Abstract
Brassinosteroids (BRs) are steroidal plant hormones that are essential for growth and development. Although insights into the functions of BRs have been provided by recent studies of biosynthesis and sensitivity mutants, the mode of action of BRs is poorly understood. With the use of DNA microarray analysis, we identified BR-regulated genes in the wild type (WT; Columbia) of Arabidopsis and in the BR-deficient mutant, det2. BR-regulated genes generally responded more potently in the det2 mutant than in the WT, and they showed only limited response in a BR-insensitive mutant, bri1. A small group of genes showed stronger responses in the WT than in the det2. Exposure of plants to brassinolide and brassinazole, which is a specific inhibitor of BR biosynthesis, elicited opposite effects on gene expression of the identified genes. The list of BR-regulated genes is constituted of transcription factor genes including the phytochrome-interacting factor 3, auxin-related genes, P450 genes, and genes implicated in cell elongation and cell wall organization. The results presented here provide comprehensive view of the physiological functions of BRs using BR-regulated genes as molecular markers. The list of BR-regulated genes will be useful in the characterization of new mutants and new growth-regulating compounds that are associated with BR function.
Exogenous application of brassinosteroids (BRs) at nanomolar to micromolar concentrations to plants induces a wide range of physiological effects, including promotion of cell elongation and division, tracheary element differentiation, gravitropic tissue bending, ethylene biosynthesis, and stress resistance as well as retardation of abscission (Mandava, 1988; Clouse and Sasse, 1998; Sasse, 1999). BR-deficient or -insensitive mutants of Arabidopsis, pea (Pisum sativum), or tomato (Lycopersicon esculentum) exhibit pleiotropic phenotypes such as dwarfism, dark green leaves, reduced fertility, prolonged life span, and abnormal skotomorphogenesis (Szekeres and Koncz, 1998; Clouse and Feldmann, 1999; Müssig and Altmann, 1999; Bishop and Yokota, 2001). Recent molecular studies thus have indicated that promotion of cell expansion and regulation of photomorphogenic responses are among the most important roles of BRs. The mechanisms by which BRs exert these actions, however, are poorly understood. This study addresses both of these issues.
The expression of several genes has been shown to be regulated by BRs, especially within relatively short periods after BR treatment (Xu et al., 1995, 1996; Szekeres et al., 1996; Goetz et al., 2000; Hu et al., 2000; Müssig et al., 2000; Jiang and Clouse, 2001). To gain insight into the mechanism of BR action at the molecular level, we have now screened for genes in Arabidopsis that respond relatively rapidly to BR exposure. We exposed a BR-deficient mutant, det2, or wild type (WT) to either 10 nm brassinolide (BL) or mock treatment and then used a highly reproducible oligonucleotide-based array, the Arabidopsis Genome Array (Affymetrix, Santa Clara, CA; Lockhart and Winzeler, 2000) to compare the abundance of transcripts corresponding to >8,000 genes between the two treatment groups. Our results represent the most comprehensive study to date of the BR-regulated genes.
RESULTS AND DISCUSSION
Identification of BR-Regulated Genes in the det2 Mutant and WT
We exposed the BR-deficient mutant, det2, to either 10 nm BL or a mock treatment for both 15 min and 3 h and then compared the abundance of >8,000 gene transcripts between the two treatment groups using the Affymetrix Arabidopsis Genome Array. Hybridization was performed with biotin-labeled cRNA samples prepared from different plant samples in three independent experiments. The fold change (FC) values, which represent ratios of hybridization signals (the average differences [AvDf]) between mock- and BL-treated plants, were calculated using Microarray Suite software (Affymetrix). Down-regulated genes (Table I) and up-regulated genes (Table II) are listed with their FC values, which were obtained with or without signal amplification using an anti-streptavidin antibody. No genes responded in a reproducible manner to a 15-min BR treatment.
Table I.
Genes down-regulated by BRs in the det2 mutant
| Gene Name | Accession No. | Function or Comment | Affymetrix No. |
det2
|
WT
|
||
|---|---|---|---|---|---|---|---|
| Amplified
|
No-Amp.
|
Amplified
|
No-Amp.
|
||||
| FC ± se | FC ± se | FC ± se | FC ± se | ||||
| DWF4/CYP90B1 | AF044216 | BR biosynthesis (P450) | 13870_at | −7.5 ± 0.76 | −3.4 ± 0.29 | −2.8 ± 0.40 | −1.8 ± 0.10 |
| CPD/CYP90A1 | X87367 | BR biosynthesis (P450) | 16042_s_at | −2.5 ± 0.66 | −3.7 ± 0.37 | −4.0 ± 1.30 | −4.0 ± 0.40 |
| ROT3/CYP90C1 | AB008097 | Leaf polar elongation (P450) | 16535_s_at | −4.5 ± 0.45 | −3.8 ± 0.17 | −2.1 ± 0.10 | −2.3 ± 0.05 |
| CYP71B3 | D78602 | Expression in older leaves (P450) | 17039_at | −2.6 ± 0.20 | −2.3 ± 0.49 | −2.3 ± 0.80 | −1.3 ± 0.25 |
| PIF3 | AF100166 | PIF3 | 14630_s_at | −2.1 ± 0.07 | −2.1 ± 0.09 | −1.1 ± 0.05 | −0.1 ± 1.05 |
| NIA1/NR1 | X13434 | Nitrate reductase (cytokinin-regulated type) | 14240_s_at | −3.0 ± 0.62 | −4.0 ± 0.32 | −4.2 ± 0.15 | −4.6 ± 0.05 |
| XTR7 | U43489 | Putative xyloglucan endotransglycosylase | 15178_s_at | −3.1 ± 0.46 | −3.5 ± 0.55 | −1.7 ± 0.05 | −1.5 ± 0.00 |
| AAP3 | X77499 | Amino acid transporter (His uptake complement) | 12372_at | −4.5 ± 0.59 | −4.5 ± 0.99 | −2.0 ± 0.10 | −2.1 ± 0.05 |
| AAP4 | X77500 | Amino acid transporter (His uptake complement) | 16522_at | −2.3 ± 0.12 | −2.4 ± 0.35 | −1.7 ± 0.10 | −2.0 ± 0.05 |
| AtKUP1 | AF029876 | High-affinity potassium transporter | 16119_s_at | −6.5 ± 0.56 | −6.3 ± 0.50 | −2.3 ± 0.00 | −2.2 ± 0.30 |
| AKT2 | U40154 | Potassium channel | 16163_s_at | −4.4 ± 0.71 | −2.6 ± 0.23 | −2.5 ± 0.10 | −1.7 ± 0.00 |
| AtNAS1 | AB021934 | Nicotianamine synthase | 20547_at | −3.3 ± 0.12 | −3.3 ± 0.31 | −2.2 ± 0.20 | −2.3 ± 0.25 |
| AtmybL2 | Z68157 | Leaf-specific myb-related protein | 20362_at | −2.9 ± 0.42 | −2.5 ± 0.33 | −2.1 ± 0.70 | −1.9 ± 0.55 |
| Athb-5 | X67033 | Athb family Leu-zipper protein | 20389_at | −2.4 ± 0.12 | −2.2 ± 0.31 | −2.4 ± 0.25 | −2.1 ± 0.00 |
| MS2-like protein | X99923 | Fatty acyl CoA reductase homolog | 19177_at | −2.9 ± 0.43 | −3.3 ± 0.35 | −8.7 ± 2.60 | −4.6 ± 2.10 |
| CYP78A6 | AC006418/F13A10.19 | P450 monooxygenase | 18190_at | −7.3 ± 0.73 | −2.9 ± 0.59 | −3.7 ± 0.50 | −1.5 ± 0.30 |
| CYP81H1 | Z99707/C7A10.50 | P450 monooxygenase | 20271_at | −3.5 ± 0.18 | −3.1 ± 0.59 | −2.6 ± 0.00 | −1.5 ± 0.35 |
| CYP710A | AC004481/F13P17.32 | P450 monooxygenase | 14856_at | −4.1 ± 0.20 | −4.4 ± 0.50 | −3.9 ± 0.05 | −3.4 ± 0.60 |
| CYP96A1 | AC002391/T20D16.19 | P450 monooxygenase | 19281_i_at | −3.2 ± 0.12 | −2.5 ± 0.65 | −2.0 ± 0.40 | −1.9 ± 0.55 |
| CYP96A9 | AL078620/F23D16.110 | P450 monooxygenase | 19730_at | −2.5 ± 0.23 | −2.2 ± 0.38 | −1.5 ± 0.00 | −1.6 ± 0.00 |
| PAP1/IAA26 | AF088281 | IAA gene family/putative phytochrome-associated protein | 16078_at | −3.4 ± 0.49 | −2.7 ± 0.07 | −2.6 ± 0.20 | −2.7 ± 0.25 |
| iaglu | U81293 | Putative indole-3-acetate β-d-glucosyltransferase | 16603_s_at | −3.1 ± 0.42 | −2.9 ± 0.24 | −1.3 ± 0.20 | −1.4 ± 0.15 |
| PIN7 | AF087820 | Putative auxin efflux carrier protein | 17576_at | −2.2 ± 0.06 | −2.2 ± 0.17 | −1.7 ± 0.00 | −1.6 ± 0.00 |
| MYB55 | AF176000 | MYB-like gene | 17977_at | −5.0 ± 0.37 | −4.5 ± 1.00 | −3.7 ± 0.95 | −2.4 ± 0.05 |
| BRD1 | AL049746/T23J7.130 | Aldose 1-epimerase like protein | 12998_at | −2.5 ± 0.20 | −2.3 ± 0.38 | −2.0 ± 0.00 | −1.7 ± 0.10 |
| BRD2 | AC006836/F19B11.20 | Putative sulfotransferase | 19127_at | −2.5 ± 0.29 | −2.5 ± 0.29 | −2.0 ± 0.10 | −2.2 ± 0.35 |
| BRD3 | AL049171/T25K17.30 | Putative caffeoyl-CoA O-methyltransferase | 19348_at | −2.3 ± 0.25 | −2.3 ± 0.26 | −1.2 ± 0.05 | 0.0 ± 1.15 |
| BRD4 | AL035528/F18A5.290 | Leu-rich repeat protein | 14110_i_at | −4.0 ± 0.35 | −1.9 ± 0.38 | −2.0 ± 0.10 | −1.3 ± 0.10 |
| BRD5/SAUR36a | AC002387/F4L23.28 | Auxin-inducible SAUR gene homolog | 14448_at | −4.4 ± 0.85 | −2.9 ± 0.70 | −3.0 ± 0.20 | −1.7 ± 0.25 |
| BRD6 | AL049659/T29H11.120 | Putative DNA-binding protein (BTB domain) | 19977_at | −4.1 ± 0.47 | −3.8 ± 0.50 | −2.1 ± 0.30 | −2.3 ± 0.50 |
| BRD7 | AC002335/T01O24.18 | MYB-like gene | 18780_at | −4.2 ± 0.82 | −2.7 ± 0.15 | −2.2 ± 0.45 | −1.2 ± 0.05 |
| BRD8 | AC007659/T14P1.14 | GATA-type zinc finger protein | 13168_i_at | −2.2 ± 0.13 | −2.2 ± 0.06 | −1.6 ± 0.05 | −1.7 ± 0.35 |
| BRD9 | Z99708/C7A10.580 | Unknown | 19221_at | −3.2 ± 0.15 | −3.1 ± 0.23 | −2.4 ± 0.40 | −2.4 ± 0.15 |
| BRD10 | AL035605/F19F18.30 | Unknown | 19398_at | −2.9 ± 0.18 | −2.9 ± 0.15 | −2.2 ± 0.05 | −2.3 ± 0.10 |
| BRD11 | AC006224/MFL8.8 | Unknown | 20174_at | −5.2 ± 0.27 | −4.2 ± 0.46 | −3.6 ± 0.45 | −2.4 ± 0.15 |
| BRD12 | AC003033/T21L14.19 | Unknown | 19889_at | −2.6 ± 0.22 | −2.5 ± 0.28 | −0.1 ± 1.35 | −0.1 ± 1.15 |
When genes have names on publications or on database, we adopted their names. Otherwise, we designated genes from BRD1 to BRD12. Amplified and No-Amp., Before and after signal amplification with antibody, respectively.
