Abstract
Behaviors are often highly heritable, polygenic traits. To investigate molecular mediators of behavior, we analyzed gene expression patterns across seven brain regions (amygdala, basal ganglia, cerebellum, frontal cortex, hippocampus, cingulate cortex, and olfactory bulb) of 10 different inbred mouse strains (129S1/SvImJ, A/J, AKR/J, BALB/cByJ, BTBR T+ tf/J, C3H/HeJ, C57BL/6J, C57L/J, DBA/2J, and FVB/NJ). Extensive variation was observed across both strain and brain region. These data provide potential transcriptional intermediates linking polygenic variation to differences in behavior. For example, mice from different strains had variable performance on the rotarod task, which correlated with the expression of >2000 transcripts in the cerebellum. Correlation with this task was also found in the amygdala and hippocampus, but not in other regions examined, indicating the potential complexity of motor coordination. Thus we can begin to identify expression profiles contributing to behavioral phenotypes through variation in gene expression.
INVESTIGATIONS into the genetics of behavioral traits, from alcohol preference to depression to cognitive ability, have revealed that behavior is highly heritable and likely influenced by many genes (Winterer and Goldman 2003; Oroszi and Goldman 2004; Hamet and Tremblay 2005). This genetic complexity has led to difficulty in identifying genes involved in psychiatric disorders as well as those contributing to general behavioral characteristics. To understand better how genotype influences behavioral phenotype, we performed a detailed analysis of expression profiles throughout the brain to determine which transcripts vary by genetic background and correlate with behavior. Recent catalogs of the mouse transcriptome indicate that there may be <30,000 protein-coding genes, but that alternate splicing, alternative start and stop sites, and microRNAs can add substantially to genetic complexity (Carninci et al. 2005). This makes the dissection of gene expression, an intermediate between polymorphic DNA sequence and variable phenotype, a logical choice to investigate relationships connecting genotype to complex phenotypes like behavior.
Previous studies have examined gene expression profiles in the brain by microarray analysis. Zapala et al. (2005) have shown that regional differences in gene expression in the adult brain are largely reflective of the developmental origin of a particular region. Investigations into strain-related differences have led to estimates that 1–2% of the genes may vary in expression between six brain regions of C57BL/6 and 129SvEv mice (Sandberg et al. 2000; Pavlidis and Noble 2001).
To extend previous studies and gain a more accurate picture of transcriptional variation, we measured gene expression in seven different regions of the mouse brain: amygdala, basal ganglia, cerebellum, frontal cortex, hippocampus, cingulate cortex, and olfactory bulb. These regions all play roles in behavior, and they encompass a range of neurocognitive functions, including locomotion, emotion, sensation, learning, and memory. Furthermore, the gene expression profile from each region was examined in 10 different inbred mouse strains: 129S1/SvImJ, A/J, AKR/J, BALB/cByJ, BTBR T+ tf/J, C3H/HeJ, C57BL/6J, C57L/J, DBA/2J, and FVB/NJ. Taking advantage of the diversity of both brain region and strain, we found that 57% of all transcripts assayed show variation across region and/or genetic background, a marked increase over previous reports (Sandberg et al. 2000; Pavlidis and Noble 2001; Zapala et al. 2005). This diversity is due to the inclusion of more distantly related strains and is a tool to focus on the molecular causes for the phenotypic diversity observed among these strains.
Performance on the accelerating rotarod is a common motor coordination task, utilized with genetically and pharmacologically modified mouse models. Comparison of this strain-specific phenotype to gene expression serves as a clear proof of principle for our approach relating expression to strain-specific phenotypes. Striking correlation was found between this task and gene expression in the cerebellum, a region involved with motor coordination. Surprisingly, correlation was also found to a lesser extent in the amygdala and hippocampus, suggesting the involvement of fear response and learning and memory in this task. No significant correlations were found in the other brain regions. These findings demonstrate the power of using gene expression profiles as an intermediate molecular phenotype to link underlying genetic variation to a behavioral phenotype.
MATERIALS AND METHODS
Mice:
Male mice, 3–4 weeks of age, were purchased from the Jackson Laboratories. Upon arrival at the University of North Carolina, mice were housed four to five per cage under standard specific pathogen free conditions. After acclimating for 1 week, mice were killed by cervical dislocation and specific brain regions were dissected. Animals used for the rotarod task were as previously described (Nadler et al. 2004).
