Abstract
Sweet basil (Ocimum basilicum) peltate glandular trichomes produce a variety of small molecular weight phenylpropanoids, such as eugenol, caffeic acid, and rosmarinic acid, that result from meta hydroxylation reactions. Some basil lines do not synthesize eugenol but instead synthesize chavicol, a phenylpropanoid that does not contain a meta hydroxyl group. Two distinct acyltransferases, p-coumaroyl-coenzyme A:shikimic acid p-coumaroyl transferase and p-coumaroyl-coenzyme A:4-hydroxyphenyllactic acid p-coumaroyl transferase, responsible for the production of p-coumaroyl shikimate and of p-coumaroyl 4-hydroxyphenyllactate, respectively, were partially purified and shown to be specific for their substrates. p-Coumaroyl-coenzyme A:shikimic acid p-coumaroyl transferase is expressed in basil peltate glands that are actively producing eugenol and is not active in glands of noneugenol-producing basil plants, suggesting that the levels of this activity determine the levels of synthesis of some meta-hydroxylated phenylpropanoids in these glands such as eugenol. Two basil cDNAs encoding isozymes of cytochrome P450 CYP98A13, which meta hydroxylates p-coumaroyl shikimate, were isolated and found to be highly similar (90% identity) to the Arabidopsis homolog, CYP98A3. Like the Arabidopsis enzyme, the basil enzymes were found to be very specific for p-coumaroyl shikimate. Finally, additional hydroxylase activities were identified in basil peltate glands that convert p-coumaroyl 4-hydroxyphenyllactic acid to its caffeoyl derivative and p-coumaric acid to caffeic acid.
Basil (Ocimum basilicum) plants synthesize significant quantities of phenylpropanoid derivatives that contain an hydroxyl group at the meta position on the aromatic ring. Several of these compounds, such as eugenol, rosmarinic acid, and caffeic acid (Fig. 1), are found in high concentrations in specialized structures that are located on the surface of the aerial parts of the plant and are known as peltate glandular trichomes (glands; Gang et al., 2001). These specialized metabolites have been found in other plant species as well, but the nature of the enzymes that catalyze the meta hydroxylation has so far remained poorly understood.
Figure 1.
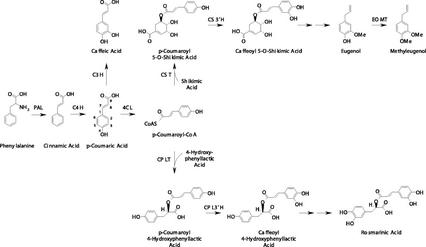
Proposed biosynthetic pathways to meta hydroxylated phenylpropanoids in sweet basil. Single arrows indicate verified transformations; double arrows indicate potential transformations. Enzymes are as follows: PAL, Phe ammonia lyase; C4H, cinnamate 4-hydroxylase; C3H, p-coumaric acid 3-hydroxylase; CPL3′H, p-coumaroyl hydroxyphenyllactate 3′-hydroxylase; CS3′H, p-coumaroyl shikimate 3′-hydroxylase; EOMT, eugenol O-methyltransferase.
One of the difficulties in identifying the enzymes catalyzing the 3-hydroxylations in the phenylpropanoid pathways is that they appear to be found at very low abundance in the tissues examined (Petersen, 1997). We have recently shown (Gang et al., 2001) that, as in peppermint (Mentha × piperita; Gershenzon et al., 1992; McCaskill and Croteau, 1995), the basil gland cells can be removed from the plant and studied in isolation from the rest of the plant, greatly facilitating biochemical and molecular investigations of a single, fully differentiated cell type (Gershenzon et al., 1992; McCaskill et al., 1992; McCaskill and Croteau, 1995; Lange et al., 2000; Gang et al., 2001). Furthermore, because these gland cells are so highly specialized for production of plant-specialized metabolites, the enzymes in these metabolic pathways are highly expressed compared with other tissues (Gang et al., 2001, 2002).
Another advantage of the basil system is the availability of different breeding lines that synthesize different sets of specialized metabolites, including phenylpropenes. For example, basil line SW synthesizes mostly eugenol, a phenylpropene with a meta hydroxyl group, whereas line EMX-1 does not synthesize eugenol and instead synthesizes methylchavicol, a phenylpropene with no meta hydroxyl group (Gang et al., 2001). We have exploited the availability of these varieties to identify the enzymatic activities in basil peltate glandular trichomes that are responsible for the differential synthesis of some meta-hydroxylated compounds.
