Abstract
NifS-like proteins catalyze the formation of elemental sulfur (S) and alanine from cysteine (Cys) or of elemental selenium (Se) and alanine from seleno-Cys. Cys desulfurase activity is required to produce the S of iron (Fe)-S clusters, whereas seleno-Cys lyase activity is needed for the incorporation of Se in selenoproteins. In plants, the chloroplast is the location of (seleno) Cys formation and a location of Fe-S cluster formation. The goal of these studies was to identify and characterize chloroplast NifS-like proteins. Using seleno-Cys as a substrate, it was found that 25% to 30% of the NifS activity in green tissue in Arabidopsis is present in chloroplasts. A cDNA encoding a putative chloroplast NifS-like protein, AtCpNifS, was cloned, and its chloroplast localization was confirmed using immunoblot analysis and in vitro import. AtCpNIFS is expressed in all major tissue types. The protein was expressed in Escherichia coli and purified. The enzyme contains a pyridoxal 5′ phosphate cofactor and is a dimer. It is a type II NifS-like protein, more similar to bacterial seleno-Cys lyases than to Cys desulfurases. The enzyme is active on both seleno-Cys and Cys but has a much higher activity toward the Se substrate. The possible role of AtCpNifS in plastidic Fe-S cluster formation or in Se metabolism is discussed.
NifS-like proteins are pyridoxal 5′ phosphate (PLP)-dependent enzymes with sequence similarity to the Cys desulfurase encoded by nifS of Azotobacter vinelandii (Zheng et al., 1993). These proteins have been found in most organisms tested, where they play a role in S or Se metabolism (Mihara et al., 1997). NifS-like proteins catalyze the breakdown of Cys to form Ala and elemental S, or they may act on related substrates such as seleno-Cys to form Ala and elemental Se (Mihara et al., 1997). The nifS of A. vinelandii is required under nitrogen fixation conditions for the formation of Fe-S clusters in nitrogenase (Zheng et al., 1993). A. vinelandii NIFS is present in a gene cluster with several other genes (nifU, nifA, and cysE) all thought to be involved in Fe-S cluster formation. A second NifS-like protein of A. vinelandii, IscS, has a housekeeping function in the formation of other cellular Fe-S proteins (Zheng et al., 1993). Interestingly, iscS is present in a gene cluster that contains paralogs of the nif genes (iscU and iscA), thus, the nif and isc clusters share a similar organization (Zheng et al., 1998). Homologs of the nif/isc genes, all thought to play a role in cellular Fe-S cluster formation have been discovered in several other bacteria including in Escherichia coli (Zheng et al., 1998). In the eukaryotes, Fe-S clusters are essential cofactors for mitochondrial respiration, as well as for many cytosolic proteins. Recent work has suggested that in yeast and in mammals, all Fe-S clusters are made in the mitochondria (for review, see Lill and Kispal, 2000). Fe-S cluster formation in the mitochondria of eukaryotes involves homologs of the genes encoded by the nif/isc clusters of bacteria (Kispal et al., 1999). It has been demonstrated in yeast that Fe-S cluster formation of enzymes present in the cytosol requires a machinery in the mitochondrial matrix, which includes yeast NifS and other homologs of the prokaryotic nif cluster. Interestingly, a transporter in the mitochondrial inner membrane of yeast is required for assembly of cytosolic Fe-S clusters (Kispal et al., 1999), and there is evidence that a similar situation may be present in other eukaryotes including plants (Kushnir et al., 2001).
In chloroplasts, Fe-S clusters play a key role in photosynthesis. They are required for the function of the cytochrome B/F complex, photosystem-I, and ferredoxin and, thus, are directly required for linear electron transport in the thylakoids (Raven et al., 1999). Some studies have been done to characterize the assembly of the 2Fe-2S cluster in plant ferredoxin by using a native gel electrophoresis system, which can distinguish the apo- and holo-forms of ferredoxin (Takahashi et al., 1986, 1990; Li et al., 1990). Cys was found to be the source of S for Fe-S cluster formation/regeneration into spinach (Spinacia oleracea) ferredoxin in isolated intact chloroplasts or lysed chloroplasts, and this process also required the presence of light or ATP and reduced NADPH (Takahashi et al., 1986, 1990). Cluster assembly into radiolabeled freshly imported ferredoxin precursor obtained by in vitro translation was shown in vitro with isolated intact chloroplasts (Li et al., 1990). Microgram amounts of a purified ferredoxin precursor obtained the 2Fe-2S cluster after in vitro import into intact chloroplasts in the absence of cytosol, as measured by high-resolution ion-exchange chromatography (Pilon et al., 1995). These experiments all strongly suggest the presence of an Fe-S cluster formation machinery in chloroplasts. Because Cys was identified as a source for Fe-S formation, a NifS-like protein with Cys desulfurase activity could well be involved in this process. At this point, still much is to be learned on how Fe-S clusters are assembled in chloroplasts (for a review on cofactor assembly in the photosynthetic machinery, see Merchant and Dreyfuss, 1998), and this will be an interesting field of study for the near future.
NifS-like proteins from various sources have higher activity with seleno-Cys than with Cys (Mihara et al., 1999, 2000a; Kato et al., 2000). In the reaction with seleno-Cys (seleno-Cys lyase activity), the NifS-like proteins produce elemental Se and Ala. In bacteria and likely also in mammals, the reaction of NifS-like proteins with seleno-Cys may have an important function in the formation of specific selenoproteins, several of which are involved in oxidative stress resistance (Lacourciere and Stadtman, 1998; Lacourciere et al., 2000). Examples of NifS-like proteins implied in this process are E. coli Cys sulfinate desulfinase (Csd) B and various mammalian seleno-Cys lyases (Mihara et al., 1999, 2000a). These proteins play a role in selenoprotein synthesis by providing a substrate for selenophosphate synthase (Lacourciere et al., 2000). This latter molecule is then used as a substrate for the formation of Se-Cys-tRNA, which can be used in the translation of UGA opal codons in specific mRNAs encoding seleno-Cys-containing enzymes (Boeck et al., 1991). Until recently, there was no evidence for the presence of specific selenoenzymes in the plant branch of the eukarya, but recently a specific seleno-form of glutathione peroxidase has been found in the unicellular algae Chlamydomonas reinhardtii (Fu et al., 2002). This raises the intriguing possibility that plants could also have selenoproteins and, thus, that Se is an essential nutrient for plants.
In plants, the S assimilation pathway leads up to the synthesis of Cys. This pathway is localized in the chloroplast and involves the reduction of sulfate to sulfite and next to S2−, which depends on electrons donated by ferredoxin (Leustek and Saito, 1999). The chemical similarity of S and Se allows the incorporation of Se into analogous compounds of the S assimilation pathway (Stadtman, 1990). Overexpression of the sulfate-activating chloroplast enzyme ATP sulfurylase was shown to lead to enhanced selenate reduction and Se accumulation in plants (Pilon-Smits et al., 1999). Thus, seleno-Cys may be formed in the chloroplast in plants. The possible formation of the NifS substrate seleno-Cys in chloroplasts and the importance of Fe-S cluster formation to photosynthesis prompted us to investigate the presence of a NifS-like protein in chloroplasts and to analyze its catalytic activity toward Cys and seleno-Cys.