Tissue samples:
Brains were placed in RNAlater (Ambion, Austin, TX) immediately upon removal and remained in the solution during dissection under low magnification (see supplemental Figure 1 at http://www.genetics.org/supplemental/). After removing the cerebellum and the olfactory bulb, the frontal cortex region was taken from the most anterior cortical area, avoiding any remaining olfactory bulb. At this point, a cut was made at the level of the optic chiasm to reveal the basal ganglia region anterior to the cut and leaving the amygdala, cingulate cortex, and hippocampus in the posterior section. The basal ganglia region was removed from the anterior portion, avoiding any cortical tissue. From the posterior portion, the cingulate cortex region was removed from both sides of the midline, above the hippocampus, which was visible from the original cut. The amygdala region was taken from both sides of the brain. Finally, the hippocampus was blunt dissected from the remaining posterior portion.
Tissues from three mice were pooled by region, such that the tissue sample contained representatives from each home cage. Tissues were stored overnight in RNAlater at 4° before transferring into TRIzol Reagent (Invitrogen, San Diego) for homogenization with a Kinematica AG (Brinkmann, Westbury, NY). The homogenate was stored at −80° until sample preparation.
RNA samples:
RNA was prepared from the pooled samples using the TRIzol protocol. Following extraction, the RNeasy miniprep cleanup protocol (QIAGEN, Valencia, CA) was used. RNA was quantitated by a spectrophotometer and its quality visualized using the Bioanalyzer Lab-on-a-chip (Agilent).
Microarray:
The strategy for microarray hybridization was as follows. Each tissue preparation consisted of a pooled sample from three animals, hybridized to a single array. Three pools were prepared for each strain-by-region condition, except C57BL/6J × frontal cortex, C3H/HeJ × cerebellum, and C3H/HeJ × hippocampus, which had four pools each. Pools were derived from independent sets of mice. Each pool was hybridized to a single array. Therefore, each strain-by-region condition had three biological replicates from which to calculate mean expression and variation. Furthermore, no two biological replicate samples were hybridized concurrently. Arrays were hybridized in batches of six to eight. Microarray analysis was performed using the Agilent mouse platform (G4121A). Fifteen micrograms of total RNA from each sample was labeled with Cy3 using the Agilent fluorescent direct label kit. Similarly, 15 μg of Stratagene mouse universal reference RNA (740100) was labeled with Cy5. Differentially labeled RNAs were cohybridized using the Agilent protocol overnight at 60°. Microarrays were washed using the Agilent SSPE/solution 3 protocol and scanned using an Agilent scanner. Raw data were collected using feature extraction software (Agilent).
Rotarod task:
Twenty mice per strain were tested on an accelerating rotarod (Ugo Basile) to assess motor coordination. Each subject was given two trials, with 45 sec between trials. Revolutions per minute (rpm) were initially 3 rpm, with a progressive increase to a maximum of 30 rpm across 5 min (maximum trial length). Measures were taken of latency to fall from the top of the rotating barrel.
Statistical analysis:
Data were normalized by the lowess function using the SMA package in R (http://stat-www.berkeley.edu/users/terry/zarray/Software/smacode.html) and further scale normalized. The log ratio of background-subtracted Cy3 signal to background-subtracted Cy5 signal was calculated for each spot and used for subsequent analyses. The Excel plug-in of the SAM package (recoded in R to handle large data sets) was used to identify significant changes in gene expression and to correlate expression in each region with the strain values on the rotarod task as described below (Tusher et al. 2001). The false discovery rate was controlled at 0.01 for differential expression hypotheses (2000 permutations) and at 0.05 for correlation with rotarod performance (500 permutations).
Further, two-way analyses of variance were performed for each transcript, to evaluate main effects of strain, region, and strain-by-region interactions. To assess the relative contribution of strain and region effects, we performed multiple regression analysis of expression values with strain and region as main-effect predictors, using the glm function in R. Contributions of strain and tissue to the overall multiple R2 were determined for each gene separately.
Hierarchical cluster analysis was performed using the hclust routine in R (http://www.r-project.org/), using the Pearson correlation metric. Separate region clustering was performed within each strain, and strains were clustered within each region. In addition, for each spot, an average expression value for each strain was obtained across all brain regions. These average values were used for an overall averaged cluster analysis of the strains. Similarly, an averaged cluster analysis was performed for brain regions, where averages were taken across strains for each brain region.
Pairwise comparisons between individual regions within strains involved too few arrays for SAM analysis, and for these comparisons we used two-sample t-tests assuming unequal variances and the Benjamini–Hochberg FDR-controlling procedure (Benjamini and Hochberg 1995).