RESULTS
Partial Purification and Properties of Basil Hydroxycinnamoyl Acyltransferases
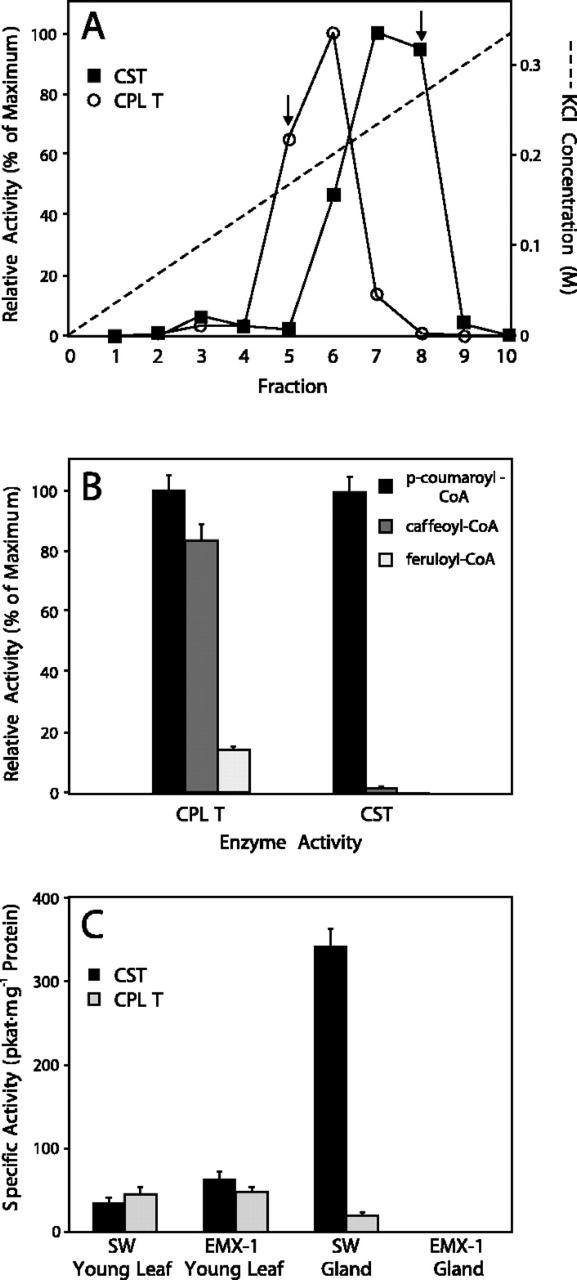
A p-coumaroyl-CoA:shikimic acid p-coumaroyl transferase (CST) and a p-coumaroyl-CoA:4-hydroxyphenyllactic acid p-coumaroyl transferase (CPLT) had been identified in other plant species (Ulbrich and Zenk, 1980; Petersen and Alfermann, 1988; Petersen, 1991; Lofty et al., 1992). Therefore, we examined the enzymatic activities present in our basil lines that can synthesize p-coumarate esters such as p-coumaroyl shikimate and p-coumaroyl 4-hydroxyphenyllactic acid. Crude protein extracts from whole young leaves of both of these basil lines contained hydroxycinnamoyl acyltransferase activities that transferred a p-coumaroyl group to hydroxyl functional groups on either (−)-shikimic acid or 4-hydroxyphenyllactic acid (Fig. 2). No extracts of leaves of any age or of isolated peltate glands from either line were able to perform similar conversions with quinic acid as the acyl acceptor (CQT activity). A partial separation of CST and CPLT activities was obtained on a weak anion-exchange column (DE53 cellulose). This indicated that CST and CPLT enzyme activities are catalyzed by two separate proteins in basil, as had been demonstrated before for CST and CQT in apple (Malus domestica) and date fruits (Lofty et al., 1992) and in Stevia raubaudiana cell suspension cultures (Ulbrich and Zenk, 1979). After the active fractions of each enzyme from the DE53 column were combined, diluted, and applied to a strong anion-exchange (MonoQ) column, the CST and CPLT activities were able to be separated (Fig. 3A) so that fractions with only one of the activities were obtained, even though the two activities overlapped significantly.
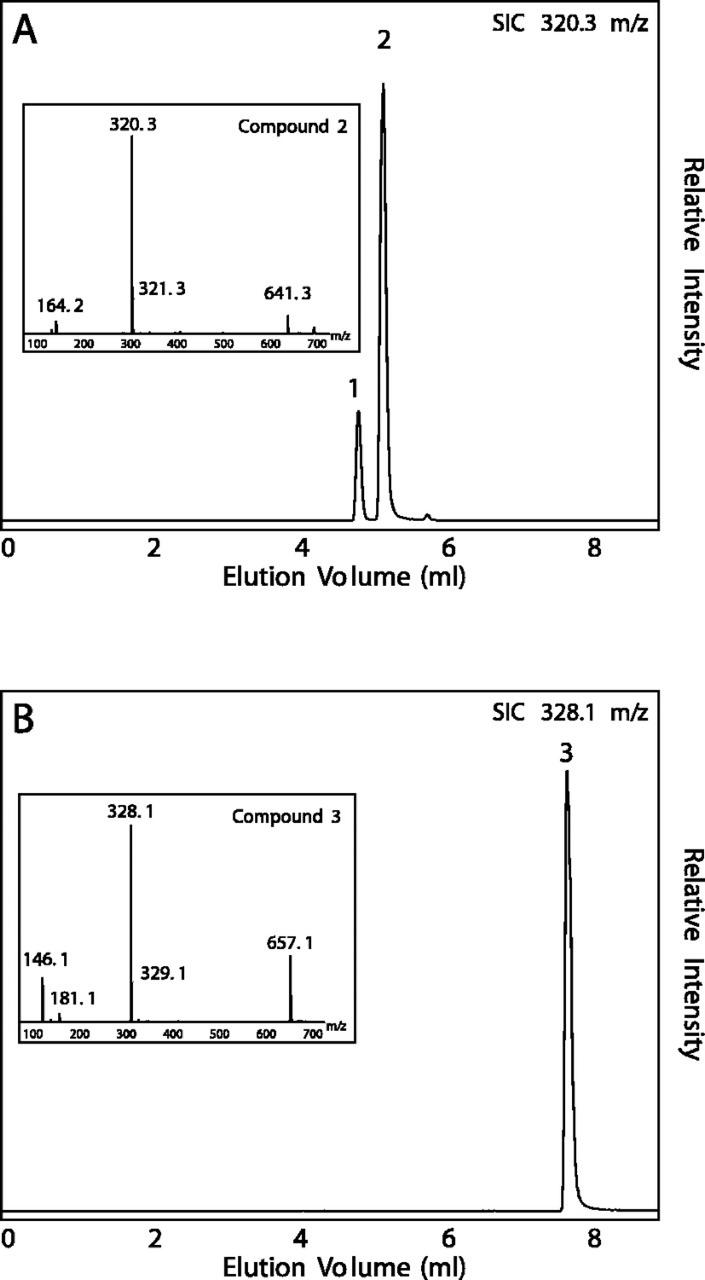
Figure 2.
Liquid chromatography (LC)/mass spectrometry (MS) analysis of acyltransferase assays with whole-leaf protein extracts from basil line SW. A, Assay with [8-13C]-p-coumaroyl-CoA and shikimic acid as substrates. Elution trace is the selected ion chromatogram at 320.3 mass-to-charge ratio (m/z). Compound 1, [8′-13C]-p-coumaroyl-4-O-shikimate. Inset, Electrospray ionization negative mode mass spectrum of compound 2 ([8′-13C]-p-coumaroyl-5-O-shikimate). B, Assay with [8-13C]-p-coumaroyl-CoA and 4-hydroxyphenyllactic acid as substrates. Elution trace is the selected ion chromatogram at 328.1 m/z. Inset, Electrospray ionization negative mode mass spectrum of compound 3 ([8′-13C]-p-coumaroyl-4-hydroxyphenyllactate).
Figure 3.