RESULTS
Detection of NIFS Activity in Arabidopsis Chloroplasts
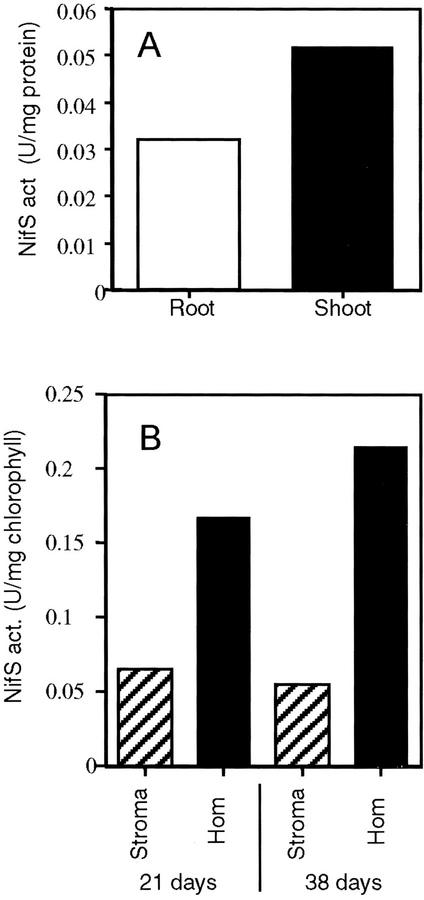
NifS-like proteins catalyze the formation of elemental S and Ala from Cys or of elemental Se and Ala from seleno-Cys. The activity is higher when seleno-Cys is used as a substrate for most NifS-like proteins, allowing for more sensitive detection of NifS activity. We therefore used seleno-Cys as a substrate to analyze the presence of NifS activity in Arabidopsis. NifS activity was detected in both root and shoot homogenates of Arabidopsis seedlings (Fig. 1A). To analyze the presence of possible NifS activity in the chloroplast of Arabidopsis, we compared the NifS activity in total rosette leaf homogenate with that in isolated chloroplasts (Fig. 1B). These chloroplasts had been obtained by a procedure that includes centrifugation on silica sol (Percoll) density gradients (Rensink et al., 1998). The specific activity of the cytosolic marker enzyme phosphoenolpyruvate carboxylase (PEPC) in the chloroplast fractions was on average less than 2% of that in total homogenate, indicating that the chloroplast fractions were essentially free of cytosolic contamination. NifS activity was detected in the total homogenate and in the stroma of isolated chloroplasts. The activity in chloroplasts accounted for 25% to 30% of the total activity found in the leaf material. Similar ratios were found for leaves from younger (3-week-old plants before flowering) and more mature plants (5.5 weeks old, after the beginning of flowering). These results suggest the presence of a NifS-like enzyme with seleno-Cys lyase activity in chloroplasts of Arabidopsis.
Figure 1.
NifS activity in plant samples. Protein extracts were obtained as outlined in “Materials and Methods,” and NifS activity was measured using 10 mm seleno-Cys as a substrate. A, NifS activity in roots and shoots. B, NifS activity in the chloroplast stroma and leaf homogenate (hom) from which the chloroplasts were isolated. All bars represent the average of three measurements. Note that the data in A are normalized based on protein and in B based on chlorophyll, which was needed to allow comparison of the latter fractions and because of the presence of bovine serum albumin in the grinding buffer in which these homogenates are made.
Cloning of a cDNA Encoding a Precursor of a NifS-Like Protein from Arabidopsis
To identify genes that encode possible chloroplast-localized NifS-like proteins, we performed database searches. Blast searches in The Arabidopsis Information Resource and National Center for Biotechnology Information (NCBI) databases (see “Materials and Methods”) revealed the presence of two possible NIFS-like genes in the genome of Arabidopsis. TargetP predictions (see “Materials and Methods”) suggest that one gene, which we named AtCpNIFS (At1g08490), may encode a chloroplast protein with a 35-amino acid long chloroplast transit sequence (TargetP score 0.925, reliability class 2). A second gene, which we named AtMtNIFS (At5g65720), may encode a precursor of a mitochondrial protein based on TargetP predictions.
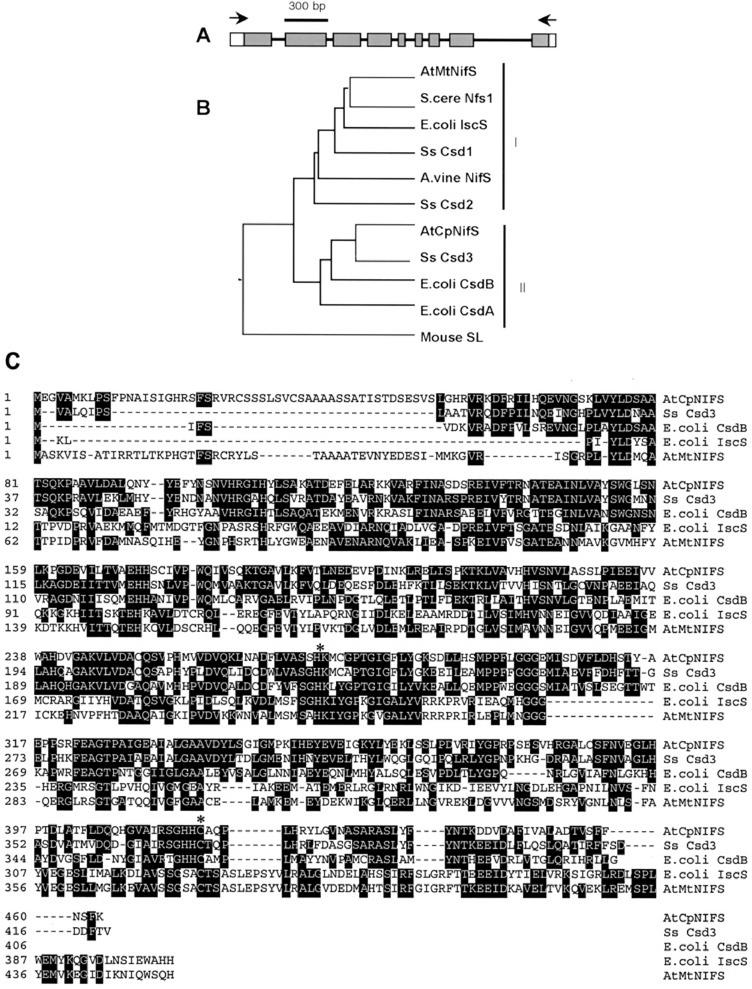
We wanted to obtain a cDNA to facilitate the analysis of the putative chloroplastic NifS-like protein encoded by AtCpNIFS. We employed reverse transcription followed by PCR amplification (reverse transcriptase-PCR) with primers that flank the coding sequence to obtain a cDNA. As a template for PCR, we used cDNA obtained by reverse transcription of the poly(A) mRNA that was isolated from 2-week-old Arabidopsis seedlings. A single PCR product was obtained. A second round of PCR using nested primers was performed to confirm the identity of the first PCR product and to introduce restriction sites for subcloning in the pET11d expression vector. Sequence analysis of this cDNA clone showed the presence of an open reading frame encoding a polypeptide of 463 amino acids. Alignment of the cDNA sequence with the published genomic sequence indicates the presence of nine exons and eight introns in the genomic sequence (see Fig. 2A). The GenBank accession number for the sequence of AtCpNIFS is AF419347.