Gene name for SAUR36 is from Hagen and Guilfoyle (2002).
Table II.
Genes up-regulated by BRs
| Gene Name | Accession No. | Function or Comment | Affymetrix No. |
det2
|
WT
|
||
|---|---|---|---|---|---|---|---|
| Amplified
|
No-Amp.
|
Amplified
|
No-Amp.
|
||||
| FC ± se | FC ± se | FC ± se | FC ± se | ||||
| IAA3/SHY2 | U18406 | Auxin-inducible nuclear protein/suppressor of phyB | 13301_at | 2.4 ± 0.42 | 2.7 ± 0.29 | 1.3 ± 0.10 | 1.3 ± 0.10 |
| IAA5 | U18407 | Auxin-inducible nuclear protein | 13660_i_at | 16.8 ± 4.64 | 7.8 ± 1.02 | 3.3 ± 0.35 | 1.8 ± 0.00 |
| SAUR-AC1/−15 | S70188 | Small auxin up RNA from Arabidopsis Columbia | 12608_i_at | 9.9 ± 0.55 | 9.6 ± 1.22 | 3.3 ± 0.00 | 3.7 ± 0.25 |
| IAA19 | U49075 | Member of IAA gene family/IAA1 binding protein | 13296_at | 3.1 ± 0.25 | 2.9 ± 0.15 | 1.8 ± 0.00 | 2.1 ± 0.20 |
| BAS1/CYP72B1 | AC003105/F18A8.8 | Activation-tagged suppressor of phyB | 12543_at | 3.8 ± 0.27 | 4.2 ± 1.16 | 2.7 ± 0.05 | 3.1 ± 0.00 |
| TCH2 | AF026473 | Touch-induced gene 2/calmodulin-related protein | 18300_at | 2.5 ± 0.19 | 2.7 ± 0.24 | 1.9 ± 0.15 | 1.8 ± 0.05 |
| TCH4 | AF051338 | Touch-induced gene 4/xyloglucan endotransglycosylase | 16620_s_at | 8.8 ± 0.81 | 16.8 ± 4.62 | 6.5 ± 0.35 | 8.2 ± 0.75 |
| XTR6 | U43488 | Xyloglucan endotransglycosylase-related protein 6 | 17533_s_at | 9.0 ± 1.10 | 5.7 ± 0.96 | 5.5 ± 0.45 | 1.4 ± 2.50 |
| MSS3 | AC004450/F14B2.23 | Suppressor of snf4/calmodulin-like protein (EF-hand) | 20689_at | 3.2 ± 0.35 | 3.8 ± 0.58 | 1.6 ± 0.15 | 1.8 ± 0.25 |
| ZAT7 | X98676 | Zinc finger protein | 15778_at | 4.2 ± 0.47 | 2.0 ± 0.15 | 1.4 ± 0.15 | 0.1 ± 1.20 |
| ATPA2 | X99952 | Root-specific peroxidase | 12356_at | 4.0 ± 2.09 | 3.0 ± 0.43 | 1.4 ± 0.20 | 1.7 ± 0.40 |
| KCS1 | AF053345 | Fatty acid elongase 3-ketoacyl-CoA synthase 1 | 17961_at | 2.7 ± 0.28 | 3.0 ± 0.29 | 1.3 ± 0.05 | 1.3 ± 0.10 |
| OPR1 | U92460 | 12-Oxophytodienoate reductase (jasmonate biosynthesis) | 18253_s_at | 2.0 ± 0.03 | 2.1 ± 0.03 | 1.4 ± 0.10 | 1.4 ± 0.10 |
| AtPMEpcrB | AF033206 | Pectin methylesterase homolog | 19267_s_at | 4.1 ± 0.32 | 4.4 ± 0.46 | 3.3 ± 0.15 | 3.5 ± 0.10 |
| ATP24a | Y11788 | Peroxidase | 18946_at | 2.3 ± 0.15 | 2.4 ± 0.30 | 0.0 ± 1.05 | 1.1 ± 0.05 |
| CYP94C1 | AC005824/F15K20.21 | P450 monooxygenase | 19288_at | 10.1 ± 3.32 | 4.0 ± 0.97 | 2.0 ± 0.20 | 1.7 ± 0.25 |
| CYP86A2 | AF013293/A_IG005I10.21 | P450 monooxygenase | 17966_at | 2.0 ± 0.09 | 2.2 ± 0.15 | 1.3 ± 0.00 | 1.3 ± 0.00 |
| BRU2/SAUR-25 | AL035528/F18A5.180 | Auxin-inducible SAUR gene family | 13395_at | 5.3 ± 0.63 | 2.6 ± 0.38 | 1.6 ± 0.05 | 1.6 ± 0.50 |
| BRU3/SAUR-9 | AL022373/T19K4.240 | Auxin-inducible SAUR gene family | 12947_at | 9.0 ± 0.21 | 9.2 ± 0.65 | 4.9 ± 0.35 | 4.9 ± 0.20 |
| BRU4/SAUR16 | AL035656/T9A14.140 | Auxin-inducible SAUR gene family | 13322_at | 5.8 ± 0.68 | 6.5 ± 1.00 | 2.2 ± 0.05 | 2.5 ± 0.30 |
| BRU5/SAUR-10 | AC006201/T27K22.12 | Auxin-inducible SAUR gene family | 13781_at | 3.0 ± 0.66 | 3.0 ± 0.06 | 1.6 ± 0.15 | 1.7 ± 0.20 |
| BRU6/GH3-2a | AL035601/F6G17.40 | Auxin-inducible GH3 gene family | 13565_at | 2.3 ± 0.17 | 2.2 ± 0.15 | 1.3 ± 0.05 | 1.8 ± 0.35 |
| BRU7/GH3-10 | AC005275/F4C21.36 | Auxin-inducible GH3 gene family | 13812_at | 2.2 ± 0.21 | 2.3 ± 0.15 | 1.7 ± 0.20 | 1.8 ± 0.40 |
| BRU8 | AC004512/T8F5.9 | Putative xyloglucan endotransglycosylase (TCH4 homolog) | 17960_at | 7.7 ± 0.48 | 8.0 ± 0.43 | 2.1 ± 0.35 | 2.0 ± 0.30 |
| BRU9 | AC007067/T10O24.17 | Putative endoxyloglucan transferase (EXGT-A3 homolog) | 19490_at | 22.0 ± 9.17 | 10.9 ± 2.54 | 5.2 ± 0.10 | 3.7 ± 0.00 |
| AtExp8 | AC002336/T2P4.4 | Putative expansin (expansin2-homolog) | 19660_at | 3.3 ± 0.57 | 3.7 ± 0.57 | 1.3 ± 0.05 | 1.3 ± 0.05 |
| BRU11 | AL035539/F22I13.170 | Putative expansin (pollen allergen protein homolog) | 19976_at | 6.2 ± 0.42 | 7.2 ± 0.92 | 2.0 ± 0.00 | 2.1 ± 0.10 |
| BRU12 | AC005169/F6F22.19 | Putative extensin with Leu-rich repeat (LRX1 homolog) | 18525_at | 3.3 ± 0.09 | 2.4 ± 0.45 | 1.3 ± 0.20 | −0.1 ± 1.10 |
| BRU13 | AC005698/T3P18.1 | Putative extensin with Leu-rich repeat (LRX1 homolog) | 18998_at | 2.2 ± 0.15 | 2.3 ± 0.29 | 2.0 ± 0.40 | 2.0 ± 0.50 |
| BRU14 | AL049608/T9E8.80 | Putative extensin with Leu-rich repeat (LRX1 homolog) | 20537_at | 2.8 ± 0.15 | 3.1 ± 0.15 | 1.6 ± 0.00 | 1.8 ± 0.00 |
| BRU15 | AC003027/F21M11.20 | Putative arabinogalactan protein (fasciclin-domain) | 16438_at | 0.6 ± 0.80 | 2.2 ± 0.10 | 1.6 ± 0.10 | 1.7 ± 0.10 |
| BRU16 | AL035601/F6GT17.100 | Putative arabinogalactan protein | 14947_at | 1.5 ± 0.28 | 2.5 ± 0.18 | 1.8 ± 0.00 | 1.8 ± 0.00 |
| BRU17 | AF060874 | Putative arabinogalactan protein (AtAGP4 homolog) | 15107_s_at | 1.7 ± 0.37 | 2.4 ± 0.24 | 1.5 ± 0.00 | 1.5 ± 0.00 |
| BRU18 | AF069298/T14P8.14 | Putative pectinesterase | 14612_at | 4.0 ± 0.18 | 4.2 ± 0.26 | 3.3 ± 0.40 | 3.6 ± 0.55 |
| BRU19 | AL022580/T5K18.200 | Putative pectinacetylesterase | 19905_at | 3.0 ± 0.28 | 3.6 ± 0.73 | 2.5 ± 0.45 | 3.3 ± 0.95 |
| BRU20 | AC004077/T31E10.15 | Endo-1,4-β-xylanase homolog | 15271_at | 2.7 ± 0.17 | 2.9 ± 0.12 | 1.5 ± 0.05 | 1.6 ± 0.10 |
| BRU21 | AL030978/M4122.170 | Ca2+-binding protein homolog | 15431_at | 3.1 ± 0.25 | 3.6 ± 0.49 | 2.3 ± 0.10 | 2.2 ± 0.00 |
| BRU22 | AL022198/F6I18.90 | Putative calmodulin-binding protein | 19857_at | 2.7 ± 0.20 | 2.7 ± 0.17 | 1.7 ± 0.10 | 2.0 ± 0.20 |
| BRU23 | AC005970/T6P5.14 | Putative protein kinase | 14030_at | 2.2 ± 0.10 | 2.4 ± 0.33 | 1.5 ± 0.15 | 1.7 ± 0.15 |
| BRU24 | AC002339/T11A7.8 | Putative receptor kinase (CLV1 homolog) | 14112_at | 3.4 ± 0.90 | 2.4 ± 0.32 | 1.7 ± 0.25 | 1.8 ± 0.10 |
