Abstract
A 268-kb chromosomal segment containing sorghum (Sorghum bicolor) genes that are orthologous to the maize (Zea mays) Rp1 disease resistance (R) gene complex was sequenced. A region of approximately 27 kb in sorghum was found to contain five Rp1 homologs, but most have structures indicating that they are not functional. In contrast, maize inbred B73 has 15 Rp1 homologs in two nearby clusters of 250 and 300 kb. As at maize Rp1, the cluster of R gene homologs is interrupted by the presence of several genes that appear to have no resistance role, but these genes were different from the ones found within the maize Rp1 complex. More than 200 kb of DNA downstream from the sorghum Rp1-orthologous R gene cluster was sequenced and found to contain many duplicated and/or truncated genes. None of the duplications currently exist as simple tandem events, suggesting that numerous rearrangements were required to generate the current genomic structure. Four truncated genes were observed, including one gene that appears to have both 5′ and 3′ deletions. The maize Rp1 region is also unusually enriched in truncated genes. Hence, the orthologous maize and sorghum regions share numerous structural features, but all involve events that occurred independently in each species. The data suggest that complex R gene clusters are unusually prone to frequent internal and adjacent chromosomal rearrangements of several types.
Disease resistance (R) genes in plants provide a major mode of defense against a wide variety of pathogens and pests. The most abundant class of R genes encodes proteins with a nucleotide-binding site (NBS) and a Leu-rich repeat (LRR) region. The genome of the model plant, Arabidopsis, contains more than 120 NBS-LRR genes (Arabidopsis Genome Initiative, 2000), but the possible R specificities of these candidate R genes are only known for a handful of loci. From the few NBS-LRR genes that have received functional analysis, it appears that the LRR region provides the specificity for recognition of a pathogen gene product (Ellis et al., 2000a), thereby leading to the initiation of a signal transduction cascade that activates several defense pathways.
Most of the R gene loci in plants are highly complex, containing numerous nearby R genes (Ellis et al., 2000b). Unequal recombination within an R gene cluster can result in variation in the size and organization of a complex locus. New R genes with novel pathogen-recognition specificities can be generated by this unequal recombination process (Richter et al., 1995). Some R genes appear to be the obvious products of unequal crossing over and/or conversion between R genes in the same tandem array, thereby providing a new “chimeric” locus with a possible new recognition specificity. Hence, complex R gene loci can undergo rapid internal reorganization. In addition, R gene loci often show a lack of synteny in closely related species (Leister et al., 1998), suggesting that they are also unstable in their gross chromosomal locations. For instance, neither the Lr1 R gene of wheat (Triticum aestivum) nor the Rpg1 rust resistance gene of barley (Hordeum vulgare) appears to have an orthologous locus in rice (Oryza sativa; Kilian et al., 1997; Gallego et al., 1998; Brueggeman et al., 2002). In contrast, most of the other types of genes in rice, wheat, and barley are found in colinear order on syntenic chromosomes (Gale and Devos, 1998; Bennetzen, 2000).
Because pathogens can easily become virulent by loss of features that allow their recognition, it is expected that R genes must evolve rapidly to keep pace with evolving pathogen populations (Bennetzen and Hulbert, 1992). Under disease pressure from a pathogen that has evolved to escape detection, selection on the plant for the few progeny with a new and appropriate R specificity would be very strong. Most of the R genes studied in plants exhibit the effects of diversifying selection, where non-synonymous substitutions exceed synonymous substitutions within an LRR region (Parniske et al., 1997; Song et al., 1997; Meyers et al., 1998). The rapid evolution of R genes is also suggested by the high degree of haplotype diversity within a species, as shown in sequence analyses of Rpm1 and Rps2 alleles from different ecotypes of Arabidopsis (Caicedo et al., 1999; Stahl et al., 1999).
In addition to selected changes in gene sequence and in the number, order, or chimeric origins of R loci, other structural variations have been detected in R gene clusters. Transposable elements are often inserted within complex R loci, sometimes causing insertional inactivation of an R gene (Song et al., 1997; Multani et al., 1998; Noel et al., 1999; Dodds et al., 2001; Ramakrishna et al., 2002b). In addition, inserted transposons can provide new regulatory sequences for any adjacent gene (Wessler et al., 1995). Moreover, two nearby transposons belonging to the same family could provide the homology needed for R gene duplication or deletion by unequal exchange. Numerous small insertions or deletions are often seen within R genes or adjacent sequences, but their mode(s) of origin and biological significance are not clear. Because so few complex R loci have been analyzed for structural variation (Parniske and Jones, 1999; Noel et al., 1999), we do not know the relative importance of these numerous rearrangement mechanisms for generating new R gene capabilities.
Rp1 is a complex R locus in maize (Zea mays) conferring race-specific resistance to a fungal pathogen, common leaf rust (Puccinia sorghi). Rp1 is an exceptionally unstable locus, even for complex R genes (Bennetzen et al., 1988; Sudupak et al., 1993; Richter et al., 1995). Both unequal crossing over and unequal conversion have been associated with changes in R specificity in the region (Sudupak et al., 1993; Hu and Hulbert, 1994; Richter et al., 1995). Small DNA segments containing most of the Rp1 homologs from the maize Rp1-D haplotype have been cloned and sequenced, providing evidence for chimeric gene generation and diversifying selection (Collins et al., 1999; Sun et al., 2001). We have recently sequenced 95- and 99-kb segments of the maize Rp1 cluster from the rust-susceptible inbred B73 (Ramakrishna et al., 2002b), uncovering a complex mixture of duplicated R genes and transposable elements. Most interesting, we found that the Rp1 complex in B73 contains at least one large (43 kb) non-tandem duplication and also several clusters of truncated genes with no homology to any known R gene (Ramakrishna et al., 2002b). Rp1 gene homologs have also been cloned and sequenced from the barley genome, although their function is not known (Ayliffe et al., 2000).
For this study, we sequenced a contiguous 268 kb of the Rp1-orthologous region in sorghum (Sorghum bicolor) to determine structural variation for an R gene cluster that has diverged at least since the ancestral divergence of maize and sorghum about 15 to 20 million years ago (mya; Gaut and Doebley, 1997). Comparison with the maize Rp1 complex indicates major differences in the current structures, but also provides evidence for common processes that create diversity in these complex R gene regions.
