Abstract
Down-regulation of the cell surface display of class I MHC proteins is an important mechanism of immune evasion by human and animal viruses. Herpesviruses in particular encode a variety of proteins that function to lower MHC I display by several mechanisms. These include binding and retention of MHC I chains in the endoplasmic reticulum, dislocation of class I chains from the ER, inhibition of the peptide transporter (TAP) involved in antigen presentation, and shunting of newly assembled chains to lysosomes. Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) is a human herpesvirus strongly linked to the development of KS and to certain AIDS-associated lymphoproliferative disorders. Here we show that KSHV encodes two distinctive gene products that function to dramatically reduce cell surface MHC I expression. These viral proteins are localized predominantly to the ER. However, unlike previously described MHC I inhibitors, they do not interfere with the synthesis, translocation, or assembly of class I chains, nor do they retain them in the ER. Rather, they act to enhance endocytosis of MHC I from the cell surface; internalized class I chains are delivered to endolysosomal vesicles, where they undergo degradation. These KSHV proteins define a mechanism of class I down-regulation distinct from the mechanisms of other herpesviruses and are likely to contribute importantly to immune evasion during viral infection.
A major arm of host immunity to viral infection is the cytotoxic T lymphocyte (CTL), which recognizes viral antigens presented as peptides bound to class I MHC (MHC I) molecules on the surface of infected cells. CTL recognition of such cells can lead to direct cytolysis as well as the release of cytokines that can contribute further to antiviral action and host immune mobilization. Therefore, evasion of CTL recognition is thought to play a key role in the establishment of a systemic viral infection. Consistent with this view, many viruses have evolved complex strategies for such evasion, usually centering on the down-regulation of cell surface MHC I display. Such strategies have been most highly developed among the herpesviruses, a family of large DNA viruses that efficiently produce persistent infections and disease in their hosts. These viruses encode many proteins that are expressed predominantly in the endoplasmic reticulum (ER) and down-regulate MHC I expression by several mechanisms. Some bind class I chains and retain them in the ER (1, 2) or, after complex formation, direct them to endolysosomal vesicles for degradation (3); others target them for dislocation from the ER to cytosolic proteosomes (4). Still other viral proteins (some located in the ER membrane, others in the cytosol) inhibit the TAP peptide transporter, causing MHC I assembly defects that block export from the ER (because only MHC I chains correctly loaded with peptides can exit this organelle).
Kaposi's sarcoma-associated herpesvirus (KSHV; also called human herpesvirus 8) is a human herpesvirus strongly linked to the development of Kaposi's sarcoma (KS) and several AIDS-related lymphoproliferative disorders (5). The striking increase in the frequency of KS in KSHV-infected individuals with HIV-mediated immunodeficiency (5) suggested to us that T cells may play a central role in host control of KSHV infection. If so, it seemed likely that viral evolution would have equipped KSHV with genes designed to counter host CTL responses. Here we describe two KSHV gene products that efficiently block MHC I display by an unusual mechanism—the up-regulation of endocytosis of surface class I chains.
Materials and Methods
Cell Lines and Cell Culture.
HeLa cells, 293 cells, and Phoenix and SLK (6) cells were grown in DMEM (DME-H21) supplemented with 10% (vol/vol) FCS and penicillin—streptomycin. BJAB lymphoma cells were grown in RPMI medium 1640 supplemented as described above.
Antibodies and Reagents.
For FACS analysis all antibodies were used at a concentration of 1 μg per 106 cells. Anti-calnexin mAb was from StressGen Biotechnologies, (Victoria, Canada) and used at a dilution of 1:1,000. Anti-CD58 and anti-MHC II mAbs were from PharMingen. Anti-CD95 mAb was from Immunotech. Rabbit polyclonal anti-hemagglutinin (anti-HA) (Y-11) and goat polyclonal anti-Flag antibodies were from Santa Cruz Biotechnology and used at a dilution of 1:200. Anti-HA mAb (HA.11) was from Babco (Richmond, CA) and used at 1:500. Monoclonal anti-HLA class I antigen (W6/32) FITC conjugate was from Sigma. Purified monoclonal anti-HLA class I antigen (W6/32) was from Dako.
E64, leupeptin, concanamycin A, bafilomycin A, and chloroquine were purchased from Sigma and used at a final concentration of 100 μg/ml, 200 μM, 50 nM, 2 μM, and 100 μM, respectively. Ammonium chloride (Fisher Scientific) was used at 50 mM.
Plasmids.
