Abstract
In most species, the synthesis of ADP-glucose (Glc) by the enzyme ADP-Glc pyrophosphorylase (AGPase) occurs entirely within the plastids in all tissues so far examined. However, in the endosperm of many, if not all grasses, a second form of AGPase synthesizes ADP-Glc outside the plastid, presumably in the cytosol. In this paper, we show that in the endosperm of wheat (Triticum aestivum), the cytosolic form accounts for most of the AGPase activity. Using a combination of molecular and biochemical approaches to identify the cytosolic and plastidial protein components of wheat endosperm AGPase we show that the large and small subunits of the cytosolic enzyme are encoded by genes previously thought to encode plastidial subunits, and that a gene, Ta.AGP.S.1, which encodes the small subunit of the cytosolic form of AGPase, also gives rise to a second transcript by the use of an alternate first exon. This second transcript encodes an AGPase small subunit with a transit peptide. However, we could not find a plastidial small subunit protein corresponding to this transcript. The protein sequence of the purified plastidial small subunit does not match precisely to that encoded by Ta.AGP.S.1 or to the predicted sequences of any other known gene from wheat or barley (Hordeum vulgare). Instead, the protein sequence is most similar to those of the plastidial small subunits from chickpea (Cicer arietinum) and maize (Zea mays) and rice (Oryza sativa) seeds. These data suggest that the gene encoding the major plastidial small subunit of AGPase in wheat endosperm has yet to be identified.
The synthesis of starch takes place inside plastids (the chloroplasts of leaves and the amyloplasts of nonphotosynthetic starch-storing organs such as seeds). The substrate for starch synthesis, ADP-Glc is synthesized by the enzyme ADP-Glc pyrophosphorylase (AGPase). AGPase catalyzes the conversion of Glc-1-P and ATP to ADP-Glc and pyrophosphate and is a heterotetrameric protein composed of two sorts of subunits referred to as the small (SSU) and large subunits (LSU; for review, see Preiss, 1991). There is evidence for the existence of multiple, tissue-specific forms of the SSU and LSU of AGPase in plants. In maize (Zea mays), for example, the SSUs of the endosperm, embryo, and leaf are encoded by three separate genes (Hannah et al., 2001). However, there is also evidence that transcripts encoding particular AGPase subunits may be expressed in more than one tissue. For, example, the transcript encoding the LSU of AGPase in barley (Hordeum vulgare) leaves is also present in the endosperm (Doan et al., 1999). At present, we do not have a complete description for any species of the nature and patterns of expression of all of the genes encoding AGPase subunits.
In most species, the synthesis of ADP-Glc occurs entirely within the plastids in all tissues so far examined. However, in the endosperm of barley (Thorbjørnsen et al., 1996a), maize (Denyer et al., 1996), and rice (Oryza sativa; Sikka et al., 2001), and probably all grasses (Beckles et al., 2001), a second form of AGPase synthesizes ADP-Glc outside the plastid, presumably in the cytosol. In barley (Thorbjørnsen et al., 1996a), maize (Denyer et al., 1996), and rice (Sikka et al., 2001), cytosolic AGPase accounts for most (85%–95%) of the total AGPase activity in the endosperm. The phenotypes of the maize mutants shrunken2, brittle2, and brittle1 (sh2, bt2, and bt1) show that the synthesis of ADP-Glc in the cytosol and its import into plastids by a specific transporter (encoded at the Brittle1 locus) is necessary for starch synthesis to occur at the wild-type rate. Mutations that abolish the LSU (Shrunken2) or the SSU (Brittle2) of the cytosolic form of AGPase in maize abolish the major AGPase activity and substantially reduce the starch content of the endosperm (Tsai and Nelson, 1966; Dickinson and Preiss, 1969). Mutations that eliminate or inactivate the ADP-Glc transporter (brittle1 mutants) also reduce starch content and cause the accumulation of ADP-Glc in the cytosol (Shannon et al., 1996).
In maize endosperm, the phenotypes of the bt2 and sh2 mutants show that the cytosolic and plastidial subunits of AGPase are encoded by four separate genes, two encoding SSUs and two encoding LSUs (Giroux and Hannah, 1994). However, this may not be the case in all cereal endosperms. In barley, cytosolic and plastidial SSU mRNAs are produced from a single gene by the use of two alternate first exons (Thorbjørnsen et al., 1996b). These transcripts predict plastidial and cytosolic SSU proteins that are identical over 90% of their length and differ only at their N termini. The predicted N-terminal domain unique to the putative plastidial SSU in barley contains a transit peptide, whereas the predicted N-terminal domain of the cytosolic protein is shorter than a typical transit peptide and lacks a consensus transit peptide cleavage site.
In wheat (Triticum aestivum), there is a limited amount of information relating gene sequences to the plastidial and cytosolic forms of AGPase. As much of this work was done before it was known that the major form of AGPase in cereal endosperms is cytosolic, it was assumed that the cDNAs isolated from wheat endosperm encoded plastidial proteins. The following is a summary of information about the subunits of AGPase in wheat and the genes encoding them. A full-length cDNA clone encoding an AGPase SSU was isolated from a wheat endosperm library (Ainsworth et al., 1993). A putative transit peptide of 22 amino acids was identified based on the alignment of the wheat SSU sequence with the N terminus of the mature AGPase SSU purified from spinach (Spinacia oleracea) leaf. However, it was noted that at 22 amino acids, this putative transit peptide is short compared with known chloroplast transit peptides. The similarity of this wheat cDNA sequence to the barley cytosolic SSU (Thorbjørnsen et al., 1996b) makes it very likely that it encodes a cytosolic rather than a plastidial AGPase SSU. No cDNA clones encoding obvious plastidial AGPase SSUs for wheat have been identified.
Partial cDNAs (Olive et al., 1989) and, later, a full-length cDNA (Ainsworth et al., 1995) encoding a LSU of AGPase were cloned from wheat endosperm. This cDNA encoded a small, putative transit peptide of 22 amino acids. A partial cDNA encoding a different LSU of AGPase was also cloned from wheat leaves (Olive et al., 1989). With the limited sequence data available at the time, Olive et al. (1989) could not conclude whether it encoded an LSU or an SSU. However, later comparisons suggested that it did encode an LSU (Smith-White and Preiss, 1992). Thus, two different cDNAs encoding LSUs have been described for wheat.
In summary, some but not all, of the cDNAs encoding subunits of wheat AGPase have been identified. The patterns of expression of the identified genes and their relationships to plastidial and cytosolic AGPase proteins are unknown. Comparison of information thus far available for barley and maize indicates that within the grasses, there may be considerable variation in the way in which AGPase subunits in the endosperm are encoded, but there is insufficient detailed information to allow general conclusions to be drawn. The aim of our work was to shed further light on this problem by providing a complete picture of AGPase transcripts and proteins in wheat endosperm. We wished to establish the identity of the genes encoding the large and small subunits of AGPase in wheat endosperm by purifying the cytosolic and plastidial forms of AGPase from this tissue; whether the cytosolic and plastidial SSUs are encoded by separate genes, as in maize, or by a single gene encoding two alternative N-termini, as in barley; the subcellular distribution of AGPase activity in wheat endosperm; and the tissue-specific pattern of expression and, for the endosperm, the temporal pattern of expression of the AGPase transcripts and proteins.
RESULTS
Sequence Analysis
The predicted amino acid sequences of subunits of plant AGPase (Smith-White and Preiss, 1992 and Fig. 1) can be divided into two groups corresponding to the LSUs and SSUs. Comparison of the N- terminal portions of the SSU sequences allows this group to be further divided into two subgroups (Thorbjørnsen et al., 1996b; Hannah et al., 2001). One group contains the plastidial AGPase SSUs from cereals and noncereals. The other group contains cereal sequences that are proven or putative cytosolic AGPase SSU sequences.