Partial purification, properties and tissue specific activity of basil acyltransferases. A, Elution of basil acyltransferases CPLT (white circles) and CST (black squares) from a strong anion-exchange analytical FLPC column (MonoQ). Arrows indicate fractions for each enzyme activity that are essentially free from the opposing acyltransferase activity. Relative activity at maximum (100%) corresponds to 196 and 108 pkat mg−1, respectively, for CPLT and CST. B, Substrate specificity of acyltransferases partially purified from basil line SW. Bars are relative activities for each respective enzyme preparation with the substrates p-coumaroyl-CoA (black), caffeoyl-CoA (dark gray), and feruloyl-CoA (light gray). Error bars are se. C, Specific enzyme activities for CST (black bars) and CPLT (light-gray bars) in crude protein extracts from selected tissues from basil lines SW and EMX-1. Error bars are se.
Both CST and CPLT, separated from each other and purified approximately 10-fold over the MonoQ column, preferred p-coumaroyl-CoA as a substrate over the other hydroxycinnamoyl-CoA esters (Fig. 3B), as was found for the related enzyme from Coleus blumei (Petersen et al., 1993). This was especially true for the activity that transferred hydroxycinnamoyl groups to shikimate (CST). The activity of CST with caffeoyl-CoA was 100-fold lower than the activity with p-coumaroyl-CoA. Feruloyl-CoA did not serve as a substrate for CST, and it was a poor substrate for CPLT.
Both CST and CPLT were found to be extremely stable enzymes at room temperature and in the freezer. Incubation for days at room temperature, multiple freeze/thaw cycles, and holding at −20°C for several months, with or without the addition of 10% (w/v) glycerol, led to no loss of activity. The enzymes appeared to be extremely stable at 37°C as well. However, temperatures above 40°C did lead to rapid and irreversible loss of activity. Therefore, all assays were carried out at 37°C. The basil CST and CPLT enzymes were most active at pH 7 in 50 mm potassium phosphate buffer. They showed approximately 50% lower activity in non-phosphate buffers. The approximate molecular mass for CST was determined on a calibrated Superose 12 column to be 48,000 D. The molecular mass of CPLT could not be clearly determined because this activity eluted in a very broad range from the Superose 12 column.
Differences between SW and EMX-1 Lines in the Activities of CST and CPLT
Next, we evaluated the specific activities of CST and CPLT in young leaves and in isolated peltate glands from basil lines SW and EMX-1 (Fig. 3C). CST activity was about 2-fold higher in leaves of the EMX-1 line than in SW leaves, but the glands of the EMX-1 line showed no CST activity, although they demonstrated good hydroxylase activities (Fig. 4). On the other hand, CST activity was greatly elevated in peltate glands of the SW line (about 10-fold higher compared with CST activity in SW leaves). The activity of CPLT was almost identical in leaves of both basil lines. However, CPLT activity was about 3-fold lower in the SW peltate glands when compared with SW leaves, and glands of the EMX-1 line did not have any detectable CPLT activity.
Figure 4.
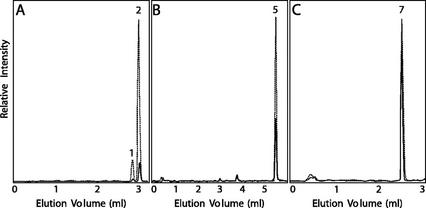
Comparison of 3′- and 3-hydroxylase activities in glands isolated from young leaves on mature plants from basil lines SW (dotted lines) and EMX-1 (solid lines). A, Representative LC/MS-selected ion chromatograms (336 m/z) for products of glands incubated with [8′-13C]-p-coumaroyl-5-O-shikimate. B, Representative LC/MS-selected ion chromatograms (344 m/z) for products of glands incubated with [8′-13C]-p-coumaroyl 4-hydroxyphenyllactate. C, Representative LC/MS-selected ion chromatograms (180 m/z) for products of glands incubated with [8-13C]-p-coumaric acid. The same amount of glands, as measured by total protein concentration and the same amount of substrate (0.6 mm), was added in each incubation for both basil lines. All assays were initiated and terminated at the same time, when approximately 5% conversion was observed for the assays in C. The peaks numbered were identified as follows: 1, [8′-13C]-caffeoyl-4-O-shikimate; 2, [8′-13C]-caffeoyl-5-O-shikimate; 5, [8′-13C]-caffeoyl 4-hydroxyphenyllactate; and 7, [8-13C]-caffeic acid. Relative activity levels are for each type of reaction, but scales are not comparable between panels.
Identification of 3-Hydroxylation Activities in Basil Glands
Peltate glands isolated from both basil lines (SW and EMX-1) possessed the ability to form caffeoyl 5-O-shikimate, caffeoyl 4-hydroxyphenyllactate, and caffeic acid from p-coumaroyl-5-O-shikimate, p-coumaroyl 4-hydroxyphenyllactate, and p-coumaric acid, respectively (see Fig. 4). For the transformation of p-coumaric acid to caffeic acid, both lines appeared to possess about the same level of 3-hydroxylase activity. However, the 3-hydroxylases that form caffeoyl 5-O-shikimate and caffeoyl 4-hydroxyphenyllactate were 9- and 3-fold, respectively, more active in line SW than in line EMX-1.
Identification of a cDNA Encoding a 3-Hydroxylase from Basil Peltate Glands
The evidence presented above indicates that glands of both SW and EMX-1 lines are able to perform 3-hydroxylation on at least three different phenylpropanoids. To determine whether the glands possess different 3-hydroxylating enzymes with unique substrate specificities, we analyzed our basil (line EMX-1) peltate glandular trichome expressed sequence tag (EST) database (Gang et al., 2001) for genes that could potentially serve as the 3-hydroxylases. Potential candidates included a 2-oxoglutarate dependent dioxygenase and several cytochromes P450. We expressed the 2-oxoglutarate dependent dioxygenase in Escherichia coli, with excellent yield of soluble recombinant protein (data not shown), but this enzyme did not serve as the catalyst for the 3-hydroxylation of the potential substrates p-coumaroyl-5-O-shikimate, p-coumaroyl 4-hydroxyphenyllactate, and p-coumaric acid or other potential intermediate in the pathways to caffeic acid, rosmarinic acid, or caffeoyl shikimate.