Figure 2.
AtCpNIFS: gene structure and protein alignment. A, Schematic genomic organization of AtCpNIFS. Exons are indicated by boxes, the coding sequence is in gray. Introns are indicated by solid lines. The arrows represent the areas where PCR primers used for initial amplification hybridize to the cDNA. B, Phylogenetic tree (Megalign program, DNASTAR, Inc.) of NifS-like proteins encoded by genes of Arabidopsis, E. coli, Synechocystis sp. 6803, A. vinelandii, and mouse. The sequences are: AtCpNifS, At1g08490 (this paper); AtMtNIFS, At5g65720, encoding the putative Arabidopsis mitochondrial NifS; A. vine NIFS, A. vinelandii NifS; E. coli CsdA; E. coli CsdB, seleno-Cys lyase; E. coli IscS, Cys desulfurase; mouse Sl, murine seleno-Cys lyase. Ss Csd1, Synechocystis sp. PCC6803 slr0387; Ss Csd2, Synechocystis sp. PCC6803 sll0704; Ss Csd3, Synechocystis sp. PCC6803 slr0077. S. cere Nfs1, Brewer’s yeast (Saccharomyces cerevisiae) mitochondrial NifS. C, Predicted protein sequence present in the cDNA of CpNIFS and Clustal alignment (DNASTAR software) with four other NifS-like protein sequences. The conserved Lys residue for cofactor binding and the active site Cys residue are indicates by asterisks. The arrow indicates the predicted transit sequence cleavage site in the AtCpNifS precursor.
NifS-like proteins are characterized by the presence of several conserved amino acid sequence elements, most notably a conserved His-Lys motif required for PLP-cofactor binding and an essential Cys residue at the active site (Zheng et al., 1993). To determine whether these elements are present in AtCpNIFS, this sequence and the putative AtMtNifS were aligned with two well-characterized NifS-like protein sequences from Escherichia coli, IscS and CsdB, and with a NifS-like sequence from Synechocystis sp. PCC6803 (Fig. 2C). The sequence alignment shows that both conserved elements are present in the Arabidopsis proteins. Relative to the bacterial proteins the plant proteins have N-terminal extensions, which may function in organelle targeting. NifS-like proteins can be divided into two classes based on sequence similarity (Mihara et al., 1997). The E. coli IscS protein is a typical class 1 protein, whereas the E. coli CsdB protein belongs to class 2. Interestingly, the alignment in Figure 2C and a grouping based on sequence similarity of various NifS-like sequences (Fig. 2B) indicate that AtCpNifS is much more related in sequence to class 2 NifS-like proteins such as E. coli CsdB than it is to class 1 NifS-like proteins and to the putative mitochondrial NifSp from Arabidopsis. The two Arabidopsis proteins have only 21% sequence similarity. In contrast, AtCpNifSp has 45% sequence similarity with the E. coli CsdB protein and 60% sequence similarity with a thus far biochemically uncharacterized cyanobacterial sequence, SsCsd3 (Kato et al., 2000).
AtCpNifS Contains Chloroplast Targeting Information
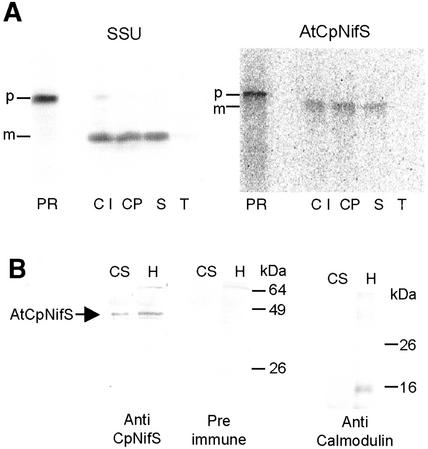
AtCpNIFS encodes a protein predicted to be active in the chloroplast stroma. To investigate directly whether the AtCpNifS precursor contains chloroplast targeting information, we performed in vitro chloroplast uptake experiments. Radiolabeled precursor protein was produced by in vitro transcription of the cloned cDNA and subsequent translation of the synthetic mRNA in the presence of 35S-Met. Translation resulted in a single radiolabeled protein of about 47 kD. This precursor was incubated with purified intact chloroplasts in the light and in the presence of ATP. Import and intra-organellar routing was monitored by re-isolation of the chloroplasts, protease treatment, and fractionation of chloroplasts into a soluble-stroma-fraction and membranes (Fig. 3A). The precursor of the small subunit of Rubisco, a stromal enzyme, which is translated and imported with high efficiency in vitro, was used as a positive control. Chloroplasts incubated with the precursor encoded by AtCpNIFS accumulated a protein of about 43 kD in size. Protease treatment and fractionation experiments showed that this protein was present in the stromal fraction. Import was dependent on the presence of a transit sequence, because removal of the 35-amino acid transit sequence resulted in no detectable import (not shown). Quantitation of the bands in Figure 3A indicated that 20% of the added AtCpNifS precursor was imported in this assay, compared with 25% for the Rubisco small subunit control precursor. We conclude that the AtCpNifS precursor contains plastid-targeting information. Immunoblot data (Fig. 3B) confirmed that the AtCpNifS protein is in the stroma.
Figure 3.
Chloroplast targeting of AtCpNifS. A, In vitro import of radiolabeled precursors. SSU, Precursor of the small subunit of ribulose-1,5-bisphosphate carboxylase oxygenase. CpNifS, Precursor of AtCpNifS. PR, Added precursor, 20% of the amount added to each of the lanes CI through T. Lanes CI, Chloroplasts recovered after import; CP, protease-treated chloroplasts; S, stromal fraction; and T, thylakoid fraction. Radiolabeled proteins were separated by SDS-PAGE and visualized with a PhosphorImager. Arrows indicate precursor (p) and mature (m) proteins. B, Western-blot analysis of chloroplast stroma (CS) and total leaf homogenate (H) of Arabidopsis. Protein loading was normalized based on the amount of chlorophyll in each fraction. Each lane corresponds to 1 μg of chlorophyll. Proteins were decorated as indicated with anti-AtCpNifS antiserum, preimmune serum, or antibodies against the cytosolic protein calmodulin as a control for cytosolic contamination.