Figure 1.
Comparison of the derived amino acid sequences of subunits of ADP-Glc pyrophosphorylase (AGPase). The dendrogram was generated using the PileUp program of the Wisconsin Package Version 10.1 (Genetics Computer Group, Madison, WI), which creates a multiple sequence alignment from a group of related sequences using progressive, pair-wise alignments. The tree shows the clustering relationships for subunits of AGPase. With one exception (AAD31613), the plant subunits of AGPase divide into two major groups, representing the LSU (pale gray) and SSU (dark gray). Presence of a transit peptide (TP) was predicted using the ChloroP server (Emanuelsson et al., 1999). y, Yes; n, no; ?, not determined due to incomplete sequence; Tissue, tissue from which transcript was isolated.
Comparison of the LSU sequences (Fig. 1) shows that these can also be divided into two major subgroups. One contains only cereal LSUs (cereal group), and the other mainly noncereal LSUs. The cereal group contains proteins expressed only in nonphotosynthetic parts of the plant such as the endosperm and embryo. None are expressed in leaves. Unlike the SSUs, comparison of the entire LSU sequences or just the N-terminal regions does not show a clear division of these sequences into ones that are proven or probable cytosolic sequences (e.g. maize endosperm Sh2, accession no. AAB24191; Bhave et al., 1990) and ones that are probably plastidial (e.g. maize embryo LSU, accession no. P55234; Giroux et al., 1995). Almost all of the sequences in this cereal group do not have clearly defined transit peptides identified by the ChloroP 1.1 Prediction Server. Thus, we cannot determine with certainty from the predicted amino acid sequence whether a LSU sequence from a cereal seed, such as that from wheat endosperm (accession no. P12299; Ainsworth et al., 1995; Olive et al., 1989), encodes a cytosolic or plastidial protein. Therefore, this must be determined experimentally.
Within the second major group of LSU sequences, which are mainly from dicots, there is one cereal sequence encoding a barley leaf LSU (accession no. T06194; Eimert et al., 1997). Unlike the LSU sequences in the cereal group, this sequence does have a clearly recognizable transit peptide of 55 amino acids (ChloroP 1.1 Prediction Server). The partial sequence of a transcript from wheat leaves (Olive et al., 1989; accession no. P12298) also falls within this group. These cereal sequences are most closely related to the dicot leaf LSU sequences. Thus LSUs expressed in cereal leaves show more similarity at the amino acid level to those of dicot leaves than to those of cereal seeds.
Identification of Transcripts in the Endosperm Encoding Cytosolic and Plastidial Forms of AGPase SSU
In wheat endosperm, the previously identified transcript encoding AGPase SSU (Ainsworth et al., 1993) is very similar to that encoding the putative cytosolic SSU in barley endosperm (Thorbjørnsen et al., 1996b) (Fig. 2a). The gene encoding this cytosolic SSU in barley also encodes a putative plastidial SSU that is produced by the use of an alternate first exon (Fig. 2b). To investigate whether a similar plastidial transcript is present in wheat endosperm, we designed primers based on the wheat and barley sequences to amplify the 5′ regions unique to the putative cytosolic and plastidial forms.
Figure 2.
Partial sequences of the 5′ ends of cDNAs encoding the AGPase SSUs of barley and wheat. The PCR products shown (AGP.S.1a and AGP.S.1b) were isolated from wheat cDNA (cv Frame) and are compared with the previously identified wheat cDNA (cv Chinese Spring; Ainsworth et al., 1993) and the barley cDNA encoding the putative cytosolic SSU (Thorbjørnsen et al., 1996b). The sequences of PCR products from two separate PCR reactions were identical. The sequences used to design the 5′ and the 3′ primers are indicated. The transcription start codon (ATG) is shown in bold and is underlined. Identical bases are shown in white on black. a, Comparison of partial cDNAs encoding the cytosolic SSU of AGPase from wheat and barley. b, Comparison of partial cDNAs encoding the putative plastidial SSU of AGPase from wheat and barley. c, Comparison of partial cDNAs encoding the cytosolic and putative plastidial SSU of AGPase from wheat. The start of the sequence common to both cDNAs (encoded by exon 2) is indicated by a star.
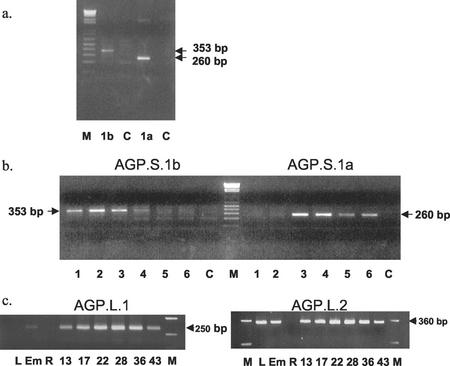
Fragments of the predicted sizes were amplified from RNA from isolated endosperms (Fig. 3a), indicating that there are two transcripts encoding SSUs in wheat endosperm. We called the transcript that gave rise to the smaller 260-bp fragment, AGP.S.1a (encoding a putative cytosolic SSU), and the transcript that gave rise to the larger 353-bp fragment, AGP.S.1b (encoding a putative plastidial SSU). The AGP.S.1a sequence was identical to that previously isolated by Ainsworth et al. (1993) (Fig. 2a). The AGP.S.1b sequence was almost identical to the barley plastidial AGPase SSU (Fig. 2b) and clearly different from AGP.S.1a (Fig. 2c) and the previously identified wheat transcript (Ainsworth et al., 1993) at the 5′ end.
Figure 3.
Analysis of AGPase transcripts. Details of the primers and PCR conditions are given in “Materials and Methods.” M, Molecular mass standards; C, control reactions (contained primers but no cDNA). Estimated sizes of the products are indicated. a, Reverse transcriptase (RT)-PCR analysis of transcripts in developing wheat endosperm using specific primer pairs for AGP.S.1a (1a) and AGP.S.1b (1b). Endosperms were dissected from grains of approximately 15 to 30 mg fresh weight. b, RT-PCR analysis of transcripts in developing wheat endosperm from grains of various developmental stages using specific primer pairs for AGP.S.1a and AGP.S.1b. The average fresh weights of the grains in each sample were 1, 4.8 mg; 2, 8.5 mg; 3, 17.5 mg; 4, 26.1 mg; 5, 42.9 mg; and 6, 64.4 mg. c, RT-PCR analysis of transcripts in leaves (L), embryos (Em), roots (R), and developing endosperms of different ages (mass of grains [mg] used is indicated). Specific primer pairs for AGP.L.1 (endosperm LSU; left, 25 ng of total RNA per reaction) and AGP.S.2 (leaf LSU; right, 400 ng of total RNA per reaction) were used. Each reaction was repeated three or four times and the results of a typical experiment are shown. To test for DNA contamination of the RT-PCR reactions, control RNA samples were first treated with DNase-free RNase before the cDNA was synthesized (Promega). In all of these control reactions, no PCR products were amplified.
