Abstract
The ACT2 gene, encoding one of eight actin isovariants in Arabidopsis, is the most strongly expressed actin gene in vegetative tissues. A search was conducted for physical defects in act2-1 mutant plants to account for their reduced fitness compared with wild type in population studies. The act2-1 insertion fully disrupted expression of ACT2 RNA and significantly lowered the level of total actin protein in vegetative organs. The root hairs of the act2-1 mutants were 10% to 70% the length of wild-type root hairs, and they bulged severely at the base. The length of the mutant root hairs and degree of bulging at the base were affected by adjusting the osmolarity and gelling agent of the growth medium. The act2-1 mutant phenotypes were fully rescued by an ACT2 genomic transgene. When the act2-1 mutation was combined with another vegetative actin mutation, act7-1, the resulting double mutant exhibited extensive synergistic phenotypes ranging from developmental lethality to severe dwarfism. Transgenic overexpression of the ACT7 vegetative isovariant and ectopic expression of the ACT1 reproductive actin isovariant also rescued the root hair elongation defects of the act2-1 mutant. These results suggest normal ACT2 gene regulation is essential to proper root hair elongation and that even minor differences may cause root defects. However, differences in the actin protein isovariant are not significant to root hair elongation, in sharp contrast to recent reports on the functional nonequivalency of plant actin isovariants. Impairment of root hair functions such as nutrient mining, water uptake, and physical anchoring are the likely cause of the reduced fitness seen for act2-1 mutants in multigenerational studies.
Actin is found in all eukaryotes as a principal and indispensable structural component of the cytoskeleton. In plants, the basal actin cytoskeleton plays many important roles in development and growth at the organismal level and in routine cellular functions. Actin is essential or at least implicated in diverse cellular processes such as cytoplasmic streaming, organelle orientation, establishment of cell polarity, cell shape and division plane determination, cell wall deposition, and tip growth (Mascarenhas, 1993; Meagher et al., 1999b). Because polarity, the orientation of cell division, and cell wall deposition are presumed to be the governing factors of organ shape and pattern formation in plants, actin is a central element in plant development (Meagher and Williamson, 1994). Different isovariants of actin often have different expression patterns, and biochemical and complementation studies indicate that not all actin isovariants are equivalent (Kumar et al., 1997; Fyrberg et al., 1998; Meagher et al., 1999a; Kandasamy et al., 2002). For example, ectopic expression of plant and animal actins can cause sever organismal phenotypes (Fyrberg et al., 1998; Kandasamy et al., 2002). This manuscript provides evidence that ACT2, one of eight actin genes expressed in Arabidopsis, is required for normal root hair elongation, but that diverse plant actin isovariants can perform the necessary protein functions.
On the basis of protein coding sequences, plant actin genes are monophyletic in origin and split to form two ancient classes encoding reproductive and vegetative actins early in land plant evolution (Hightower and Meagher, 1986; Meagher and McLean, 1990; McDowell et al., 1996b; Kandasamy et al., 1999). The vegetative class of the actin multigene family in Arabidopsis is divided into two distinct subclasses, one composed of ACT7 and the other composed of ACT2 and ACT8 (McDowell et al., 1996b). ACT2 and ACT8 are most closely related, differing by only one amino acid residue, yet their high level of silent nucleotide substitution differences indicates that they have not shared a common ancestral gene for 30 to 60 million years (McDowell et al., 1996b). ACT2 expression is virtually constitutive in vegetative tissues, whereas ACT8 expression is weaker than ACT2, especially in young tissue. In addition, ACT8 is expressed in only a fraction of the tissues with ACT2 expression (An et al., 1996b). ACT7 is expressed in the early developmental stages of nearly all vegetative tissues. ACT2, ACT7, and ACT8 are all well expressed in roots and root hairs.