RESULTS
Isolation and Mapping of Rp1-Orthologous Clones from Sorghum
Ten bacterial artificial chromosome (BAC) clones that hybridized to a probe from the 5′ end of the Rp1-D gene (Collins et al., 1999) were identified from the sorghum BTx623 BAC library (http://hbz.tamu.edu/bacindex.html). Restriction enzymes MluI, NcoI, and NotI were used to make a physical map based on overlapping restriction fragments. Gel-blot hybridization analysis with the Rp1 5′ probe was used to confirm the physical map, generating a contiguous series (contig) of overlapping clones. The number of Rp1 homologs in the contig was determined by hybridization of NcoI-digested BAC DNAs with the Rp1 5′ probe. Sun et al. (2001) previously suggested that the number of NcoI-hybridizing fragments should correspond to the number of Rp1 homologs in maize, but we found that some Rp1 homologs within a cluster are so similar in sequence that they give identical fragment sizes with most restriction enzymes (Ramakrishna et al., 2002b). In addition, more than one hybridizing band per gene would be observed if there were an NcoI restriction site in the region of Rp1 covered by the probe. Taking these factors into account, we identified five Rp1 homologs that mapped to a 27-kb region in a 350-kb contig (Fig. 1). This contig mapped to linkage group H of sorghum (P.E. Klein, personal communication). Two other fingerprinted BACs that hybridized to the probe from the 5′ region of Rp1-D revealed one unique band on an NcoI gel blot. However, these clones did not hybridize to a probe from the Rp1 3′ region, suggesting that the sequences were 3′-truncated paralogs and/or not closely related to Rp1. In agreement with this prediction, these two clones were unlinked to the Rp1-orthologous region, forming a contig that mapped to linkage group G of sorghum (P.E. Klein, personal communication; Klein et al., 2000).
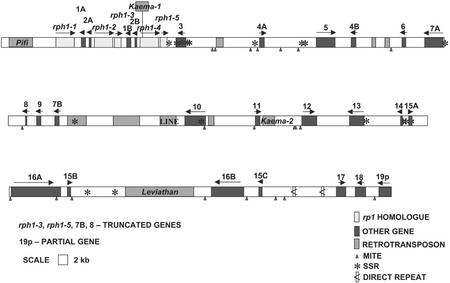
Figure 1.
Physical map of the rph1 region from sorghum inbred BTx623. Each circle represents an Rp1 homolog (i.e. rph1 gene). Physical distances and names of BACs are represented by lines above and below the positions of the rph1 genes. The names of BACs in parentheses are from the naming convention used in fingerprint analysis (Klein et al., 2000; P.E. Klein, personal communication).
Sequence Analysis of Sorghum Rp1-Orthologous BACs
The genomic organization at the Rp1-orthologous region in sorghum was studied by sequencing two sorghum BACs, Sb95A23 and Sb98N08. The two clones that were chosen contain all five Rp1 homologs, and more than 200 kb of sorghum DNA that is 3′ to the duplicated Rp1 homologs (Fig. 1). These two BACs were fully sequenced by our standard shotgun approach (Dubcovsky et al., 2001; Song et al., 2001; Ramakrishna et al., 2002a). Sb95A23 (97, 616 bp) and Sb98N08 (237, 576 bp) overlap by 67,759 bp and together cover 268,433 bp (accession no. AY144442) of the sorghum genome.
Sequence analysis by our standard approach, using a combination of gene-prediction programs, homology searches, and manual annotation indicates that this 268-kb region contains 31 candidate genes, including four that appear to be truncated. Numerous mobile DNAs are also found in the region (Fig. 2). Identified retrotransposon sequences constitute about 20% of the region, a relatively high percentage for the sorghum genome. We recently sequenced a different sorghum region of 425 kb that contains only 4% retrotransposon sequences (Song et al., 2002).
Figure 2.
Sequence organization of the sorghum genomic segment as represented by overlapping BACs Sb95A23 and Sb98N08. The five Rp1 homologs were named rph1-1 to rph1-5. All other genes are given simplex numbers from 1 to 19. Arrows indicate genes or truncated genes, plus their size and proposed direction of transcription. A through C, Multiple copies of the same gene family. Asterisks, SSRs that contain tandem repeat numbers of eight or more. Retrotransposons are indicated by rectangles without arrows on top. Novel full-length LTR-retrotransposons were named as described by SanMiguel et al. (2002). MITEs are indicated by small triangles. Arrowheads with bars between genes 15C and 17 indicate direct repeat sequences of 468 and 470 bp that are 91% identical to each other and separated by 6,112 bp. Gene 19p is given its unique designation because the sequenced region only contains the 3′ portion (p) of the gene.
Sequences annotated as genes on these BACs are described in Table I. Genes were classified as truncated when homology was detected only to a part of a protein entry in the GenBank database using the BLASTX program. The homology in these truncated genes was limited only to portions of the protein-encoding regions that was confined to either N-terminal, C-terminal, or central regions of the putative protein.
Table I.
Predicted genes on sorghum BACs Sb95A23 and Sb98N08
| Gene | Homology | Accession No. | e Value | Sorghum ESTs (Identity %) |
|---|---|---|---|---|
| rph1-1 | Rust resistance-like protein, RP1–3 (maize) | AAM03018 | 0 | AW923446 (100%) |
| AW923525(100%) | ||||
| 1A | BE362870 (93%) | |||
| 1B | BE362870 (90%) | |||
| rph1-2 | Rust resistance RP1D-like protein (maize) | AAK18308 | 0 | AW923446 (99%) |
| AW923525 (100%) | ||||
| rph1-3a | Rust resistance-like protein, RP1–2 (maize) | AAM03016 | e−148 | AW923446 (91%) |
| AW923525 (95%) | ||||
| 2Ab | ||||
| 2Bb | ||||
| rph1-4 | Rust resistance-like protein, RP1 (maize) | AAM03014 | 0 | AW923446 (93%) |
| AW923525 (98%) | ||||
| rph1-5a | Rust resistance protein (maize) | AAD47197 | e−158 | AW923446 (93%) |
| AW923525 (96%) | ||||
| 3 | Signal transducer of phototropic response, RPT2 (Arabidopsis) | AAF33112 | 2e−62 | BM324312 (94%) |
| BE596595 (94%) | ||||
| 4A | Hypothetical protein (Arabidopsis) | AAG51364 | 9e−32 | BG241140 (84%) |
| 5 | Hypothetical protein (Arabidopsis) | AAG51355 | e−167 | |
| 4B | Hypothetical protein (Arabidopsis) | AAG51364 | 1e−36 | |
| 6 | BE599243 (100%) | |||
| 7A | Xa1-like (rice) | AAK52518 | e−179 | |
| 8a | Putative protein (Arabidopsis) | CAB96668 | 3e−25 | |
| 9 | Putative protein (Arabidopsis) | CAB82970 | 2e−48 | BE356244 (94%) |
| 7Ba | Xa1-like (rice) | AAK52518 | 8e−20 | |
| 10 | Expressed protein (Arabidopsis) | AAB63084 | 4e−42 | BE599243 (86%) |
| 11 | Cysteine proteinase (Arabidopsis) | CAB41164 | 6e−17 | BG053332 (99%) |
| BF480815 (97%) | ||||
| BG051257 (96%) | ||||
| 12 | Cytochrome P450 (sorghum) | AAC49659 | 0 | BF657281 (99%) |
| (Obtusifoliol 14-α demethylase) | BM323786 (100%) | |||
| BG240113 (100%) | ||||
| BF656939 (100%) | ||||
| 13 | Cyclin-dependent kinase | AAG01533 | 7e−47 | BG487760 (98%) |
| (Tobacco [Nicotiana tabacum]) | BE360020 (100%) | |||
| 14b | ||||
| 15A | Hypothetical protein (rice) | BAB62578 | 3e−32 | BE125790 (99%) BE359797 (100%) |
| 16A | Ankyrin-like protein (Arabidopsis) | AAF04911 | 2e−76 | |
| 15B | Hypothetical protein (rice) | BAB62578 | 1e−30 | BM032442 (97%) AW680740 (82%) |
| 16B | Ankyrin-like protein (Arabidopsis) | AAF04911 | 4e−69 | |
| 15C | Hypothetical protein (rice) | BAB62578 | 1e−33 | BE125790 (92%) |
| BE359797 (90%) | ||||
| 17 | ATPase-like (Arabidopsis) | BAA96915 | 2e−54 | BG047811 (100%) BM330200 (99%) |
| 18 | Hypothetical protein (Arabidopsis) | NP_178129 | 4e−14 | |
| 19-p | Unknown protein (rice) | BAB93427 | 1e−26 | AW283956 (100%) BE358200 (99%) |
Truncated genes.