Identification of a Wheat Genomic Clone Encoding AGPase SSU
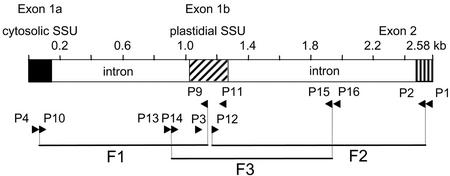
The fact that AGP.S.1a and AGP.S.1b differed at their 5′ ends but were identical at their 3′ ends (Fig. 2c) suggested that in wheat, as in barley, these two transcripts are probably encoded by a single gene. To identify this gene in wheat, we used primers designed to amplify the DNA encoding the 5′ region in three overlapping fragments (Fig. 4). We called this partial wheat AGPase gene Ta.AGP.S.1 (GenBank accession no. AF536819). It encodes two exons, 1a and 1b, corresponding to the 5′ sequences of AGP.S.1a and AGP.S.1b respectively, and the first part of exon 2, which is common to both transcripts. The arrangement of the exons is identical to those described for the barley AGPase SSU gene (Thorbjørnsen et al., 1996b), and the sequence of exons and introns in these genes are very similar (88% identity).
Figure 4.
Analysis of AGPase genes. A diagrammatic representation of a genomic clone of Ta.AGP.S.1. Introns are shown in white, and exons are shaded. Primers (P1–P16) and their orientation are indicated by arrowheads. The clone was isolated as three overlapping fragments (F1–F3).
Developmental Pattern of Expression of AGP.S.1a and AGP.S.1b
We investigated the patterns of expression of AGP.S.1a and AGP.S.1b during wheat grain development (Fig. 3b) using the specific primers shown in Figure 2 and reverse transcriptase-PCR. The RT-PCR products corresponding to AGP.S.1a and AGP.S.1b differed in their developmental pattern of expression (two experiments with mRNA from different batches of grain). The product corresponding to AGP.S.1a was detected in endosperms undergoing rapid starch synthesis, but not in very young endosperms when starch synthesis was minimal. The product corresponding to AGP.S.1b was detected only in young endosperms at the beginning of their starch-synthesizing period. The RT-PCR product corresponding to the constitutively expressed cytosolic isoform of glyceraldehyde 3-P dehydrogenase was present at all developmental stages tested, showing that the cDNA was intact (data not shown).
Pattern of Expression of Transcripts Encoding AGPase LSUs
Two wheat cDNAs encoding AGPase LSUs were identified previously (accession nos. Z21969 and X14348). These transcripts will be referred to as AGP.L.1 and AGP.L.2, respectively. The tissue-specific and, in the endosperm, temporal patterns of expression of the genes encoding AGP.L.1 and AGP.L.2 were examined using specific primers and RT-PCR (Fig. 3c). In all reactions, the RT-PCR products were of the predicted length and sequencing of the product amplified from endosperm at 16 d postanthesis and for leaf confirmed that these were amplified fragments of the AGPase cDNAs to which the primers were designed (data not shown). AGP.L.1 was expressed in endosperms at each developmental stage tested (Fig. 3c) and could also be detected in embryos and to a lesser extent roots when higher concentrations of RNA were used (100 ng, data not shown). No AGP.L.1 transcript could be detected in leaves. AGP.L.2 was expressed in leaves, endosperms, and embryos. No AGP.L.2 transcripts were detected in roots at the concentration of RNA used in these experiments.
The Relative Amounts of Plastidial and Cytosolic AGPase in the Endosperm
To examine the subcellular location of AGPase activity and proteins, we used a mechanical method to isolate plastids from developing wheat endosperm. A pellet fraction was obtained that was enriched in plastidial marker enzymes (soluble starch synthase and alkaline pyrophosphatase) relative to cytosolic marker enzymes (alcohol dehydrogenase and Suc synthase [SuSy]). The distribution of AGPase between pellet and supernatant was very similar to that of the cytosolic marker enzymes. This suggested that most of the activity of AGPase in wheat endosperm is extraplastidial. The activities in the plastid-enriched pellets as a percentage of the total activity recovered in the supernatant plus pellet (means ± se from measurements of six separately prepared batches of plastids) were 1.6 ± 0.2 (AGPase), 1.1 ± 0.2 (alcohol dehydrogenase), 1.3 ± 0.1 (SuSy), 14.6 ± 0.8 (soluble starch synthase), and 14.3 ± 1.1 (alkaline pyrophosphatase).
To determine how much of the total AGPase activity was plastidial, we compared the activities of AGPase and marker enzymes in aliquots of the plastid preparations that were deliberately contaminated with differing amounts of cytosolic enzymes (using a method described in Denyer and Smith, 1988). The subcellular distribution of AGPase activity calculated from these data showed that the plastidial AGPase activity was much less than the cytosolic AGPase activity. Endosperms from a range of developmental stages (grains of 4–30 mg fresh weight) were tested, and in no experiments was the activity in the plastids more than 7% of the total activity.
Identification of AGPase SSU Using Specific Antisera
Using specific antiserum to the Bt2 SSU of maize AGPase (Giroux and Hannah, 1994), we examined the relative amounts of the plastidial and cytosolic SSU proteins present in developing wheat endosperm at three developmental stages (Fig. 5). As with barley (Thorbjørnsen et al., 1996a; Beckles et al., 2001), the cytosolic and plastidial SSUs are of different molecular mass (approximately 54 and 51 kD, respectively). The plastidial and cytosolic proteins were present at all three developmental stages. Although there was some variation between samples, in general, the amount of the cytosolic SSU per endosperm (and possibly the amount of plastidial SSU also) increased with developmental age.
Figure 5.
The abundance of AGPase SSUs in endosperms of different ages. Three replicate extracts of endosperms from grains 12, 17, or 21 d after anthesis (DAA) were subjected to SDS-PAGE on 7.5% (w/v) gels and were then immunoblotted and probed with Bt2 antiserum at a concentration of 1/5,000 as described in “Materials and Methods.” The average grain weights were 10.8 mg (12 DAA), 18.9 mg (17 DAA), and 43.3 mg (21 DAA). Each track contains proteins from an equivalent proportion of an endosperm. C, Cytosolic small subunit; P, plastidial small subunit. Proteins of unknown identity that were smaller than the SSUs also cross-reacted with the antiserum (not shown).
Separation of Cytosolic and Plastidial Forms Using Anion-Exchange Chromatography
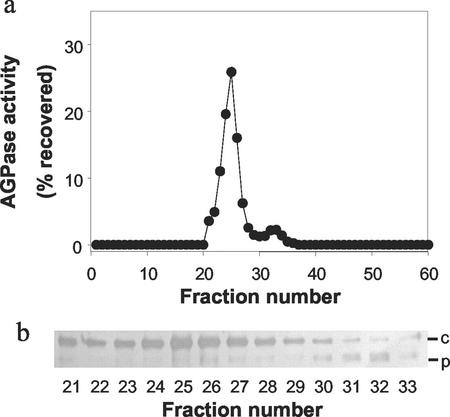
In addition to the estimation of the subcellular distribution of AGPase activity by fractionation (above), we used ion-exchange chromatography to separate and quantify the plastidial and cytosolic AGPase activities. In extracts of developing wheat endosperm, we observed two peaks of activity (Fig. 6). In western blots probed with the Bt2 antiserum, the cytosolic SSU could be detected in fractions from the major peak and the plastidial SSU could be detected in fractions from the minor peak. This suggested that the two peaks represented the separated cytosolic and plastidial forms of AGPase. The relative activities of the two peaks of AGPase were used to estimate the relative abundance of these forms in the endosperm. In a typical experiment (Fig. 6), the plastidial AGPase (second peak) accounted for approximately 6% of the total AGPase activity.
Figure 6.