Next, we evaluated the ESTs that represented cytochromes P450 more closely and found that one EST encoded an enzyme (CYP98A13) that was very closely related to other cytochromes P450 whose sequences had been obtained as a result of EST production projects with sorghum (Sorghum bicolor; CYP98A1; Bak et al., 1998) and soybean (Glycine max; CYP98A2) and as a result of the Arabidopsis genome sequencing project (CYP98A3).
Based on this EST, we used a combined 5′-RACE/genome walking approach (see “Materials and Methods”) to obtain full-length cDNAs for two basil cytochrome P450 proteins with 99% identity to each other. Interestingly, all 5′-RACE experiments with basil line EMX-1 yielded a product that was truncated at the same position observed in the EST originally identified, whereas 5′-RACE with line SW yielded products of several different lengths, one of which was almost full length. Genome walking with both basil lines EMX-1 and SW revealed that the truncation observed in the original EST and the 5′-RACE experiments with line EMX-1 occurred at a splice junction. Thus, the truncated mRNAs appear to have resulted from incomplete transcript processing.
The two closely related basil cytochromes P450 proteins have been designated CYP98A13v1 and CYP98A13v2 (GenBank accession nos. AY082611 and AY082612, respectively). To determine the enzymatic activity of these two isoforms, functional CYP98A13 proteins were obtained using a yeast (Saccharomyces cerevisiae) expression system. This system uses yeast strain WAT11, which expresses the Arabidopsis cytochrome P450 reductase (Urban et al., 1994), with plant cytochromes P450 being expressed from a stable self-replicating plasmid (pYeDP60).
CO difference spectra from the microsomes purified from yeast expressing the basil P450s indicated that good expression of the recombinant proteins was achieved (data not shown). We next assayed these microsomal fractions for hydroxylase activity (Fig. 5). These microsomal fractions efficiently converted [8′-13C]-p-coumaroyl shikimate substrate into [8′-13C]-caffeoyl shikimate as previously described for Arabidopsis CYP98A3 (Schoch et al., 2001). Two major isomers of [8′-13C]-p-coumaroyl shikimate, the [8′-13C]-p-coumaroyl 5-O-shikimate and [8′-13C]-p-coumaroyl 4-O-shikimate esters, were present in the substrate mixture added to these assays. Formation of the [8′-13C]-p-coumaroyl 4-O-shikimate isomer is the result of rearrangement at physiological pH of the [8′-13C]-p-coumaroyl 5-O-shikimate substrate. This isomerization is a well-characterized property of these substrates (Kühnl et al., 1987). Both CYP98A13v1 and CYP98A13v2 converted >97% of both p-coumaroyl shikimate esters to the corresponding caffeoyl esters under these conditions (see “Materials and Methods”). p-Coumaroyl-quinate was also a substrate for these enzymes, resulting in the formation of caffeoyl-quinate (Table I).
Figure 5.
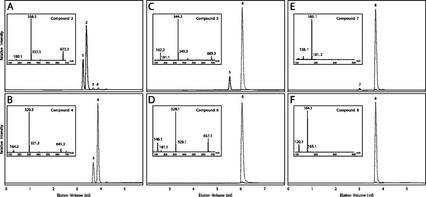
Enzymatic activity of basil CYP98A13 in purified yeast microsomes. A, C, and E, Assays with recombinant CYP98A13v1. B, D, and F, Assays with WAT11 control. A and B, Assays with [8′-13C]-p-coumaroyl shikimate as substrate. C and D, Assays with [8′-13C]-p-coumaroyl 4-hydroxyphenyllactate as substrate. E and F, Assays with [8-13C]-p-coumaric acid as substrate. Solid lines, Selected ion chromatograms for major ion (m/z) of eluting product peak. Dotted lines, Selected ion chromatograms for major ion (m/z) of eluting substrate peak. Insets, Electrospray ionization negative mode mass spectra for selected peaks. Peaks were identified as follows: 1, [8′-13C]-caffeoyl-4-O-shikimate; 2, [8′-13C]-caffeoyl-5-O-shikimate; 3, [8′-13C]-p-coumaroyl-4-O-shikimate; 4, [8′-13C]-p-coumaroyl-5-O-shikimate; 5, [8′-13C]-caffeoyl 4-hydroxyphenyllactate; 6, [8′-13C]-p-coumaroyl 4-hydroxyphenyllactate; 7, [8-13C]-caffeic acid; and 8, [8-13C]-p-coumaric acid.
Table I.
Comparison of some kinetic parameters for selected potential substrates with recombinant basil CYP98A13 proteins
| Substrate | Enzyme | Km | Kcat | Kcat/Km |
|---|---|---|---|---|
| μm | min−1 | μm−1 min−1 | ||
| p-Coumaroyl-shikimate | CYP98A13v1 CYP98A13v2 | 2.7 ± 0.5 2.9 ± 0.4 | 109.0 ± 3.1162.8 ± 3.7 | 4156 |
| p-Coumaroyl-quinate | CYP98A13v1 CYP98A13v2 | 27.5 ± 4.7 19.7 ± 6.8 | 91.4 ± 7.4189.0 ± 2.4 | 3.3 9.6 |
| p-Coumaric acid | CYP98A13v1 CYP98A13v2 | 5,100 ± 1005,100 ± 300 | n.d.an.d. | n.d. n.d. |
n.d., Not determined.
In contrast to the high conversion of the p-coumaroyl shikimate esters, other metabolites found to be hydroxylated in basil glands and leaves were poor substrates for these recombinant enzymes (Fig. 5; Table I). For example, only about 15%, 2.5%, and 0.5% conversions were observed, respectively, for p-coumaroyl 4-hydroxyphenyllactate, p-coumaric acid, and p-coumaroyl-CoA, when near 100% conversion of the p-coumaroyl shikimate esters to the caffeoyl forms had been achieved. When assays were run for a period of time such that only 5% to 10% of the p-coumaroyl 5-O-shikimate substrate was converted to the caffeoyl shikimate derivative, no detectable conversion of the other potential substrates could be observed. Apparent Km values for p-coumaroyl shikimate and p-coumaric acid were about 3 and >5,000 μm, respectively (Table I). Thus, it appears that the substrate specificity of the basil CYP98A13 enzymes is similar to the substrate specificity of the recently characterized Arabidopsis homolog (Schoch et al., 2001) and that these enzymes are responsible for formation of caffeoyl shikimate in basil peltate glands.