| BRU25 | AC004238/F19I3.16 | Leu-rich repeat domain | 12251_at | 4.5 ± 0.67 | 4.7 ± 0.72 | 3.5 ± 0.85 | 2.6 ± 0.25 |
| BRU26 | AC002354/F6P23.7 | NAM/NAC gene homolog | 13806_at | 2.9 ± 0.44 | 3.0 ± 0.70 | 1.8 ± 0.30 | 1.6 ± 0.05 |
| BRU27 | AC006533/F20M17.23 | Ethylene-Regulated protein ER33 homolog | 15403_s_at | 3.7 ± 0.25 | 3.8 ± 0.29 | 1.9 ± 0.15 | 2.1 ± 0.20 |
| BRU28 | AF128396/T3H13.3 | Phosphate-induced gene phl-1 homolog | 14077_at | 3.2 ± 0.35 | 5.8 ± 0.29 | 3.6 ± 0.30 | 3.5 ± 0.30 |
| BRU29 | AL021711/F13C5.140 | Pro-rich protein (APG) homolog (GDSL-motif) | 16434_at | 2.8 ± 0.15 | 3.0 ± 0.03 | 2.4 ± 0.40 | 2.4 ± 0.35 |
| BRU30 | AL035353/F16A16.110 | Pro-rich protein (APG) homolog (GDSL-motif) | 17196_at | 3.9 ± 0.38 | 4.7 ± 0.48 | 1.9 ± 0.20 | 2.3 ± 0.25 |
| BRU31 | AC005917/F3P11.22 | Pollen-specific protein (SF21) homolog | 15892_at | 2.4 ± 0.18 | 2.7 ± 0.32 | 1.8 ± 0.15 | 1.8 ± 0.25 |
| BRU32 | AC005679/F9K20.9 | Carrot (Daucus carota) EP1 homolog | 17440_i_at | 2.7 ± 0.32 | 3.2 ± 0.55 | 1.6 ± 0.20 | 1.6 ± 0.25 |
| BRU33 | AC007060/T5I8.14 | Phosphatidylinositol/phosphatidylcholine transfer protein homolog | 12046_at | 3.6 ± 0.34 | 3.8 ± 0.25 | 1.9 ± 0.10 | 1.9 ± 0.15 |
| BRU34 | AP007269/A_IG002N01.14 | Unknown | 12953_at | 3.1 ± 0.35 | 3.3 ± 0.49 | 1.6 ± 0.05 | 1.8 ± 0.05 |
| BRU35 | AC005169/F6F22.17 | Unknown | 13916_at | 2.2 ± 0.12 | 3.6 ± 0.20 | 3.6 ± 0.20 | 3.8 ± 0.45 |
| BRU36 | AC005106/T25N20.6 | Unknown | 14913_at | 3.7 ± 0.34 | 3.4 ± 0.47 | 0.1 ± 1.05 | 0.0 ± 1.10 |
| BRU37 | AL049481/T5L19.20 | Unknown | 14951_at | 2.6 ± 0.26 | 2.8 ± 0.32 | 2.0 ± 0.10 | 2.2 ± 0.20 |
| BRU38 | AC002535/T30B22.26 | Unknown | 19880_at | 2.6 ± 0.15 | 2.9 ± 0.25 | 1.6 ± 0.20 | 1.6 ± 0.10 |
| BRU39 | AC007138/T7B11.21 | Unknown | 19992_at | 3.0 ± 0.21 | 3.4 ± 0.32 | 1.6 ± 0.10 | 1.6 ± 0.05 |
| BRU40 | AL022224/F1C12.90 | Unknown | 12027_at | 2.8 ± 0.32 | 2.5 ± 0.54 | 1.2 ± 0.05 | 0.0 ± 1.05 |
| BRU41 | AC000348/T7N9.22 | Unknown | 14443_at | 2.1 ± 0.17 | 2.1 ± 0.12 | 1.8 ± 0.20 | 2.0 ± 0.25 |
| BRU42 | AC006921/F2H17.17 | Unknown | 18885_at | 2.0 ± 0.12 | 2.1 ± 0.12 | 2.0 ± 0.00 | 2.2 ± 0.10 |
| BRU43 | AC004521/F411.31 | Unknown | 12584_at | 2.1 ± 0.15 | 2.8 ± 0.57 | 1.4 ± 0.10 | 1.5 ± 0.45 |
When genes have names on publications or on database, we adopted their names. Otherwise, we designated genes from BRU2 to BRU43. Ampified and No-Amp., Before and after signal amplification with antibody, respectively.
Gene names for SAUR and GH3 genes are from Hagen and Guilfoyle (2002).
A single GeneChip experiment, for example for the 3-h exposure experiment, revealed that 1,009 of 8,300 genes (12%) were up- or down-regulated by BL, as defined by a more than 2-fold difference in the FC values (before or after signal amplification). We used absolute call and difference call values calculated by the Microarray Suite software to exclude false-positive signals that resulted from cross-hybridization or noise. Taking into account these two parameters, 655 of 1,009 genes (65%) were excluded from the list for this experiment. Genes that were reproducibly regulated by BL in three independent experiments were compiled into a final list of 59 up-regulated genes and 36 genes down-regulated genes (Tables I and II). On the other hand, we also processed raw signal data (AvDf) from triplicate GeneChip analysis using Welch's approximate t test. This analysis identified 155 BL-regulated genes (as defined by more than 2-fold difference in the mean AvDf values) at a significance level of P < 0.05. Given that 84 of these 155 genes were included in the list of 95 BL-regulated genes that were identified in our initial analysis, we consider that our threshold is reliable and more stringent than the conventional approach based on Welch's approximate t test.
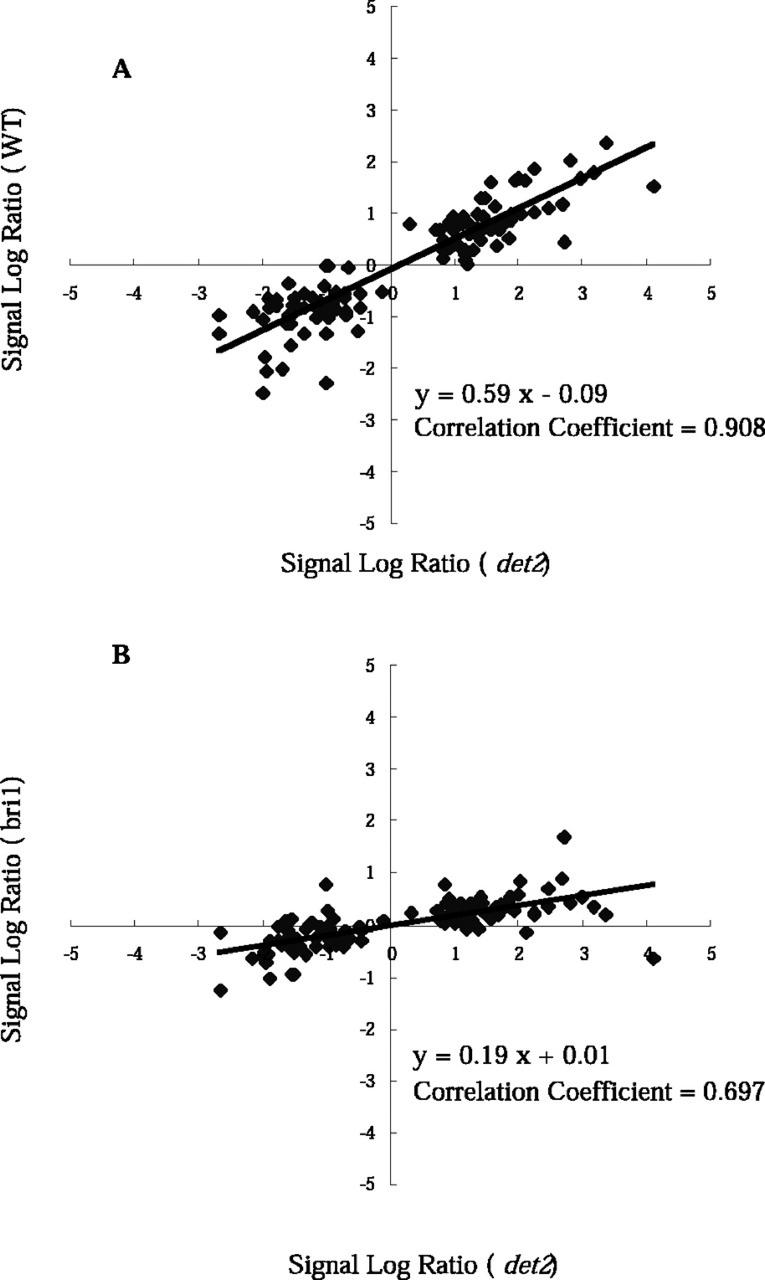
We also exposed a WT plant to either 10 nm BL or a mock treatment for 3 h and then compared the abundance of transcripts between the two treatment groups using the Affymetrix GeneChips. Fifty-one genes were identified as the BR-regulated genes in the WT. Fifteen of these identified as novel BR-regulated loci, because they did not appear in the BR-regulated genes in Table I or II. Interestingly, all of these genes were down-regulated by BL, and these genes generally showed milder responses to BL in the det2 mutant background than in the WT background (Table III). The signal log ratio values, which represent ratios of hybridization signals (AvDf values) between mock- and BL-treated plants on a log scale (base 2), were calculated using Microarray Suite software. A signal log ratio of 1, for example, indicates a 2-fold increase in the transcript level and −1 indicates a 2-fold decrease. The signal log ratio values from the WT experiments were plotted against those from the det2 mutant experiments for all of the genes listed in Tables I to III (Fig. 1A). In general, most of the genes responded to BL similarly in the WT and det2 mutant, although some genes responded poorly in the WT. The correlation coefficient for the WT and det2 experiments was 0.908, which indicates a strong positive correlation between the WT and det2 experiments. The inclination of the regression line in Figure 1A is 0.59. We therefore conclude that BR-regulated genes generally respond to BL in a similar fashion in the WT and det2 but that the det2 response to BL is more sensitive than that of the WT. We speculate that the det2 mutant has a stronger response to exogenous BL than WT, because the det2 mutant accumulates lower levels of endogenous BRs (Fujioka et al., 1997).
Table III.