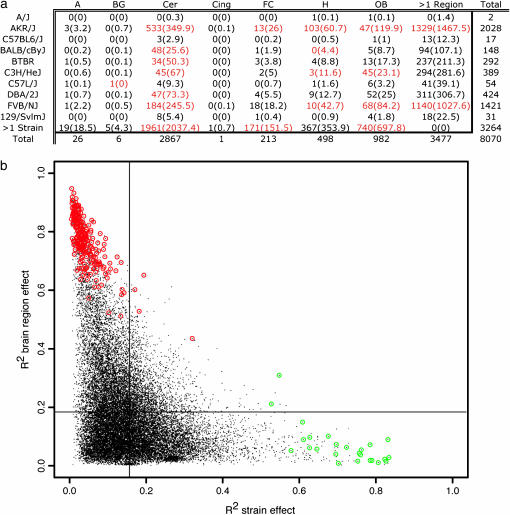
For the table showing transcripts with expression levels specific to a single strain-by-region combination (Figure 3a), the independence of strain and region was assessed via a standard χ2-contingency table statistic. We subjected the entries to 10,000 random permutations to obtain an empirical P-value.
Figure 3.—
Gene expression is influenced by strain and region. (a) The number of genes whose variation is attributed to a particular brain region-by-strain identity. This is the intersection of genes in the first column of Figures 1a and 2a. Cells in red indicate significant departure from that expected. A, amygdala. BG, basal ganglia. Cer, cerebellum. Cing, cingulate cortex. FC, frontal cortex. H, hippocampus. OB, olfactory bulb. Observed (expected) values are shown. (b) Each point represents a single gene with variable expression across strain or region. The x-axis indicates the proportion of expression variation due to strain, the y-axis indicates variation due to brain region. Lines indicate the mean R2-value for each axis. Genes from the last bar of Figure 1a are circled in red. Genes from the last bar of Figure 2a are circled in green.
RESULTS
Gene expression profiles were generated for seven brain regions in 10 inbred strains of mice. Three arrays were hybridized for each strain-by-region condition. Each array consisted of pooled RNA from three individual animals, for a total of nine animals per strain-by-region condition. Samples were hybridized against a reference RNA (740100; Stratagene, La Jolla, CA) on the Agilent mouse oligo platform (G4121A, Agilent), which contains features corresponding to 20,871 transcripts. Of these, 14,003 are annotated genes, 4974 are ESTs or hypothetical genes, and 1893 are unknown. These microarray data are publicly available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). A total of 11,884 transcripts showed variation across brain region and/or strain. The effects of strain and region on expression were analyzed using the significance analysis of microarrays (SAM) (Tusher et al. 2001) and supported by analogous two-way ANOVA analyses including strain-by-region interaction effects (supplemental data 1 at http://www.genetics.org/supplemental/).
Widespread gene expression differences across seven mouse brain regions:
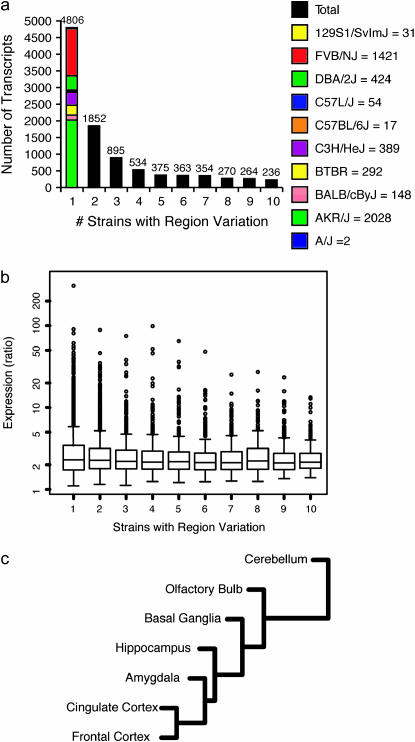
SAM analyses were performed within each strain to identify transcripts exhibiting significant regional variation with a false discovery rate (FDR) <0.01. A total of 9949 transcripts showed statistically significant regional expression differences in ≥1 of the 10 strains. These transcripts were grouped by the number of strains in which they exhibited regional variation (Figure 1a, supplemental data 2 at http://www.genetics.org/supplemental/). For example, 4806 transcripts showed regional variation in a single strain, the majority of which appeared in AKR/J or FVB/NJ. A total of 236 transcripts exhibited regional variation in all 10 strains. Expression profiles of these 236 transcripts show, with few exceptions, that the profile of gene expression across regions is similar in all strains (supplemental data 3 at http://www.genetics.org/supplemental/). For example, Nts expression is higher in the amygdala, basal ganglia, and hippocampus of all 10 strains.