The act2-1 mutant examined in this study was isolated from an Arabidopsis T-DNA insertion library using a sequence-based screening technique (McKinney et al., 1995). The act2-1 allele contains a T-DNA insertion at the beginning of the first protein coding exon (exon 1/2) as shown in Figure 1a. The insertion replaces 16 nucleotides of ACT2 DNA spanning this intron/exon junction. Close inspection of homozygous act2-1 lines grown on soil revealed no phenotypic distinction from wild-type plants. However, a multigenerational population study demonstrated the act2-1 mutation acts as a deleterious mutation that can be detected in the 2n sporophytic generation (Gilliland et al., 1998), which is consistent with the vegetative expression pattern of the ACT2 gene. Homozygous mutant plants have only 70% of the fitness of wild-type plants in each generation and even heterozygous plants have slightly lowered fitness. The kinetics by which the act2-1 mutant allele was lost over several generations were consistent with requirements in vegetative growth and viability but were not consistent with any requirement for ACT2 during meiotic development (Asmussen et al., 1998). Asmussen et al. (1998) estimated that the act2-1 allele would be lost from a large population in 20 generations, making it effectively lethal. Because natural selection should act on phenotypic variation (Lewontin, 1974), the lack of an obvious outward physical phenotype associated with the strong negative selection potential of the act2-1 allele appeared paradoxical.
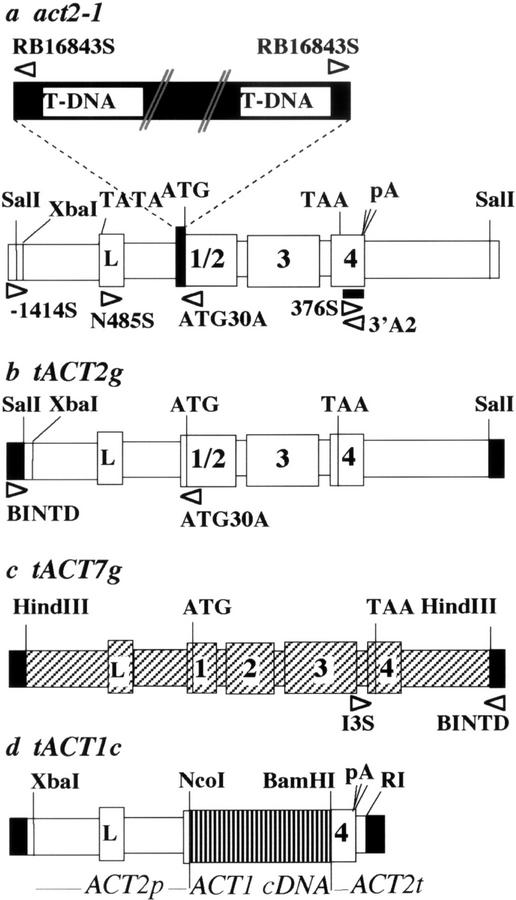
Figure 1.
Map of act2-1 mutant allele and complementing transgenes. a, The act2-1 allele contains a T-DNA insertion (black) separating most of the ACT2 5′-UTR (white) from the body of the ACT2 actin coding sequence (white rectangles, with exons drawn larger than introns or flanking sequence). The T-DNA insertion event deleted the acceptor splice junction including 12 nucleotides of the leader (5′-UTR) intron and four noncoding nucleotides of exon one-half of the act2-1 gene, leaving just five nucleotides between the T-DNA and the act2-1 start codon. Because the insertion is flanked on either side by a right border sequence, the allele contains two or more T-DNAs in tandem at this site. b, The genomic ACT2 transgene (tACT2g) used to complement the act2-1 mutant was contained on a 4-kb SalI fragment (white) flanked by T-DNA sequences (black). c, The genomic actin ACT7 transgene (tACT7g) used to complement the act2-1 mutant was contained on a 4-kb HindIII fragment (diagonal lined rectangles). d, The cDNA transgene, tACT1c, used to complement the act2-1 mutant, contained the actin ACT1 protein coding sequence (vertical lined rectangles), under control of the ACT2 (white) promoter (ACT2p) and terminator (ACT2t) sequences (Kandasamy et al., 2002). The white arrowheads refer to positions and orientations of PCR primers and the thick horizontal black bar (a) refers to the location of the 3′-UTR DNA probe used for northern analysis.
A certain amount of functional redundancy is expected between the eight highly similar actin proteins in Arabidopsis. However, each of these actin genes presumably must have its own essential properties to be selectively maintained through tens or in some cases hundreds of millions of years of evolution (Meagher et al., 1999a, 1999b). To distinguish the functional roles of the predominantly vegetatively expressed actins, the possible physical phenotype(s) of the act2-1 mutant was explored further in the following study. A highly shortened root hair phenotype was observed. This phenotype was directly attributable to the loss of ACT2 and is the probable cause of low variability in multigenerational studies. The data presented herein suggest a high level of functional redundancy among the actin isovariants in their ability to direct root hair elongation, but a distinct role is nevertheless indicated for ACT2 gene regulation in this process.