Genes down-regulated by BRs in WT
| Gene Name | Accession No. | Function or Comment | Affymetrix No. |
det2
|
WT
|
||
|---|---|---|---|---|---|---|---|
| Amplified
|
No-Amp.
|
Amplified
|
No-Amp.
|
||||
| FC ± se | FC ± se | FC ± se | FC ± se | ||||
| CYP79B2 | AF069495 | Cytochrome P450 | 20479_i_at | −0.4 ± 0.70 | −0.4 ± 0.68 | −1.6 ± 0.35 | −2.2 ± 0.05 |
| CYP81D8 | AL035601/F6G17.20 | Cytochrome P450 | 14032_at | −2.0 ± 0.50 | −0.7 ± 1.07 | −3.7 ± 0.20 | −1.9 ± 0.15 |
| 4HPPD | AF047834 | 4-Hydroxyphenylpyruvate dioxygenase | 15669_s_at | −2.4 ± 0.30 | −2.3 ± 0.30 | −6.9 ± 1.60 | −3.0 ± 0.30 |
| CHI | M86358 | Chalcone isomerase | 20413_at | −1.4 ± 0.15 | −1.3 ± 0.12 | −2.2 ± 0.05 | −2.3 ± 0.45 |
| BRI1 | AF017056 | BRI1 | 20209_at | −1.8 ± 0.06 | −1.9 ± 0.13 | −2.0 ± 0.05 | −2.0 ± 0.20 |
| Eli3-1 | X67816 | Cinnamyl-alcohol dehydrogenase | 13242_at | −1.9 ± 0.25 | −1.9 ± 0.15 | −1.7 ± 0.00 | −2.1 ± 0.05 |
| BRD13 | AL021713/T9A21.60 | Putative protein | 13514_s_at | −1.9 ± 0.74 | −1.8 ± 0.03 | −4.0 ± 0.50 | −2.0 ± 0.35 |
| BRD14 | AC006085/F11M15.3 | Putative Ser/Thr protein kinase | 16878_at | −3.3 ± 0.56 | −1.8 ± 0.74 | −4.0 ± 0.10 | −1.8 ± 0.00 |
| BRD15 | AC012562/F17O14.23 | Osmotic stress-inducible kinase like-protein | 13246_at | −1.8 ± 0.39 | −0.6 ± 0.83 | −3.8 ± 1.10 | −1.9 ± 0.70 |
| BRD16 | AL049480/F14M19.60 | Putative pathogenesis-related protein | 18755_at | −2.1 ± 0.38 | −2.0 ± 0.68 | −2.6 ± 0.50 | −2.0 ± 0.25 |
| BRD17 | AL033545/F7K2.50 | Extensin-like protein | 12115_at | −2.2 ± 0.19 | −1.8 ± 0.10 | −2.4 ± 0.25 | −2.3 ± 0.05 |
| BRD18 | AC005662/F13H10.7 | Putative embryo-abundant protein | 14083_at | −2.0 ± 0.15 | −1.8 ± 0.27 | −2.4 ± 0.15 | −2.2 ± 0.15 |
| BRD19 | Y12776 | Late embryogenesis-abundant protein like-protein | 18594_at | −1.5 ± 0.09 | −1.6 ± 0.09 | −2.1 ± 0.05 | −2.0 ± 0.00 |
| BRD20 | AL021811/F10M6.70 | Putative protein | 14574_at | −2.8 ± 0.38 | −2.4 ± 0.52 | −1.8 ± 0.15 | −2.2 ± 0.15 |
| BRD21 | AC007661/T8P21.9 | Putative protein | 15487_at | −1.6 ± 0.12 | −1.6 ± 0.29 | −1.8 ± 0.55 | −2.1 ± 0.05 |
When genes have names on publications or on database, we adopted their names. Otherwise, we designed genes from BRD13 to BRD21. Amplified and No-Amp., before and after signal amplification with antibody, respectively.
Figure 1.
Comparison of BL treatment on the WT, bri1, and det2 seedlings with the use of GeneChip. A, The distribution of the signal log ratio values for WT (y axis) and det2 (x axis) are shown for the BR-regulated genes, listed in Tables I to III. B, The distribution of signal log ratio values for bri1 (y axis) and det2 (x axis) are shown for the BR-regulated genes, listed in Tables I to III. The signal log ratio represents the ratio of hybridization signals between BL- and mock-treated samples using a log (base 2) scale. A signal log ratio of 1 represents a gene that shows a 2-fold increase in expression by BL treatment; a signal log ratio of −1 represents a gene that shows a 2-fold reduction by BL treatment.
Analysis of BR Response in the bri1 Mutant
It has been shown that BRI1 is a critical component of the BR receptor (Wang et al., 2001). We have examined the effect of BR on a bri1-5 mutant, a weak mutant allele of BR-insensitive 1 (Noguchi et al., 1999). The bri1-5 seedlings were exposed to either 10 nm BL or a mock treatment for 3 h, and transcript abundance was analyzed using the Affymetrix GeneChip. Only three genes were identified to respond to BR in bri1-5 mutant, namely DWF4/CYP90B1, AtKUP1, and MS2-like genes. These genes are among the genes with the strongest response in the det2 and WT plants (Table I). All three genes showed weaker responses in the bri1 than in det2 background (data not shown). The signal log ratio values of the bri1 experiments were plotted against those of the det2 experiments for all the genes listed in Tables I to III (Fig. 1B). In the bri1 mutant, most of the genes responded to BL in a similar fashion to those in the det2 mutant. The correlation coefficient for the bri1 and det2 experiments is 0.697, indicating a positive correlation between the bri1 and det2 experiments. The inclination of the regression line in Figure 1B is 0.19. These results indicate that BR-regulated genes generally respond to BL similarly in the bri1 and det2, but the responses in bri1 are weaker than those in det2 for all BR-regulated genes. This observation provides good evidence that the BL responses of the genes listed in Tables I to III are dependent on the BRI1 gene. However, this dose not necessarily mean that the listed genes are all regulated via a BRI1-dependent-signaling pathway. It may be the case that the bri1 mutant is less sensitive to exogenously applied BRs, because the bri1 mutant accumulates enormous amounts of endogenous BRs, i.e. BL, castasterone, and typhasterol (Noguchi et al., 1999). Hu et al. (2000) suggested the existence of a BRI1-independent-signaling pathway. Moreover, there are several bri1 homologs in the Arabidopsis genome. However, the existence of a BRI1-independent-signaling pathway has yet to be verified.
Evaluation of Effects of Brassinazole (Brz), a BR-Specific Biosynthesis Inhibitor
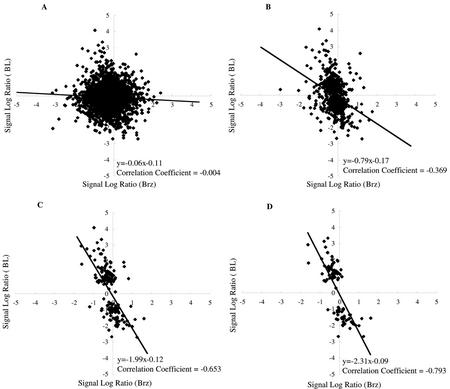
Brz, a synthetic chemical developed in our laboratory, is a triazole-type BR biosynthesis inhibitor that induces dwarfism (Asami and Yoshida, 1999). We analyzed BR-regulated genes to evaluate the effects of newly developed Brz, Brz220 (Sekimata et al., 2002), which has the strongest and the most specific effects on Arabidopsis among the Brz inhibitors (our unpublished data). WT seedlings were exposed to either 3 × 10−6 m Brz or a mock treatment for 3 h, and the abundance of transcripts was compared using GeneChip. The signal log ratio values from the Brz experiments were plotted against the BL experiments for all genes (Fig. 2A) or for BR-regulated genes listed in Tables I and II (Fig. 2D). The correlation coefficient for the Brz and BL experiments was −0.004 for all genes (n > 8,000), indicating that there was no significant correlation between the BL and Brz treatments. On the other hand, a correlation coefficient of −0.793 was calculated for the BR-regulated genes, suggesting a strong inverse correlation between the BL and Brz treatments. These observations indicate that our list of BR-regulated genes presented here is useful to evaluate new mutants as well as new growth-regulating compounds that are related to BR function. Our observations contrarily suggest that a short (3 h) and moderate concentration (3 × 10−6 m) of Brz treatment decreased the levels of endogenous active BRs within 3 h. This observation, together with our previous observation that Brz selectively interacts with a putative BR 22-hydroxylase (DWF4; Asami et al., 2001), also suggests rapid depletion of the BR-precursor pool, i.e. the BR precursors are metabolized to BL and/or degraded to the inactive form during the 3-h treatment of Brz.
Figure 2.
Comparison of BL and Brz treatment with the use of GeneChip experiment. The distribution of signal log ratio values for treatments with BL (y axis) and Brz (x axis) are shown. A, All of the genes (>8,000) on the GeneChip are plotted. B, Genes that are induced or reduced more than 2-fold in a single GeneChip experiment are plotted. C, Genes that are induced or reduced more than 2-fold in two of the three GeneChip experiments are plotted. D, The BR-regulated genes (induced or reduced more than 2-fold in three GeneChip experiments, listed in Tables I and II) are plotted. The signal log ratio represents the ratios of hybridization signals using a log (base 2) scale. A signal log ratio of 1 represents a gene whose expression is increased 2-fold by treatment with either BL or Brz, and a signal log ratio of −1 represents a gene whose expression is reduced 2-fold by treatment with either BL or Brz.
Modulation of the PIF3 (Phytochrome-Interacting Factor 3) Gene
Given that they are fixed in space, plants have evolved highly flexible programs to adapt to the changing environment. Light is one of the most important environmental signals regulating the programs of growth and development. Although interactions between light and hormonal signals have been studied extensively (von Arnim and Deng, 1996; Fankhauser and Chory, 1997; Kraepiel and Miginiac, 1997), the molecular mechanisms that underlie connectivity between the respective signaling pathways remain unclear. In addition, plant hormones have been widely regarded as signaling molecules that act downstream of light signal. BR-deficient or -insensitive mutants and plants treated with Brz, which is an inhibitor of BR biosynthesis, exhibit abnormal skotomorphogenesis or dark green leaves when grown in light (Fankhauser and Chory, 1997; Szekeres and Koncz, 1998; Asami and Yoshida, 1999; Clouse and Feldmann, 1999; Müssig and Altmann, 1999; Nagata et al., 2000). These observations have led to the proposal that light might alter either the concentration of BRs or the responsivity of cells to these steroids (Fankhauser and Chory, 1997; Kraepiel and Miginiac, 1997).
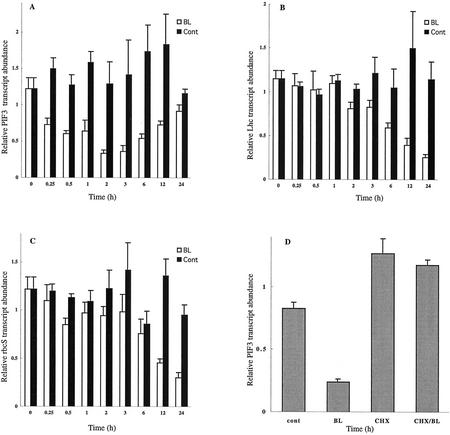
Our microarray analysis has now demonstrated that BL down-regulates the gene for PIF3 (Table I), a well-characterized transcription factor that functions at the upstream end of the light-signaling pathway (Ni et al., 1998). PIF3 is a basic helix-loop-helix protein that is localized to the nucleus and both interacts with the biologically active-type photoreceptors, Pfrs (Ni et al., 1999; Fairchild et al., 2000; Zhu et al., 2000), and binds to light-regulated promoters through the G-box sequence motif (CACGTG; Friedrichsen et al., 2000; Martinez-Garcia et al., 2000). Quantitative reverse transcription (RT)-PCR analysis revealed that repression of PIF3 expression was apparent within 15 min after BL treatment, and the expression was reduced by two-thirds within 2 to 3 h (Fig. 3A). Dose-response analysis revealed that inhibition of PIF3 expression by BL was apparent at 0.1 nm BL and was maximal at 100 nm BL (data not shown), indicating that BL regulates the expression of this gene at physiological concentrations.
Figure 3.
Regulation of PIF3, Lhcb1.3, and rbcS-1A gene expression by BL. Time courses of the inhibition of PIF3 (A), Lhcb1.3 (B), and rbcS-1A (C) gene expression by BL. Light-grown det2 seedlings were treated with 10 nm BL or were mock treated (cont) for the indicated times, and transcript abundance was analyzed by quantitative Taq-Man RT-PCR. D, Effects of cycloheximide (CHX) on PIF3 expression. Seedlings were treated with 100 μm CHX or mock treatment for 1 h, and then for an additional 3 h with 10 nm BL or mock treatment in the continued presence of CHX. The amount of PIF3 mRNA was determined by quantitative Taq-Man RT-PCR analysis. Transcript abundance levels are presented as relative values that are normalized with respect to the levels of 18S ribosomal RNA. Data are means ± se from three different plant samples.