Figure 1.—
Gene expression varies with brain region. (a) Number of genes with variable expression across brain regions in one or more strains. Column 1, genes with variable expression across brain regions in a single strain, is divided by which strain has variation (supplemental data 2 at http://www.genetics.org/supplemental/). (b) Range of expression level as a ratio of maximum expression to minimum expression. Columns correspond to a. (c) Cluster of brain regions using strain-averaged gene expression profiles.
For each transcript showing significant regional variation within a strain, we calculated the range of expression across the regions. The ratio of extreme values (highest expression among all regions divided by lowest expression among regions) was averaged across all strains in which the transcript showed variation. Interestingly, the distribution of these average expression ratios did not seem to depend greatly on the number of strains showing regional variation, with most values falling in the range of two to three (Figure 1b).
The relationship between brain regions is illustrated by clustering the regions using strain-averaged gene expression (Figure 1c). The cerebellum is the most unique and clusters away from the telencephalic regions. In the telencephalon, olfactory bulb and basal ganglia are distinctive with amygdala and hippocampus more closely related to each other. The frontal cortex and cingulate cortex are the most closely related regions examined. These data are consistent with those of Zapala et al. (2005), who found that regional cluster analysis recapitulated the development of the embryonic brain. Expression data from each individual strain were also clustered separately to examine the differences in relationship between brain regions from strain to strain (supplemental data 4 at http://www.genetics.org/supplemental/). Two pairs of strains, A/J-FVB/NJ and 129/SvImJ-C57L/J, showed identical region clusters, the latter of which is very similar to the strain-averaged cluster. The brain region clusters of BTBR, BALB/cByJ, and C57BL/6J differ only in the placement of a single branch. In 8 of the 10 strains, frontal and cingulate cortex cluster most closely together. Within each strain we performed all pairwise comparisons of the seven regions and report transcripts with FDR <0.05 (supplemental data 9 at http://www.genetics.org/supplemental/).
Extensive variation in gene expression among inbred mouse strains:
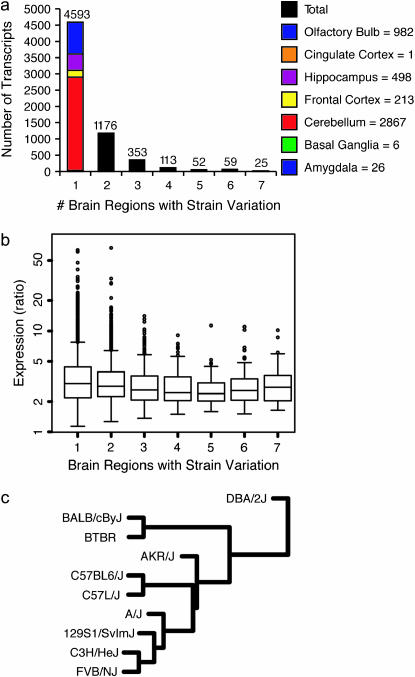
SAM analyses were performed within each brain region to identify transcripts exhibiting significant strain variation at FDR <0.01. A total of 6371 transcripts showed statistically significant expression differences by strain in one or more brain regions, with the majority being specific to a single brain region (Figure 2a). Transcripts showing strain variation in only one brain region are variably expressed most often in the cerebellum (supplemental data 5 at http://www.genetics.org/supplemental/). For the 25 transcripts showing significantly variable expression across strains in all brain regions, variation is due to expression differences in the same strains in all regions (supplemental data 6 at http://www.genetics.org/supplemental/). For example, Chi3l3 is expressed at a higher level in C3H/HeJ than in the other strains in all seven regions.
Figure 2.—
Gene expression is quantifiably different between strains. (a) Number of genes with variable expression across strains in one or more brain regions. Column 1, genes with variable expression across strains in a single region, is divided by which region has variation (supplemental data 5 at http://www.genetics.org/supplemental/). (b) Range of expression level as a ratio of maximum expression to minimum expression. Columns correspond to a. (c) Cluster of strains using region-averaged gene expression profiles.
For each transcript showing significant variation within a region, we calculated the range of expression across the strains. The ratio of extreme values was averaged across all regions in which the transcript showed variation. Similar to the results for region, the distribution of these ratios does not depend upon the number of regions showing strain variation, with most values around two to three (Figure 2b).