KSHV genes (K genes) were amplified by PCR using λ clones spanning the entire KSHV genome as template and inserted into pCR3.1 vector (Invitrogen) and into pLNCX (CLONTECH). The HA and Flag epitope tags were inserted by PCR downstream of the first methionine of each ORF; the resulting HA-K3 and Flag-K5 were cloned into pCR3.1 vector. From pCR3.1-K5, K5 was inserted into pCDEF3 (7) and into pIRES2-EGFP (CLONTECH). The resulting vectors were named pCDEF3-K5 and pIRES2-K5, respectively. From pCR3.1-K3, K3 was inserted into pCEP4 (Invitrogen) to obtain pCEP4-K3. pcDNA3 vector expressing the wild-type dynamin or the K44E dynamin-1 trans-dominant-negative were kindly provided by R. B. Vallee (8).
Transfection and Retroviral Infection.
Adherent cells were transfected by using the Fugene-6 product according to the manufacturer's instructions (Boehringer Mannheim). BJAB transfection has been described (9). For transient transfection analysis, the cells were used 36–48 h after transfection.
On transfection of pLNCX vector, the Phoenix packaging cell line produces replication-defective viral particles that have been used for stable gene transfer and expression in HeLa cells.
Flow Cytometry Analysis.
Adherent cells were detached by using enzyme-free/PBS-based cell dissociation buffer (GIBCO/BRL) according to the manufacturer's instructions. Cells were washed in PBS/1% BSA and incubated with specific mAbs (1 μg per 106 cells) for 30 min at 4°C. Bound mouse antibodies were revealed by a fluorescein or R-phycoerythrin-conjugated goat anti-mouse antibody. Cell surface fluorescence was analyzed with a Becton Dickinson FACScalibur. For intracellular staining, cells were fixed in 4% paraformaldehyde before incubations with specific antibodies in a buffer containing 0.02% saponin (Sigma).
For kinetics of MHC I surface internalization, cells were incubated for 60 min at 4°C with saturating amounts of W6/32 mAb (12 μg/ml) in DMEH-21 supplemented with 2% FCS, washed, and incubated at 37°C for different periods of time. To stop endocytosis, cells were returned to 4°C. W6/32-bound MHC I surface molecules were revealed by a fluorescein-conjugated goat anti-mouse antibody, and cells were analyzed by flow cytometry. The data were collected in a logarithmic mode, and the geometric mean of fluorescence intensity was calculated. To measure export of MHC I molecules to the cell surface, cells were stained with a saturating amount of W6/32 mAb (as described above for endocytosis) followed by a second step of incubation with a saturating amount of R-phycoerythrin-conjugated goat anti-mouse IgG (1:50). After extensive washing, cells were incubated at 37°C for various periods of time in the presence of mAb W6/32 that was fluorescein-conjugated. Cells were analyzed by flow cytometry. The data were collected in a logarithmic mode, and the geometric mean of fluorescence intensity was calculated.
Indirect Immunofluorescence and Confocal Microscopy.
Subconfluent layers of HeLa cells were grown on glass coverslips and transfected with Fugene-6. Thirty-six hours after transfection cells were rinsed with PBS and fixed 10 min with 1% formalin in PBS. After washing with PBS, cells were permeabilized with 0.05% saponin in PBS/2% BSA before 1 h of incubation with combinations of primary antibodies raised in different species. After extensive washing, the cells were incubated with fluorescent secondary antibodies (1:200) for 1 h. After washing, coverslips were mounted on glass slides with Vectashield (Vector Laboratories). The mounted cells were analyzed by using a Zeiss Axiovert and a MRC 1000 laser-scanning confocal microscope.
Metabolic Labeling, Immunoprecipitation, and Endoglycosidase H (Endo H) Digestion.
Cells (5 × 106) were preincubated with methionine-free RPMI medium 1640 supplemented with 10% (vol/vol) FCS and penicillin–streptomycin for 1 h at 37°C. The cells were labeled for 20 min with 200 μCi of Trans35S-Label (ICN) in a final volume of 2 ml. After washes, cells were chased for the indicated time in 50 ml of complete RPMI medium. At the end of chase periods, cells were lysed in 500 μl of PBS containing 1% Nonidet P-40 and protease inhibitor mixture (Sigma) on ice for 30 min. Nuclei and insoluble debris were removed by centrifugation. The remaining extract was incubated with 4 μg of W6/32 mAb for 1 h at 4°C. The mixture was supplemented with BSA to a final concentration of 1% and incubated overnight with 40 μl of protein A/G beads (Santa Cruz Biotechnology) at 4°C. Protein A/G beads were washed extensively, and the bound proteins were analyzed by electrophoresis in an SDS/12.5% polyacrylamide gel. For endo H treatment, samples were denatured in 40 μl of 0.3% SDS/150 mM 2-mercaptoethanol/50 mM sodium acetate, pH 5.5, at 100°C for 5 min and incubated with 4 milliunits of endo H (Boehringer Mannheim) or without enzyme overnight at 37°C. Gels were fixed, dried, and analyzed by using a PhosphorImager Storm 860 (Molecular Dynamics).