AtCpNIFS mRNA Is Expressed in All Tissues
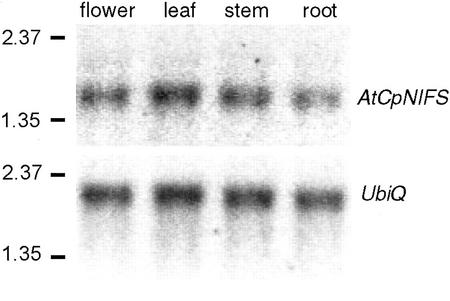
To analyze in which tissues AtCpNIFS is expressed at the mRNA level, we analyzed transcript levels in roots, stems, leaves, and flowers by northern blots (Fig. 4). Equal amounts of poly(A+) mRNA from each tissue was loaded on gel. The full AtCpNIFS coding sequence was used as a probe. A probe specific for the constitutively expressed ubiquitin was used as a control. A single transcript of about 1.8 kb is detected in all tissues. There were no apparent differences in AtCpNIFS mRNA levels between different tissues. These results indicate that AtCpNIFS is expressed in all major plant tissues.
Figure 4.
Northern analysis of AtCpNIFS expression. Equal amounts of poly(A+) mRNA were separated on an agarose gel and blotted to nitrocellulose. The blot was probed with an AtCpNIFS-specific probe, and hybridized bands were visualized with a PhosphorImager. The blot was stripped to remove the probe and reprobed with a probe for the UbiQ (ubiquitin) gene.
AtCpNIFS Encodes a PLP-Dependent Enzyme with Seleno-Cys Lyase and Cys Desulfurase Activities
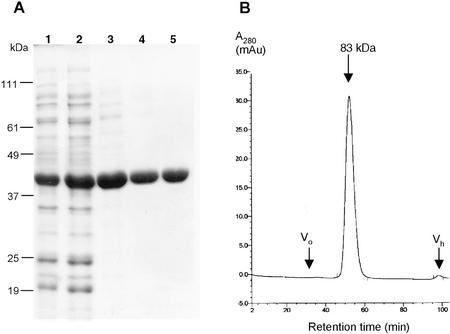
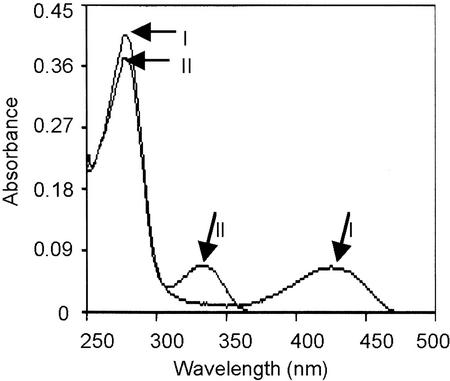
To facilitate the biochemical characterization of AtCpNifSp, the mature protein without its transit sequence was expressed in E. coli. The mature sequence was cloned under the control of the T7 promoter in expression vector pET11d. The resulting vector pMNFS-8 was introduced into codon+ E. coli cells. Induction of this strain grown in SOB (20 g L−1 tryptone, 5 g L−1 yeast extract, 0.5 g L−1 NaCl, 2.5 mm KCl, 10 mm MgCl2, and 10 mm MgSO4)medium with isopropyl-β-d-thiogalactopyranoside resulted in the production of a protein of about 43 kD. We estimate by comparing the staining intensities on Coomassie Brilliant Blue-stained protein gels that this protein constituted about 10% of the total cellular protein. However, upon disruption of the cells followed by a centrifugation at 12,000g, the majority of this protein was present in the pellet. Enzyme activity assays showed only a 2- to 3-fold increase in NifS activity in the lysate from cells overproducing AtCpNifS compared with strains harboring a control plasmid (the activity in control strains is attributed to IscS, which is constitutively expressed in E. coli). However, addition of 1 mm PLP to cultures before induction resulted in almost quantitative recovery of the overproduced protein in the soluble fraction and resulted in a 12- to 15-fold increase in NifS activity in the cell lysate compared with control strains. The recombinant protein was purified to homogeneity by a combination of ammonium sulfate fractionation and hydrophobic interaction and anion-exchange chromatography (Fig. 5A). Seleno-Cys lyase activity was used to monitor the purification (see Table I). Amino-terminal sequencing of the first six residues of the purified protein produced the sequence AAAASS, which matches the expected amino-terminal sequence of mature AtCpNifS. This result and the observation that IscS has a higher mobility on SDS-PAGE (not shown) identified the purified protein as recombinant AtCpNifS. The purified protein eluted with an apparent molecular mass of 83 kD from a calibrated S200 gel filtration column, suggesting that the protein is a dimer. Recombinant AtCpNifS had an absorption spectrum with a peak around 420 nm, characteristic of PLP-dependent enzymes (Fig. 6). Reduction of the protein with sodium borohydride resulted in a marked change in the absorption spectrum; the peak at 420 nm disappeared and at the same time an increase was seen in the A335. These data indicate the presence of a PLP cofactor in AtCpNifS (Mihara et al., 1997).
Figure 5.
Purification of AtCpNifS. A, Coomassie Brilliant Blue staining of an SDS-PAGE (12.5% [w/v]) gel with the following fractions from the protein purification. 1, Total cell lysate; 2, 70% (w/v) ammonium sulfate pellet; 3, phenyl-Sepharose-FF column eluate; 4, Hi-Prep Q16/10 column eluate; 5, Resource-Phe column eluate. B, Elution profile A280 of purified AtCpNifS from a calibrated S-200 Hi-prep 16/60 column. The void volume (V0) and hold-up (VH) volume are indicated with arrows. The buffer used was 50 mm KPi and 150 mm NaCl.
Figure 6.
Absorption spectra of AtCpNifS. The absorption spectrum of the protein at a concentration of 100 μg mL−1 was measured in 25 mm phosphate buffer at pH 7.5. Line I, Native protein; line II, 5 min after the addition of 1 mm sodium borohydride.
The specific activity of the purified enzyme (as measured by determining Se0 production, using 10 mm seleno-Cys as a substrate) was 3.7 units mg−1, which is comparable with other NifS-like proteins (Mihara et al., 1999). The enzyme had a comparable activity over a broad pH range from pH 7 to 9. The substrate specificity of AtCpNifS for both seleno-Cys and Cys substrates was determined by measuring Ala production, which is more sensitive than the lead acetate method. Activity was measured over a wide range of substrate concentrations. Apparent Km values were determined over the substrate range of 0.01 to 10 mm and are presented together with the specific activity at 10 mm substrate (see Table II). However, it should be noted that with AtCpNifS, we observed deviation from standard Michaelis-Menten kinetics at Cys concentrations above 10 mm. Non-Michaelis-Menten kinetics have also been observed with other NifS-like proteins (Mihara et al., 2000b), and this precludes the determination of a true Km and Vmax for such enzymes. AtCpNifS can use both substrates to produce Ala. However, somewhat surprisingly, purified AtCpNifS has a relatively low activity toward Cys. The observed discrimination factor for activity on seleno-Cys relative to Cys is 283. This is more comparable with that of CsdB (290-fold), the E. coli enzyme thought to function as a seleno-Cys lyase, than that of IscS (8.2-fold), the E. coli enzyme thought to function as a Cys desulfurase (Mihara et al., 1999).