DISCUSSION
In the work presented here, we were able to identify two distinct acyltransferases in basil leaves. These enzymes, CST and CPLT, are very specific for their substrates and are expressed differentially in basil tissues: CST activity is much higher in tissues that are actively producing eugenol (i.e. in the peltate glands of basil lines producing eugenol) than in tissues that are not, and CPLT activity is higher in whole leaf tissue than in peltate glands. Similar acyltransferase activities, which transfer a p-coumaroyl group to quinic acid, have been identified in other species (Ulbrich and Zenk, 1979; Lofty et al., 1992). Quinic acid is a component of chlorogenic acid (=caffeoyl-3-O-quinate), an aromatic acid that is relatively abundant in the plant kingdom. Chlorogenic acid has been implicated as a possible intermediate of the phenylpropanoid pathway (Schoch et al., 2001), along with caffeoyl shikimate. However, no quinic acid esters (including chlorogenic acid) were found in extracts from leaves or peltate glandular trichomes from either basil line. Furthermore, no CQT activity was found in leaf or gland extracts from either basil line.
We also showed that the presence or absence of the two distinct acyltransferases (CST and CPLT) in basil is not sufficient to explain the production of different 3-hydroxylated compounds in this plant. The basil CYP98A13 enzymes appear to have about the same level of catalytic efficiency as their Arabidopsis counterpart (CYP98A3) in the conversion of p-coumaroyl 5-O-shikimate to caffeoyl 5-O-shikimate, with turnover numbers between 100 and 600 per minute (between 6,000 and 36,000 per hour) and apparent Km values between 2.5 and 7 μm (Schoch et al., 2001). In addition, the basil CYP98A13 isozymes, like the orthologous Arabidopsis enzyme, were capable of meta-hydroxylating p-coumaroyl-quinate. However, the basil enzymes were 5- to 10-fold more efficient with the shikimate ester, whereas the Arabidopsis enzyme is about 4-fold more efficient with the shikimate ester than with the quinate ester (Schoch et al., 2001). On the other hand, the characterization of basil CYP98A13v1 and CYP98A13v2 showed that these enzymes do not readily hydroxylate p-coumaroyl 4-hydroxyphenyllactate or p-coumaric acid. The latter observation was also reported by Franke et al. (2002), who measured a turnover number for Arabidopsis CYP98A with p-coumaric acid of three per hour and an apparent Km value around 2,500 to 5,000 μm, values that clearly indicate that p-coumaric acid is not a physiological substrate for this enzyme.
Therefore, our results indicate that the formation of rosmarinic acid and free caffeic acid in basil leaves and peltate glands proceeds through the action of separate hydroxylases, which are not part of the more general phenylpropanoid pathway that includes the activity of the CYP98A enzymes. Such additional hydroxylase activities were identified in peltate glands from basil plants (Fig. 4), but these enzymes are yet to be purified and characterized. Nevertheless, NADPH and O2 were required for their activity, and the enzymes were fully functional in the presence of reducing agents such as dithioerythritol (DTE; which was included in the extraction and assay buffers). This suggests that these hydroxylases in sweet basil peltate glands and leaves are not likely to be nonspecific phenolases (Patil and Zucker, 1965), nor are they likely to be the potential hydroxylases that have been reported to require FAD (Kamsteeg et al., 1981) or Zn2+ and ascorbate (Kneusel et al., 1989) for activity. It seems likely that the enzyme(s) responsible for the 3′-hydroxylation of p-coumaroyl 4-hydroxyphenyllactate in sweet basil will be related to the analogous enzyme in C. blumei (Petersen et al., 1993; Petersen, 1997). This enzyme, not purified to date, is also very specific for its substrate, and p-coumaric acid cannot be utilized as a substrate (Petersen, 1997). It will be interesting to discover whether these alternative hydroxylases evolved from CYP98A homologs, or from other unrelated enzymes.
MATERIALS AND METHODS
Plant Material
Two lines of basil (Ocimum basilicum) designated EMX-1 and SW were grown as described elsewhere (Gang et al., 2001).
Reagents
Unless specified otherwise, all solvents and reagents were molecular biology grade or reagent grade or better and were obtained from Sigma (St. Louis), Aldrich (Milwaukee, WI), or Fisher Scientific (Pittsburgh). [U-13C]-l-Phe (>99.99% 13C) was purchased from Isotec (Miamisburg, OH). [8-14C]p-Coumaric, [8-14C]caffeic, and [8-14C]ferulic acids were synthesized at 0.2 mmol scale from [2-14C]malonic acid (Amersham Biosciences, Piscataway, NJ) and the corresponding substituted 4-hydroxybenzaldehyde, as previously described (Gang et al., 2001). Stable isotope-labeled hydroxycinnamic acids ([8-13C]-p-coumaric, [8-13C]-caffeic, and [8-13C]-ferulic acids) were synthesized using the same procedure at 1 mmol scale, using [2-13C]-malonic acid, >99.9% 13C at the 2 position (Isotec, Inc.). Identity of the synthesized compounds was confirmed by comparison with authentic standards for matching elution volume, UV spectra and mass spectra in LC/MS analysis, and retention time and mass spectra in gas chromatography (GC)/MS analysis.
Instrumentation
Protein purification was performed using FPLC (Pharmacia Biotech, Piscataway, NJ). GC/MS was performed on a QP-5000 GC/MS system (Shimadzu, Columbia, MD) equipped with an Econo-Cap SE-54 capillary column (30-m × 0.32-mm i.d., 1.0-μm film thickness, Alltech, Deerfield, IL), as previously described (Gang et al., 2001).