Separation of isoforms of AGPase by ion-exchange chromatography. Wheat endosperms were extracted and the homogenate applied to a Q-Sepharose column as described in “Materials and Methods.” a, The activity of AGPase in each fraction is expressed as a percentage of the total AGPase activity recovered in all fractions from the column. b, Samples of fractions 21 through 33 (containing AGPase activity) were subjected to SDS-PAGE, blotted onto nitrocellulose, and probed with Bt2 antiserum at a dilution of 1/5,000. c, Cytosolic SSU; P, plastidial SSU.
Purification of Cytosolic and Plastidial AGPase from Developing Wheat Endosperm
The cytosolic and plastidial activities of AGPase in extracts of developing wheat endosperms were separated using ion-exchange chromatography (as shown in Fig. 6). Gel-filtration chromatography of each form of AGPase on Superose 200 gave a single peak of activity with an estimated mass of between 250 and 300 kD. This mass is consistent with the idea that the cytosolic and plastidial enzymes are heterotetramers of two small and two large subunits, as suggested by Fu et al. (1998) for the potato (Solanum tuberosum) tuber AGPase. The purification scheme we devised (Table I) is rapid and recovers approximately 45% of the initial AGPase activity. Therefore, it is unlikely that any major forms of AGPase were lost during purification.
Table I.
Purification of cytosolic and plastidial AGPase from developing wheat endosperm
| Step | Volume | Total Activity | Recovery of Activity | Total Protein | Purification Relative to Crude Homogenate | Specific Activity |
|---|---|---|---|---|---|---|
| mL | mmol min−1 | % crude homogenate | mg | -fold | mmol min−1 | |
| Crude homogenate | 6 | 52.4 ± 1.54 | 100 | 238 ± 9.17 | 1 | 0.22 ± 0.01 |
| Q Sepharose (cytosolic) | 20 | 36.2 ± 1.77 | 69 | 4.53 ± 0.16 | 36.3 | 7.99 ± 0.56 |
| Q Sepharose (plastidial) | 15 | 1.68 ± 0.16 | 3.2 | 0.18 ± 0.02 | 43.6 | 9.60 ± 1.15 |
| Sephadex 200 (cytosolic) | 12 | 23.1 ± 1.12 | 44 | 1.02 ± 0.09 | 103 | 22.7 ± 2.85 |
| Sephadex 200 (plastidial) | 9 | 0.47 ± 0.04 | 0.9 | 0.015 ± 0.001 | 143 | 32.4 ± 3.64 |
The plastidial and cytosolic forms of AGPase were simultaneously purified from 500 endosperms of 14 to 25 mg fresh wt. Values are means (±se) of measurements made in six (cytosolic) or four (plastidial) separate experiments. The final specific activities of the purified cytosolic and plastidial AGPase from wheat endosperm were similar to the specific activities of AGPase partially purified from rice endosperm (42.9 mmol min−1 mg protein−1; Nakamura et al., 1992) and maize endosperm (33.6 mmol min−1 mg protein−1; Plaxton and Preiss, 1987) and considerably greater than that of AGPase purified from wheat endosperm (2.4 mmol min−1 mg protein−1) by Gómez-Casati and Iglesias (2002).
Analysis of the Purified Cytosolic AGPase Proteins
When analyzed by SDS-PAGE (Fig. 7A), the final preparation of cytosolic AGPase contained major proteins of approximately 50 kD (proteins a and b), the expected size for subunits of AGPase, as well as major contaminating proteins of 90 kD (protein c) and 105 kD (protein d) and other minor proteins (identity unknown). All visible proteins of between 40 and 60 kD were excised from the gel and were subjected to digestion with trypsin, and the masses of the resulting fragments were analyzed using MALDI-ToF. Analysis of protein b (Fig. 7A) showed that it was an SSU of AGPase. Nine fragments (covering 30% of the protein) were identified that matched the putative cytosolic SSU from wheat endosperm (accession no. P30523; Ainsworth et al., 1993) encoded in part by AGP.S.1a. One fragment from the purified protein was the same mass as a fragment expected from the unique N terminus encoded by AGP.S.1a.
Figure 7.
SDS PAGE of purified cytosolic (A) and plastidial (B) AGPase. AGPase was purified as described in “Materials and Methods,” and was subjected to SDS-PAGE on a 7.5% (w/v) acrylamide gel (A) or a 4% to 12% (w/v) acrylamide gradient gel (B). Masses in kilodaltons of Mr marker proteins are indicated. Protein bands labeled a through d and a through c were excised and subjected to matrix-assisted laser-desorption ionizing time-of-flight mass spectrometry (MALDI-ToF) analysis. Proteins identified by MALDI-ToF analysis are indicated.
Analysis of a group of four proteins ranging in size from 48 to 55 kD (protein a) showed that they were fragments of the wheat endosperm LSU encoded by AGP.L.1. For the largest of the four proteins, 13 fragments (covering 39% of the protein) were identified.
MALDI-ToF analysis of the major contaminating proteins of 90 and 105 kD (proteins c and d) in the AGPase showed that they were SuSy 1 (accession no. CAA04543) and SuSy 2 (accession no. CAA03935), respectively.
Analysis of the Purified Plastidial AGPase Proteins
When analyzed by SDS-PAGE (Fig. 7B), the final preparation of plastidial AGPase contained major proteins of approximately 50 kD (protein a), the expected size for subunits of AGPase, as well as major contaminating proteins of approximately 110 and 150 kD (proteins b and c) and other minor proteins (identity unknown). Proteins a, b, and c (Fig. 7B) were excised from the gel, subjected to digestion with trypsin, and the masses of the resulting protein fragments were analyzed using MALDI-ToF. This analysis showed that the region of the gel labeled a contained two types of protein. One was a SSU of AGPase and the other was a LSU. Thirteen fragments (covering 30% of the protein) were identified that matched an AGPase SSU from chickpea (Cicer arietinum; accession no. AAK27720). The fragment sizes did not match as closely to those predicted for the barley homolog of AGP.S.1b, the putative plastidial SSU (accession no. P55238).
For the LSU in protein a, the best matches of fragment masses in two separate experiments were to those predicted for the cytosolic LSU from wheat endosperm, AGP.L.1. The total number of fragments obtained from the two experiments combined covered 46% of the protein. In both experiments, a fragment corresponding to the predicted N terminus of the LSU was obtained, showing that the protein had not been processed to remove a transit peptide.
MALDI-ToF analysis of the major contaminating proteins showed that protein b was a heat shock protein. The closest match of fragment sizes was to HSP80–2 from wheat (accession no. X98582). Analysis of protein c did not reveal its identity.
To obtain protein sequence information for the purified plastidial SSU protein, we used a quadrupole time-of-flight (Q-ToF) mass spectrometer (MS). The AGPase SSUs that matched most closely to the two fragments of protein sequence obtained for the purified wheat protein were those from rice seed, maize embryo, chickpea, and the predicted sequence of wheat AGP.S.1b (Fig. 8). However, the amino acid sequence obtained by Q-ToF did not exactly match any of these sequences, including that of wheat AGP.S.1b. It differed from these sequences in at least six out of the 26 residues.
Figure 8.
Comparison of the sequence of purified plastidial AGPase SSU with the predicted sequences of other AGPase SSUs. The purified plastidial SSU protein from wheat endosperm was subjected to MALDI analysis, and the sequence of two fragments was obtained by Q-ToF analysis. These sequences were compared with those of other SSUs available in databases. The corresponding sequences of the four SSUs that matched most closely to those of the purified plastidial SSU protein from wheat are shown. The positions of the amino acids in the wheat endosperm AGPase SSU are indicated. Leu (L) and Ile (I) have the same mass and are therefore indistinguishable by Q-ToF. For L and I in the Q-ToF sequence, the amino acid most likely to be present was determined by comparison with other deduced sequences and is shown on the top line with the alternative, less likely amino acid shown below in parenthesis. Rice, Rice seed (AAK27313); Maize, maize embryo (AAK39640); Chickpea, unknown source tissue (AAK27720); Wheat (AGP.S.1b), wheat endosperm (P30523).
