APX gene expression and protection against chilling injury in rice seedlings. J Exp Bot. 2001;52:145–151. [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. [PubMed] [Google Scholar]
- Storozhenko S, De Pauw P, Van Montagu M, Inzé D, Kushnir S. The heat-shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol. 1998;118:1005–1014. doi: 10.1104/pp.118.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZR, Gantt E, Cunningham FX. Cloning and functional analysis of the beta-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem. 1996;271:24349–24352. doi: 10.1074/jbc.271.40.24349. [DOI] [PubMed] [Google Scholar]
- Terashima I, Noguchi K, Itoh-Nemoto T, Park YM, Kubo A, Tanaka K. The cause of PSI photoinhibition at low temperatures in leaves of Cucumis sativus, a chilling-sensitive plant. Physiol Plant. 1998;103:295–303. [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Thijs G, Lescot M, Marchal K, Rombauts S, De Moor B, Rouze P, Moreau Y. A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics. 2001;17:1113–1122. doi: 10.1093/bioinformatics/17.12.1113. [DOI] [PubMed] [Google Scholar]
- Utsugi S, Sakamoto W, Murata M, Motoyoshi F. Arabidopsis thaliana vegetative storage protein (VSP) genes: gene organization and tissue-specific expression. Plant Mol Biol. 1998;38:565–576. doi: 10.1023/a:1006072014605. [DOI] [PubMed] [Google Scholar]
- Vallelian-Bindschedler L, Schweizer P, Mosinger E, Metraux JP. Heat-induced resistance in barley to powdery mildew (Blumeria graminis f.sp. hordei) is associated with a burst of active oxygen species. Physiol Mol Plant Pathol. 1998;52:185–199. [Google Scholar]
- Wang WJ, Ghosh S, Guo SW. Quantitative quality control in microarray image processing and data acquisition. Nucleic Acids Res. 2001;29:U32–U39. doi: 10.1093/nar/29.15.e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Wu CH, Capar T, Browse J, Lindquist S, Somerville C. Characterization of an HSP70 cognate gene family in Arabidopsis. Plant Physiol. 1988;88:731–740. doi: 10.1104/pp.88.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto HY, Chichester CO, Nakayama TO. Studies on light and dark interconversions of leaf xanthophylls. Arch Biochem Biophys. 1962;97:168–173. doi: 10.1016/0003-9861(62)90060-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.