On the basis of these observations, we examined the effects of BL on the abundance of transcripts derived from Lhcb1.3 (Jansson, 1999) and rbcS-1A (Krebbers et al., 1998), which encode light-harvesting chlorophyll a/b-binding protein (Lhc) and the small subunit of ribulose-1,5-bisphosphate carboxylase (rbcS), respectively. The promoter regions of both Lhcb1.3 and rbcS-1A contain G-box sequence motifs, which bind to PIF3 protein. The expression of each of these genes was repressed by BL treatment after a lag period of approximately 1 h; transcript levels were reduced by about two-thirds after 24 h (Fig. 3, B and C), at which time the chlorophyll content of BL-treated plants was about one-half that of mock-treated plants, without a significant change in the chlorophyll a/b ratio (Table IV). The relative temporal patterns of the expression profiles of PIF3 and Lhcb1.3 coincided with those of regulatory genes and their putative output genes in former microarray studies (e.g. Seki et al., 2001; Tepperman et al., 2001). Because light-induced expression of Lhcb1.3 (CAB3) was reduced in plants expressing PIF3 antisense RNA (Ni et al., 1998), our results suggest that the BL-induced repression of Lhcb1.3 expression is mediated, at least in part, by the effect of this BR on PIF3 expression. Our data also suggest that BRs modulate the light-regulated plant development by affecting PIF3 expression. In other words, BRs may act as regulators of the light-signaling pathway, in addition to or rather than functioning as downstream mediators of light signal transduction. On the other hand, it has been controversial whether BR-induced photomorphogenic responses, such as de-etiolation and dark green leaves, are secondary effects of the retarded cell elongation in BR-deficient plants (Bishop and Yokota, 2001). It is noteworthy that repression of PIF3 gene is apparent in 15 min of BR treatment. The speed of this response is similar to that of BR-biosynthetic genes and quicker than that of genes implicated in cell elongation or cell wall organization (discussed below).
Table IV.
Chlorophyll contents after BL treatment
| Chl a | Chl b | Chl a/b | |
|---|---|---|---|
| mg g−1 fresh wt. | |||
| BL | 0.26 ± 0.04 | 0.08 ± 0.01 | 3.22 ± 0.04 |
| Cont | 0.50 ± 0.03 | 0.16 ± 0.01 | 3.19 ± 0.06 |
Data are means ± se. Chl, Chlorophyll.
To determine whether the down-regulation of PIF3 by BL is dependent on de novo protein synthesis, we examined the effect of CHX, an inhibitor of cytosolic protein synthesis. The amount of PIF3 mRNA was unexpectedly moderately increased by treatment of plants with 100 μm CHX for 1 h (Fig. 3D). Furthermore, in plants exposed to CHX, PIF3 expression did not respond to subsequent treatment with BL. These results suggest that CHX might prevent the de novo synthesis of a short-lived repressor of PIF3 expression that acts at the transcriptional or posttranscriptional level. The results further suggest that BL might function upstream of this putative repressor. Recent studies have indicated the importance of short-lived repressors, whose degradation is mediated by the ubiquitin system (Estelle, 2001; Schwechheimer and Deng, 2001). Early auxin-inducible genes have been suggested to be regulated by the ubiquitin-mediated system (Gray and Estelle, 2000; Leyser, 2001; Ramos et al., 2001). The possible involvement of such regulatory system in BR-signaling pathway is presented in the following section.
Modulation of Early Auxin-Inducible Genes
Some effects of BRs have been thought to be related to auxin action (Mandava, 1988; Sasse, 1999). For example, the application of BRs together with auxins in the rice lamina inclination test or hypocotyl elongation test results in synergistic responses (Mandava, 1988; Sasse, 1999). However, although numerous physiological studies have addressed the interactions between BRs and auxins, little is known about the underlying molecular mechanisms. Clouse and colleagues performed comparisons of the physiological effects of BRs and auxins, as well as molecular analyses of auxin-inducible genes and auxin-insensitive mutants in soybean (Glycine max) and tomato. They concluded that the promotion of cell elongation by BRs is not likely mediated by the auxin-signaling pathway (Clouse et al., 1992, 1993; Zurek et al., 1994). On the other hand, our comprehensive microarray analysis, which encompasses most of the known auxin-inducible genes and various homologs of these genes, revealed a marked overlap in the BR- and auxin-signaling pathways in Arabidopsis.
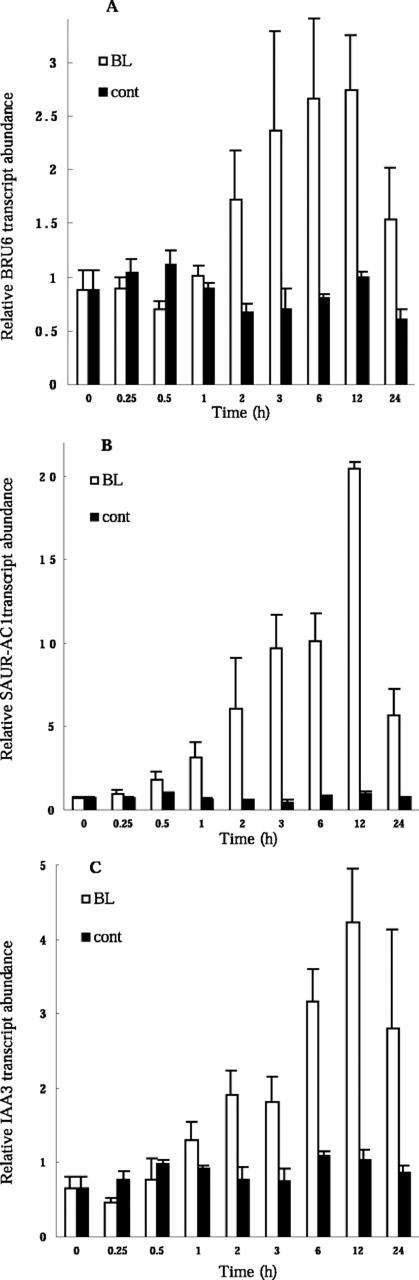
BL induced the expression of the early auxin-inducible genes indole-3-acetic acid (IAA)3, IAA5 (Abel et al., 1995), and SAUR-AC1 (Gil et al., 1994) and that of homologs IAA, GH3, and SAUR genes (Table II). Although the functions of the proteins encoded by early auxin-inducible genes remain controversial, these proteins are thought to serve as important factors in auxin signaling (Abel and Theologis, 1996; Gray and Estelle, 2000; Reed, 2001). Because the BL-induced genes include members of the three most well-characterized families of early auxin-inducible genes (Abel and Theologis, 1996), we chose one member from each family, namely, IAA3, SAUR-AC1, and a GH3 homolog (BRU6 in Table II), for further analysis. Before the analysis of the BL-induced expression, we confirmed that these genes exhibited a more than 2-fold increase in gene expression within 15 min of exposure to auxin (Abel et al., 1995; our unpublished data).
RT-PCR analysis revealed similar kinetics for the induction by BL of all three genes; the maximal level of expression was apparent between 6 and 12 h (Fig. 4, A–C). The IAA3 and GH3 homolog (BRU6) genes showed limited induction within 1 h of BL treatment, whereas SAUR-AC1 showed significant induction within 30 min. Because none of the genes was BR-regulated within 15 min, we conclude that SAUR-AC1 is one of the early BR-inducible genes. Clouse and colleagues (1992) reported that members of the SAUR and GH3 gene family were not rapidly induced during BR-promoted cell expansion but were induced by BR at later time points with different kinetics than when treated with auxin (Zurek et al., 1994). They concluded that these genes were not required for initial elongation in BR-treated tissue. On the other hand, we demonstrate here that some members of these gene families are induced more quickly by BL than previously described. The BL-induction kinetics of these genes are similar to those of genes involved in cell elongation and cell wall organization (see following section). It is not currently clear whether BRs activate auxin biosynthesis or activate a part of the auxin-signaling pathway or whether these genes represent shared signaling components of the auxin- and BR-signaling pathways.
Figure 4.
Induction of early auxin-inducible genes by BL. Kinetics of the induction of a GH3 homolog, BRU6 (A), SAUR-AC1 (B), and IAA3 (C) by BL are shown. Light-grown det2 seedlings were treated with 10 nm BL or were mock treated (cont) for the indicated times, after which transcript abundance was analyzed by Taq-Man RT-PCR. Transcript abundance levels are presented as relative values that are normalized with respect to the levels of 18S ribosomal RNA. Data are means ± se from three different plant samples.
In contrast to the BL induction of some early auxin-inducible genes, none of the late auxin-inducible genes is significantly induced by BL within the 3-h treatment period, even though a number of probes for such genes are present on the Arabidopsis Genome Array. It remains to be determined whether longer exposure to BRs results in the activation of late auxin-inducible genes. On the other hand, neither IAA1 nor IAA2, both of which are early auxin-inducible genes (Abel et al., 1995), was induced by exposure of Arabidopsis to BL for 3 h. The expression of PAP1/IAA26 and a SAUR homolog (BRD5), both of which are homologs of auxin-inducible genes, was repressed by BL treatment (Table I); the responses of these genes to auxin have not been known. These observations indicate that members of the IAA, GH3, and SAUR gene families can be classified into at least four groups: (a) those that are induced specifically by auxin (such as IAA1 and IAA2); (b) those that are induced both by auxin and by BRs (IAA3, IAA5); (c) those that are induced by BRs, but not by auxins (GH3 homolog [BRU7; auxin-insensitive data not shown]); and (d) those that are repressed by BRs (PAP1, SAUR homolog [BRD5]). Further analysis is needed to enable classification of members of the auxin-inducible gene families, depending on their dose- or time-dependent responses to auxin and BRs. Such classification should provide insight into functions of these genes, and it will further extend understanding involvement of these genes in the regulation of growth and development, including photomorphogenesis (Reed, 2001). Functional characterization of these genes should also provide insights into the functional divergence between auxins and BRs.