DISCUSSION
Identification of Genes Encoding the LSU and SSU of AGPase in Wheat Endosperm
To identify the genes encoding the LSU and SSU of AGPase in wheat endosperm, we purified the cytosolic and plastidial forms of the enzyme and used MALDI-ToF and Q-ToF MS analysis to match the purified proteins to previously characterized cDNAs. This analysis showed that the LSU of the cytosolic form of AGPase is encoded by the cDNA identified by Ainsworth et al. (1995), and the SSU of the cytosolic AGPase is encoded by the cDNA identified by Ainsworth et al. (1993). Both cDNAs were previously thought to encode plastidial AGPase subunits. However, our results show that the proteins encoded by these transcripts are not imported into the plastids and are therefore processing by removal of part of their N-terminal sequences would not be expected.
Purification and sequencing of plastidial AGPase from wheat endosperm suggested that some, or all, of the plastidial SSU is encoded by a gene that has yet to be identified. The MALDI-ToF and Q-ToF analyses of fragments of the purified protein showed that the wheat endosperm plastidial SSU was more similar to AGPase SSUs from chickpea, maize embryo, and rice seeds than to the cytosolic SSU of wheat or to the putative plastidial SSU from barley endosperm (Thorbjørnsen et al., 1996a). The isolation of the gene in wheat encoding this protein is in progress.
Purification of the plastidial AGPase did not result in the identification of the gene encoding the plastidial LSU from wheat endosperm. We obtained sequence information from the purified protein that matched the cytosolic LSU. Possible explanations for this are that the cytosolic and plastidial LSUs are encoded by the same gene; that our plastidial AGPase preps were contaminated with some cytosolic AGPase; or that there was some exchange of subunits between the plastidial and cytosolic enzymes prior to their separation on Q-Sepharose. We consider the first of these possibilities to be very unlikely. A MALDI fragment matching the extreme N terminus of the protein was found that would indicate that no processing of the protein to remove a transit peptide had occurred. It is not likely that a plastidial LSU protein would lack a transit peptide (Vothknecht and Soll, 2000). Consistent with the second and/or third possibilities, we saw a small amount of the plastidial SSU in the (mainly) cytosolic AGPase peak after Q-Sepharose (Fig. 6, Peak 1), suggesting that a limited amount of contamination or subunit exchange had occurred.
A Single SSU Gene Encodes Two Transcripts, But Neither Corresponds to the Plastidial SSU Protein
Our experiments suggested that in wheat, as in barley, two different mRNAs are produced from a single SSU gene. We identified two transcripts, AGP.S.1a and AGP.S.1b, in developing wheat endosperm that were almost identical to two previously identified barley endosperm transcripts (bepsF1 and blps14; Thorbjørnsen et al., 1996a). The gene in wheat (Ta.AGP.S.1) encoding these transcripts was partially sequenced and it showed an arrangement of the exons encoding the N termini of the two alternative AGPase SSUs similar to that of the AGPase SSU gene in barley (Thorbjørnsen et al., 1996b). The existence of such a gene in wheat was predicted by Thorbjørnsen et al. (1996b) based on the similarity between the deduced N-terminal sequences of the wheat and barley SSU proteins.
The smaller of the two transcripts (AGP.S.1a) produced from gene Ta.AGP.S.1 corresponds to the cDNA identified by Ainsworth et al. (1993) and encodes the cytosolic SSU of AGPase. The larger of the two transcripts (AGP.S.1b) produced from gene Ta.AGP.S.1 encodes a putative protein predicted to have a transit peptide. However, as no plastidial protein was found that matched the predicted sequence of this transcript, its significance is unknown.
As no proof in the form of protein sequence from purified plastidial AGPase is available to support the idea that the barley equivalent of AGP.S.1b, blps14 encodes some or all of the plastidial SSU in barley endosperm, we cannot exclude the possibility that another, yet to be discovered, gene may encode at least some of the plastidial AGPase SSU in barley as well as in wheat. This question will be addressed in a future publication from our group.
Most of the AGPase Activity in Wheat Endosperm Is Cytosolic
Two different approaches were used to assess the subcellular distribution of AGPase activity in wheat endosperm: preparation of fractions enriched in plastids and separation of cytosolic and plastidial forms of AGPase using ion-exchange chromatography. Both approaches showed that that the cytosolic form accounts for most of the total AGPase activity in the endosperm during the phase of rapid accumulation of starch. There is a second, minor form of AGPase in developing endosperm that is plastidial. The activity of the plastidial AGPase was variable. In our experiments, it was generally very low (<2%–7% of the total AGPase activity in the endosperm).
AGPase Subunits Have Different Temporal and Spatial Patterns of Expression
We investigated the tissue-specific pattern of expression of the endosperm cytosolic LSU (AGP.L.1) and the major leaf LSU (AGP.L.2) and, for the endosperm, the temporal pattern of expression of these LSUs and the two SSU transcripts (AGP.S.1a and AGP.S.1b) encoded by gene Ta.AGP.S.1. We also compared the pattern of expression of AGP.S.1a and AGP.S.1b with changes in the relative amounts of protein and activities of the plastidial and cytosolic forms of AGPase during endosperm development.
The AGP.L.1 transcript was expressed strongly only in the endosperm. This confirms the results of others for wheat (Ainsworth et al., 1995) and for the corresponding transcript in barley (Villand et al., 1992; Thorbjørnsen et al., 1996b; Eimert et al., 1997; Doan et al., 1999). The gene may also be expressed in tissues other than the endosperm. In general agreement with others, we found a low level of expression of the AGP.L.1 transcript in embryos and roots, but none in leaves. This may indicate that the cytosolic form of AGPase is not confined to the endosperm. However, it is more likely that the probes used in these experiments were not gene specific and may have crosshybridized with transcripts encoding other forms of LSU or that, as suggested by Doan et al. (1999), very low levels of these largely endosperm-specific transcripts may be present in other tissues due to promoter leakage, but are unlikely to encode the bulk of the LSU protein in those tissues. Purification and sequencing of the AGPase proteins from other tissues, as well as the endosperm, may be necessary to resolve these controversies.
The AGP.L.2 transcript was expressed in leaves, endosperms, and embryos, but not in roots. This suggests that the LSU protein encoded by this transcript may be present, presumably in the plastids, of leaves, endosperms, and embryos. Similar patterns of expression of the leaf LSU, blpl, were observed in barley (Doan et al., 1999). However, as we were unable to identify the plastidial LSU protein in this study, we do not know whether the AGP.L.2 transcript or another, as yet unidentified, transcript is responsible for the plastidial LSU in wheat endosperm.
The transcripts encoding the LSU and SSU of cytosolic AGPase increase in abundance during the early to middle grain-filling period. Similar results were obtained for barley transcripts encoding cytosolic subunits of AGPase (Doan et al., 1999). This expression pattern correlates with the increase in total (mainly cytosolic) AGPase activity (reaching a peak at approximately 20 DAA; Pilling 2001) and the abundance of the cytosolic SSU protein (our results and Reeves et al., 1986).
The putative plastidial SSU transcript AGP.S.1b and the plastidial SSU protein did not show a coordinate pattern of expression through endosperm development. The transcript AGP.S.1b was most abundant in very young endosperms, whereas the abundance of the plastidial SSU protein differed very little in endosperms of all stages examined. This is consistent with the idea that the transcript AGP.S.1b encoded by gene Ta.AGP.S.1 may not be responsible for the major plastidial SSU of AGPase in wheat endosperm.