Predicted as a gene by two or more gene prediction programs but does not have significant homology to any entries in EST or protein databases of GenBank.
The Sorghum rph1 Gene Cluster
Because the sequenced genes are highly homologous to the Rp1 genes of maize and because they map on sorghum linkage group H at a syntenic position with the short arm of maize chromosome 10 where Rp1 is found (Peng et al., 1999), we have named these genes rph1-1 through rph1-5 (Fig. 2). The portion of the Rp1-orthologous gene cluster that we sequenced covers approximately 27 kb. This region contains three apparent full-length Rp1 homologs (rph1-1, rph1-2, and rph1-4) and two truncated Rp1 homologs (rph1-3 and rph1-5, truncated at exactly the same nucleotide). Four other predicted genes are found within the sequenced Rp1-orthologous region. These four predicted genes are present as duplicated pairs, with two between rph1-1 and rph1-2 (genes 1A and 2A) and the other two between rph1-3 and rph1-4 (genes 1B and 2B). One cDNA has been sequenced from sorghum that has 93% and 90% identities to genes 1A and 1B, respectively, but homology was not identified for either gene 1 or 2 in any other species, nor can any function be predicted for these genes.
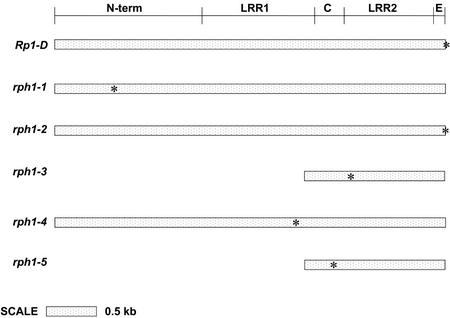
Although genes rph1-1, rph1-2, and rph1-4 are all largely intact, only rph1-2 appears to encode a predicted full-length RP1 protein like that encoded by the maize Rp1-D gene (Fig. 3). Gene rph1-1 contains a stop codon at the position corresponding to amino acid 197 in Rp1-D. A novel LTR retrotransposon, Kaema-1, is inserted 752 bp downstream of the predicted translation initiation codon of rph1-4, which should also result in a truncated protein product. The 5′ regions that would encode the N-terminal 840 amino acids are deleted from rph1-3 and rph1-5. This presumably indicates that the deletion was a single event that predated the duplication of rph1-3 and rph1-5 from a common ancestral gene. This interpretation is supported by the observation that the retained sequences of rph1-3 and rph1-5 are more closely related to each other than to the same region in any of the other rph1 candidate genes in this region.
Figure 3.
Comparison of Rp1 and rph1 gene structures. N-term is the N-terminal region that contains the NBS, LRR1 is LRR region 1 and LRR2 is LRR region 2. The region between LRR1 region and LRR2 region is domain C. The region from the end of LRR2 region to the position of the stop codon predicted for the maize Rp1-D gene is domain E. Asterisks, Stop codons. Genes rph1-3 and rph1-5 are missing their N termini and most of their LRR1 regions.
A 4,077-bp region from the end of rph1-1 to the start of rph1-2 and a 4,256-bp region from the end of rph1-3 to the start of rph1-4 exhibit 96.2% sequence identity, ignoring insertions and deletions (indels). These are the regions that contain the predicted genes 1A, 2A, 1B, and 2B. The very high sequence identity within this area suggests that the region was duplicated within the last three million years, probably by unequal recombination sited within the rph1 genes themselves.
Non-Tandem Gene Duplications Near the rph1 Cluster
We identified 31 possible genes within the more than 268 kb of genomic DNA that we sequenced from sorghum. Five of these genes are Rp1 homologs. Duplicated genes 1A and 1B are predicted by three independent gene-finding programs (FGENESH, GeneMark.hmm, and GENSCAN), but only part of gene 1 has similarity to a cDNA sequence (BE362870: 90% identity over a 260-bp region). Duplicated genes 2A and 2B are annotated as genes based only on gene prediction programs. The degrees of homology and predicted functions (based exclusively on these homologies) for the other 22 predicted genes in this region are provided in Table I. These genes encode a wide variety of predicted proteins, ranging from ankyrin-like proteins, to a cytochrome P-450 enzyme, to a protein that is involved in signal transduction of the phototropic response.
Many of the genes in this region are duplicated, but none are duplicated in a simple tandem manner. Genes 4A and 4B are in an inverted orientation, with predicted gene 5 in between. Gene 7A is homologous to a Xa1-like putative disease R gene from rice and has a truncated copy (gene 7B) upstream of gene 9. Gene 7B has an N-terminal deletion compared with gene 7A (3,855 bp) and the rice Xa1-like gene (4,206 bp; OSJNBa0056A20.14), leaving only about 0.9 kb of gene-like sequence. Gene 15 has three copies (genes 15A–15C) with predicted protein products that show homology to a rice hypothetical protein (BAB62578). Comparison of gene 15A with gene 15C shows 91% identity over a 681-bp region. In contrast, gene 15B has respective 73% and 69% identities with genes 15A and 15C. This suggests that genes 15A and 15C are the more recent duplication products. An alternative hypothesis, that gene 15B is evolving more rapidly, seems unlikely because the Ka to Ks ratios for all three gene 15 comparisons are very similar (0.57–0.67; data not shown). Gene 16 also has duplicated copies, genes 16A and 16B, that are 84% identical. Genes 16B and 15C are in an inverted orientation compared with genes 15A, 16A and 15B, suggesting that a minimum of four rearrangements (duplication of 15A/16A and 15B/16B, followed by inversion of 16B, insertion of Leviathan, and duplication/insertion of 15C from 15A) would have been required to generate the current genomic structure in this region. A more complex series of rearrangements is more likely, however, given the apparent relative dates of the duplication events.