HPLC was performed using a Discovery HS C18 column (15-cm × 2.1-mm i.d., Supelco, Bellefonte, PA) attached to a 2690 HPLC system (Waters, Milford, MA) with in-line degasser, autosampler, sample incubator, and column heater. Compound elution was monitored (200–400 nm) with a Waters 996 UV/Visible photodiode array detector. Complete baseline separation of all phenylpropene and phenylpropanoid compounds was achieved at a flow rate of 0.25 mL min−1 with the column incubated at a constant temperature of 40°C. Solvent A was 0.05% (v/v) formic acid and 5 mm ammonium acetate in water; solvent B was 100% (v/v) acetonitrile. The column was pre-equilibrated with 5% (v/v) B in A. After injection of up to 25 μL of aqueous sample, the column was washed with 0.5 mL of pre-equilibration solvent. Phenylpropanoids and phenylpropenes were eluted from the column with a linear gradient from 5% to 66% (v/v) B over 13.75 mL. The column was then washed by increasing B to 100% (v/v; linear gradient in 0.75 mL) and holding at 100% (v/v) B for 0.75 mL. The column was then re-equilibrated by returning the column to 5% (v/v) B (over 0.75 mL), followed by a 2.5-mL wash with this solvent. Total run time was 70 min. A shortened gradient (5% to 66% [v/v] B in 6 mL) was also used when only separation of compounds with widely separated elution volumes was required.
LC/MS was performed on HPLC eluents using a Micromass Quattro LCZ triple quadrupole mass spectrometer (Micromass, Inc., Beverly, MA). Flow splitting (10:1) after the Waters 996 photodiode array detector resulted in an inlet flow rate into the mass spectrometer electrospray source (ESI Z-Spray) of 20 to 25 μL min−1. This dramatically enhanced the signal when compared with non-split samples. Ionization of target molecules in negative ion mode was achieved with a capillary voltage of −3.2 kV and a cone voltage of 30 V. For positive ionization mode, the capillary voltage was set at 3.25 kV. The desolvation and cone gases were set at 450 and 50 L h−1, respectively, and the desolvation and source temperatures were 250°C and 120°C, respectively. Mass detection was performed in scanning mode, at 450 atomic mass units s−1, with 0.1-s interscan delay. All other electrospray source and instrument parameters were set as recommended by the instrument manufacturer. Data analysis was performed using MassLynx computer software (Micromass, Inc.).
Protein Extracts
Small basil leaves (<2 cm long, 20 g total) were harvested and ground in liquid nitrogen. Protein extraction buffer (50 mm BIS-Tris [pH 7], 1% [w/v] polyvinylpyrrolidone [Mr of 40,000], and 10 mm DTE), was added at a ratio of 10:1 (v/w) buffer:tissue, and the samples were kept on ice for 30 min. After filtration through Miracloth (Calbiochem, San Diego) and centrifugation (25,000g for 30 min), the crude extracts were either used immediately for production of 13C-labeled compounds, used immediately for purification of enzyme activities, or supplemented with 10% (w/v) glycerol and stored at −80°C for later enzyme assays. Basil peltate glandular trichomes, isolated as previously described (Gang et al., 2001), were also used for crude protein extracts. Isolated glandular trichomes were resuspended in the same protein extraction buffer as was used for leaves. The glandular trichome cells were lysed by sonication (30 s) on ice, and the cellular debris was removed by centrifugation at 20,000g in a cooled microcentrifuge. The supernatant was supplemented with 10% (w/v) glycerol and stored at −80°C for later enzyme assays.
Partial Purification of Acyltransferases from Basil Leaves
Crude protein extracts (up to 45 mL) obtained from whole young leaves were applied to a freshly prepared DE53 Cellulose column (8-mL bed volume), pre-equilibrated in buffer A (25 mm BIS-Tris [pH 7.0] and 10% [w/v] glycerol). After washing to remove unbound proteins, the acyltransferase activities were eluted using a linear salt gradient from 0 to 0.5 m KCl in buffer A. Fractions containing CST and CPLT activities, which overlapped significantly, were pooled and diluted five times. This mixture was then applied to a MonoQ analytical column (Pharmacia), and the two acyltransferase activities, which bound to the column under these conditions, were separated by a linear gradient of 0 to 0.5 m KCl (elution around 200 mm). Active fractions were desalted into buffer A and concentrated (Nanosep microconcentrators, Pall Filtron, Pall Corporation (East Hills, NY), and then applied to a Superose 12 column (Pharmacia) that had been calibrated using cytochrome c (Mr of 12,400), carbonic anhydrase (Mr of 29,000), bovine serum albumin (Mr of 66,000), and β-amylase (Mr of 200,000) in buffer A.
Radiolabeled Acyltransferase Assay
To monitor protein elution from columns during purification, we developed a radiochemical assay for CST and CPLT activities that used [8-14C]p-coumaroyl-CoA as substrate. [8-14C]p-Coumaroyl-CoA, [8-14C]caffeoyl-CoA, and [8-14C]feruloyl-CoA were produced enzymatically from CoA and the corresponding [8-14C]-labeled hydroxycinnamic acid using recombinant tobacco (Nicotiana tabacum) 4-coumarate:CoA ligase (4CL) as described by Beuerle and Pichersky (2002). The enzymatic hydroxycinnamoyl-CoA ester synthesis proceeded at room temperature in Tris-HCl buffer (50 mm, pH 7.5) or in potassium phosphate buffer (50 mm, pH 7) with near equal efficiency. Because the acyltransferase assays were much more efficient in the potassium phosphate buffer, this buffer was used for all syntheses. The reactions consisted of 50 mm potassium phosphate buffer (pH 7), 0.5 mm ascorbate (Na+ salt), 2.5 mm ATP, 1 mm CoA, 1,000 cpm μL−1 [8-14C]hydroxycinnamic acid in ethanol (approximately 5,500 cpm nmol−1 for all three), and approximately 0.02 mg mL−1 purified recombinant 4CL. After incubation at room temperature for several hours, two to four were usually sufficient for 100% conversion of free hydroxycinnamic acid to the corresponding CoA ester. The synthesis mixture was used directly as substrate/buffer for acyltransferase assays. These 50-μL assays were initiated by addition to 35 μL of [8-14C]hydroxycinnamoyl-CoA substrate/buffer mixture, of 5 μL of 20 mm acyl acceptor (shikimic acid, CQT, or 4-hydroxyphenyllactic acid), and 10 μL of enzyme to test. Assays were performed at 37°C for up to 30 min and quenched by addition of 4 μL of 6 n HCl. Products were then extracted with 100 μL of ethyl acetate and monitored either by radio-thin-layer chromatography (Polygram SIL G/UV24 silica gel plates, 20 × 20 cm, Machery-Nagel, Duren, Germany; solvent system of 20:1 [v/v] diethyl ether:methanol with 10 drops acetic acid for 120 mL; detection with BioScan 200 Imaging Scanner), or by scintillation counting. Background counts for these assays were typically less than 1%.