MATERIALS AND METHODS
Plant Material
Grains of wheat (Triticum aestivum) were from the John Innes Germplasm Collection (cv Bobwhite), the Waite Institute (Adelaide, Australia; cv Frame), or from the Montana Agricultural Experimental Station (Bozeman; cv HiLine). Plants were grown in individual pots in a greenhouse at a minimum temperature of 12°C, with supplementary lighting in winter to give a 16-h day. In an alternate manner, plants were grown in a controlled environment room at 15°C/12°C day/night and with 16 h of light per day. Leaves and roots were harvested from 14-d-old plants grown in vermiculite, endosperm was dissected from grains at various stages of development, and embryos were dissected from grains at 20 to 25 DAA. Tissues were used immediately or were harvested directly into liquid nitrogen and stored at −80°C prior to use.
Isolation of Total RNA and cDNA Synthesis
Total RNA was extracted from developing grain using a commercially available phenol/guanidine isothiocyanate procedure (Trizol reagent; Invitrogen, Paisley, UK). For experiments using the cultivar Frame, cDNA was prepared from 2 μg of total RNA with Thermoscript RT (Invitrogen, Paisley, UK) and an oligo-dT20 primer at a temperature of 55°C according to the manufacturer's instructions. For experiments using the cultivar HiLine, cDNA was prepared from 0.025 or 0.4 μg of total RNA with Moloney-murine leukemia virus RT (Applied Biosystems, Foster City, CA) and 1 or 2 μm of the respective antisense oligonucleotide at a temperature of 42°C according the manufacturer's instructions.
PCR Amplification of cDNAs
For experiments using primers designed to AGPase SSUs, a 2-μL aliquot of single-stranded cDNA from the cultivar Frame was used as a template for the PCR amplification of each product using standard PCR procedures. Control PCR reactions with GAPDH primers were carried out as described in Burton et al. (1999). PCR reactions were performed in a total volume of 50 μL with 0.2 μL of primers at 2 μm. Cycling parameters were 35 cycles of 94°C for 40 s, 50°C for 40 s, and 72°C for 60 s. The cDNA fragment encoding part of the cytosolic AGPase SSU was amplified with the sense primer 5′-CGGCAATGGATGTGCCTTTGG-3′ and the antisense primer 5′-CAGACAATTACTGACAGG-3′. The cDNA fragment encoding part of the plastidial AGPase SSU was amplified with the sense primer 5′-CATCCACCTCAATGGCGATGGC-3′ and the antisense primer 5′-CAATAAGCCTGTAGTTGGCACCCAATGGCA-3′. Single PCR products were isolated from gels and purified using Geneclean (Geneworks, Adelaide, Australia) and were cloned into the pGEM TEASY vector (Promega, Madison, WI). Each product was completely sequenced using the Applied Biosystems 373 DNA sequencer.
For experiments using primers designed to AGPase LSUs, PCR was primed with 0.2 μm (LSU.L.1) or 0.4 μm (LSU.L.2) primers using an RNA PCR kit (Roche Molecular Biochemicals, Summerville, NJ) following the manufacturer's instructions, except that PCR was done using GC-Advantage Polymerase 2 PCR system (CLONTECH, Palo Alto, CA). Cycling parameters were 95°C for 5 min, 25 cycles of 95°C for 1 min, 55°C for 2 min, 72°C for 3 min, and an extension time of 72°C for 7 min. AGPase LSU.1 was amplified with primers 5′-GTGACGGGTTCTGCGACA-3′and 5′-GTTTGTTTGCTCGCTGCC-3′. AGPase LSU.2 was amplified with primers 5′-TCTGTTGCTTGCCTATTGATGG-3′ and the 5′-CTGTTCAGCAAGGGC AAGATTT-3′. Single PCR products were isolated from gels and were purified using a QIAquick kit (Qiagen) and sequenced directly with each oligonucleotide primer using the Applied Biosystems ABI 3700 capillary Sequencer with the ABI BigDye terminator chemistry (PE Applied Biosystems, Foster City, CA).
The Cloning and Sequencing of a Gene Encoding AGPase SSU (Ta.AGP.S.1)
To clone Ta.AGP.S.1, we used nested PCR on DNA prepared from developing grains harvested 10 to 13 d after fertilization. The PCR reactions were carried out using the high-fidelity Taq polymerase Elongase (Invitrogen, Paisley, UK) and 10% (v/v) dimethyl sulfoxide. The conditions were 4 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 50°C, 1 min at 72°C, and a final extension time of 10 min at 72°C. For the first-round amplification of fragment 1, the primer sequences were CGGCAATGGATGTGCCTTTGG (P4) and GAGACAGGTCTGGGAGCTCTTC (P11) and for the second, nested round, they were AGCGTGAACAATGCAACATTG (P10) and GCGGAGGGATCAGGATCTTGG (P9). For the first-round amplification of fragment 2, the primer sequences were CATCCACCTCAATGGCGATGGC (P3) and CAGACAATTACTGACAGG (P1) and for the second, nested round, they were ACGTGCCGTGTCAGACTCCAAGA (P12) and CAATAAGCCTGTAGTTGGCACCCAATGGCA (P2). For the first-round amplification of fragment 3, the primer sequences were GTTAAGTTACTCAGCCACACTG (P13) and GATGTCTCAATCCAGCAATTCC (P16) and for the second, nested round, they were CAGTGGGTGTCAGGCGTATCTC (P14) and TGCAGCTGAGACATGATCCAAG (P15). PCR products to be analyzed were cloned into the pGEM-T Easy vector (Promega) and were sequenced using an ABI 3700 capillary sequencer.
Purification of Cytosolic and Plastidial AGPase from Developing Wheat Endosperm
All steps were performed at 4°C or on ice. Five hundred wheat endosperms (each of 14–25 mg fresh weight) from cv Bobwhite plants grown in a controlled environment room were excised into 20 mL of homogenization medium (50 mm HEPES, pH 7.4, 2 mm MgCl2, 2 mm EDTA, 5 mm NaCl, 5% [v/v] ethanediol, and 0.1 μg mL−1 protease inhibitors [Sigma Complete; Sigma, St. Louis]). The endosperms were homogenized using a mortar and pestle, and were then centrifuged for 20 min at 10,000g. The pellet was discarded and the supernatant was loaded onto a HiLoad 16/60 FPLC column containing Q-Sepharose (Amersham Pharmacia Biotech, Buckinghamshire, UK) pre-equilibrated with 50 mm BisTris propane, pH 7.4, 10 mm NaCl, and 5% (v/v) ethanediol. Protein was eluted from the column by applying a linear gradient of 10 to 500 mm NaCl at a flow rate of 2.5 mL min−1. Fractions were assayed and those containing cytosolic AGPase activity (peaks 1) were pooled separately from those containing plastidial AGPase activity (peak 2). Both fractions were concentrated to 200 μL using a Centriplus YM-100 (Millipore, Bedford, MA) concentrator (100 kD cut-off). Each of the concentrated samples was then applied to a Sephadex-200 16/60 gel-filtration column and eluted with 50 mm HEPES, pH 7.2, and 100 mm NaCl at a flow rate of 0.5 mL min−1. Fractions containing AGPase activity were pooled and concentrated as above to a volume of 30 to 150 μL prior to SDS-PAGE.