Truncated Genes
Several of the predicted genes in the sequenced region appear to be truncated relative to more intact copies in this same region, and sometimes also to other homologs in the databases. Genes rph1-3, rph1-5, and 7B mentioned above all contain deletions that remove a major portion of their single coding exons. Gene 8 has only a single copy in this region but also appears to be truncated, from both the 5′ and 3′ ends. The predicted protein that could be encoded by gene 8 shows homology to a part (amino acids 183–252) of an Arabidopsis putative protein (CAB96668). Gene 8 also shows 87% nucleotide identity, over the identical 210-bp region, to the interior portion of a wheat expressed sequence tag (BG313327).
Mobile DNAs and Other Structural Features
Five largely intact retroelements were identified in the 268-kb sorghum region. Four of these elements are retrotransposons with long terminal repeats (LTRs), whereas one element is a LINE. Only one of the LTR-retrotransposons, Leviathan, had been previously identified in sorghum (Bennetzen, 1996; Chen et al., 1998). We discovered and named two new elements (Kaema and Pifi). These retroelements were given designations using the approach described by SanMiguel et al. (2002). Pifi was found upstream of rph1-1. About 123 kb from the region where Kaema-1 is inserted in rph1-4, there is another copy of this retrotransposon between genes 11 and 12. A full-length Leviathan retrotransposon of 14,522 bp is present between genes 15B and 16B. In addition, there are many truncated retrotransposons in this region. Miniature inverted-repeat transposable elements (MITEs) were abundant (at least 21) in the sorghum BACs. There were no large inverted repeat (class II) transposable elements found in this region. In comparison with the rph1 region, the 425-kb sorghum region described earlier has only three retroelements (Song et al., 2002).
Several simple sequence repeats (SSRs) were annotated in the region. Most of these SSRs were found within or near genes, but one was identified within an LTR retrotransposon fragment. These hypervariable sequences can serve as excellent markers to follow the segregation of this region, including in future tests to see whether any disease resistance genes in sorghum might map at this location.
Molecular Analysis of Rp1 Homolog Origins and Evolution
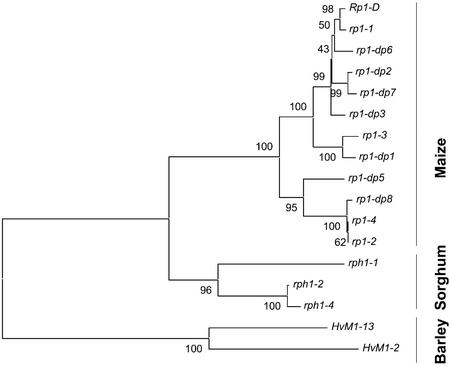
To investigate the evolutionary history of the Rp1-like genes in the rph1 region, all of the Rp1 homologs sequenced in the present study and those in the GenBank database (Collins et al., 1999; Ayliffe et al., 2000; Sun et al., 2001; Ramakrishna et al., 2002b) were used to construct a neighbor-joining tree. Because the Rp1 homologs show diversifying selection mostly in the LRR region, where non-synonymous substitutions exceed synonymous substitutions (Sun et al., 2001; Ramakrishna et al., 2002b), synonymous substitutions were used to construct gene trees. Neighbor-joining trees with different topologies were obtained for different domains, indicative of mosaics of shared gene segments (chimeric genes) that could have resulted from unequal crossing over and/or gene conversion events. However, most of the mosaic patterns were observed in the C-terminal region of the gene (Ramakrishna et al., 2002b). Therefore, the N-terminal regions were used to make neighbor-joining trees using Kimura two-parameter distance estimates and synonymous substitutions separately (Fig. 4). Trees with similar topologies were obtained. Maize, barley and sorghum Rp1 homologs were organized in separate clades. This suggests that either a single ancestral Rp1 gene gave rise to the known Rp1 homologs by repeated duplications in their respective lineages after their divergence from common ancestors and/or that ectopic conversion has homogenized the Rp1 gene family within each species.
Figure 4.
A neighbor-joining tree of barley, maize, and sorghum Rp1 homologs. A neighbor-joining tree was constructed based on synonymous substitutions in the N-terminal region of all the barley, maize, and sorghum Rp1 gene family members from the present study and those present in the GenBank database. Bootstrap values based on 1,000 replicates are indicated at the nodes.
Retrotransposon Insertion Times
The insertion times of full-length sorghum retrotransposons was estimated from the divergence of their LTRs (Table II) as described by SanMiguel and coworkers (2002). The synonymous substitution rate for adh1 and adh2 genes of grasses (6.5 × 10−9 substitutions year−1 site−1) was used to estimate the insertion times of retrotransposons. Under these criteria, the analyses indicated that three of four sorghum LTR-retrotransposons inserted within the last one million years. Our analyses indicated that Kaema-2 is the most ancient insertion in this region, but it still dates back to only about 2.5 mya. The Kaema-1 insertion time is very recent, presumably after the duplication of the segments between the rph1 genes that also duplicated genes 1 and 2. By this same reasoning, it appears that Leviathan also inserted very recently (<0.5 mya), long after the duplications of genes 15 and 16.
Table II.
Predicted retrotransposon insertion times
| Name | Sites | K | se | Mya | se |
|---|---|---|---|---|---|
| Pifi | 235 | 0.009 | 0.006 | 0.66 | 0.47 |
| Kaema-1 | 457 | 0.002 | 0.002 | 0.17 | 0.17 |
| Kaema-2 | 377 | 0.024 | 0.008 | 1.87 | 0.63 |
| Leviathan | 3,922 | 0.004 | 0.001 | 0.33 | 0.08 |
K, No. of substitutions per site calculated as per Kimura two-parameter method; Mya, the estimated insertion time of the retrotransposon in million years.
Comparison of Orthologous rp1/rph1 Loci from Maize and Sorghum
Two segments of the maize genome containing Rp1 homologs have been sequenced, and these 95- and 99-kb maize BAC inserts (AF466931 and AF466932) mapped to the syntenic location with the sorghum rph1 cluster (Ramakrishna et al., 2002b). The entire Rp1 R gene cluster was mapped to two segments of 250 and 300 kb (separated by an unknown distance), containing approximately 15 Rp1 homologs. In sorghum, the orthologous locus is much smaller (about 27 kb) and contains only five Rp1 homologs, rph1-1 through rph1-5. Most of the difference in regional size appears to be attributable to the greater number of LTR retrotransposons inserted in the maize region (Ramakrishna et al., 2002b), but one such mobile DNA (Kaema-1) is found in the sorghum region. The recent insertion times of both the maize and sorghum LTR retrotransposons in these regions indicates that they landed at these locations more than 10 million years after the divergence of maize and sorghum from a common grass ancestor.