DISCUSSION
AtCpNifS is the first characterized NifS-like protein from higher plants. The isolated protein forms a dimer and has activity toward both seleno-Cys and Cys, but the activity toward seleno-Cys is 283-fold higher for the purified enzyme. The absorption spectrum and the observed increased yield of soluble active protein when expressed in the presence of PLP indicate the presence of this cofactor in the enzyme. Dimer formation, the presence of a PLP cofactor, and activity toward both Cys and seleno-Cys are all properties of NifS-like proteins (Zheng et al., 1993). The cDNA for AtCpNIFS encodes a precursor with a functional N-terminal cleavable transit sequence. The protein was detected in the stroma of isolated chloroplasts. Judged from northern blotting, AtCpNIFS is expressed in all major tissue types. Consistent with that observation, the protein was detected in both roots and shoots by western blots (not shown). We assume that the protein expressed in roots is also present in plastids.
Mihara et al. (1997) classified NifS-like proteins in two groups based on sequence similarity. AtCpNifS is grouped in class 2 (Fig. 2B). It shares 60% sequence similarity with a close homolog, SsCsd3 in the cyanobacterium Synechocystis sp. PCC6803, one of three NifS-like proteins from this organism thought to be related to an evolutionary predecessor of the chloroplast. To our knowledge, the biochemical function of SsCsd3 has not been analyzed to date, although the gene has been reported to be essential for viability in this organism (Seidler et al., 2001). Other class 2 proteins include the E. coli CSD encoded by csdA and E. coli CsdB, a protein that seems to be a counterpart of the mammalian seleno-Cys lyase.
Another NifS-like protein (At5g65720) is most likely present in Arabidopsis mitochondria (Kushnir et al., 2001). This second NifS is a class 1 protein. Other well-characterized class 1 proteins include A. vinelandii NifSp (Zheng et al., 1993) and Iscs (Zheng et al., 1998), E. coli IscS (Zheng et al., 1998; Mihara et al., 2002), two proteins from the cyanobacterium Synechocystis sp. PCC6803 (Kato et al., 2000), and mitochondrial NifS-like proteins from mammals and yeast (Kispal et al., 1999). Each of these proteins seems to function as a Cys desulfurase, and for each of these proteins, a possible role in providing elemental S for Fe-S formation has been demonstrated.
Fe-S cluster formation can take place in isolated intact chloroplasts (Li et al., 1990; Pilon et al., 1995). Because Cys was identified as a source for Fe-S formation (Takahashi et al., 1986, 1990), a Cys desulfurase could well be involved in this process. The AtCpNifS identified in this paper has Cys desulfurase activity, albeit low in comparison with class 1 NifS-like proteins. However, the chloroplast harbors the key enzymes of the S assimilation pathway and is the site of Cys formation in plants (Leustek and Saito, 1999). It is feasible that the observed relatively low Cys desulfurase activity of the purified protein reflects a need for regulation of AtCpNifS with respect to available Cys if the protein serves a role as a Cys desulfurase for Fe-S formation. In bacteria, such regulation of the Cys pool may be achieved by the coordinated expression of the NIF/ISC genes with the expression of CysE genes localized close to the NIF/ISC clusters (Zheng et al., 1998). CysE genes encode the enzymes that catalyze the formation of O-acetyl-Ser, a direct precursor of Cys (Evans et al., 1991). NifU-like proteins may also play a regulatory role by accepting the S from the NifS, as was recently shown for E. coli IscU and IscS (Urbina et al., 2001; Kato et al., 2002). Interestingly, four genes (At4g01940, At5g49940, At4g25910, and At1g51390) that encode proteins with similarity to cyanobacterial NifU-like proteins and that possess putative chloroplast targeting domains are present in the genome of Arabidopsis (http://www.Arabidopsis.org). Reverse transcriptase-PCR analysis for some of these genes (not shown) and the presence of expressed sequence tag clones in the database indicate that these NIFU-like genes are expressed in Arabidopsis.
The activity toward Cys and seleno-Cys of purified AtCpNifS is most comparable with the activity of E. coli CsdB, the counterpart of mammalian seleno-Cys lyase, and a protein that may play a role in selenoprotein synthesis by providing a substrate for selenophosphate synthase (Mihara et al., 1999; Lacourciere et al., 2000). This latter molecule is then used as a substrate for the formation of Se-Cys-tRNA, which can be used in the translation of UGA opal codons in specific mRNAs encoding seleno-Cys-containing enzymes. Until recently, there was no evidence for the presence of specific selenoenzymes in the plant branch of the eukarya, but a specific selenoform of glutathione peroxidase has recently been found in the unicellular algae Chlamydomonas reinhardtii (Fu et al., 2002). This glutathione peroxidase is most likely active in the mitochondria but is encoded by a nuclear gene (Fu et al., 2002) and, thus, is translated in the cytosol. This situation raises the intriguing possibility that AtCpNifS could play a role in selenoprotein synthesis. A dual role in both S and Se metabolism has recently been shown for the E. coli IscS (Mihara et al., 2002).
In vivo approaches such as the analysis of plant lines expressing antisense RNA to silence the gene and the analysis of T-DNA insertion lines may shed light on the role of AtCpNifs in Fe-S formation and possibly Se metabolism in plants.
MATERIALS AND METHODS
Plant Growth and Fractionation
Arabidopsis (Ecotype Columbia-0) plants were routinely grown in a greenhouse on soil with supplementary light on a 15-h-light/9-h-dark cycle. For experiments requiring root tissue, plants were grown in sterile containers on one-half-strength Murashige and Skoog (1962) agar. Plant root and shoot protein samples used for the experiments in Figure 1A were obtained by grinding 1 g of plant tissue in liquid nitrogen with mortar and pestle, after which 2 mL of extraction buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol [DTT], and 0.5% [v/v] Triton X-100) was added. After 5 min of incubation on ice with occasional mixing, the mixture was centrifuged for 5 min at 12,000g, and the supernatant fraction was brought up to 10% (w/v) glycerol and frozen in aliquots until further analysis. Homogenates and intact chloroplasts used in Figure 1B were isolated from rosette leaves of plants as described by Rensink et al. (1998). The extent of cytosolic contamination of all chloroplast fractions used in Figures 1 and 3 was checked by measuring the specific activity of the cytosolic enzyme PEPC in both total leaf homogenate and stromal fractions as described (Pilon-Smits et al., 1990). The specific activity for PEPC in chloroplasts was at the detection limit and on average was less than 2% of the specific activity observed in homogenates. Therefore, it was concluded that the chloroplast fractions were free of cytosolic contamination. A stromal protein fraction was obtained from the chloroplasts essentially as described by Smeekens et al. (1986). Chloroplasts equivalent to 100 μg of chlorophyll were precipitated and lysed by resuspension in 300 μL of 10 mm Tris-HCl (pH 8). After 2 min on ice, an equal volume of 660 mm sorbitol and 100 mm HEPES-KOH (pH 8) was added. The thylakoid membranes were removed by centrifugation at 10,000g for 5 min and glycerol was added to the supernatant to a final concentration of 10% (w/v) before freezing in liquid nitrogen.