Stable Isotope Acyltransferase Assay
To determine specific activity and substrate specificity and to verify the identity of assay products, we used a nonradioactive assay. This was performed very similarly to the radiolabeled assays. Instead of [8-14C]-labeled hydroxycinnamic acids used in CoA ester synthesis with recombinant 4CL, [8-13C]-labeled hydroxycinnamic acids were used. The buffer system, pH, temperature, and assay time, as well as quenching with 6 n HCl and ethyl acetate extraction, remained the same as for the radiolabeled assays. The ethyl acetate was then removed under dry nitrogen, and the assay products resuspended in ethanol before LC/MS analysis. Alternatively, the assays were quenched by addition of 4 μL of 6 n HCl and, after centrifugation to remove precipitated protein, 25 μL of the remaining assay mixture was injected directly into the LC/MS. Products were identified by HPLC elution volume, UV spectrum, and mass spectrum. Quantification of assay products was performed by comparing peak area of samples to a standard curve made from known concentrations of standard compounds.
2-Oxoglutarate-Dependent Dioxygenase Assay
Assays (100-μL final volume) for 2-oxoglutarate-dependent dioxygenase catalyzed hydroxylation of phenylpropanoid intermediates consisted of 50 mm Bis-Tris-propane (pH 7.5), 8 mm 2-oxoglutarate, 8 mm ascorbic acid, 0.05 mm Fe(NH2)2(SO4)2·6H2O, 5 mm dithiothreitol, 1 unit catalase, 2 mg mL−1 bovine serum albumin, 2 mm substrate, and 30 μg of crude protein (basil protein extracts or lysates from overexpressed recombinant protein). After incubation at 23°C for 1 h, the assays were stopped by addition of 10 μL of glacial acetic acid. After centrifugation (14,000g for 10 min), the samples were analyzed by LC/MS as described above.
Isolation of the cDNA Encoding CYP98A13
A preliminary EST database was previously constructed by sequencing random clones from a peltate glandular trichome cDNA library (Gang et al., 2001) obtained from basil line EMX-1. Several cytochromes P450 were identified in the EST database, including transcripts representing cinnamate-4-hydroxylase and a related gene that possessed high similarity to sequences in GenBank from sorghum (Sorghum bicolor; CYP98A1), soybean (Glycine max; CYP98A2), and Arabidopsis (CYP98A3). The basil CYP98A13 sequence was truncated; therefore, RACE (Chenchik et al., 1996; Matz et al., 1999) and genome walking (Siebert et al., 1995a, 1995b) were used to obtain the sequence missing from the 5′ end of the gene. The full-length cDNA was then amplified from first strand cDNA, which had been obtained from total RNA from peltate glands of basil line SW using the First Strand cDNA Synthesis Kit (Amersham-Pharmacia Biotech, Uppsala), using the Advantage cDNA Polymerase (CLONTECH Laboratories, Palo Alto, CA) with primers designed for the 5′ and 3′ ends of the coding region (5′ primer, 5′-CAACCAGCCATGGCAGCTCTCC; and 3′ primer, 5′-CGCCATTTACAAGTCCACAGCAATACG). The resulting 1,500-bp band was subcloned into a TA cloning vector (pCRT7/CT TOPO-TA, Invitrogen, Carlsbad, CA). Several individual transformants were sequenced completely on both strands.
Expression of CYP98A13 in Yeast (Saccharomyces cerevisiae)
Yeast strain WAT11, a derivative of the W303-B strain that expresses the Arabidopsis cytochrome P450 reductase ATR1 (Pompon et al., 1996), was used for expression of the basil CYP98A13 proteins. The cDNAs encoding the basil CYP98A13s were amplified via PCR with the same primers (5′ primer, 5′-AAAGATCTATGGCAGCTCTCCTCCTCCT; and 3′ primer, 5′-GGGGTACCTTACAAGTCCACAGCAATACG) that introduced BglII and KpnI restriction sites (underlined), respectively, to the 5′ and 3′ ends of the cDNA. The amplification products were then subcloned into pCRT7/CT TOPO-TA cloning/expression plasmid and resequenced to ensure that no base pair substitutions had been introduced during PCR. After digestions with the appropriate restriction enzymes, the fragments were directionally ligated into the expression cassette of the plasmid pYeDP60. Plasmids containing the correct inserts were transformed into WAT11 cells, grown, and induced for expression as previously described (Pompon et al., 1996). Microsomes containing expressed CYP98A13 and the Arabidopsis cytochrome P450 reductase were isolated (Pompon et al., 1996) for enzyme assays. Yields from 300 mL of liquid culture after 20 h of induction in Gal medium were typically 30 to 40 mg of microsomal protein.
Cytochrome P450 Assay
Assays for 3-hydroxylase activity with a variety of substrates, including p-coumaric acid, p-coumaroyl-CoA, p-coumaroyl shikimate, and p-coumaroyl 4-hydroxyphenyllactate, were performed and analyzed in a manner similar to the acyltransferase assays. For these assays, [8-13C]-labeled compounds were used, to differentiate from any possible contamination from endogenous substrate. Each 100-μL assay consisted of 50 mm potassium phosphate buffer (pH 7.1), 0.6 mm substrate (10 μL of 6 mm substrate in 10% [v/v] ethanol), 1 mm NADPH, and 0.1 mg of yeast microsomes or basil leaf protein extracts (which contained DTE). Alternatively, whole isolated peltate glandular trichomes were used instead of protein extracts. The assays were incubated at 28°C with shaking to ensure proper oxygenation and were quenched after 1 h by addition of 4 μL of 6 n HCl. After centrifugation to remove precipitated proteins, the resulting assay products were analyzed by LC/MS.
Footnotes
This work was supported by the U.S. Department of Agriculture National Research Initiative (competitive grant no. 0003497), by a Margaret and Herman Sokal Fellowship in the Sciences (to D.R.G.), and by the Deutscher Akademischer Austauschdienst (Gemeinsames Hochschulsonderprogramm III von Bund und Ländern, Germany; fellowship to T.B.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.007146.