In both maize and sorghum, many of the Rp1-homologs appear to be nonfunctional. However, despite the absence of any known R phenotype specified by either the maize rp1 region (from inbred B73) or the sorghum rph1 region, cDNAs have been identified in their respective species, indicating that at least one gene in the region is expressed.
As in the rp1 complex of maize, there are additional genes within the rph1 R gene cluster. None of these genes has any known association with disease resistance activity. Despite the presence of potentially functional non-R genes in each cluster, the same “extra” genes found within the rp1 complex were not seen in the sorghum rph1 complex, nor were the extra genes from the rph1 complex observed in the maize rp1 complex.
DISCUSSION
The Evolution of R Gene Clusters
Many previous studies have shown that comparative analysis of DNA sequences from orthologous chromosomal regions can provide important and unique insights into genome evolution (for review, see Bennetzen, 2000). Loci containing R genes have not been subjected to this analysis, partly because the first few attempts have yielded a lack of conserved R gene presence/absence between the compared species (Kilian et al., 1997; Feuillet and Keller, 1999; Brueggeman et al., 2002). This is not an entirely surprising result, given that comparative recombinational mapping in grass genomes indicated a rapid reorganization of R gene homologs, often leading to a complete lack of synteny for these candidate R genes (Leister et al., 1998).
In this study, we have determined the sequence of the complex locus that we call rph1, plus more than 200 kb of contiguous downstream sequence. The rph1 complex maps in the orthologous position relative to the complex Rp1 locus of maize, and contains duplicated copies of Rp1-like genes. Comparison of the gene sequences suggests that the Rp1-homologous loci from maize, sorghum, and barley have all arisen independently from a single Rp1-like gene that was present in their common ancestor. However, this conclusion may be incorrect because the high frequency of known and predicted unequal recombination events in this region (Sudupak et al., 1993) can cause the R genes within a cluster to evolve as a concerted set. The exception to this prediction of concerted evolution is present in the LRR region, where natural selection has apparently enriched for sequence diversity in this pathogen-recognition component.
Compared with the maize rp1 complex from maize inbred B73, the rph1 cluster of sorghum inbred BTx623 has a significantly lower number of Rp1-homologous loci (15 versus five). However, various haplotypes of Rp1 in maize contain different numbers of Rp1 homologs, ranging from possibly as few as 1 to as many as 20 in known alleles (Collins et al., 1999; Sun et al., 2001; Ramakrishna et al., 2002b; Webb et al., 2002). Hence, the sorghum rph1 complex of BTx623 is within the range of maize Rp1 variability. In both maize and sorghum, it is expected that unequal interchromatid exchange will cause exactly as many decreases in Rp1 homolog number as increases, and that only selection for gene function within the duplicated regions will allow maintenance of a high copy number. It is not known whether the gene fragments might have any role, but they could serve as a reservoir of R gene variability for later use in creating a new Rp1 gene by unequal exchange or conversion.
Unlike most other complex disease resistance loci, the maize Rp1 region contains at least two apparently intact genes within the cluster that have no obvious role in disease resistance (Ramakrishna et al., 2002b). One of these genes exhibits homology to Arabidopsis and rice hypothetical proteins with no similarity to any known cDNAs. The second gene shows homology to rice unknown and hypothetical proteins with 91% and 96% identity to maize cDNAs (H89387 and AW424864) over a 440-bp region. We did not find homologs of these genes in the sorghum rph1 complex. In fact, only the Rp1 homologs are conserved between the sequenced maize and sorghum BACs. The sorghum rph1 region does contain some non-R genes, and these were apparently duplicated as part of an unequal recombination event that duplicated rph1 genes.
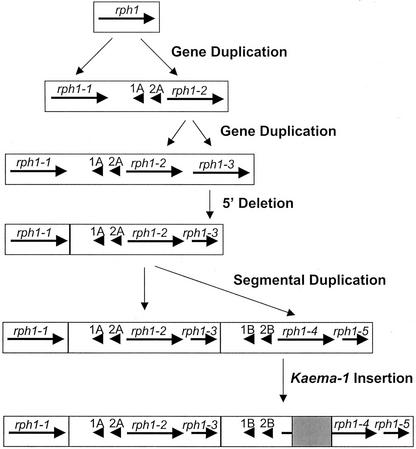
The precise orders of the events that gave rise to the current rph1 complex is ambiguous, given that multiple events have apparently occurred. The simplest series of events that we can predict would require a minimum of five steps. The first step would have been an ancient duplication of a single-copy rph1 gene to give rise to rph1-1 and rph1-2, either by a mechanism that allowed them to be separated by two predicted genes (1A and 2A), or followed by insertions of these two genes in this region. It may seem unlikely that genes frequently insert into new locations, but we have determined that the adh1 gene was inserted as a single functional locus onto a new chromosome in a common ancestor of maize and sorghum (Tikhonov et al., 1999) and that two genes were inserted near the adh2 locus of sorghum after its divergence from a common ancestor with maize (K. Ilic and J. Bennetzen, unpublished data). The second step would have been the direct duplication of the rph1-3 gene from rph1-2, to create a [rph1-1-gene 1A-gene 2A-rph1-2-rph1-3] complex. The third step would have been the 5′ truncation of rph1-3. We know that this step preceded the next duplication because the identical 5′ truncation is seen in rph1-5. The fourth step was the direct tandem duplication of the [gene 1A-gene 2A-rph1-2-rph1-3] portion of the complex to create the current order and content of R gene homologs. This may have occurred by unequal recombination, apparently sited just 3′ to rph1-1 and 3′ to rph1-3. We do not see any mobile DNAs or other repeats at these sites now that may have provided homology for such unequal events, but the rapid decay of repeats in higher plants makes it likely that such legacies would no longer be detectable (SanMiguel et al., 1998; Devos et al., 2002). One additional structural rearrangement must then have occurred, the insertion of Kaema-1. The predicted nature and order of these events are depicted in Figure 5. We cannot predict the precise timing of these proposed events. However, the degree of sequence divergence between rph1-1, rph1-2, and rph1-4 (Fig. 4) can be used to predict approximate dates for the duplications of these genes. Comparing the maize and sorghum Rp1 orthologs indicates that the synonymous substitution rate at this locus is 2-fold higher than the synonymous substitution rate (6.5 × 10−9 substitutions year−1 site−1) at the adh locus of grasses (Gaut et al., 1996). Taking this into account, the initial rph1 duplication into rph1-1 and rph1-2 would have occurred about 8 to 10 mya, and the duplication of the [rph1-1-gene 1A-gene 2A-rph1-2-rph1-3] complex would have taken place within the last two million years.