The relationship between the strains examined is illustrated by cluster analysis using expression levels averaged across brain region (Figure 2c). DBA/2J shows the most distinctive pattern of expression of the 10 strains. BALB/cByJ and BTBR form a group more distinct from the rest of the strains than from each other. Among the remaining strains, AKR/J forms a branch by itself, while C57L/J and C57BL/6J are more similar to each other. C3H/HeJ and FVB/NJ also cluster together. When the strains are clustered using each brain region individually, the structure of the clusters is markedly different (supplemental data 7 at http://www.genetics.org/supplemental/). DBA/2J remains the outlier in three of the seven regions. A/J and AKR/J are closely related in four of the seven regions, consistent with a recent single-nucleotide polymorphism (SNP) analysis (Petkov et al. 2005). None of the regional clusters recapitulates the region-averaged cluster structure.
Interaction between strain- and region-specific gene expression profiles:
Transcripts appearing in the last column of Figure 1a exhibit regional variation in all 10 strains. Similarly, the transcripts in the last column of Figure 2a exhibit characteristic variation in strain-specific expression in all seven brain regions. We next considered transcripts appearing in the first columns of Figures 1a and 2a. Such transcripts show regional variation in only 1 strain and strain variation in only one region and thus exhibit expression levels largely specific to a single strain-by-region combination. In Figure 3a, the table of transcripts with expression levels specific to strain-by-region combinations shows that certain combinations appear to be over- or underrepresented (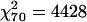 , permutation P = 0.0033). Strain-by-region combinations AKR/J × cerebellum, AKR/J × hippocampus, BALB/cByJ × cerebellum, and C3H/HeJ × olfactory bulb have an overrepresentation of variable transcripts, as do multiple regions of BTBR and FVB/NJ. Strain-by-region combinations involving the cerebellum, hippocampus, and olfactory bulb of multiple strains, C57L/J × basal ganglia, and multiple regions of AKR/J have an underrepresentation of variable transcripts.
, permutation P = 0.0033). Strain-by-region combinations AKR/J × cerebellum, AKR/J × hippocampus, BALB/cByJ × cerebellum, and C3H/HeJ × olfactory bulb have an overrepresentation of variable transcripts, as do multiple regions of BTBR and FVB/NJ. Strain-by-region combinations involving the cerebellum, hippocampus, and olfactory bulb of multiple strains, C57L/J × basal ganglia, and multiple regions of AKR/J have an underrepresentation of variable transcripts.
The relative contribution of strain or region to the variation in expression of each transcript can be described by the portion of the multiple regression R2 attributable to strain and region as main effects. A slightly larger average R2-value is observed for brain region effect than for strain effect (Figure 3b). This tendency is more pronounced for higher R2-values. For example, ninefold more transcripts exhibit R2 > 0.6 for brain region effect (716) than for strain effect (78). However, there are some transcripts whose explained variation is due more to strain than to brain region. The transcripts at the extremes are often those from the last columns of Figures 1a and 2a.
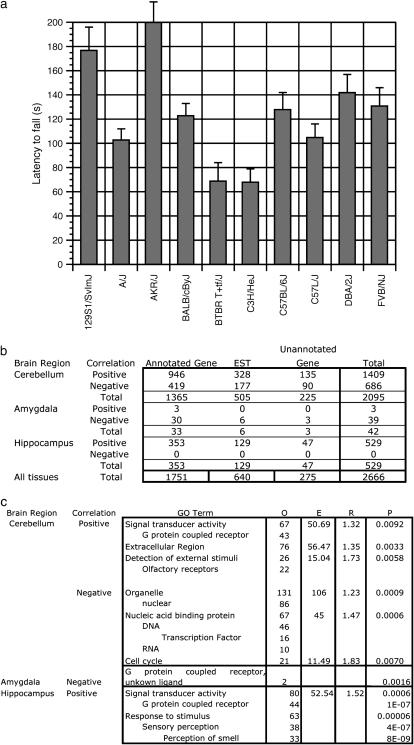
Gene expression is correlated with motor task performance in the cerebellum, amygdala, and hippocampus:
A further goal of quantifying expression variation is to investigate the genetic contribution to biological function. Inbred mouse strains differ in many physiological and behavioral phenotypes (Bogue and Grubb 2004). One behavioral phenotype showing strain-specific variation is performance on the rotarod task (Figure 4a). This task assays motor coordination by requiring a mouse to walk on a rotating rod as its rotation increases; latency to fall is a measure of motor performance. In the cerebellum, transcripts were tested for correlation with rotarod performance using linear regression analysis in the SAM package. At an FDR of 0.05, 1409 transcripts showed a positive correlation with rotarod performance, while 686 showed a negative correlation (Figure 4b, supplemental data 8 at http://www.genetics.org/supplemental/). Of the 500 transcripts most significantly positively correlated with an increase in rotarod performance, several gene ontology (GO) term categories were overrepresented (Zhang et al. 2004). These included signal transducer activity, the extracellular region, and detection of external stimuli. Of the 500 most negatively correlated transcripts, significant GO term categories included the organelle, nucleic acid binding proteins, and the cell cycle (Figure 4c).