Control of Cell Elongation and Cell Wall Organization
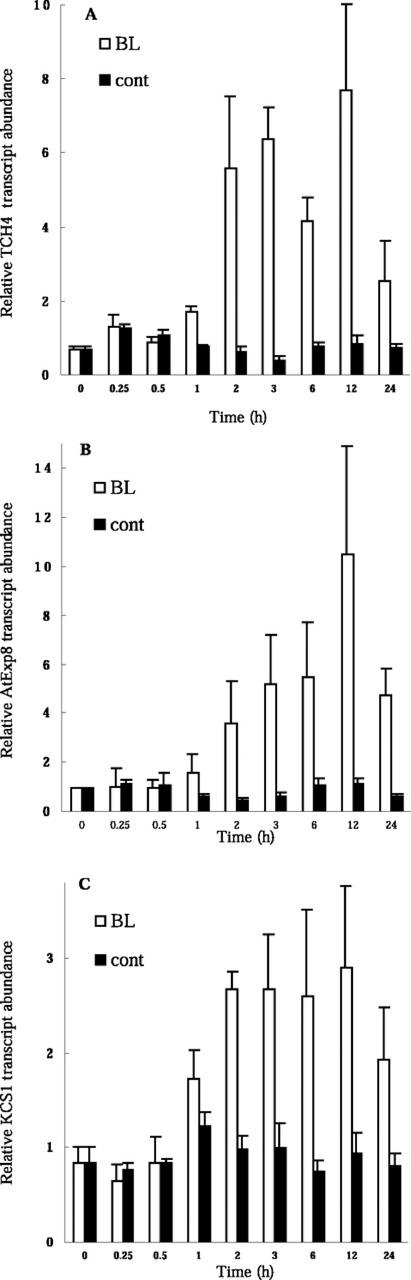
Microarray analysis revealed that BL induces the expression of various genes implicated in cell elongation or cell wall organization (Table II). As far as we are aware, this list contains the largest number of genes in this class shown to be regulated by a single signaling molecule. Most of these genes have not previously been shown to be under the control of any hormone. We chose three of the genes for further analysis of their induction kinetics: TCH4, which encodes xyloglucan endotransglycosylase (XET; Xu et al., 1995); a putative expansin gene (AtExp8) that is highly homologous to ZmExp2 (Im et al., 2000); and KCS1, which encodes a fatty acid elongase 3-ketoacyl-CoA synthase that functions in wax biosynthesis (Todd et al., 1999). RT-PCR analysis revealed that the induction of these three genes was apparent after a lag period of 30 min to 1 h, with maximal expression observed between 6 and 12 h (Fig. 5, A–C). The kinetics of induction of these genes were thus similar to those of BL-induced expression of the early auxin-inducible genes (Fig. 4) and to those of BR-induced cell elongation (Sasse, 1999), i.e. the lag period for the response to BRs is generally longer than that for responses to auxins. The lag period of BL-induced TCH4 expression was similar to that demonstrated previously (Xu et al., 1995).
Figure 5.
BL induction of genes that are implicated in cell elongation or cell wall organization. Kinetics of induction by BL of TCH4 (A), a putative expansin gene AtExp8 (B), and KCS1 (C) are shown. Light-grown det2 seedlings were treated with 10 nm BL or were mock treated (cont) for the indicated times, after which transcript abundance was analyzed by Taq-Man RT-PCR. Transcript abundance levels are presented as relative values that are normalized with respect to the levels of 18S ribosomal RNA. Data are shown as means ± se from three different plant samples.
On the other hand, the expression of XTR7, a member of the XET/XTH gene family, was down-regulated by BL (Table I), whereas the expression of this gene was previously shown to be independent of BR and auxin regulation (Xu et al., 1996). We demonstrated that BL reduced the abundance of XTR7 transcripts in a dose-dependent manner between 10−10 and 10−5 m by RT-PCR, but 10−6 m auxin increased the levels of XTR7 transcripts (data not shown). Our observations thus suggest the existence of multiple BR-signaling pathways that regulate XET/XTH gene family: One regulates the TCH4 gene in the manner observed for auxin induction, and the other regulates the XTR7 gene in a manner that is opposite to that observed with auxin. These observations pave the way for the exploration of the complex interactions between auxin- and BR-signaling pathways.
P450s and BR Metabolic Enzymes
In contrast to animals and yeast, plants possess a large family of P450 genes that contribute to the synthesis of secondary metabolites. The Arabidopsis genome thus contains nearly 300 P450 genes (Arabidopsis Genome Initiative, 2000). Recent studies into the specific functions of the proteins encoded by these genes have revealed that they are involved in the biosynthesis of a number of signaling molecules in plants. CYP85A (Bishop et al., 1999; Shimada et al., 2001), CYP90A (Szekeres et al., 1996), and CYP90B (Choe et al., 1998) participate in BR biosynthesis, CYP701A3 (Helliwell et al., 1998) and CYP88A3 (Helliwell et al., 2001) in gibberellin (GA) biosynthesis, and CYP79B2 (Hull et al., 2000) and CYP83B1 (Barlier et al., 2000; Bak et al., 2001) in auxin metabolism. It is important to know how BRs regulate P450 genes because it will help elucidate signaling network cross-talk between BRs and other signaling molecules. Furthermore, the oxygenation steps in the BR biosynthetic pathway, which are probably catalyzed by P450 enzymes, have yet to be characterized. Using the DNA array analysis, we examined the effects of BL on the expression of P450 genes, including those that contribute to BR metabolic pathways. Many P450 genes were regulated by BL (Fig. 6A). Such information will be useful in the future characterization of their specific functions. BL represses the expression of CYP79B2 and CYP81D8 (Table III). The CYP79B2 has shown to convert Trp to indole-3-acetaldoxime, the first step of Trp-independent IAA biosynthesis. We have recently identified that the CYP81D8 is the only auxin-inducible P450 gene by the comprehensive screening of early auxin-inducible genes using DNA microarray (S. Sawa and our unpublished data). These P450 genes may account for the physiological interactions between auxin and BRs.
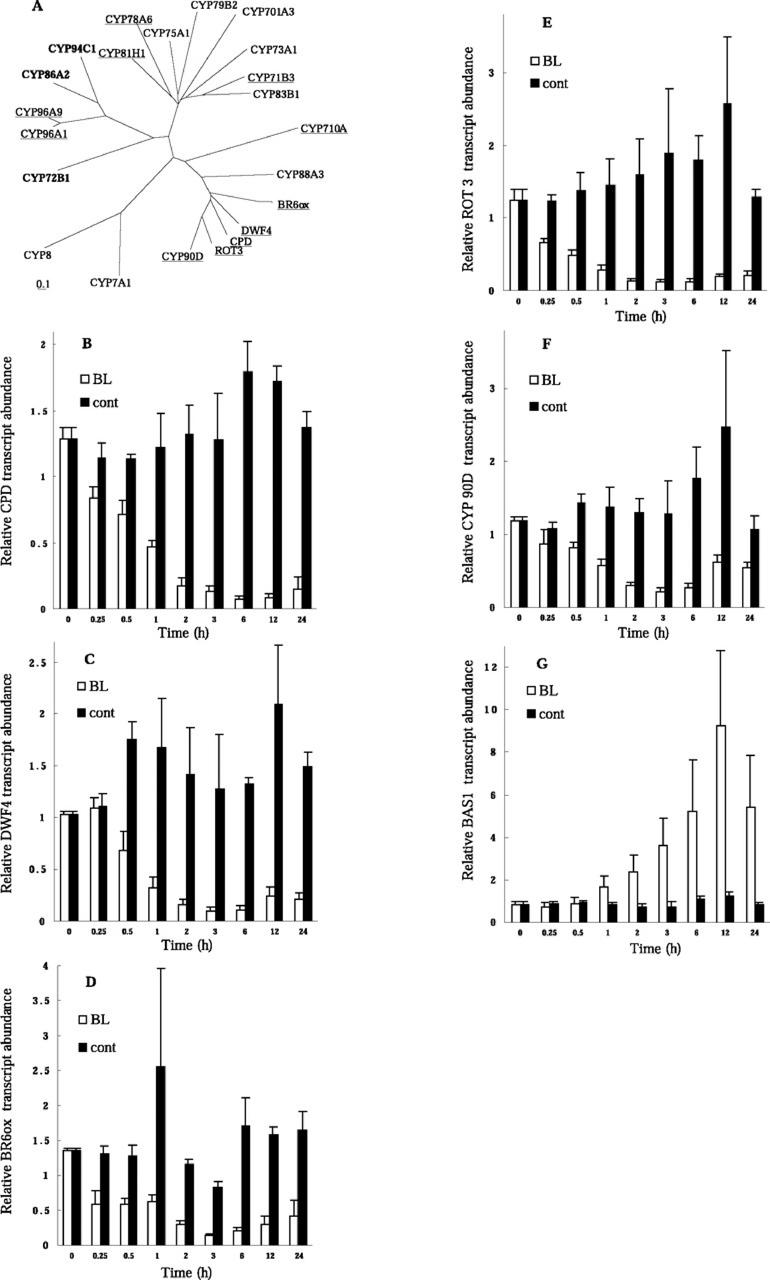
Figure 6.
Regulation of P450 genes by BL. A, Phylogenetic relationships among BR-regulated and other P450 genes. Proteins whose genes were shown to be up- or down-regulated by BL are indicated in bold and underlined, respectively (Tables I and II). BR6ox (Arabidopsis, CYP85A, AB035868), CPD (Arabidopsis, CYP90A, X87367), ROT3 (Arabidopsis, CYP90C, AB008097), DWF4 (Arabidopsis, CYP90B, AF044216), and CYP90D (Arabidopsis, AB066286) belong to the CYP85 or CYP90 families. CYP72B1 (Arabidopsis, BAS1, AC003105) is a suppressor of the phyB mutant. CYP88A3 (Arabidopsis, AtKAO1, AF318500) and CYP701A3 (Arabidopsis, GA3, AF047720) participate in GA biosynthesis. CYP83B1 (Arabidopsis, D78598) and CYP79B2 (Arabidopsis, AF069495) participate in auxin metabolism. The genes for CYP73A1 (Helianthus sp., cinnamate 4-hydroxylase, Z17369) and CYP75A1 (Petunia sp., flavonoid-3′, 5′-hydroxylase, D14588) were the first P450 genes to be identified functionally in higher plants and belong to the higher plant-specific group A of P450 genes. The accession numbers of the other P450 family members are D30718 (CYP8) and M93133 (CYP7A1). Kinetics of regulation of the CPD (B), DWF4 (C), BR6ox (D), ROT3 (E), CYP90D (F), CYP72, CYP85, CYP90, and BAS1 (G) genes by BL are shown. Light-grown det2 seedlings were treated with 10 nm BL or were mock treated (cont) for the indicated times, after which transcript abundance was analyzed by Taq-Man RT-PCR. Transcript abundance levels are presented as relative values that are normalized with respect to the levels of 18S ribosomal RNA. Data are shown as means ± se from three different plant samples.
Among P450 genes, a cluster consisting of the CYP85 and CYP90 families attracted our interest. Three members of these families have been shown to contribute to BR biosynthesis, and we found that the expression of the clustered genes was all repressed by BL treatment (Fig. 6, B–F). The gene cluster included ROT3/CYP90C, which functions in polar cell elongation in leaf cells (Kim et al., 1998), and CYP90D, a novel P450 gene (the full-length sequence of which has determined and has been submitted to GenBank; accession no. AB066286). These data suggest that all of the genes in the cluster encode BR-related enzymes. The observation that BL represses the expression of several BR biosynthetic genes indicates that the BR biosynthetic pathway is subject to feedback regulation at multiple points to ensure homeostasis of endogenous BRs. The rate of repression of CPD gene in Figure 4B was slower than that described in a previous report (Mathur et al., 1998), probably because of the low concentration of BL used in our experiments. On the other hand, our microarray experiments revealed that the expression of genes for enzymes that function upstream in the pathway of BR biosynthesis, including DWF7/STE1 (Choe et al., 1999), DIM/DWF1 (Klahre et al., 1998), and DET2 (Li et al., 1996), was not significantly affected by BL treatment (data not shown). The results indicate that the genes encoding upper step enzymes are less likely to be regulated by BL in a feedback regulatory manner. Furthermore, we observed that BAS1/CYP72B1, which encodes a repressor of a phyB mutant, was induced by BL treatment (Fig. 6G). Although CYP72B1 has been shown to catalyze C26 hydroxylation of BL (Neff et al., 1999), its physiological function is not fully understood. Our results imply that the expression of this gene was induced to promote the catabolism of the exogenously applied BL.