Enzyme Assays
NifS activity toward l-seleno-Cys and l-Cys was measured in 0.12 m Tricine-NaOH, pH 7.9, 50 mm DTT, and 0.2 mm PLP; incubation was at 37°C. Freshly dissolved substrate was used for every reaction. The substrate concentration was 10 mm unless noted otherwise. Elemental Se or S produced was measured with lead acetate as described (Esaki et al., 1982). Production of Ala from l-seleno-Cys and l-Cys was determined with a high performance amino acid analyzer (7300, Beckman Coulter, Fullerton CA). The specific activities are expressed in units per milligram of protein with 1 unit of enzyme defined as the amount that catalyzes the formation of 1 μmol of product in 1 min. Because very low activities were measured for purified AtCpNifS with the Cys substrate, we performed positive controls for the reactions with the same substrate mixtures by using the purified IscS enzyme from Escherichia coli, and these measurements confirmed the high activity on Cys previously reported for this enzyme (Mihara et al., 1999).
Database Searches and Sequence Alignments
Blast searches in the sequenced genomes of Arabidopsis and cyanobacteria were performed using E. coli IscS as an initial query sequence using the TBLASTN program on the NCBI web site (http://www.ncbi.nlm.nih.gov:80/BLAST/). Several sequences with significant similarity to IscS from cyanobacteria and one sequence from Arabidopsis were used in a second round of blast searches to find sequences encoding NifS-like proteins in Arabidopsis both in the NCBI and The Arabidopsis Information Resource (http://www.Arabidopsis.org/) databases. The Clustal program (Megalign program, DNASTAR, Inc., Madison, WI) was used to align all Arabidopsis sequences found in the first and second blast search with known and well-characterized group I and II NifS-like protein sequences from E. coli, Azotobacter vinelandii, Brewer's yeast (Saccharomyces cerevisiae), Synechocystis sp., and Mus musculus (Mihara et al., 1997). Only two Arabidopsis sequences could be grouped with the NifS-like proteins, and these contained the conserved Cys and Lys residues characteristic of NifS-like proteins. Subcellular localization and the transit peptide cleavage site were predicted with the TargetP program (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2000).
Cloning Procedures and Northern Blots
General cloning procedures were used (Ausubel et al., 1998). Total RNA was isolated from 2-week-old Arabidopsis seedlings using the Plant RNeasy kit (Qiagen USA, Valencia, CA) according to manufacturer's instructions. Ten micrograms of RNA was treated with DNaseI (Roche Diagnostics, Palo Alto, CA), and cDNA was synthesized using Moloney murine leukemia virus-reverse transcriptase (Promega, Madison, WI), according to instructions. The Expand polymerase enzyme kit (Roche Diagnostics) was used to amplify the Arabidopsis chloroplastic NIFS (AtCpNIFS) sequence from this cDNA by PCR. The following primers were used: NFS-out1 (5′-ACTTTGAAGACTCACTCTTGTTCATTCGT-3′) and NFS-out2, (5′-AGATCCAGCAGGAAGGTGTTCCACTTAT-3′). The annealing temperature was 64°C. The resulting 1.6-kb PCR product was isolated and used as template in a second round of PCR using again primer NFS-out1 and a nested primer (NFS-B2, 5′-TCGCCGGATCCACTTATTTGAAAGAGTTGAA-3′), which introduced a BamHI restriction site downstream of the coding sequence to clone the precursor sequence in the NcoI/BamHI sites of vector pET11d (Studier et al., 1990) producing plasmid pPNFS-a2. To subclone the mature sequence in pET11d, PCR was performed with another nested primer NFS-mat (5′-TCACCTCCATGGCCGCCGCCGCCTCCTC-3′), introducing an NcoI site, and NFS-B2; the PCR product was subcloned to produce plasmid pMNFS-8. The inserts in both plasmids were sequenced by the dideoxy dye termination method.
For northern analysis, total RNA was isolated from different tissues (roots, stems, leaves, and flowers) by the TRIzol reagent method (Invitrogen, Carlsbad, CA) and poly(A+) RNA was prepared using the oligotex mRNA kit (Qiagen USA) according to the manufacturer's instructions. Twelve micrograms of poly(A+) RNA was electrophoresed on a 1% (w/v) agarose gel containing 4% (w/v) formaldehyde, transferred to a nylon membrane, and probed with a 32P-labeled 1,300-bp AtCpNIFS cDNA. Prehybridization and hybridization were performed at 65°C in a solution containing 0.5 m sodium phosphate and 0.7% (w/v) SDS. After hybridization, the membrane was washed with 0.1× SSC and 1% (w/v) SDS at 65°C, and radiolabeled bands were visualized and quantified in a PhosphorImager (STORM, Molecular Dynamics, Sunnyvale, CA). The hybridized probe was removed by washing the membrane three times at 65°C, 5 min each, with stripping buffer (0.1× SSC, 1% [w/v] SDS, and 40 mm Tris, pH 7.5) containing 50% (v/v) formamide and once with stripping buffer without formamide according to Ausubel et al. (1998). The membrane was reprobed with a 32P-labeled 2-kb cDNA fragment specific for Arabidopsis ubiquitin. Prehybridization, hybridization, and washing were performed as before.
In Vitro Import Experiments
Plasmid pPNFS-a2 was linearized with BamHI before in vitro transcription using T7-polymerase (Epicenter Technologies, Madison, WI) according to the manufacturer's instructions. Plasmid pSP8114 containing the sequence for the precursor of the small subunit of ribulose bisphosphate carboxylase oxygenase (Lubben and Keegstra, 1986) was linearized with PstI before transcription with SP6 polymerase. Radiolabeled precursors were synthesized in a wheat germ lysate system in the presence of 35S-Met (specific activity 1,300 Ci mmol−1, Amersham Biosciences Inc., Piscataway, NJ) according to suggested protocols (Promega). Chloroplasts for import experiments were isolated from 10-d-old pea (Pisum sativum cv Little Marvel) seedlings and incubated with radiolabeled precursors as described (Pilon et al., 1992). The postimport thermolysin treatment, re-isolation of intact chloroplasts through 40% (v/v) Percoll cushions, and fractionation into the stroma and the thylakoid membranes were performed as described (Smeekens et al., 1986). The proteins in fractions from import experiments equivalent to 10 μg of chlorophyll were separated by SDS-PAGE (15% [w/v] gel). The gel was fixed in 7% (v/v) acetic acid, 25% (v/v) methanol, and dried, and radiolabeled proteins were visualized and quantified using a STORM PhosphorImager.
Expression and Purification of AtCpNifS from E. coli
The E. coli strain codon+ (Stratagene, La Jolla, CA) was used. This strain is optimized for the translation of eukaryotic genes by overexpressing tRNA species that match codons that are frequent in eukaryotes but rare in E. coli. Because the strain is a derivative of BL21(DE3), it can be induced to produce the T7 RNA polymerase by isopropyl-β-d-thiogalactopyranoside induction, resulting in expression of the gene under control of the T7 promoter (Studier et al., 1990). Cells harboring plasmid pMNFS-8 were grown at 30°C with vigorous shaking in SOB medium supplemented with 1 mm PLP and 50 μg mL−1 ampicillin. When the culture reached an OD600 nm of 0.5, isopropyl-β-d-thiogalactopyranoside was added to 0.4 mm to induce expression. After 16 h of further incubation, the culture was chilled on ice, and the cells were collected by centrifugation for 5 min at 5,000g at 4°C. From here on, all procedures were performed at 4°C except where mentioned.