Results
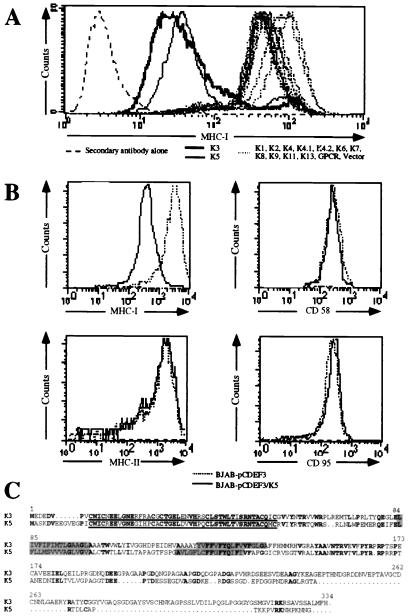
We systematically screened the KSHV genome for viral products that might mediate down-regulation of class I MHC molecules. The KSHV genome contains at least 15 ORFs (ORFs K1–K15) that are not conserved in other herpesviruses. We reasoned that potential regulators of MHC I likely would fall into this class of genes, which are suspected to encode specialized functions not required for replication in vitro but which play roles in pathogenesis or spread in vivo. We therefore generated retroviral expression vectors for most of these coding regions and used them to stably transduce HeLa cells; cell surface levels of MHC I heavy chains then were determined by flow cytometry. As shown in Fig. 1A, cells expressing two of these genes—ORFs K3 and K5—displayed strong (ca. 20- to 30-fold) reductions of surface MHC I levels.
Figure 1.
(A) Detection of MHC I molecules in HeLa cells stably transduced with retrovirus vectors expressing the different K genes. After infection, cells were selected in the presence of geneticin for 2 weeks; colonies were pooled and stained by flow cytometry with a monoclonal anti-HLA A,B,C antibody directly conjugated to fluorescein. Results presented correspond to the intensity of fluorescein fluorescence of the HeLa cells analyzed by flow cytometry. (B) BJAB cells stably transfected with pcdef3-K5 (solid lines) or the pcdef3 vector alone (dashed lines) were stained with mAbs specific for MHC I (Upper Left), CD58 (Upper Right), MHC II (Lower Left), or CD95 (Lower Right) and analyzed by flow cytometry. (C) Alignment of K3 and K5. Bold characters correspond to identity, rectangles represent the two putative transmembrane domains predicted by the program psort (10), and underlined sequences represent the putative ring finger motif.
To determine whether this effect was specific for MHC I, we stably transfected a human B cell line (BJAB) with a vector expressing K5 (or with an empty vector) and assayed by flow cytometry for surface levels of MHC I, MHC II, CD58, and CD95. As shown in Fig. 1B, cells expressing the K5 protein had reduced levels of MHC class I but normal surface levels of MHC class II chains and CD58 and CD95 polypeptides. Thus, the down-regulation of surface MHC I is selective and does not reflect a nonspecific process affecting all surface constituents.
Fig. 1C shows the predicted amino acid sequences of the K3 and K5 gene products. The two proteins have no homologs in GenBank, but are clearly related to one another: 40% of K5's residues are identical to those of K3. No N-terminal signal sequence is predicted for either protein, but both have two strongly hydrophobic stretches that likely represent transmembrane domains. The primary sequence reveals few other clues as to their potential function(s).
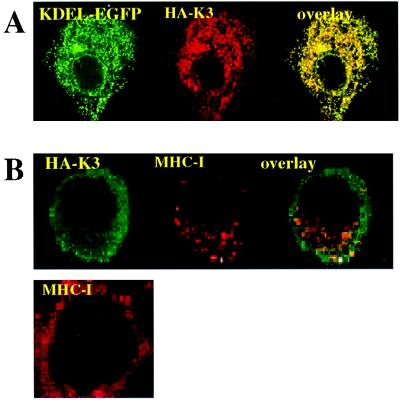
To determine the subcellular localization of the proteins, we tagged them with N-terminal epitopes derived from influenza HA (for K3) or Flag (for K5). These tagged versions of both K3 and K5 were fully functional for MHC I down-regulation (data not shown). We expressed the tagged versions in HeLa cells, together with an expression vector for KDEL-EGFP, encoding an ER-localized form of green fluorescent protein (GFP). The transfected cells then were examined by immunofluorescence with a mAb specific for the epitope tag. As shown in Fig. 2A for K3, a lacy reticular pattern of HA staining was observed in the cytoplasm, typical of that expected for the ER. This pattern was identical with that observed for the ER marker (GFP-KDEL) in the same cells, and confocal imaging revealed striking colocalization of the two polypeptides. Staining of K3-expressing cells with antibody to an endogenous ER resident protein, calnexin, confirmed that the proteins indeed colocalized with this ER marker (data not shown). Identical results were obtained for K5 expression (data not shown). We found no colocalization of K3 or K5 with markers of more distal compartments of the secretory or endocytotic pathways, including the Golgi, trans-Golgi network, or lysosomes (not shown). Thus, at steady state the bulk of the K3 and K5 chains are localized within the ER (see also ref. 11). We cannot exclude that a small subpopulation of K3/K5 chains (<5% of the total) may reach the plasma membrane, although attempts to demonstrate such surface chains biochemically or by flow cytometry have not been consistently successful (see Discussion).