CONCLUSIONS
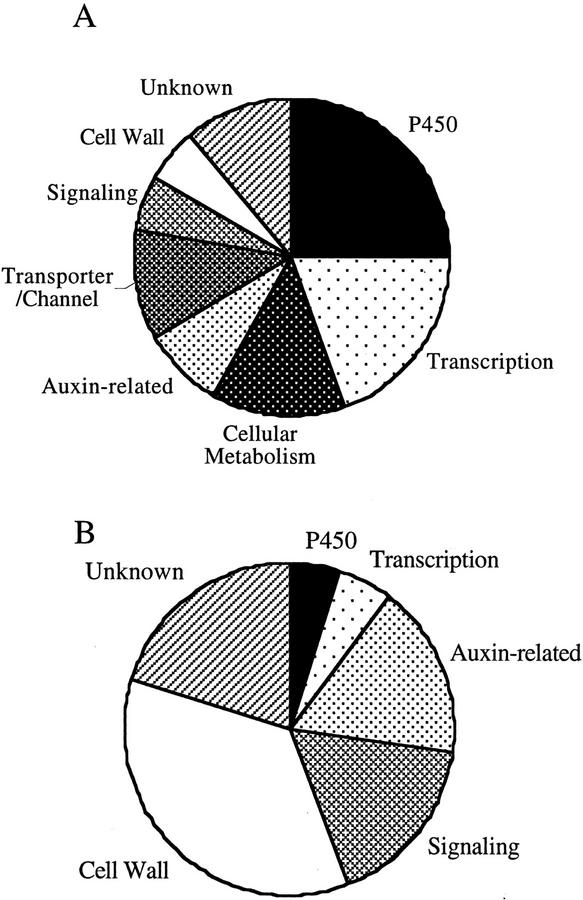
Very recently, three independent groups reported microarray analysis of BR-regulated genes (Hu et al., 2001; Müssig et al., 2002; Yin et al., 2002). The presented list of BR-regulated genes have significant differences with each other. Such differences may be attributable to different experimental conditions, but further studies may be required to account for the discrepancy. We present here the largest list of the BR-regulated genes, which may provide comprehensive view of the physiological functions of BRs using BR-regulated genes as molecular markers. P450 genes and transcription factor genes predominated among down-regulated genes (Fig. 7A). Auxin-related genes and genes that are implicated in cell elongation or cell wall organization predominated among up-regulated genes (Fig. 7B). We focused some of these genes for further characterization (Figs. 3–6). We did not discuss all of the genes shown to be regulated by BL in our microarray analysis (Tables I and II). These additional genes also provide insight into the actions of BRs and their underlying mechanisms. For example, the microarray results suggest possible mechanisms through which BRs interact with auxin. The expression of PIN7, a homolog of the PIN1 and PIN2 genes for putative auxin-efflux carrier proteins (Galweiler et al., 1998; Muller et al., 1998), was repressed by BL treatment, suggesting that BL might control auxin efflux. The expression of iaglu, which encodes a putative indole-3-acetate β-d-glucosyltransferase, was also repressed by BL, suggesting that BL might regulate auxin metabolism. BR signaling also appears to exhibit cross-talk with that of other plant hormones, including jasmonic acid (OPR1 in Table II; Biesgen and Weiler, 1999) and an unidentified ligand of a putative receptor kinase (BRU23 in Table II). Some of the remaining genes encoded proteins with Leu-rich repeats that lacked kinase domains, either with or without an extensin-like domain (Tables I and II). Genes that potentially related to signaling molecules, such as calmodulin-related proteins and their putative binding proteins, were regulated by BL (Tables I and II). Finally, BL controlled the expression of genes potentially regulating cell elongation, such as a K+ channel, a K+ transporter, and other transporters (Table I). BRs exert a wide variety of effects on both growth and development in plants. It is likely that many BR-regulated genes, presented here, contribute to these pleiotropic effects. Further analysis of the functions of these genes will provide insights into BR activities and will facilitate the understanding functions of the steroidal hormone in plants.
Figure 7.
Frequencies of up-regulated and down-regulated genes. A, Distribution of BR-down-regulated genes that are listed in Table I. B, Distribution of BR-up-regulated genes that are listed in Table II. The BR-regulated genes are classified into the functional categories based on their established or putative functions.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis ecotype Columbia was used as WT in this study. The Arabidopsis mutant det2-1 (Chory et al., 1991) was used as BR-deficient mutant. Seedlings were grown for 7 d at 22°C under continuous light in one-half-strength Murashige and Skoog (1962) liquid medium (Invitrogen, Carlsbad, CA) supplemented with 1.5% (w/v) Suc. The seedlings were then treated with 10 nm BL or mock-treated with dimethyl sulfoxide (final concentration, 0.1%, v/v), after which they were immediately frozen in liquid nitrogen and stored at −80°C until RNA isolation.
DNA Microarray Analysis
Total RNA was isolated from seedlings by the acid-guanidinium-phenol-chloroform method (Sambrook et al., 1989). Polyadenylated RNA was purified with the Oligotex-dT30 kit (Roche Diagnostics, Indianapolis) and converted into double-stranded cDNA with the use of a Super Script Choice cDNA Synthesis kit (Invitrogen) and with an oligo(dT)24 primer containing a T7 polymerase promoter site at its 3′ end (Amersham Biosciences AB, Uppsala). Biotin-labeled cRNA was generated from the double-stranded cDNA using the BioArray High Yield RNA transcript labeling kit (Enzo Diagnostics, New York) and was then purified with the use of the RNeasy RNA purification kit (Qiagen USA, Valencia, CA). Each cRNA sample (20 μg) was fragmented by incubation for 35 min at 94°C in fragmentation buffer (40 mm Tris-acetate [pH 8.1], 100 mm potassium acetate, and 30 mm magnesium acetate). The hybridization mixture comprised 15 μg of fragmented cRNA in 300 μL of a solution containing 100 mm MES, 1 m [Na+], 20 mm EDTA, 0.01% (v/v) Tween 20, 0.1 mg mL−1 herring sperm DNA, 0.5 mg mL−1 acetylated bovine serum albumin, and control cRNA (Eukaryotic Hybridization Control Kit, Affymetrix). Portions (200 μL) of each mixture were subjected to hybridization with the Arabidopsis Genome Array (Affymetrix) for 16 h at 45°C with rotation at 60 rpm. Each array was then washed consecutively with non-stringent wash buffer (6× SSPE [Sambrook et al., 1989], 0.01% [v/v] Tween 20, and 0.005% [v/v] Antifoam) and stringent wash buffer (100 mm MES, 0.1 m [Na+], and 0.01% [w/v] Tween 20). Hybridization complexes were then detected by consecutive exposure to phycoerythrin-streptavidin (Molecular Probes, Eugene, OR), biotinylated antibodies to streptavidin (Vector Laboratories, Burlingame, CA), and phycoerythrin-streptavidin, after which each array was washed again with non-stringent wash buffer. All washing and staining procedures were performed with a Fluidics Station 400 (Affymetrix). The array was scanned by a confocal microscope scanner (HP Genome Array Scanner, Affymetrix) at a wavelength of 570 nm.
DNA microarray technology is developing continually, but technical problems, such as a narrow signal dynamic range and aberrant detection of cross-hybridization or noise, remain to be resolved. We used the following methods to solve these problems. We chose GeneChip system (Affymetrix) as a DNA microarray system. The protocols for data analysis of the GeneChip system and the issues of sensitivity and quantitation have been described previously (Lockhart et al., 1996). In brief, each gene is represented on the array as a set of 16 oligonucleotide probes that match the sequence of the gene exactly. The specificity of hybridization is verified by inclusion on the array of the same set of probes each with a single-nucleotide mismatch in the center of its sequence. The difference between the hybridization signal obtained with the matching set of probes and that obtained with the mismatched probes is proportional to the abundance of the corresponding transcript and is calculated as the AvDf value. Analysis of absolute and differential gene expression was performed with the GeneChip software, Microarray Suite (v4.0, Affymetrix). AvDf values of highly abundant transcripts that are obtained with the standard GeneChip protocol are frequently affected by signal saturation when antibody amplification is used. To achieve a higher signal dynamic range, we scanned each chip before and after signal amplification using an anti-streptavidin antibody. Each chip was normalized relative to the sum of the AvDf values, and then each gene was compared between the control and BL-treated samples. Genes that were up- or down-regulated as reflected by a more than 2-fold difference in their AvDf values and were assigned to an “increase” or “decrease” in the difference call of the comparison analysis by Microarray Suite were identified. Furthermore, genes with an “absent” value in the absolute call of baseline data and with a decrease value in the difference call were excluded from the list. Genes with an absent value in the absolute call of experimental data and with an increase value in the difference call were conversely also excluded from the list. To ensure the reproducibility of results, we performed three independent experiments with different plant samples, and genes that showed the same responses in all three experiments were classified as genes regulated by BR (Tables I–III). The BR6ox and CYP90D genes were identified with the use of Arabidopsis GEM1 microarrays (Incyte Systems, Palo Alto, CA) containing 7,942 cDNA clones.
Taq-Man RT-PCR Analysis
Total RNAs were isolated as described above and then treated with DNase I. They were then converted to cDNAs using a Super Script first-strand synthesis system (Invitrogen). Quantitative RT-PCR was performed with the use of real-time Taq-Man technology (Holland et al., 1991) and a sequence detector (model 7700, Applied Biosystems, Foster City, CA). Gene-specific primers and Taq-Man probes (Table V) were used to analyze transcript abundance. The 18S ribosomal RNA was analyzed as an internal control and was used to normalize the values for transcript abundance. We performed three independent experiments with different plant samples. Each experiment was normalized relative to the median of the experiment, and then means and ses of three experiments were calculated.
Table V.