Figure 4.—
Gene expression as a function of motor coordination. (a) Performance on rotarod as measured by latency to fall in seconds. (b) Number of genes correlated to rotarod performance. Positive and negative correlations divided by known genes, ESTs, and unknown genes as listed on the Agilent gene list are shown. (c) Overrepresented GO term categories in the 500 most highly correlated sequences for both positive and negative correlations. O, observed. E, expected. R, enrichment. P, P-value.
Significant gene expression correlation was also found in the amygdala and hippocampus. In the amygdala, 3 genes were positively correlated and 38 were negatively correlated with rotarod performance. In the hippocampus, 529 genes showed a positive correlation between expression and rotarod performance (Figure 4b, supplemental data 8 at http://www.genetics.org/supplemental/). GO term analysis of these genes shows an overrepresentation of signal transduction activity, response to stimulus, and sensory perception (Figure 4c). When similar SAM analyses were performed using gene expression from olfactory bulb, basal ganglia, and cingulate and frontal cortices, no significant correlations were found.
DISCUSSION
Widespread gene expression variation across both brain region and strain:
Earlier studies estimated that 1% of genes show variable expression in a comparison of C57BL6 and 129SvEv (Sandberg et al. 2000). Reanalysis of these data with a refined statistical method doubled this estimate (Pavlidis and Noble 2001). Using our data, comparison of only these two strains yielded similarly low estimates (data not shown). However, using 10 inbred strains, we found that nearly 30% of transcripts exhibit strain-specific expression variation in at least one region of the brain. This increase is due, in large part, to inclusion of more distantly related strains, such as AKR/J and FVB/NJ, providing a greater opportunity to relate expression to phenotypic differences among strains. The majority of differentially expressed transcripts identified in earlier work are present in our gene lists (Sandberg et al. 2000; Pavlidis and Noble 2001). We hypothesize that many of these genes contribute directly to neurological and behavioral phenotypes, while others are influenced by genetic variation among strains without direct behavioral consequences. Therefore, the list of differentially expressed genes generated by this approach is a rich resource for the genetic dissection of behavior.
These data reveal how genetic background interacts with gene expression across brain regions. Strain-averaged cluster analysis of the relatedness of brain regions supports the finding that regional expression profile is reflective of brain development (Zapala et al. 2005). When brain regions are clustered by expression profile for each strain, different relationships emerge. Cluster analysis of region-averaged strain relatedness does not recapitulate the derivation of the strains or their relatedness based on SNP analyses (Petkov et al. 2005). In fact, the region-averaged strain cluster differs from all individual brain region clusters. The differential relatedness of strains on each brain region may be an indication of why certain strains behave similarly on some tasks, but not others. The difference between averaged and individual clusters is yet another indication of the complex relationship between the influence of genetic background and brain region on gene expression.
Prior to this experiment, it was often assumed that differences between strains would be much smaller than between regions (Zapala et al. 2005). The present analysis shows that there are more transcripts with an R2 > 0.6 for region than there are for strain. For the bulk of the transcripts, however, the mean R2 is very similar for region or strain. Furthermore, the mean range of transcript expression is similar across regions and strains. We also observed that transcripts exhibiting significant regional variation in all 10 strains tend to show remarkably similar expression profiles across different strains. These genes are likely to perform functions specific to the regions in which they are highly expressed, since this expression has been conserved in all strains. Similarly, transcripts exhibiting strain variation in all seven regions show similar profiles for different regions. The expression of these genes is dependent upon the genetic background and may be involved strain-specific phenotypic changes in multiple organ systems. The range of expression changes in these two sets of genes, however, is still quite similar. So even though the expression of a transcript varies by region in all 10 strains, the change in expression observed is not significantly larger than what is seen in a transcript with variable expression in a single strain.