Figure 2.
(A) Intracellular localization of K3. HeLa cells grown on coverslips were transfected with expression vectors for the HA-tagged version of K3 (HA-K3) plus KDEL-GFP. Cells were stained 36 h posttransfection as described in Materials and Methods. HA-K3 was detected with a rabbit polyclonal anti-HA followed by a goat anti-rabbit IgG rhodamine conjugate. (B) Localization of MHC I molecules in K3- and K5-expressing HeLa cells. HeLa cells were transfected with vector alone (Lower) or HA-tagged version of K3 (Upper). Cells were stained 36 h posttransfection. MHC I molecules were visualized by staining with the monoclonal W6/32 antibody followed by a goat anti-mouse IgG rhodamine conjugate. HA-K3 was detected with a rabbit anti-HA revealed by goat anti-rabbit fluorescein conjugate.
To determine whether class I chains were being retained in the ER by K3/K5 expression, we examined the localization of MHC I polypeptides in K3- or K5-expressing HeLa cells. As shown in Fig. 2B Lower, which illustrates cells transfected with a control vector, all MHC I staining is localized to the cell surface. However, in cells expressing HA-tagged K3 (Fig. 2B Upper) or K5 (not shown), MHC I staining is largely relocalized to small, punctate cytoplasmic structures, presumably vesicular in nature. This pattern does not resemble ER or plasma membrane staining, and costaining for K3 (Fig. 2B Upper) confirms that MHC I and K3 are not colocalized. These data suggest that despite the abundant presence of K3/K5 in the ER MHC I chains were able to access a more distal compartment of the vesicular transport pathway.
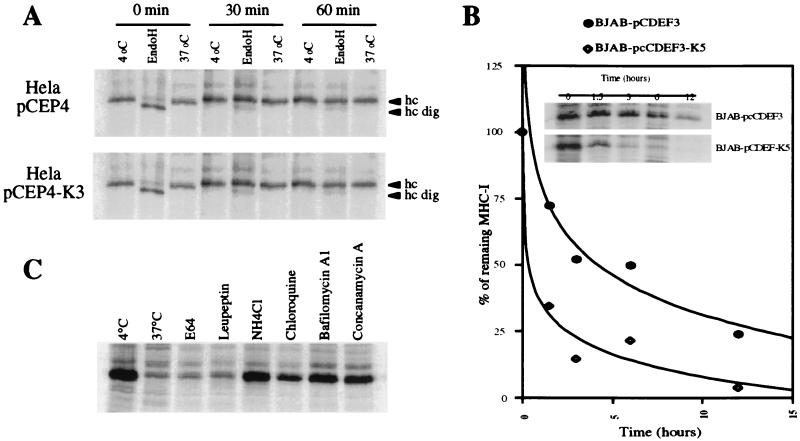
To further examine the effects of K3/K5 on class I metabolism, we conducted the pulse—chase analysis of Fig. 3A. Control HeLa cells or HeLa cells stably expressing K3 were pulse-labeled for 20 min with [35S]methionine; one aliquot of cells was harvested then, and the remainder was washed and incubated with unlabeled medium for 30 or 60 min. At each time point, extracts were prepared and incubated at either 4°C or 37°C, then immunoprecipitated with a mAb (W6/32) to MHC I. This antibody reacts principally with properly assembled MHC I heavy-chain β2-microglobulin dimers. Previous studies have shown that MHC I chains that fail to be loaded with peptide are destabilized at 37°C (but not 4°C), losing reactivity with W6/32. As shown in Fig. 3A, at each time point the MHC I chains in K3-expressing cells (Lower) behaved identically to those in control cells (Upper) expressing the vector alone—there was no early degradation of the chains (as would be expected if ER translocation had been aborted) or destabilization at 37°C (as would occur if peptide loading had been impaired). Analysis of the N-linked carbohydrates of the MHC I chains at each time point confirmed that K3 had no effect on MHC I maturation and egress from the ER. At 0 min of chase, most chains were endo H-sensitive, as expected for newly made polypeptides in the ER. But after as little as 30 min of chase, nearly all chains in both vector- and K3-expressing cells were now endo H-resistant, indicating ER export and traversal of at least the proximal Golgi complex. Identical results were obtained in K5-expressing cells (not shown).