Figure 5.
A model for the origin of the current state of the sequenced rph1 complex from a single region with only one rph gene. Horizontal arrows designate the positions, orientations, and approximate transcript sizes for the indicated genes.
In some R genes of the NBS-LRR class, unequal recombination is often sited in the repeats in the LRR region, as observed in mutants at the M locus in flax and RPP5 in Arabidopsis (Anderson et al., 1997; Noel et al., 1999). Unequal intragenic recombination has been shown to be frequent at the maize Rp1 locus (Collins et al., 1999; Sun et al., 2001), although rearrangements with breakpoints outside the Rp1 homologs may have been under-represented in these studies. Haplotypes of the Cf-4/Cf-9 region primarily show unequal recombination events sited at a specific location that is not within a R gene (Parniske et al., 1997; Parniske and Jones, 1999). Our data do not allow us to determine the precise breakpoints of the unequal recombinational events that gave rise to the current rph1 complex in BTx623, but they do suggest that the recombination occurred just 3′ to rph1-1, before gene 1. Interestingly, our analysis of the rp1 complex from maize inbred B73 detected similar end points for a large segmental duplication: just 3′ to an Rp1 homolog (Ramakrishna et al., 2002b).
Nearby Rearrangements: Non-Tandem Gene Duplication and Truncation
Of the rph1 R gene homologs, only one of them (rph1-2) contains an uninterrupted reading frame like that seen in the maize Rp1-D gene. Although sorghum does not have any R gene mapped to this region, rph1-2 does show 99% to 100% identity over a 700-bp region with cDNAs from sorghum inbred BTx623 (AW923446 and AW923525). This indicates that at least one Rp1-homologous gene in this complex is expressed in the inbred that was the source of the sequenced BACs.
Most of the Rp1 homologs in the rph1 complex are defective, two by apparent nonsense mutations and two by 5′ truncations. All of the sorghum rph1 genes that we sequenced are in the same transcriptional orientation. Several other genes downstream from the rph1 complex also show nearby duplicated copies and/or genic truncation.
Three duplicated gene pairs and one gene triplication are found in the more than 200 kb of sorghum DNA that we sequenced downstream of the rph1 cluster. One (gene 7) of the duplicated gene pairs is in direct orientation, whereas two (genes 4 and 16) are in an inverted orientation. Of the triplicated genes, 15A and 15B are directly repeated, whereas 15C is inverted relative to the two other copies. In every case, the duplicated or triplicated loci are separated by additional predicted genes. Hence, none of these rearrangements appear to have arisen from a single simple event like an unequal exchange. It seems likely that the current gene content, order, and orientation of this region is attributable to multiple independent rearrangements within the last few million years. Hence, the area adjacent to the rph complex of sorghum is unusually unstable, compared with other analyzed regions of the sorghum genome (Chen et al., 1998; Tikhonov et al., 1999; Ramakrishna et al., 2002a; Song et al., 2002).
The gene truncations that we observed in this region are, in contrast, mostly simple events. Genes rph1-3, rph1-5, and 7B all contain only 5′ deletions of the single predicted exon that they originally encoded. There are several processes that could account for these deletions, including illegitimate recombination (Devos et al., 2002). Gene 8 exhibits apparent deletions that have removed all homology to previously sequenced Arabidopsis and wheat genes from both the 5′ and 3′ ends. Only a central 210-bp segment of a single exon remains in the sequence we have generated for gene 8. Although two independent deletions are possible, it is also possible that the gene 8 exon fragment arrived in this location in the same way that similarly truncated genes were found to cluster within the rp1 complex (Ramakrishna et al., 2002b). Although the precise mechanism of gene fragment insertion is not known, illegitimate repair of a double stranded DNA break seems a likely candidate (Wessler et al., 1990; Salomon and Puchta, 1998). The gene insertions, inversions, duplications, and fragmentations observed in the rph1-adjacent region have all been seen as rare events in other regions of various plant genomes. However, they are greatly enriched in this sequenced region of sorghum. Further studies are needed to determine whether close juxtaposition to an unstable R gene complex might increase the instability of nearby loci. At the very least, it is clear that numerous genic rearrangements have occurred in this small region within the last eight million years, the approximate date of two of the gene duplication events (genes 7A/B and 15A/C) in this small chromosomal segment.
Mobile DNA Accumulation Specificities
As we and others have observed previously (Tikhonov et al., 1999; Mao et al., 2000; Ramakrishna et al., 2002a), the small class II transposable elements in the sequenced region (MITEs) are primarily associated with genes. None is inserted within a known retroelement. In contrast to other regions of the sorghum genome that have been sequenced (Tikhonov et al., 1999; Ramakrishna et al., 2002a; Song et al., 2002), there are quite a few retrotransposons in the sequenced DNA, although none are in an obvious nested organization like that seen in maize (SanMiguel et al., 1996). Many of these retroelements are also truncated, suggesting that they have undergone the same type of illegitimate recombination that has removed most of the retrotransposon sequences from Arabidopsis (Devos et al., 2002).
Orthologous rp1 and rph1 Regions of Sorghum and Maize: Similarities and Contrasts
The Rp1 region in maize has been placed near the telomere of the short arm of chromosome 10 by both cytogenetic and recombinational mapping (Rhoades, 1935; Saxena and Hooker, 1968; Hulbert and Bennetzen, 1991). Although recombination rates in the region can vary a great deal in different haplotype combinations, the complex Rp1 locus usually is found to cover about 0.4 centiMorgans. Rp1 is flanked by markers bnl3.04 and ksu3/4, separated by about 4 centiMorgans on the maize genetic map (Jiang et al., 1996). Most of the sorghum BACs harboring Rp1 homologs mapped to a 27-kb region close to marker bnl3.04 on sorghum linkage group H. Few markers near Rp1 in maize have also been mapped in sorghum, but markers bnl3.04, umc130 and rz561 flank Rp1 in maize and flank the region of linkage group H in sorghum that we sequenced (Wilson et al., 1999; Klein et al., 2000; http://www.gramene. org/cmap). Therefore, the sequenced region in sorghum appears to be syntenic to the Rp1 region mapped on the short arm of chromosome 10. Also, because no other confirmed BAC clones other than those we sequenced exhibited strong homology to Rp1 probes, all evidence suggests that rph1 and rp1 are orthologous complex loci.
In both sorghum and maize, the Rp1 homologs in these syntenic regions are arrayed as nearby duplicated copies, but with some other gene candidates found interrupting the duplicated regions. In maize, a large number of the non-R gene sequences found in the Rp1 region were present as truncated gene fragments. Thirteen of the 16 truncated genes that we found in the maize rp1 complex were in three clusters, suggesting that they arose from multiple rounds of illegitimate break repair at the same sites or from single repairs at each of these sites with multiple unlinked DNA templates (Ramakrishna et al., 2002b). We do not know how frequently genes are truncated in plants, but they tend to occur after gene amplification (Llaca and Messing, 1998). However, the frequency of truncated genes near the rph1 region of sorghum suggests that there is something about disease resistance gene clusters that attracts this type of DNA rearrangement. Further genome analysis is needed to investigate this point.