The cells were briefly washed in 150 mm NaCl, collected by centrifugation, and suspended in 50 mm potassium phosphate (KPi), pH 7.5, and 1 mm EDTA, and frozen at −80°C in 1/50 of the original culture volume until lysis. Before lysis, the cell suspension was thawed and supplemented with 20 μm PLP, 4 μg mL−1 pepstatin, 0.1 mm phenyl-methyl-sulfonyl fluoride, and 2 mm DTT. The suspension was passed twice through a French press at 8,000 psi to disrupt the cells. The lysate was centrifuged for 10 min at 12,500g. The cleared lysate was stirred, and ammonium sulfate was slowly added to 30% saturation. After 30 min, precipitated proteins were removed by centrifugation for 10 min at 12,000g. To the supernatant, ammonium sulfate was added to 70% saturation, and precipitated protein was collected by centrifugation for 20 min, at 12,500g. Precipitated proteins were suspended in a 50 mm KPi, pH 7.5, 1 mm EDTA, and 1 m (NH4)2SO4. The sample was applied to a phenyl-Sepharose FF column (2.5 × 12 cm; Amersham Biosciences Inc.) equilibrated in 50 mm KPi, pH 7.5, 1 mm EDTA, and 1 m (NH4)2SO4 at a flow rate of 4 mL min−1. The column was washed with 100 mL of 50 mm KPi, pH 7.5, 1 mm EDTA, and 1 m (NH4)2SO4 and eluted with a 500-mL linear gradient of 1 to 0 m (NH4)2SO4; 8-mL fractions were collected. Peak fractions (as measured by the OD420 nm) were concentrated by the addition of (NH4)2SO4 to 70% saturation, followed by centrifugation. Concentrated proteins were suspended in 25 mm Tris, pH 7.5, and 1 mm EDTA and dialyzed against 3 L of 25 mm Tris, pH 7.5. The dialyzed protein was filtered through a 0.2-μm filter and applied at room temperature to a Q-Sepharose Hi-Prep 16/10 column (Amersham Biosciences Inc.) equilibrated in 25 mm Tris, pH 7.5, and connected to a Summit HPLC system (Dionex, Sunnyvale, CA). The column was eluted with a linear 0 to 400 mm NaCl gradient in the same buffer at a flow rate of 2 mL min−1. Fractions of 2 mL were collected, and elution was monitored by simultaneous detection of the OD at 420, 280, and 220 nm. AtCpNifS eluted at 200 mm NaCl, and the peak fractions were pooled and adjusted to 1 m ammonium sulfate. Final purification was performed at room temperature by repetitive HPLC on a 1-mL Resource-Phe column using a gradient from 1 to 0 m (NH4)2SO4 in 50 mm KPi, pH 7.5, and 1 mm EDTA at a flow of 1 mL min−1. AtCpNifS eluted at a concentration of 400 mm ammonium sulfate. The purified protein was dialyzed against 50 mm KPi, pH 7.5, 1 mm EDTA, and 10% (w/v) glycerol and stored in small aliquots at −80°C.
Antibodies
For antibody production, the purified AtCpNifS protein was dialyzed to 25 mm sodium phosphate, pH 7.0, and 150 mm NaCl. Polyclonal antibodies were raised in chickens at a commercial facility (Aves Labs, Tigard, OR). Antibodies for calmodulin were obtained from Zymed Laboratories (South San Francisco).
General Methods
Protein was assayed according to Bradford (1976). Chlorophyll was assayed according to Bruinsma (1961). Absorption spectra were recorded in a Beckman DU 530 spectrophotometer.
Table 1.
Purification of AtCpNifS
| Step | Total Protein | Total Activitya | Specific Activity | Yield |
|---|---|---|---|---|
| mg | units | units mg−1 | % | |
| Crude extract | 645 | 207 | 0.32 | 100 |
| Ammonium sulfate | 225 | 151 | 0.67 | 73 |
| Phenyl-Sepharose | 57 | 63 | 1.1 | 30 |
| Q-Sepharose | 17 | 36 | 2.1 | 17 |
| Resource-Phe | 12 | 44b | 3.7b | 21 |
Determined with l-selenocysteine as substrate by measuring Se0 formation using lead acetate.
Measured after dialysis to remove ammonium sulfate.
Table 2.
Kinetic properties and substrate specificity of AtCpNifS
| Substrate | Apparent Kma | Specific Activityb (at 10 mm Substrate) |
|---|---|---|
| mm | units mg−1 | |
| l-Cysteine | 0.11 | 0.0142 |
| l-Selenocysteine | 2.7 | 4.03 |
Determined over the substrate range of 0.01 to 10 mm.
Determined by measuring Ala production.
Footnotes
This work was supported by the U.S. National Science Foundation (NSF; grant nos. MCB–9982432 to E.A.H.P.-S. and MCB–0091163 to M.P.). The international collaborative research was supported by the NSF (supplement to grant no. MCB9982432) and by a Grant-in-aid for Joint Research Projects between Japan and the United States from the Japan Society for the Promotion of Science (to T.K.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010280.