Figure 3.
Intracellular transport and stability of MHC I molecules. (A) HeLa cells stably transfected with pCEP4-K3 (Lower) or the pCEP4 vector alone (Upper) were metabolically labeled for 20 min and chased for the indicated time periods. After being lysed in 1% Nonidet P-40, lysates were divided into three aliquots. Two were kept on ice and one was incubated at 37°C for 1 h. Heterodimeric MHC I molecules were immunoprecipitated with mAb W6/32 and treated with endo H where indicated. (B) BJAB cells stably transfected with pcdef3-K5 or the vector alone were radiolabeled for 30 min and chased for the indicated time periods. MHC I molecules in cell lysates were immunoprecipitated with mAb W6/32. Quantitation of the data is presented as a fraction of initial MHC I levels; ⋄, K5 expressing cells; ●, control cells. (C) BJAB cells stably transfected with pcdef3-K5 were radiolabeled for 30 min and chased for 3 h in the presence or absence of the indicated inhibitors. MHC I molecules were immunoprecipitated by using mAb W6/32.
We next conducted a more extended pulse—chase analysis to determine whether MHC I chains undergo degradation in the more distal regions of the secretory pathway (for example, by being targeted to lysosomes). After 30 min of pulse labeling, K5- or vector-expressing BJAB cells were chased for 1.5, 3, 6, and 12 h, and then their MHC I chains were immunoprecipitated and examined by SDS/PAGE. As shown in Fig. 3B, MHC I chains synthesized in the presence of K5 displayed a reduced half-life at these later times when compared with those synthesized in control cells lacking K5 expression. If this instability were the result of degradation in the acidic environment of endosomes or lysosomes, it would be expected to be abrogated by lysosomotropic weak bases such as ammonium chloride or chloroquine. Fig. 3C reveals that this is indeed the case: when a similar pulse—chase experiment is conducted in the presence of NH4Cl or chloroquine, the half-life of MHC I chains is restored to nearly control levels. Similar results were obtained with the vacuolar H+-ATPase inhibitors bafilomycin A1 and concanamycin A, which also raise endolysosomal pH. These results suggest that degradation occurs within the endolysosomal compartment—a site devoid of detectable K3 or K5 chains (data not shown). The protease inhibitor E64, which is specific for lysosomal cysteine proteases, did not restore the levels of MHC I chains in K5-positive cells, suggesting that other families of proteases may be involved in the turnover of MHC proteins in this compartment.
Two possible models can be envisioned for how the chains come to be localized in internal endolysosomal vesicles. In one model, chains may go to the cell surface and then be reclaimed by enhanced endocytosis. In the second, chains exiting the trans-Golgi network may be shunted directly to such vesicles without ever arriving at the cell surface. (Of course, the two models are not mutually exclusive.)
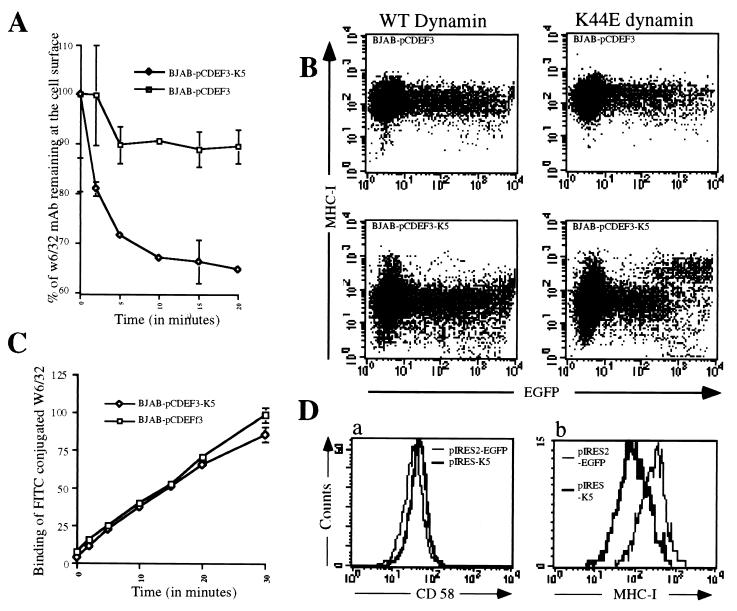
To search for enhanced endocytosis of surface MHC I chains, we incubated BJAB cells stably expressing K5 or a control vector with anti-MHC I antibody at 4°C. After washing away the free antibody, the cells were shifted to 37°C and the complement of surface MHC I chains present at various times thereafter was detected by staining with FITC-conjugated anti-IgG. As shown in Fig. 4A, expression of K5 (or K3, data not shown) led to an accelerated decline of surface MHC I staining. Together with the data of Fig. 3, this suggests an important role for enhanced endocytosis in this form of MHC I down-regulation. To exclude the possibility of accelerated MHC I degradation at the cell surface, we examined the total (intracellular plus cell surface) levels of MHC I chains prelabeled in this fashion, both before and after 20 min at 37°C. (This was done by flow cytometry in the presence and absence of saponin; data not shown.) This experiment revealed no net loss of MHC I reactivity, confirming that the loss of surface MHC I chains at 20 min (Fig. 4A) reflects internalization rather than either degradation or enhanced dissociation of MHC chains from their antibody. (The degradation of MHC I chains observed in Fig. 3B occurs subsequent to this time point.)