It is not clear why the rp1 complex of maize and the rph1 complex of sorghum have retained syntenic/orthologous locations on their respective genetic maps, given that conserved syntenic positions are often not seen with R genes (Leister et al., 1998; Feuillet and Keller, 1999; Brueggeman et al., 2002). In several other ways, the rp1 and rph1 complex loci share common properties. They have repeated R genes that are mostly in direct orientation, they have non-R genes within the complex that have been duplicated along with the R genes, they have created direct repetitions of large segments of the R gene region that contain more than one R gene with breakpoints sited just 3′ to Rp1 homologs, they have recently inserted retrotransposons within the cluster, and they have nearby truncated gene fragments. Although all of these events appear to have occurred independently in each lineage, their common general outcomes suggest that similar mechanisms and/or selective forces have continued to act on these loci for many millions of years.
MATERIALS AND METHODS
BAC Selection and Restriction Mapping
An Rp1 5′ probe, previously described (Ramakrishna et al., 2002b), was used to screen a BAC library from sorghum (Sorghum bicolor) inbred BTx623 (http://hbz.tamu.edu/bacindex.html). A total of 10 BACs with homology to this probe were identified, and these clones were mapped using different combinations of MluI, NcoI, and NotI. Fragments were size fractionated by field inversion gel electrophoresis, transferred to nylon membranes, and hybridized with the Rp1 5′ probe.
Detailed restriction maps of BACs Sb95A23 and Sb98N08 were constructed to experimentally validate sequence assembly. BACs were digested with restriction enzymes AscI, NotI, PacI, PmeI, SfiI, and SwaI. The DNAs in the gels were transferred to nylon membranes and hybridized with the Rp1 5′ probe. Fragments observed on the agarose gel were compared between BACs to identify common fragments.
Sequencing and Sequence Analysis
Shotgun libraries for BACs Sb95A23 and Sb98N08 were made as described previously (Dubcovsky et al., 2001; Song et al., 2001). For finishing the sequences of the BACs, gaps were closed as described by Ramakrishna et al. (2002a). Annotation and sequence analysis were performed as described earlier (Dubcovsky et al., 2001; Song et al., 2001; Ramakrishna et al., 2002a). FGENESH (http://www.softberry.com/berry.phtml) with the monocot training set was used for gene prediction in addition to GENSCAN (http://genes.mit.edu/GENSCAN.html) and GeneMark.hmm (http://opal.biology.gatech.edu/GeneMark/eukhmm.cgi). The criteria used to define a gene were, first, a match to a sequence in a protein database using BLASTX (Altschul et al., 1997), second, a match to cDNAs or, third, prediction as a gene by two or more gene prediction programs. These criteria are used after excluding identified transposons.
All of the Rp1 homologs and retrotransposons were aligned using CLUSTALX (Thompson et al., 1997). Rates of nucleotide substitution were estimated using the distance measures of Nei and Gojobori (1986) and the Jukes-Cantor correction as implemented in the MEGA2 (molecular evolutionary genetic analysis) package (Kumar et al., 2001). Neighbor-joining trees using synonymous substitution rates were constructed as implemented in MEGA2. Insertion times of retrotransposons and divergence times of genes were estimated as described previously (Ramakrishna et al., 2002b, 2002a).
Footnotes
This work was funded by the National Science Foundation Plant Genome Program (grant no. 9975618).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.014951.
LITERATURE CITED
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PA, Lawrence GJ, Morrish BC, Ayliffe MA, Finnegan EJ, Ellis JG. Inactivation of the flax rust resistance gene Massociated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell. 1997;9:641–651. doi: 10.1105/tpc.9.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Ayliffe MA, Collins NC, Ellis JG, Pryor A. The maize rp1rust resistance gene identifies homologues in barley that have been subjected to diversifying selection. Theor Appl Genet. 2000;100:1144–1154. [Google Scholar]
- Bennetzen JL. The contributions of retroelements to plant genome organization, function and evolution. Trends Microbiol. 1996;4:347–353. doi: 10.1016/0966-842x(96)10042-1. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell. 2000;12:1021–1029. doi: 10.1105/tpc.12.7.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Hulbert SH. Organization, instability and evolution of plant disease resistance genes. Plant Mol Biol. 1992;20:575–578. [PubMed] [Google Scholar]
- Bennetzen JL, Qin MM, Ingels S, Ellingboe AH. Allele-specific and mutator-associated instability at the rp1disease-resistance locus of maize. Nature. 1988;332:369–370. [Google Scholar]
- Brueggeman R, Rostoks N, Kudrna D, Kilian A, Han F, Chen J, Druka A, Steffenson B, Kleinhofs A. The barley stem rust-resistance gene Rpg1is a novel disease-resistance gene with homology to receptor kinases. Proc Natl Acad Sci USA. 2002;99:9328–9333. doi: 10.1073/pnas.142284999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo AL, Schaal BA, Kunkel BN. Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:302–306. doi: 10.1073/pnas.96.1.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, SanMiguel P, Bennetzen JL. Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice. Genetics. 1998;148:435–443. doi: 10.1093/genetics/148.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T. Molecular characterization of the maize Rp1-Drust resistance haplotype and its mutants. Plant Cell. 1999;11:1365–1376. doi: 10.1105/tpc.11.7.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JKM, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Ellis JG. Contrasting modes of evolution acting on the complex Nlocus for rust resistance in flax. Plant J. 2001;27:439–453. doi: 10.1046/j.1365-313x.2001.01114.x. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Ramakrishna W, SanMiguel P, Busso C, Yan L, Shiloff B, Bennetzen J. Comparative sequence analysis of colinear barley and rice BACs. Plant Physiol. 2001;125:1342–1353. doi: 10.1104/pp.125.3.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Dodds P, Pryor T. Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol. 2000a;3:278–284. doi: 10.1016/s1369-5266(00)00080-7. [DOI] [PubMed] [Google Scholar]
- Ellis J, Dodds P, Pryor T. The generation of plant disease resistance gene specificities. Trends Plant Sci. 2000b;5:373–379. doi: 10.1016/s1360-1385(00)01694-0. [DOI] [PubMed] [Google Scholar]