Primers and TaqMan probes used for RT-PCR
| Gene Name | Forward Primer Sequence | Reverse Primer Sequence | TaqMan Probe Sequence |
|---|---|---|---|
| CPD | CCCAAACCACTTCAAAGATGCT | GGGCCTGTCGTTACCGAGTT | TCTGCCATCTCCAAGGGTTGAAAGTGC |
| DWF4 | GTGATCTCAGCCGTACATTTGGA | CACGTCGAAAAACTACCACTTCCT | CAGCAAAACAACGGAGCGTCATCG |
| ROT3 | ATTGGCGCGTTCCTCAGAT | CAAGACGCCAAAGTGAGAACAA | CTCACCTCAAAGACCGGATCACTCGAGA |
| BR6ox | TGGCCAATCTTTGGCGAA | TCCCGTATCGGAGTCTTTGGT | ACCGAGTTTCTCAAACAAGGCCCCAAC |
| CYP90D | CTCATTACCCTTGCCGTCAAA | CAGCTTCATGTTTTCTTCCGTTAG | CCTCTCTGATTCTCCTGCTGCCCTCAAT |
| BAS1 | TTGGCTTCATACCGTTTGGC | TTACAGCGAGTGTCAATTTGGC | CGGAGTTCGTACATGCATTGGTCAGAATC |
| Lhcb1.3 | GGAGCTCAAGAACGGAAGATTG | GGTTCTCTATCGGTCCCTTACCA | TGGATTCTTCGTTCAAGCCATCGTCA |
| rbcS1A | GCTCTCTTCCGCTACTATGGTTG | AAGGCAGCGGAGGACTTAAGT | TCAGGCCACTATGGTCGCTCCTTTCA |
| TCH4 | CAAGAACATGGAGTCTCTAGGCACT | GTGAAAGGAGCTTTAGACCAATCG | ACTCGAGTCTTTGGAACGCTGATGATTGG |
| PIF3 | TCAGGCTCACCAAAGCTAAGC | CACACCAGCTCCACAACTTCA | AATCTGCTCAAGACAGGAACCCTTCTCCAC |
| GH3 homolog (BRU6) | CGTATATCCAACGGCGATTGT | CCAGCAGATGTTCCTGAGCTT | TCTTCTCACCCCATCACCGAGTTTCTCA |
| SAUR-AC1 | GAGGATTCATGGCGGTCTATG | GTTAAGCCGCCCATTGGAT | TGGTGCCGGTTTCATACTTAAACCAGCCT |
| IAA3 | AGCCTAAACCTTTGGCTTCTGA | GGTGATTGGATGCTCATTGGT | CTTGCACGTACATATGAACATCTCCCATGG |
| XTR7 | TCGACGAGTTTGACCTCACTTG | GTCCAGCGACAAAGACAGCATA | ACCACAGAGGCAAAATCTTCAACGGAGG |
| AtExp8 | CAACCATCACCGTCACAGCTA | TGAAGAGGAGGATTGCACCAA | AAACTTTTGCCCACCTAACCCTGGCCT |
| KCS1 | GCTCAAATACGTGAAGCTTGGA | CAGCACGGTTCCGGTTAAA | CAACTCTTGCAACGTGACCACCATTCTCT |
| 18S ribosomal RNA | CGGCTACCACATCCAAGGAA | GCTGGAATTACCGCGGCT | TGCTGGCACCAGACTTGCCCTC |
Sequence Analysis
DNA sequences were determined with an automated DNA sequencer (model 373A DNA Sequencing System, Applied Biosystems). Nucleotide sequences were compiled and analyzed with GENETYX-Mac software (Software Development Co. Ltd., Tokyo). The BLAST program (Altschul et al., 1990) was used to search for entries of homologous sequences in the DNA data bank of Japan. The ClustalW program on the server at DNA data bank of Japan was used to align amino acid sequences and to derive phylogenetic relations based on the neighbor-joining method (Saitou and Nei, 1987).
Determination of Chlorophyll Content
Acetone was added to the seedlings to bring the final concentration to 80%. After homogenization, the solution was centrifuged at 10,000g for 5 min, and chlorophyll was determined according to the method of Arnon (1949). Chlorophyll contents were normalized with respect to the fresh weight of the seedlings.
ACKNOWLEDGMENTS
We thank Drs. Joanne Chory and Kenneth A. Feldmann for providing mutants, Dr. Kiyotaka Okada for careful reading of the manuscript, Narumasa Miyauchi for technical assistance with quantitative RT-PCR and other molecular biological analyses, and Katsuhiko Sekimata for assistance with Brz preparation.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.011254.
LITERATURE CITED
- Abel S, Nguyen MD, Theologis A. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol. 1995;251:533–549. doi: 10.1006/jmbi.1995.0454. [DOI] [PubMed] [Google Scholar]
- Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts: polyphenol-oxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Mizutani M, Fujioka S, Goda H, Min YK, Shimada Y, Nakano T, Takatsuto S, Matsuyama T, Nagata N et al. Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J Biol Chem. 2001;276:25687–25691. doi: 10.1074/jbc.M103524200. [DOI] [PubMed] [Google Scholar]
- Asami T, Yoshida S. Brassinosteroid biosynthesis inhibitors. Trends Plant Sci. 1999;4:348–353. doi: 10.1016/s1360-1385(99)01456-9. [DOI] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R. CYP83B1, a cytochrome P450 at the metabolic branch paint in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell. 2001;13:101–111. doi: 10.1105/tpc.13.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA. 2000;97:14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesgen C, Weiler EW. Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta. 1999;208:155–165. doi: 10.1007/s004250050545. [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JD, Kamiya Y. The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA. 1999;96:1761–1766. doi: 10.1073/pnas.96.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Yokota T. Plants steroid hormones, brassinosteroids: current highlights of molecular aspects on their synthesis/metabolism, transport, perception and response. Plant Cell Physiol. 2001;42:114–120. doi: 10.1093/pcp/pce018. [DOI] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22a-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE et al. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of Det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Feldmann KA. Molecular genetics of brassinosteroid action. In: Sakurai A, Yokota T, Clouse S, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 163–190. [Google Scholar]
- Clouse SD, Hall AF, Langford M, McMorris TC, Baker ME. Physiological and molecular effects of brassinosteroids on Arabidopsis thaliana. J Plant Growth Regul. 1993;12:61–66. [Google Scholar]
- Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Zurek DM, McMorris TC, Baker ME. Effect of brassinolide on gene expression in elongating soybean epicotyls. Plant Physiol. 1992;100:1377–1383. doi: 10.1104/pp.100.3.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle M. Proteases and cellular regulation in plants. Curr Opin Plant Biol. 2001;4:254–260. doi: 10.1016/s1369-5266(00)00169-2. [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CAP, Li JM, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1255. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Li JM, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J et al. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galweiler L, Guan CH, Muller A, Wisman E, Mendgen K, Yephremov A, Palme K. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- Gil P, Liu Y, Orbovic V, Verkamp E, Poff KL, Green PJ. Characterization of the auxin-inducible Saur-Ac1 gene for use as a molecular-genetic tool in Arabidopsis. Plant Physiol. 1994;104:777–784. doi: 10.1104/pp.104.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Godt DE, Roitsch T. Tissue-specific induction of the mRNA for an extracellular invertase isoenzyme of tomato by brassinosteroids suggests a role for steroid hormones in assimilate partitioning. Plant J. 2000;22:515–522. doi: 10.1046/j.1365-313x.2000.00766.x. [DOI] [PubMed] [Google Scholar]
- Gray WM, Estelle M. Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem Sci. 2000;25:133–138. doi: 10.1016/s0968-0004(00)01544-9. [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–385. [PubMed] [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA. 2001;98:2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Sheldon CC, Olive MR, Walker AR, Zeevaart JAD, Peacock WJ, Dennis ES. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA. 1998;95:9019–9024. doi: 10.1073/pnas.95.15.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain-reaction product by utilizing the 5′→3′ exonuclease activity of thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YX, Bao F, Li JY. Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 2000;24:693–701. doi: 10.1046/j.1365-313x.2000.00915.x. [DOI] [PubMed] [Google Scholar]
- Hu YX, Wang ZK, Wang YH, Bao F, Li N, Peng ZH, Li JY. Identification of brassinosteroid responsive genes in Arabidopsis by cDNA array. Sci China Ser C Life Sci. 2001;44:637–643. doi: 10.1007/BF02879358. [DOI] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im KH, Cosgrove DT, Jones AM. Subcellular localization of expansin mRNA in xylem cells. Plant Physiol. 2000;123:463–470. doi: 10.1104/pp.123.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Jiang JR, Clouse SD. Expression of a plant gene with sequence similarity to animal TGF-beta receptor interacting protein is regulated by brassinosteroids and required for normal plant development. Plant J. 2001;26:35–45. doi: 10.1046/j.1365-313x.2001.01007.x. [DOI] [PubMed] [Google Scholar]
- Kim GT, Tsukaya H, Uchimiya H. The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 1998;12:2381–2391. doi: 10.1101/gad.12.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepiel Y, Miginiac E. Photomorphogenesis and phytohormones. Plant Cell Environ. 1997;20:807–812. [Google Scholar]
- Krebbers E, Seurinck J, Herdies L, Cashmore AR, Timko MP. Four genes in two diverged subfamilies encode the ribulose-1,5-bisphosphate carboxylase small subunit polypeptides of Arabidopsis thaliana. Plant Mol Biol. 1998;11:745–759. doi: 10.1007/BF00019515. [DOI] [PubMed] [Google Scholar]
- Leyser O. Auxin signalling: the beginning, the middle and the end. Curr Opin Plant Biol. 2001;4:382–386. doi: 10.1016/s1369-5266(00)00189-8. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Dong HL, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang CW, Kobayashi M, Horton H et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Mandava NB. Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:23–52. [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C et al. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998;14:593–602. doi: 10.1046/j.1365-313x.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- Muller A, Guan CH, Galweiler L, Tanzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998;17:6903–6911. doi: 10.1093/emboj/17.23.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant. 1962;15:473–498. [Google Scholar]
- Müssig C, Altmann T. Physiology and molecular mode of action of brassinosteroids. Plant Physiol Biochem. 1999;37:363–372. [Google Scholar]
- Müssig C, Biesgen C, Lisso J, Uwer U, Weiler EW, Altmann T. A novel stress-inducible 12-oxophytodienoate reductase from Arabidopsis thaliana provides a potential link between brassinosteroid-action and jasmonic-acid synthesis. J Plant Physiol. 2000;157:143–152. [Google Scholar]
- Müssig C, Fischer S, Altmann T. Brassinosteroid-regulated gene expression. Plant Physiol. 2002;129:1241–1251. doi: 10.1104/pp.011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N, Min YK, Nakano T, Asami T, Yoshida S. Treatment of dark-grown Arabidopsis thaliana with a brassinosteroid-biosynthesis inhibitor, brassinazole, induces some characteristics of light-grown plants. Planta. 2000;211:781–790. doi: 10.1007/s004250000351. [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S et al. BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indole acetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell. 2001;13:2349–2360. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 2001;6:420–425. doi: 10.1016/s1360-1385(01)02042-8. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sasse J. Physiological actions of brassinosteroids. In: Sakurai A, Yokota T, Clouse SD, editors. Brassinosteroids: Steroidal Plant Hormones. Tokyo: Springer-Verlag; 1999. pp. 137–161. [Google Scholar]
- Schwechheimer C, Deng XW. COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol. 2001;11:420–426. doi: 10.1016/s0962-8924(01)02091-8. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1,300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimata K, Han SY, Yoneyama K, Takeuchi Y, Yoshida S, Asami T. A specific and potent inhibitor of brassinosteroid biosynthesis possessing a dioxolane ring. J Agric Food Chem. 2002;50:3486–3490. doi: 10.1021/jf011716w. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S. Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 2001;126:770–779. doi: 10.1104/pp.126.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Koncz C. Biochemical and genetic analysis of brassinosteroid metabolism and function in Arabidopsis. Plant Physiol Biochem. 1998;36:145–155. [Google Scholar]
- Szekeres M, Nemeth K, KonczKalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J, Post-Beittenmiller D, Jaworski JG. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999;17:119–130. doi: 10.1046/j.1365-313x.1999.00352.x. [DOI] [PubMed] [Google Scholar]
- von Arnim A, Deng XW. Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:215–243. doi: 10.1146/annurev.arplant.47.1.215. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2002;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- Xu W, Campbell P, Vargheese AK, Braam J. The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J. 1996;9:879–889. doi: 10.1046/j.1365-313x.1996.9060879.x. [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. Arabidopsis Tch4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YH, Wang ZY, Mora-Garcia S, Li JM, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Zhu YX, Tepperman JM, Fairchild CD, Quail PH. Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc Natl Acad Sci USA. 2000;97:13419–13424. doi: 10.1073/pnas.230433797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek DM, Rayle DL, McMorris TC, Clouse SD. Investigation of gene-expression, growth-kinetics, and wall extensibility during brassinosteroid-regulated stem elongation. Plant Physiol. 1994;104:505–513. doi: 10.1104/pp.104.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]