RESULTS
Root Hair Phenotypes of act2-1 Mutants
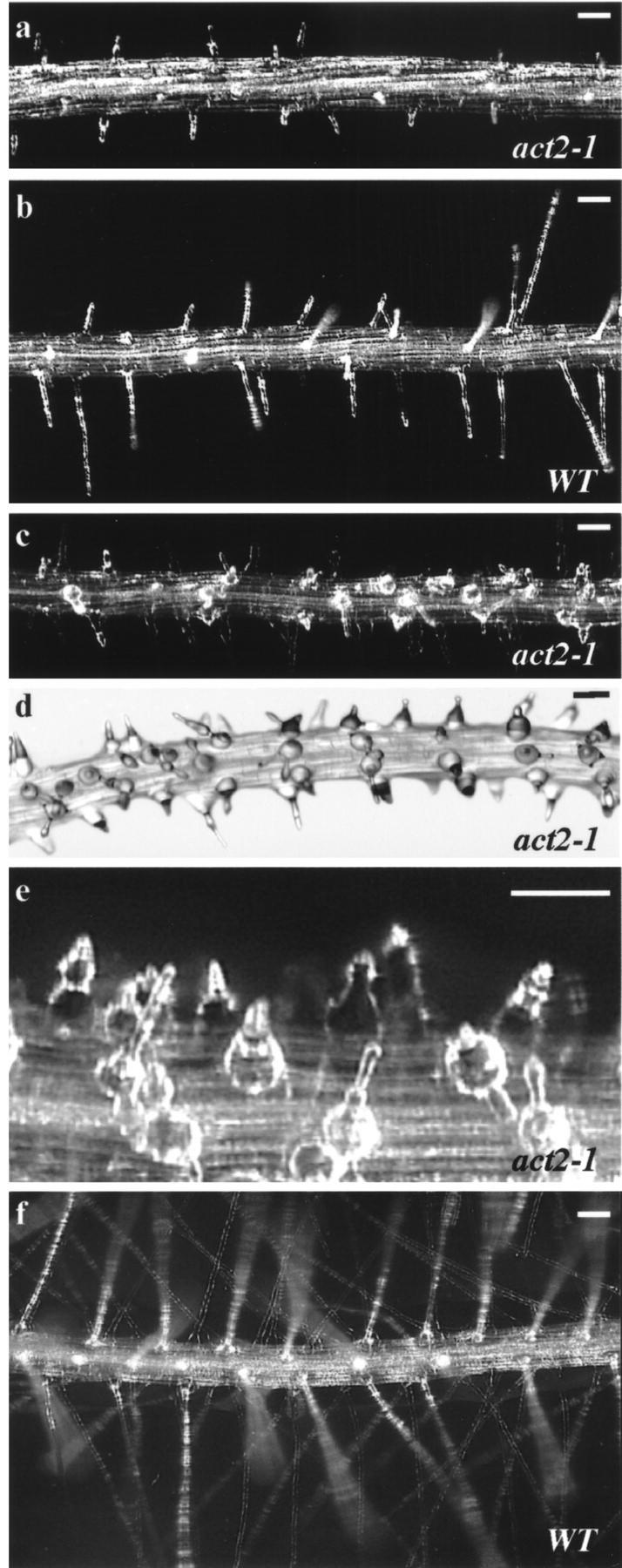
A previous multigenerational population study (Gilliland et al., 1998) demonstrated a significant drop in act2-1 allele frequencies relative to the wild-type allele within two generations, suggesting that the act2-1 mutation is a highly deleterious allele. The strong negative selection potential of the act2-1 allele on soil-grown plants implied a phenotypic defect that was not apparent from physical examination of these plants. Close inspection of act2-1 homozygous mutants roots on different defined growth media revealed a strong phenotype in root hairs. The root hairs of act2-1 mutants were much shorter than wild-type root hairs as show in Figure 2. Mutant root hairs averaged 20% the length of those on wild-type plants when grown on solid phytagar (Invitrogen, Carlsbad, CA)-Murashige and Skoog medium. Mutant root hairs were 60% the length of those on wild-type plants when grown in liquid Murashige and Skoog culture (data not shown). These root hairs, although shorter, still resembled wild-type root hairs in shape, number, and placement. However, when the act2-1 mutants were grown on medium containing Phytagel (Sigma-Aldrich, St. Louis) and Suc, the root hairs are distinctly bulbous (Fig. 2, c–e). The mutant root hairs (Fig. 2, d–e) are wider and rounder than wild type at their base and become more narrow toward the tip, exhibiting a pear-like shape. Occasional branching was also observed. The bulbous root hairs (Fig. 2c) averaged one-seventh (14%) the length of the wild-type filamentous root hairs (Fig. 2f). The length of root hairs was quantitatively compared between wild type and mutant as shown in Figure 3d (left two bars). ACT2/act2-1 heterozygotes appear fully wild type (data not shown), indicating that act2-1 is a recessive allele. The pear-like phenotype is maintained when mannitol, a non-metabolizable sugar, is substituted for Suc on the phytagel plates, indicating the bulbous base of the root hair is modulated by osmolarity rather than nutrient response or metabolism of Suc. Without either sugar included in the plates, the root hairs produced the short spiked configuration. The number and position of the root hairs is not altered under any conditions, indicating the act2-1 mutant allele does not effect root hair determination or initiation.