Isolation of Plastids from Developing Wheat Endosperm
Plastids were isolated from wheat endosperm at approximately 10 to 12 DAA essentially according to the method of Tetlow et al. (1993). Approximately 2 g of wheat endosperm was excised into a glass petri dish containing 10 mL of amyloplast isolation medium (AIM; 50 mm HEPES, pH 7.4, and 0.8 m sorbitol) on ice. All subsequent steps were performed at 4°C. The AIM was removed and 10 mL of plasmolysis buffer (50 mm HEPES, pH 7.4, 0.8 m sorbitol, 1 mm EDTA, 1 mm KCl, and 2 mm MgCl2) was added to the endosperms that were left on ice for 90 min. Plasmolysis buffer was removed and replaced by 10 mL of 50 mm HEPES, pH 7.4, 0.8 m sorbitol, 1 mm EDTA, 1 mm KCl, 2 mm MgCl2, and 1% (w/v) bovine serum albumin. Endosperms were gently chopped using a new razor blade, and the resultant homogenate was filtered through two layers of Miracloth (Calbiochem, San Diego). The filtrate was centrifuged at 50g for 5 min and the pellet enriched in plastids was resuspended in a suitable volume of AIM (typically 1 mL). To break the plastids and recover stromal enzymes, the suspension of organelles was mixed vigorously, squirted 10 times through a fine-bore syringe needle, and then centrifuged at 12,000g for 5 min. The supernatant was assayed for enzyme activity.
Identification of AGPase Subunits by MALDI-ToF and Q-ToF
Fractions containing AGPase activity were subjected to SDS-PAGE and the separated proteins were stained with Coomassie Brilliant Blue R-250. Protein bands were excised and subjected to tryptic digestion according to Speicher (2000) followed by analysis by MALDI-ToF MS. Mass fragment sizes were used to query the National Center for biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) nonredundant database using the MASCOT search tool (http://www.matrixscience.com). All matched sequences showed mass errors of <50 μL L−1.
For Q-ToF analysis, tryptic peptides for each sample were separated using C18 reverse-phase HPLC (75 μm i.d. × 150 mm column, with 3 μm C18 100Å PepMap packing, LC Packings, Amsterdam) and were eluted directly into the nanoelectrospray ion source of a Q-ToF 2 MS (Micromass, Manchester, UK) at a flow rate of 300 nL min−1 using a CapLC (Waters, Milford, MA). After an initial wash for 3 min at 5% (w/v) B (100% [w/v] acetonitrile and 0.1% [w/v] formic acid), a binary gradient consisting of 5% to 80% (w/v) solvent B (100% [w/v] acetonitrile and 0.1% [w/v] formic acid) was developed (t = 0 min, 5% [w/v] B; t = 3, 5% [w/v] B; t = 3.5, 15% [w/v] B; t = 15, 40% [w/v] B; t = 22, 60% [w/v] B; and t = 25, 80% [w/v] B). Spectra were acquired in the data-dependent mode throughout the HPLC gradient, switching between a full survey mass spectrum (m/z range 400–1,600; 1.2 s scan time; MS to MS/MS switch above threshold intensity of 5 counts/s) and two MS/MS spectra (m/z range 50–3,000; 1.2 s scan time; charge-state recognition for 2+, 3+, and 4+ ions; argon collision gas; switch back to MS survey after 6 s or below threshold of 2 counts/s; dynamic exclusion time of 25 s) recorded sequentially on the two most abundant ions present in the survey mass spectrum. All of the MS/MS spectra recorded for each sample were searched against the NCBI nonredundant database using the MASCOT search tool. MS/MS spectra that were not identified by searching against the database were analyzed using the PepSeq de novo sequencing tool (Micromass).
SDS-PAGE and Immunoblotting
SDS-PAGE was on 7.5% (w/v) acrylamide gels according to Beckles et al. (2001) or on Novex (San Diego) preprepared 4% to 12% (w/v) acrylamide gradient gels run in the MOPS buffer system according to the manufacturer's instructions (Invitrogen, Groningen, The Netherlands). Gels were stained with Coomassie Brilliant Blue R-250. Immunoblotting was according to Denyer et al. (1997).
Localization of Wheat Endosperm AGPase Activity
The AGPase activity of wheat endosperm was localized to the plastid or extraplastidial compartments essentially according to the method of Denyer and Smith (1988) except that the extraplastidial compartment was the cytosol rather than the mitochondria.
Enzyme Activities
For AGPase, the activity reported was dependent upon the presence in the assay of all of the appropriate substrates and cofactors and also upon extract concentration within the range used to make the measurements. The concentrations of components of each of the assays and their pH values were optimized to give the maximum rate. The rate of the reaction was linear with respect to time for at least 4 min. Reaction mixtures were as follows: AGPase: as in Smith et al. (1989, assay 2b), except that 100 mm HEPES, pH 7.9, 0.4 mm NAD, 1 mm ADP-Glc, 1.5 mm NaPPi, and 5 U phosphoglucomutase were used; SuSy: As in Craig et al. (1999), except that the buffer was 82 mm AMPSO {(3-[1, 1-dimethyl-2-hydroxyethyl] amino)-2-hydroxy-propanesulfonic acid}, pH 9.0; soluble starch synthase: As in Jenner et al. (1994), the resin method, except that 0.5 mg of potato amylopectin, 2 mm ADP[U-14C] Glc at 2.3 GBq mol−1, and 10 μL of extract were used; Alkaline inorganic pyrophosphatase: As in Gross and ap Rees (1986), except that 50 mm Bicine, pH 8.9, 20 mm MgCl2, and 1.25 mm NaPPi were used; and Alcohol dehydrogenase: as in Cossins et al. (1968), except that the buffer was 85 mm glycyl-Gly (pH 8.6), and 1.3 mm NAD and 100 mm ethanol were used.
ACKNOWLEDGMENTS
We thank Dr. Alison M. Smith and Dr. David Laurie for support, encouragement, and useful discussions throughout the course of this work and for constructive criticism of the manuscript, and Dr. Anne Edwards for advice and help with sequence comparisons.
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (UK; competitive strategic grant to the John Innes Centre), by DuPont Agricultural Products, and by the Australian Grains Research and Development Corporation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010363.