Some transcripts exhibit expression levels largely specific to single strain-by-region combinations, and the pattern of strain-by-region specificity is complex. Transcripts with this property may be of particular interest for phenotypes likely to involve specific brain regions. For example, the cerebellum of an AKR/J mouse has more variable transcripts than can be accounted for by transcripts differentially regulated in all AKR/J tissues. This indicates that AKR/J may perform significantly differently on a motor task. Similarly, overrepresentation of variable genes in the AKR/J hippocampus may contribute to a differential learning and memory phenotype.
Gene expression is correlated with behavior:
Although region-specific expression in the brain may provide clues to function, genetic background-specific expression is required to explain behavioral differences among inbred strains. A gene whose expression does not vary between strains cannot contribute to a difference in behavioral phenotype. Gene expression profiles can be used as an intermediary in efforts to understand phenotypic variation. To demonstrate the feasibility of such an approach, we correlated gene expression data with a behavioral task showing strain-specific variation. In the cerebellum, 2095 transcripts (10%) showed significant correlation between expression level and performance on the rotarod motor performance task across inbred strains. This is indicative of the complexity of motor coordination, as well as of the central role of the cerebellum in motor activity. Many known genes were identified in this analysis, several of which have been shown to affect rotarod performance, such as Rac3 (Corbetta et al. 2005). Furthermore, a proportion of the genes identified are of unknown function, providing a rich resource for investigation of the genetic control of motor coordination.
Examination of GO terms in the correlation lists revealed functions associated with transcription, such as signal transduction activity and nucleic acid-binding proteins (Zhang et al. 2004). GO analysis also shows unexpected results, such as an overrepresentation of transcripts annotated as olfactory receptors in regions other than olfactory bulb and cell cycle genes in the postmitotic cerebellum. These unexpected results could reflect the incompleteness of GO annotation, regional specificity in gene function, or the indirect coexpression of genes involved in motor function with other genes in these brain regions. Additional clues to biological function may be revealed by analysis of genes that are targeted by known transcription factors.
While the cerebellum is thought to be involved in motor tasks, regions such as olfactory bulb have not been shown to exhibit similar involvement (Mauk et al. 2000; Nixon 2003; Ohyama et al. 2003). Analysis of gene expression in the olfactory bulb yielded no significant correlations with rotarod performance, supporting our general approach. Furthermore, no significant correlations were found in the basal ganglia or cingulate or frontal cortices with this task, whereas significant correlations were observed in the amygdala and hippocampus. Relatively few genes were found to be correlated with motor function in these regions, perhaps indicating that the major influence on motor coordination is from the cerebellum, with some level of mediation by the other two brain regions, even though the basal ganglia are involved with other aspects of motor function, such as motor planning (Glover 2004). Furthermore, transcripts with significant correlations in the amygdala and hippocampus are not merely a subset of transcripts identified in the cerebellum. This indicates that different mechanisms function in each brain region and suggests that the experiment is not merely identifying the gene expression signature of a particular genetic background in each region examined. There is evidence for a hippocampal role in locomotion (Bast and Feldon 2003), and the marked improvement with repeated trials typically observed in this procedure suggests a contribution of learning and memory to rotarod performance. The amygdala is not immediately implicated in the rotarod task, but there is potential that differential fear response could be a factor in the strength of the motivation to remain on the rotarod and could vary with gene expression in the amygdala. These results indicate that performance on the rotarod is a combination of neurocognitive processes involving more than simply cerebellar function.
It is clear that regions of the brain, having specific biological functions, express a unique suite of genes to perform these functions. Many of the genes showing significant variation in specific brain regions in this data set have also been identified in other published reports on regional specificity. For example, we found significant variation in Foxp1, Actn2, and Rgs9 in the basal ganglia, in accordance with previous studies (Dunah et al. 2000; Ferland et al. 2003; Rahman et al. 2003; Tamura et al. 2003, 2004; Zapala et al. 2005). However, our data also expand previous studies and show that variation in gene expression in the brain is substantially higher than previously reported.
This catalog of gene expression is a useful tool for generating hypotheses about the genetic basis of any phenotype showing variation across inbred mouse strains, encompassing behavioral, physiological, or pharmacological studies. These data can also be informative in choosing strains likely to be sensitive to manipulations of certain pathways and thus suitable for a particular experiment. By defining the expression differences between inbred mouse strains, we can examine the effect of genotype on molecular phenotype and ultimately the effect of genetic background on behavior.
Acknowledgments
We thank Antonio Perez and Nancy Young for their work on the rotarod phenotyping. This work was supported by the University of North Carolina Studies to Advance Autism Research and Treatment U54 MH66418, 5P30HD003110, and EPA RD83272001.