- Feuillet C, Keller B. High gene density is conserved at syntenic loci of small and large grass genomes. Proc Natl Acad Sci USA. 1999;96:8265–8270. doi: 10.1073/pnas.96.14.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego F, Feuillet C, Messmer M, Penger A, Graner A, Yano M, Sasaki T, Keller B. Comparative mapping of the two wheat leaf rust resistance loci Lr1 and Lr10in rice and barley. Genome. 1998;41:328–336. doi: 10.1139/g98-024. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA. 1997;88:2060–2064. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Hulbert SH. Evidence for the involvement of gene conversion in meiotic instability of the Rp1rust resistance genes of maize. Genome. 1994;37:742–746. doi: 10.1139/g94-105. [DOI] [PubMed] [Google Scholar]
- Hulbert SH, Bennetzen JL. Recombination at the rp1locus of maize. Mol Gen Genet. 1991;226:377–382. doi: 10.1007/BF00260649. [DOI] [PubMed] [Google Scholar]
- Jiang JM, Hulbert SH, Gill BS, Ward DC. Interphase fluorescence in situhybridization mapping: a physical mapping strategy for plant species with large complex genomes. Mol Gen Genet. 1996;252:497–502. doi: 10.1007/BF02172395. [DOI] [PubMed] [Google Scholar]
- Kilian A, Chen J, Han F, Steffenson B, Kleinhofs A. Towards map-based cloning of the barley stem rust resistance genes Rpg1 and rpg4using rice as an intergenomic cloning vehicle. Plant Mol Biol. 1997;35:187–195. [PubMed] [Google Scholar]
- Klein P, Klein RR, Cartinhour S, Ulanch P, Dong J, Obert J, Morishige DT, Schlueter S, Childs K, Ale M. A high-throughput AFLP-based method for constructing integrated genetic and physical maps: progress toward a sorghum genome map. Genome Res. 2000;10:789–807. doi: 10.1101/gr.10.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Leister DM, Kurth J, Laurie DA, Yano M, Sasaki T, Devos KM, Graner A, Schulze-Lefert P. Rapid reorganisation of resistance gene homologues in cereal genomes. Proc Natl Acad Sci USA. 1998;95:370–375. doi: 10.1073/pnas.95.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llaca V, Messing J. Amplicons of maize zein genes are conserved within genic but expanded and constricted in intergenic regions. Plant J. 1998;15:211–220. doi: 10.1046/j.1365-313x.1998.00200.x. [DOI] [PubMed] [Google Scholar]
- Mao L, Wood TC, Yu Y, Budiman MA, Tomkins J, Sung-Sick W, Sasinowski M, Presting G, Frisch D, Goff S et al. Rice transposable elements: a survey of 73,000 sequence-tagged-connectors. Genome Res. 2000;10:982–990. doi: 10.1101/gr.10.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW. Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell. 1998;10:1833–1846. doi: 10.1105/tpc.10.11.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multani DS, Meeley RB, Paterson AH, Gray J, Briggs SP, Johal GS. Plant-pathogen microevolution: molecular basis for the origin of a fungal disease in maize. Proc Natl Acad Sci USA. 1998;95:1686–1691. doi: 10.1073/pnas.95.4.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Noel L, Moores TL, van der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JDG. Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell. 1999;11:2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BBH, Jones JDG. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9locus of tomato. Cell. 1997;91:821–832. doi: 10.1016/s0092-8674(00)80470-5. [DOI] [PubMed] [Google Scholar]
- Parniske M, Jones JDG. Recombination between diverged clusters of the tomato Cf-9plant disease resistance gene family. Proc Natl Acad Sci USA. 1999;96:5850–5855. doi: 10.1073/pnas.96.10.5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Schertz KF, Cartinhour S, Hart GE. Comparative genome mapping of Sorghum bicolor(L.) Moench using an RFLP map constructed in a population of recombinant inbred lines. Plant Breed. 1999;118:225–235. [Google Scholar]
- Ramakrishna W, Dubcovsky J, Park Y-J, Busso C, Emberton J, SanMiguel P, Bennetzen JL (2002a) Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics (in press) [DOI] [PMC free article] [PubMed]
- Ramakrishna W, Emberton J, Ogden M, SanMiguel P, Bennetzen JL (2002b) Structural analyses of the maize Rp1 complex uncovers numerous sites and unexpected mechanisms of local rearrangement. Plant Cell (in press) [DOI] [PMC free article] [PubMed]
- Rhoades VH. The location of a gene for disease resistance in maize. Proc Natl Acad Sci USA. 1935;21:243–246. doi: 10.1073/pnas.21.5.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter TE, Pryor TJ, Bennetzen JL, Hulbert SH. New rust resistance specificities associated with recombination in the Rp1complex in maize. Genetics. 1995;141:373–381. doi: 10.1093/genetics/141.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Ramakrishna W, Bennetzen JL, Busso CS, Dubcovsky J. Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5Am. Funct Integr Genom. 2002;2:70–80. doi: 10.1007/s10142-002-0056-4. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Saxena KMS, Hooker AL. On the structure of a gene for disease resistance in maize. Proc Natl Acad Sci USA. 1968;61:1300–1305. doi: 10.1073/pnas.61.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Llaca V, Linton E, Messing J. Sequence, regulation, and evolution of the maize 22-kD α zein gene family. Genome Res. 2001;11:1817–1825. doi: 10.1101/gr.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Llaca V, Messing J. Mosaic organization of orthologous sequences in grass genomes. Genome Res. 2002;13:1549–1555. doi: 10.1101/gr.268302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W-Y, Pi L-Y, Wang G-L, Gardner J, Holsten T, Ronald PC. Evolution of the rice Xa21disease resistance gene family. Plant Cell. 1997;9:1279–1287. doi: 10.1105/tpc.9.8.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- Sudupak MA, Bennetzen JL, Hulbert SH. Unequal exchange and meiotic instability of disease-resistance genes in the rp1region of maize. Genetics. 1993;133:119–125. doi: 10.1093/genetics/133.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Collins NC, Ayliffe M, Smith SM, Drake J, Pryor T, Hulbert SH. Recombination between paralogues at the rp1rust resistance locus in maize. Genetics. 2001;158:433–438. doi: 10.1093/genetics/158.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov AP, SanMiguel PJ, Nakajima Y, Gorenstein ND, Bennetzen JL, Avramova Z. Colinearity and its exceptions in orthologous adhregions of maize and sorghum. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CA, Richter TE, Collins NC, Nicolas M, Trick HN, Pryor T, Hulbert SH. Genetic and molecular characterization of the maize rp3rust resistance locus. Genetics. 2002;162:381–394. doi: 10.1093/genetics/162.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler S, Tarpley A, Purugganan M, Spell M, Okagaki R. Filler DNA is associated with spontaneous deletions in maize. Proc Natl Acad Sci USA. 1990;87:8731–8735. doi: 10.1073/pnas.87.22.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR, Bureau TE, White SE. LTR-retrotransposons and MITES: important players in the evolution of plant genomes. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- Wilson WA, Harrington SE, Woodman WL, Lee M, Sorrells ME, McCouch SR. Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated panicoids. Genetics. 1999;153:453–473. doi: 10.1093/genetics/153.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]