LITERATURE CITED
- Bak S, Kahn RA, Nielsen HL, Møller BL, Halkier BA. Cloning of three a-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moensch by a PCR approach and identification by expression in Escherichia coli of the CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol Biol. 1998;36:393–405. doi: 10.1023/a:1005915507497. [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichersky E. Enzymatic synthesis and purification of aromatic coenzyme A esters. Anal Biochem. 2002;302:305–312. doi: 10.1006/abio.2001.5574. [DOI] [PubMed] [Google Scholar]
- Chenchik A, Zhu Y, Diatchenko L, Li R, Hill J, Siebert P. Generation and use of high-quality cDNA from small amounts of total RNA by SMART PCR. In: Larrick J, editor. RT-PCR Methods for Gene Cloning and Analysis. Natick, MA: BioTechniques Books; 1996. pp. 305–319. [Google Scholar]
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 2002;30:33–45. doi: 10.1046/j.1365-313x.2002.01266.x. [DOI] [PubMed] [Google Scholar]
- Gang DR, Lavid N, Zubieta C, Chen F, Beuerle T, Lewinsohn E, Noel JP, Pichersky E. Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell. 2002;14:505–519. doi: 10.1105/tpc.010327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang DR, Wang J, Dudareva N, Nam KH, Simon J, Lewinsohn E, Pichersky E. An investigation of the storage and biosynthesis of phenylpropenes in sweet basil (Ocimum basilicum L.) Plant Physiol. 2001;125:539–555. doi: 10.1104/pp.125.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon J, McCaskill D, Rajaonarivony JIM, Mihaliak C, Karp F, Croteau R. Isolation of secretory cells from plant glandular trichomes and their use in biosynthetic studies of monoterpenes and other gland products. Anal Biochem. 1992;200:130–138. doi: 10.1016/0003-2697(92)90288-i. [DOI] [PubMed] [Google Scholar]
- Kamsteeg J, van Brederode J, Verschuren PM, van Nigtevecht G. Identification and genetic control of p-coumaroyl-coenzyme A, 3-hydroxylase isolated from petals of Silene dioica. Z Pflanzenphysiol. 1981;102:435–442. [Google Scholar]
- Kneusel RE, Matern U, Nicolay K. Formation of trans-caffeoyl-CoA from trans-4-coumaroyl-CoA by Zn2+-dependent enzymes in cultured plant cells and its activation by an elictor-induced pH shift. Arch Biochem Biophys. 1989;269:455–462. doi: 10.1016/0003-9861(89)90129-x. [DOI] [PubMed] [Google Scholar]
- Kühnl T, Koch U, Heller W, Wellmann E. Chlorogenic acid biosynthesis: characterization of a light-induced microsomal 5-O-(4-coumaroyl)-d-quinate/shikimate 3′-hydroxylase from carrot (Daucus carota L.) cell suspension cultures. Arch Biochem Biophys. 1987;258:226–232. doi: 10.1016/0003-9861(87)90339-0. [DOI] [PubMed] [Google Scholar]
- Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R. Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Nat Acad Sci USA. 2000;97:2934–2939. doi: 10.1073/pnas.97.6.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofty S, Fleuriet A, Machiex J-J. Partial purification and characterization of hydroxycinnamoyl CoA:transferases from apple and date fruits. Phytochemistry. 1992;31:767–772. [Google Scholar]
- Matz M, Lukyanov S, Bogdanova E, Diatchenko L, Chenchik A. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill D, Croteau R. Monoterpene and sesquiterpene biosynthesis in glandular trichomes of peppermint (Mentha × piperita) rely exclusively in plastid-derived isopentenyl diphosphate. Planta. 1995;197:49–56. [Google Scholar]
- McCaskill D, Gershenszon J, Croteau R. Morphology and monoterpene biosynthetic capabilities of secretory cell clusters isolated from glandular trichomes of peppermint (Mentha × piperita) Planta. 1992;187:445–454. doi: 10.1007/BF00199962. [DOI] [PubMed] [Google Scholar]
- Patil SS, Zucker M. Potato phenolases. purification and properties. J Biol Chem. 1965;240:3938–3943. [PubMed] [Google Scholar]
- Petersen M. Characterization of rosmarinic acid synthase from cell cultures of Coleus blumei. Phytochemistry. 1991;30:2877–2881. [Google Scholar]
- Petersen M. Cytochrome p450-dependent hydroxylation in the biosynthesis of rosmarinic acid in Coleus. Phytochemistry. 1997;45:1165–1172. [Google Scholar]
- Petersen M, Alfermann AW. Two new enzymes of rosmarinic acid biosynthesis from cell cultures of Coleus blumei: hydroxyphenylpyruvate reductase and rosmarinic acid synthase. Z Naturforsch. 1988;43c:501–504. [Google Scholar]
- Petersen M, Häusler E, Karwatzki B, Meinhard J. Proposed biosynthetic pathway for rosmarinic acid in cell cultures of Coleus blumei Benth. Planta. 1993;189:10–14. [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996;272:51–64. doi: 10.1016/s0076-6879(96)72008-6. [DOI] [PubMed] [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D. CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem. 2001;276:36566–36574. doi: 10.1074/jbc.M104047200. [DOI] [PubMed] [Google Scholar]
- Siebert PD, Chen S, Kellogg DE. The Human GenomeWalker DNA Walking Kit: a new PCR method for walking in uncloned genomic DNA. CLONTECHniques. 1995a;X:1–3. [Google Scholar]
- Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995b;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich B, Zenk MH. Partial purification and properties of p-hydroxycinnamoyl-CoA:quinate hydroxycinnamoyl transferase from higher plants. Phytochemistry. 1979;18:929–933. [Google Scholar]
- Ulbrich B, Zenk MH. Partial purification and properties of p-hydroxycinnamoyl-CoA:shikimate-p-hydroxycinnamoyl transferase from higher plants. Phytochemistry. 1980;19:1625–1629. [Google Scholar]
- Urban P, Werck-Reichhart D, Teutsch H, Durst F, Regnier S, Kazmaier M, Pompon D. Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast. Kinetic and spectral properties of the major plant P450 for the phenylpropanoid pathway. Eur J Biochem. 1994;222:843–850. doi: 10.1111/j.1432-1033.1994.tb18931.x. [DOI] [PubMed] [Google Scholar]