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1998. [Google Scholar]
- Boeck A, Forschhammer K, Heider J, Baron C. Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem Sci. 1991;16:463–467. doi: 10.1016/0968-0004(91)90180-4. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bruinsma J. A comment on the spectrophotometric determination of chlorophyll. Biochim Biophys Acta. 1961;52:576–578. doi: 10.1016/0006-3002(61)90418-8. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Esaki N, Nakamura T, Tanaka H, Soda K. Selenocysteine lyase, a novel enzyme that specifically acts on selenocysteine: mammalian distribution and purification and properties of pig liver enzyme. J Biol Chem. 1982;257:4386–4391. [PubMed] [Google Scholar]
- Evans DJ, Jones R, Woodley PR, Wilborn JR, Robson RL. Nucleotide sequence and genetic analysis of the Azotobacter chroococcum nifUSVWZM gene cluster, including a new gene (nifP) which encodes a serine acetyltransferase. J Bacteriol. 1991;173:5457–5469. doi: 10.1128/jb.173.17.5457-5469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L-H, Wang X-F, Eyal Y, She Y-M, Donald LJ, Standing KG, Ben-Hayyim G. A selenoprotein in the plant kingdom: mass spectrometry confirms that an opal codon (UGA) encodes selenocysteine in Chlamydomonas reinhardtii glutathione peroxidase. J Biol Chem. 2002;277:25983–25991. doi: 10.1074/jbc.M202912200. [DOI] [PubMed] [Google Scholar]
- Kato S-I, Mihara H, Kurihara T, Takahashi Y, Tokumoto U, Yoshimura T, Esaki N. Cys-328 of IscS and Cys-63 of IscU are the sites of disulfide bridge formation in a covalently bound IscS/IscU complex: implications for the mechanism of iron-sulfur cluster assembly. Proc Natl Acad Sci USA. 2002;99:5948–5952. doi: 10.1073/pnas.082123599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S-I, Mihara H, Kurihara T, Yoshimura T, Esaki N. Gene cloning, purification, and characterization of two cyanobacterial NifS homologs driving iron-sulfur cluster formation. Biosci Biotechnol Biochem. 2000;64:2412–2419. doi: 10.1271/bbb.64.2412. [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir S, Babiychuk E, Storozhenko S, Davey MW, Papenbrock J, De Rycke R, Engler G, Stephan UW, Lange H, Kispal G et al. A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell. 2001;13:89–100. doi: 10.1105/tpc.13.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacourciere GM, Mihara H, Kurihara T, Yoshimura T, Esaki N, Stadtman TC. Escherichia coli NifS-like proteins provide selenium in the pathway for the biosynthesis of selenophosphate. J Biol Chem. 2000;275:23769–23773. doi: 10.1074/jbc.M000926200. [DOI] [PubMed] [Google Scholar]
- Lacourciere GM, Stadtman TC. The NIFS protein can function as a selenide delivery protein in the biosynthesis of selenophosphate. J Biol Chem. 1998;273:30921–30926. doi: 10.1074/jbc.273.47.30921. [DOI] [PubMed] [Google Scholar]
- Leustek T, Saito K. Sulfate transport and assimilation in plants. Plant Physiol. 1999;120:637–643. doi: 10.1104/pp.120.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-M, Theg SM, Bauerle CM, Keegstra K. Metal-ion-center assembly of ferredoxin and plastocyanin in isolated chloroplasts. Proc Natl Acad Sci USA. 1990;87:6748–6752. doi: 10.1073/pnas.87.17.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Kispal G. Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem Sci. 2000;25:352–356. doi: 10.1016/s0968-0004(00)01589-9. [DOI] [PubMed] [Google Scholar]
- Lubben TH, Keegstra K. Efficient in vitro import of a cytosolic heat shock protein into pea chloroplasts. Proc Natl Acad Sci USA. 1986;83:5502–5506. doi: 10.1073/pnas.83.15.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S, Dreyfuss BW. Posttranslational assembly of photosynthetic metalloproteins. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:25–51. doi: 10.1146/annurev.arplant.49.1.25. [DOI] [PubMed] [Google Scholar]
- Mihara H, Kato S, Lacourciere G, Stadtman TC, Kennedy RAJD, Kurihara T, Tokumoto U, Takahashi Y, Esaki N. The iscS gene is essential for the biosynthesis of 2-selenouridine in tRNA and the selenocysteine-containing formate dehydrogenase H. Proc Natl Acad Sci USA. 2002;99:6679–6683. doi: 10.1073/pnas.102176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara H, Kurihara T, Watanabe T, Yoshimura T, Esaki N. cDNA cloning, purification, and characterization of mouse liver selenocysteine lyase: candidate for selenium delivery protein in selenoprotein synthesis. J Biol Chem. 2000a;275:6195–6200. doi: 10.1074/jbc.275.9.6195. [DOI] [PubMed] [Google Scholar]
- Mihara H, Kurihara T, Yoshimura Y, Esaki N. Kinetic and mutational studies of three NifS homologs from Escherichia coli: mechanistic difference between l-cysteine desulfurase and l-selenocysteine lyase reactions. J Biochem. 2000b;127:559–567. doi: 10.1093/oxfordjournals.jbchem.a022641. [DOI] [PubMed] [Google Scholar]
- Mihara H, Kurihara T, Yoshimura T, Soda K, Esaki N. Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities: gene cloning, purification, and characterization of a novel pyridoxal enzyme. J Biol Chem. 1997;272:22417–22424. doi: 10.1074/jbc.272.36.22417. [DOI] [PubMed] [Google Scholar]
- Mihara H, Maeda M, Fujii T, Kurihara T, Hata Y, Esaki N. A nifS-like gene, csdB, encodes an Escherichia coli counterpart of mammalian selenocysteine lyase: gene cloning, purification, characterization and preliminary x-ray crystallographic studies. J Biol Chem. 1999;274:14768–14772. doi: 10.1074/jbc.274.21.14768. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:437–497. [Google Scholar]
- Pilon M, America T, van 't Hof R, de Kruijff B, Weisbeek P. Protein translocation into chloroplasts. In: Rothman SS, editor. Advances in Molecular and Cell Biology: 1. Membrane Protein Transport. Greenwich, CT: JAI Press; 1995. pp. 229–255. [Google Scholar]
- Pilon M, de Kruijff B, Weisbeek PJ. New insights into the import mechanism of the ferrodoxin precursor into chloroplasts. J Biol Chem. 1992;267:2548–2556. [PubMed] [Google Scholar]
- Pilon-Smits EAH, Hwang S, Lytle CM, Zhu Y, Tai JC, Bravo RC, Chen Y, Leustek T, Terry N. Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction and tolerance. Plant Physiol. 1999;119:123–132. doi: 10.1104/pp.119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits EAH, ‘t Hart H, van Brederode J. Phosphoenolpyruvate carboxylase in Sedum rupestre (Crassulaceae): drought-enhanced expression and purification. J Plant Physiol. 1990;136:155–160. [Google Scholar]
- Raven JA, Evans MC, Korb RE. The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosynth Res. 1999;60:111–149. [Google Scholar]
- Rensink WA, Pilon M, Weisbeek P. Domains of a transit sequence required for in vivo import in Arabidopsis chloroplasts. Plant Physiol. 1998;118:691–699. doi: 10.1104/pp.118.2.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler A, Jaschkowitz K, Wollenberg M. Incorporation of iron-sulphur clusters in membrane-bound proteins. Biochem Soc Trans. 2001;29:418–421. doi: 10.1042/bst0290418. [DOI] [PubMed] [Google Scholar]
- Smeekens S, Baurle C, Hageman J, Keegstra K, Weisbeek P. The role of the transit peptide in the routing of precursors toward different chloroplast compartments. Cell. 1986;46:365–375. doi: 10.1016/0092-8674(86)90657-4. [DOI] [PubMed] [Google Scholar]
- Stadtman TC. Selenium biochemistry. Annu Rev Biochem. 1990;59:111–127. doi: 10.1146/annurev.bi.59.070190.000551. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Mitsui A, Hase T, Matsubara H. Formation of the iron-sulfur cluster of ferredoxin in isolated chloroplasts. Proc Natl Acad Sci USA. 1986;83:2434–2437. doi: 10.1073/pnas.83.8.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Mitsui A, Matsubara H. Formation of the Fe-S cluster of ferredoxin in lysed spinach chloroplasts. Plant Physiol. 1990;95:97–103. doi: 10.1104/pp.95.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina HD, Silberg JJ, Hoff KG, Vickery LE. Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J Biol Chem. 2001;276:44521–44526. doi: 10.1074/jbc.M106907200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters: identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- Zheng L, White RH, Cash VL, Jack RF, Dean DR. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc Natl Acad Sci USA. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]