Figure 4.
(A) Kinetics of decrease of surface-bound anti-MHC I mAb in BJAB cells stably transfected with pcdef3-K5 or the pcdef3 vector. Cells were labeled at 4°C with the anti-MHC I mAb W6/32, washed, and incubated at 37°C for the indicated periods. Cells then were cooled to 4°C and stained with goat anti-mouse IgG fluorescein conjugate. Surface levels of W6/32-MHC I complexes were analyzed by flow cytometry. The initial values (t = 0) were standardized to 100, corresponding to 200 fluorescence units for BJAB pcdef3-K5 and 1100 fluorescence units for BJAB pcdef3. Data are expressed as the percentage of the initial values. □, Cells bearing pcdef3 vector alone; ⋄, pcdef3-K5-expressing cells. (B) Susceptibility of K5-induced MHC I endocytosis to a dominant-negative dynamin-1 mutant. BJAB cells (Upper) or BJAB-K5 cells (Lower) were transfected with 21 μg of dynamin-1 wild type (WT; left) and 4 μg of an EGFP expression vector or with 21 μg of the dynamin-1 trans-dominant-negative (K44E; Right) and 4 μg of an EGFP expression vector. After 36 h, cells were incubated with mAb W6/32 and W6/32-bound MHC I surface molecules were revealed by a fluorescein-conjugated goat anti-mouse antibody and analyzed by flow cytometry. (C) Kinetics of MHC I export to the cell surface. BJAB stably transfected with pcdef3-K5 (⋄) or the vector alone (□) were labeled at 4°C with an excess of MHC I mAb W6/32 to saturate the preexisting surface MHC I molecules. Cells were washed and incubated at 37°C in the presence of a MHC I mAb W6/32 fluorescein conjugate for the indicated periods of time. Cells were analyzed by flow cytometry. (D) Effect of K5 expression on preexisting surface MHC I molecules. BJAB cells were transfected with 20 μg of pIRES2-EGFP (thin lines) or pK5-IRES2-EGFP (thick lines). Immediately after transfection, cells were labeled for 1 h at 4°C with mAb W6/32 or with a mAb against CD58, washed, and returned to the 37°C incubator. Thirty-six hours after transfection, surface-labeled MHC I (b) or CD58 (a) in cells displaying GFP expression was detected by flow cytometry using a goat anti-mouse IgG R-phycoerythrin conjugate.
To further explore the possibility that K5 enhances endocytosis, we have examined the effects of transient expression of a dominant-negative mutant of dynamin (K44E) in K5-expressing cells. This dynamin mutant is known to specifically block the endocytic pathway (8). Expression vectors for wild-type or mutant dynamin were transfected into BJAB cells, along with a GFP expression vector to identify transfected cells. As shown in Fig. 4B Upper, transient expression of either wild-type dynamin (WT; Left) or K44E (Right) has little effect on surface MHC I levels in transfected normal BJAB cells (Upper). However, in BJAB cells expressing K5 (Fig. 4B Lower), the expression of dynamin K44E (Lower Right) but not wild-type dynamin (Lower Left) results in a strong inhibition of MHC I down-regulation. This is visible as a restoration of surface levels of class I chains (to levels comparable to those observed in the absence of K5) in cells expressing high levels of GFP (fluorescence intensity above 103). Because dynamin is not known to participate in the exocytotic pathway, this result indicates that most of the effect of K5 is via augmentation of the endocytosis of MHC I chains.