LITERATURE CITED
- Ainsworth C, Hosein F, Tarvis M, Weir F, Burrell M, Devos KM, Gale MD. Adenosine diphosphate glucose pyrophosphorylase genes in wheat: differential expression and gene mapping. Planta. 1995;197:1–10. doi: 10.1007/BF00239933. [DOI] [PubMed] [Google Scholar]
- Ainsworth C, Tarvis M, Clark J. Isolation and analysis of a cDNA clone encoding the small subunit of ADP-glucose pyrophosphorylase from wheat. Plant Mol Biol. 1993;23:23–33. doi: 10.1007/BF00021416. [DOI] [PubMed] [Google Scholar]
- Beckles DM, Smith AM, ap Rees T. A cytosolic ADP-glucose pyrophosphorylase is a feature of Graminaceous endosperms, but not of other starch-storing organs. Plant Physiol. 2001;125:818–827. doi: 10.1104/pp.125.2.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhave MR, Lawrence S, Barton C, Hannah LC. Identification and molecular characterization of Shrunken-2cDNA clones of maize. Plant Cell. 1990;2:581–588. doi: 10.1105/tpc.2.6.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Zhang X-Q, Hrmova M, Fincher GB. A single limit dextrinase gene is expressed both in the developing endosperm and in germinated grains of barley. Plant Physiol. 1999;119:859–872. doi: 10.1104/pp.119.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossins EA, Kopala LC, Blawacky B, Spronk AM. Some properties of higher plant alcohol dehydrogenase. Phytochemistry. 1968;7:1125–1134. [Google Scholar]
- Craig J, Barratt P, Tatge H, Déjardin A, Handley L, Gardner C, Barber L, Wang TL, Hedley C, Martin C et al. Mutations at the rug4locus alter the carbon and nitrogen metabolism of pea plants through an effect on sucrose synthase. Plant J. 1999;17:101–110. [Google Scholar]
- Denyer K, Barber LM, Edwards EA, Smith AM, Wang TL. Two isoforms of the GBSSI class of granule-bound starch synthase are differentially expressed in the pea plant (Pisum sativumL) Plant Cell Environ. 1997;20:1566–1572. [Google Scholar]
- Denyer K, Dunlap F, Thorbjørnsen T, Keeling P, Smith AM. The major form of ADP-glucose pyrophosphorylase in maize endosperm is extraplastidial. Plant Physiol. 1996;112:779–785. doi: 10.1104/pp.112.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer K, Smith AM. The capacity of plastids from developing pea cotyledons to synthesize acetyl CoA. Planta. 1988;173:172–182. doi: 10.1007/BF00403008. [DOI] [PubMed] [Google Scholar]
- Dickinson DB, Preiss J. Presence of ADP-glucose pyrophosphorylase in Shrunken-2 and Brittle-2mutants of maize endosperm. Plant Physiol. 1969;44:1058–1062. doi: 10.1104/pp.44.7.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan DNP, Rudi H, Olsen O-A. The allosterically unregulated isoform of ADP-glucose pyrophosphorylase from barley endosperm is the most likely source of ADP-glucose incorporated into endosperm starch. Plant Physiol. 1999;121:965–975. doi: 10.1104/pp.121.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimert K, Luo C, Déjardin A, Villand P, Thorbjørnsen T, Kleczkowski LA. Molecular cloning and expression of the large subunit of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) leaves. Gene. 1997;189:79–82. doi: 10.1016/s0378-1119(96)00837-2. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YB, Ballicora MA, Leykam JF, Preiss J. Mechanism of reductive activation of potato tuber ADP-glucose pyrophosphorylase. J Biol Chem. 1998;273:25045–25052. doi: 10.1074/jbc.273.39.25045. [DOI] [PubMed] [Google Scholar]
- Giroux MJ, Hannah LC. ADP-glucose pyrophosphorylase in shrunken-2 and brittle-2mutants of maize. Mol Gen Genet. 1994;243:400–408. doi: 10.1007/BF00280470. [DOI] [PubMed] [Google Scholar]
- Giroux MJ, Smith-White B, Gilmore V, Hannah LC, Preiss J. The large subunit of the embryo isoform of ADP glucose pyrophosphorylase from maize. Plant Physiol. 1995;108:1333–1334. doi: 10.1104/pp.108.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Casati DF, Iglesias AA. ADP-glucose pyrophosphorylase from wheat endosperm: purification and characterization of an enzyme with novel regulatory properties. Planta. 2002;214:428–434. doi: 10.1007/s004250100634. [DOI] [PubMed] [Google Scholar]
- Gross P, ap Rees T. Alkaline inorganic pyrophosphatase and starch synthesis in amyloplasts. Planta. 1986;167:140–145. doi: 10.1007/BF00446381. [DOI] [PubMed] [Google Scholar]
- Hannah LC, Shaw JR, Giroux MJ, Reyss A, Prioul J-L, Bae J-M, Lee J-Y. Maize genes encoding the small subunit of ADP-glucose pyrophosphorylase. Plant Physiol. 2001;127:173–183. doi: 10.1104/pp.127.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner CF, Denyer K, Hawker JS. Caution on the use of the generally accepted methanol precipitation technique for the assay of soluble starch synthase in crude extracts of plant tissues. Aust J Plant Physiol. 1994;21:17–22. [Google Scholar]
- Nakamura Y, Kawaguchi K. Multiple forms of ADPglucose pyrophosphorylase of rice endosperm. Physiol Plant. 1992;84:336–342. [Google Scholar]
- Olive MR, Ellis RJ, Schuch WW. Isolation and nucleotide sequences of cDNA clones encoding ADP-glucose pyrophosphorylase polypeptides from wheat leaf and endosperm. Plant Mol Biol. 1989;12:525–538. doi: 10.1007/BF00036967. [DOI] [PubMed] [Google Scholar]
- Pilling EM. The origins of growth rings in starch granules. PhD thesis. East Anglia, UK: University of East Anglia; 2001. [Google Scholar]
- Plaxton WC, Preiss J. Purification and properties of nonproteolytically degraded ADPglucose pyrophosphorylase from maize endosperm. Plant Physiol. 1987;83:105–112. doi: 10.1104/pp.83.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J. Biology and molecular biology of starch synthesis and its regulation. Oxford Surveys Plant Mol Cell Biol. 1991;7:59–114. [Google Scholar]
- Reeves CD, Krishnan HB, Okita TW. Gene expression in developing wheat endosperm. Plant Physiol. 1986;82:34–40. doi: 10.1104/pp.82.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC, Pien F-M, Liu K-C. Nucleotides and nucleotide sugars in developing maize endosperms. Plant Physiol. 1996;110:835–843. doi: 10.1104/pp.110.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikka VK, Choi S-B, Kavakli H, Sakulsingharoj C, Gupta S, Ito H, Okita TW. Subcellular compartmentation and allosteric regulation of the rice endosperm ADPglucose pyrophosphorylase. Plant Sci. 2001;161:461–468. [Google Scholar]
- Smith AM, Bettey M, Bedford ID. Evidence that the rblocus alters the starch content of developing pea embryos through an effect on ADP glucose pyrophosphorylase. Plant Physiol. 1989;89:1279–1284. doi: 10.1104/pp.89.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-White BJ, Preiss J. Comparison of proteins of ADP-glucose pyrophosphorylase from diverse sources. J Mol Evol. 1992;34:449–464. doi: 10.1007/BF00162999. [DOI] [PubMed] [Google Scholar]
- Speicher KD. Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J Biomol Technol. 2000;11:74–86. [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Blissett KJ, Emes MJ. A rapid method for the isolation of purified amyloplasts from wheat endosperm. Planta. 1993;189:597–600. [Google Scholar]
- Thorbjørnsen T, Villand P, Denyer K, Olsen O-A, Smith AM. Distinct isoforms of ADPglucose pyrophosphorylase occur inside and outside the amyloplasts in barley endosperm. Plant J. 1996a;10:243–250. [Google Scholar]
- Thorbjørnsen T, Villand P, Kleczkowski LA, Olsen OA. A single gene encodes two different transcripts for the ADP-glucose pyrophosphorylase small subunit from barley (Hordeum vulgare) Biochem J. 1996b;313:149–154. doi: 10.1042/bj3130149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CY, Nelson OE. Starch-deficient maize mutants lacking adenosine diphosphate glucose pyrophosphorylase activity. Science. 1966;151:341–343. doi: 10.1126/science.151.3708.341. [DOI] [PubMed] [Google Scholar]
- Villand P, Aalen R, Olsen OA, Lüthi E, Lonneborg A, Kleczkowski LA. PCR amplification and sequences of cDNA clones for the small and large subunits of ADP-glucose pyrophosphorylase from barley tissues. Plant Mol Biol. 1992;19:381–389. doi: 10.1007/BF00023385. [DOI] [PubMed] [Google Scholar]
- Vothknecht UC, Soll J. Protein import: the hitchhikers guide into chloroplasts. Biol Chem. 2000;381:887–897. doi: 10.1515/BC.2000.110. [DOI] [PubMed] [Google Scholar]