References
- Bast, T., and J. Feldon, 2003. Hippocampal modulation of sensorimotor processes. Prog. Neurobiol. 70: 319–345. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate / a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57: 289–300. [Google Scholar]
- Bogue, M. A., and S. C. Grubb, 2004. The Mouse Phenome Project. Genetica 122: 71–74. [DOI] [PubMed] [Google Scholar]
- Carninci, P., T. Kasukawa, S. Katayama, J. Gough, M. C. Frith et al., 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- Corbetta, S., S. Gualdoni, C. Albertinazzi, S. Paris, L. Croci et al., 2005. Generation and characterization of Rac3 knockout mice. Mol. Cell. Biol. 25: 5763–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunah, A. W., M. Wyszynski, D. M. Martin, M. Sheng and D. G. Standaert, 2000. alpha-actinin-2 in rat striatum: localization and interaction with NMDA glutamate receptor subunits. Brain Res. Mol. Brain Res. 79: 77–87. [DOI] [PubMed] [Google Scholar]
- Ferland, R. J., T. J. Cherry, P. O. Preware, E. E. Morrisey and C. A. Walsh, 2003. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J. Comp. Neurol. 460: 266–279. [DOI] [PubMed] [Google Scholar]
- Glover, S., 2004. Separate visual representations in the planning and control of action. Behav. Brain Sci. 27: 3–78. [DOI] [PubMed] [Google Scholar]
- Hamet, P., and J. Tremblay, 2005. Genetics and genomics of depression. Metabolism 54: 10–15. [DOI] [PubMed] [Google Scholar]
- Mauk, M. D., J. F. Medina, W. L. Nores and T. Ohyama, 2000. Cerebellar function: Coordination, learning or timing? Curr. Biol. 10: R522–R525. [DOI] [PubMed] [Google Scholar]
- Nadler, J. J., S. S. Moy, G. Dold, D. Trang, N. Simmons et al., 2004. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain Behav. 3: 303–314. [DOI] [PubMed] [Google Scholar]
- Nixon, P. D., 2003. The role of the cerebellum in preparing responses to predictable sensory events. Cerebellum 2: 114–122. [DOI] [PubMed] [Google Scholar]
- Ohyama, T., W. L. Nores, M. Murphy and M. D. Mauk, 2003. What the cerebellum computes. Trends Neurosci. 26: 222–227. [DOI] [PubMed] [Google Scholar]
- Oroszi, G., and D. Goldman, 2004. Alcoholism: genes and mechanisms. Pharmacogenomics 5: 1037–1048. [DOI] [PubMed] [Google Scholar]
- Pavlidis, P., and W. S. Noble, 2001. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2: RESEARCH0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov, P. M., J. H. Graber, G. A. Churchill, K. DiPetrillo, B. L. King et al., 2005. Evidence of a large-scale functional organization of mammalian chromosomes. PLoS Genet. 1: e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, Z., J. Schwarz, S. J. Gold, V. Zachariou, M. N. Wein et al., 2003. RGS9 modulates dopamine signaling in the basal ganglia. Neuron 38: 941–952. [DOI] [PubMed] [Google Scholar]
- Sandberg, R., R. Yasuda, D. G. Pankratz, T. A. Carter, J. A. Del Rio et al., 2000. Regional and strain-specific gene expression mapping in the adult mouse brain. Proc. Natl. Acad. Sci. USA 97: 11038–11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, S., Y. Morikawa, H. Iwanishi, T. Hisaoka and E. Senba, 2003. Expression pattern of the winged-helix/forkhead transcription factor Foxp1 in the developing central nervous system. Gene. Expr. Patterns 3: 193–197. [DOI] [PubMed] [Google Scholar]
- Tamura, S., Y. Morikawa, H. Iwanishi, T. Hisaoka and E. Senba, 2004. Foxp1 gene expression in projection neurons of the mouse striatum. Neuroscience 124: 261–267. [DOI] [PubMed] [Google Scholar]
- Tusher, V. G., R. Tibshirani and G. Chu, 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterer, G., and D. Goldman, 2003. Genetics of human prefrontal function. Brain Res. Brain Res. Rev. 43: 134–163. [DOI] [PubMed] [Google Scholar]
- Zapala, M. A., I. Hovatta, J. A. Ellison, L. Wodicka, J. A. Del Rio et al., 2005. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc. Natl. Acad. Sci. USA 102: 10357–10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., D. Schmoyer, S. Kirov and J. Snoddy, 2004. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using gene ontology hierarchies. BMC Bioinformatics 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]