In keeping with this, we have examined the delivery of class I chains to the cell surface in the presence or absence of K5. BJAB cells stably expressing either K5 or a control plasmid first were incubated with excess antibody to MHC I chains at 4°C to bind all preexisting class I chains. (Control experiments revealed that this preincubation reduces the binding of FITC-labeled anti-class I antibody to background levels.) Then, cells were shifted to 37°C and newly arrived cell surface MHC I chains were scored by FACS analysis using FITC-conjugated MHC I antibody. Fig. 4C shows that, by this criterion, delivery of MHC I chains to the cell surface proceeded with similar kinetics and efficiency in the presence or absence of K5 expression.
If the effect of K5 on surface MHC I is due principally to effects on endocytosis (rather than on delivery of newly made chains to the cell surface), then it should be evident on MHC chains made and delivered to the surface in the absence of K5. To explore this possibility, we examined the effect of de novo K5 expression on preexisting MHC class I chains (Fig. 4D). BJAB cells were transfected with expression vectors encoding either GFP alone (Fig. 4D, thin lines) or both K5 plus GFP (thick lines). Immediately after transfection (i.e., before expression of the vector), the cells were incubated with mAbs to MHC I chains (Fig. 4D, a) or control CD58 chains (Fig. 4D, b) to tag the preexisting surface pools of these polypeptides. After this binding, excess free antibodies were removed by washing and cells were incubated at 37°C to allow expression of the transfected plasmids. Thirty-six hours later, surface MHC I or CD58 chains were identified by binding of phycoerythrin-labeled anti-IgG and quantified by flow cytometry. As shown in Fig. 4D, a clear reduction in surface MHC I is evident only in GFP-positive (i.e., transfected) cells expressing K5 (Fig. 4D, b). This effect is specific for MHC I, as surface levels of preformed CD58 are unaffected (Fig. 4D, a). [Note: The level of prelabeled MHC I chains remaining on the GFP-expressing cells at 36 h after antibody labeling is down 20- to 30-fold from that observed in cells labeled directly without further incubation (compare Fig. 4D with Fig. 1B); this reflects the known half-life of MHC I (see Fig. 3B). Nonetheless, K5 expression can decrease this level further.] Thus, even chains sorted to the surface in the absence of K5 expression can be down-regulated by subsequent addition of K5, clearly indicating that the molecule can function outside of the exocytotic pathway.
Discussion
These results establish that KSHV encodes two previously undescribed gene products that strongly down-regulate class I MHC expression. The mechanism by which the K3 and K5 proteins achieve this end is distinct from other previously described modes of herpesvirus-induced MHC modulation, although it can be instructively compared with them. Although, like other herpesviral immunoregulatory proteins, the K3 and K5 chains are found largely on ER membranes, their effects on MHC I metabolism are not exerted in this organelle. This distinguishes them from the ER-localized HCMV US6 protein (12, 13) [and the cytosolic herpes simplex virus ICP 47 protein (14–16)], which blocks peptide transport, and the US3 protein of cytomegalovirus (1, 2), which directly binds and retains MHC I chains in the ER. Similarly, K3 and K5 differ from the US11 and US2 proteins of human cytomegalovirus, which cause newly made MHC I chains to be dislocated from the ER and then degraded by cytosolic proteosomes (4). Rather, K3 and K5 promote the endocytosis of MHC I chains from the cell surface and their subsequent degradation within an intracellular endolysosomal compartment. We can envision two models by which such endocytosis might be promoted. First, a small subpopulation of K3/K5 chains may be sorted to the plasma membrane, where they might interact (directly or indirectly) with surface MHC I, leading to conformational or other changes in the latter that would promote interactions with the endocytic machinery. If so, the steady-state level of such surface chains must be very low. Because we do not observe colocalization of MHC I and K3/K5 in endosomal vesicles (data not shown), the fate of such putative surface K3/K5 chains is unknown; possibilities include degradation or rapid return to the ER compartment. Consistent with this model, the ability of transfected K3 and K5 vectors to down-regulate preexisting surface MHC I chains is abrogated by the drug brefeldin A, which blocks the transit of newly made chains to the cell surface (L.C., unpublished results). Alternatively, ER-resident chains of K3/K5 might mediate a signaling pathway that induces host surface proteins that mediate the effect on MHC I endocytosis. Several signaling pathways mediated by ER-resident proteins are known (17), although none has been implicated as yet in the regulation of downstream components of the vesicular transport pathway. If this model is correct, then the brefeldin result just mentioned requires that the target(s) induced by this signaling must include at least one protein that traverses the ER and Golgi. Further experiments are necessary to distinguish among these models.
The action of K3 and K5 on MHC I has features that recall two other viral modulators of MHC I. The gp48 protein of murine cytomegalovirus also targets class I chains to endolysosomes for degradation (3), but this is due to redirection of exocytosis of MHC I to these vesicles after egress from the ER and Golgi. By contrast, K3 and K5 appear not to interfere with exocytosis but rather to promote enhanced reclamation of cell surface MHC chains via endocytosis. We know of no other herpesviral protein that functions in this fashion. Interestingly, a retroviral protein (HIV nef) also has been shown recently to enhance MHC I endocytosis (18), probably by binding to PACS-1 molecule (19), but, unlike the KSHV effectors described here, nef is localized to the cytosol and the cytosolic face of the plasma membrane. It will be interesting to see whether K3 and K5 share common mechanisms of action with HIV nef in this regard.
In KSHV biology, the expression of K3 and K5 appears to be limited to the early phase of the lytic replicative cycle (20). Because MHC I molecules are required for the presentation of viral peptides to CTLs, down-regulation of surface MHC I expression likely represents a key mechanism by which KSHV attempts to evade the immune system and spread within its host.
Acknowledgments
This manuscript is dedicated to the memory of R. H. Sadler, a friend and a colleague.
Abbreviations
- ER
endoplasmic reticulum
- CTL
cytotoxic T lymphocyte
- KS
Kaposi's sarcoma
- KSHV
Kaposi's sarcoma-associated herpesvirus
- HA
hemagglutinin
- GFP
green fluorescent protein
- endo H
endoglycosidase H
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140129797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140129797
References
- 1.Ahn K, Angulo A, Ghazal P, Peterson P A, Yang Y, Fruh K. Proc Natl Acad Sci USA. 1996;93:10990–10995. doi: 10.1073/pnas.93.20.10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones T R, Wiertz E J, Sun L, Fish K N, Nelson J A, Ploegh H L. Proc Natl Acad Sci USA. 1996;93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reusch U, Muranyi W, Lucin P, Burgert H G, Hengel H, Koszinowski U H. EMBO J. 1999;18:1081–1091. doi: 10.1093/emboj/18.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiertz E J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 5.Whitby D, Boshoff C. Curr Opin Oncol. 1998;10:405–412. doi: 10.1097/00001622-199809000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Siegal B, Levinton-Kriss S, Schiffer A, Sayar J, Engelberg I, Vonsover A, Ramon Y, Rubinstein E. Cancer. 1990;65:492–498. doi: 10.1002/1097-0142(19900201)65:3<492::aid-cncr2820650320>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 7.Goldman L A, Cutrone E C, Kotenko S V, Krause C D, Langer J A. BioTechniques. 1996;21:1013–1015. doi: 10.2144/96216bm10. [DOI] [PubMed] [Google Scholar]
- 8.Herskovits J S, Burgess C C, Obar R A, Vallee R B. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro V S, Mollenauer M N, Greene W C, Weiss A. J Exp Med. 1996;184:1663–1669. doi: 10.1084/jem.184.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haque M, Chen J, Ueda K, Mori Y, Nakano K, Hirata Y, Kanamori S, Uchiyama Y, Inagi R, Okuno T, et al. J Virol. 2000;74:2867–2875. doi: 10.1128/jvi.74.6.2867-2875.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahn K, Gruhler A, Galocha B, Jones T R, Wiertz E J, Ploegh H L, Peterson P A, Yang Y, Fruh K. Immunity. 1997;6:613–621. doi: 10.1016/s1074-7613(00)80349-0. [DOI] [PubMed] [Google Scholar]
- 13.Hengel H, Koopmann J O, Flohr T, Muranyi W, Goulmy E, Hammerling G J, Koszinowski U H, Momburg F. Immunity. 1997;6:623–632. doi: 10.1016/s1074-7613(00)80350-7. [DOI] [PubMed] [Google Scholar]
- 14.Fruh K, Ahn K, Djaballah H, Sempe P, van Endert P M, Tampe R, Peterson P A, Yang Y. Nature (London) 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 15.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Nature (London) 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 16.Ahn K, Meyer T H, Uebel S, Sempe P, Djaballah H, Yang Y, Peterson P A, Fruh K, Tampe R. EMBO J. 1996;15:3247–3255. [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman R, Sidrauski C, Walter P. Annu Rev Cell Dev Biol. 1998;14:459–485. doi: 10.1146/annurev.cellbio.14.1.459. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard J M. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 19.Piguet V, Wan L, Borel C, Mangasarian A, Demaurex N, Thomas G, Trono D. Nat Cell Biol. 2000;2:163–167. doi: 10.1038/35004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukac D M, Kirshner J R, Ganem D. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]