Figure 2.
Root hair phenotypes of the act2-1 mutant and wild-type Arabidopsis plants. Root hairs of act2-1/act2-1 mutants (a) are shorter than those of wild type (b), when grown on Phytagel plates in Murashige and Skoog salts without Suc. When the same medium is supplemented with 1% (w/v) Suc (c–f), the act2-1/act2-1 mutant displays distinctive bulbous root hairs that are typically pear-shaped with occasional branching (c–e), whereas wild-type plants have long and straight root hairs (f). Bars in the dark field images (a–c, e and f) and bright field image (d) = 50 μm.
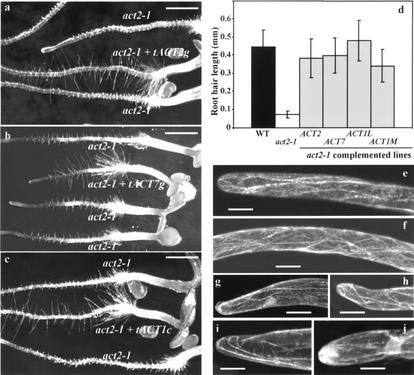
Figure 3.
Root hairs of the act2-1 mutant Arabidopsis plants. a through d, Complementation of the act2-1 root hair phenotype in plants expressing various actin transgenes. a, Seven-day-old act2-1 mutant seedlings display wild-type filamentous roots when transformed with genomic tACT2g fragment (middle seedling). Flanking seedlings are homozygous act2-1 mutant segregants that did not contain the complementing tACT2g transgene. b, Seven-day-old act2-1 mutant seedlings display wild-type filamentous roots when transformed with genomic tACT7g fragment (middle seedling). Flanking seedlings are homozygous act2-1 mutants. c, Seven-day-old act2-1 mutant seedlings display wild-type filamentous roots when transformed with tACT1c (Fig. 1d), ACT1 cDNA under expression of ACT2 regulatory regions (middle seedling). Flanking seedlings are homozygous act2-1 mutants. d, Root hair length in wild-type, act2-1, and complemented lines. n = 20 for each. ses are indicated. e though j, Immunocytochemistry of wild-type (e and f) and act2-1/act2-1 mutant Arabidopsis root hairs (g–j). Bar in a through c = 1 mm. Bars in e through j = 10 μm.
Immunocytochemical examination of the freeze-substituted root hairs labeled with the monoclonal actin antibody mAbGPa revealed several thick actin bundles oriented parallel to the longitudinal axis of the short mutant root hairs. However, in the very apex of these stunted root hairs, the actin bundles were often looping through the tip (Fig. 3, g–i) or occasionally showing dense but diffuse fluorescence labeling (Fig. 3j). Such organization of actin was never observed in the growing wild-type hairs of the identical root zone (Fig. 3, e and f). In the long wild-type root hairs, there were dense arrays of longitudinally oriented thick actin bundles and thin filaments all through the axis, with the latter extending even into the tip. Overall, there was no drastic alteration in the cytoarchitecture of the mutant root hairs except that they contained less actin filaments and showed frequent looping of the actin bundles at the tip (Fig. 3, h and i).
Complementation of the act2-1 Phenotype by Different Actin Isovariants
Although the act2-1 allele had been backcrossed to wild type and segregated with the root hair phenotype in latter generations, the possibility remained that the mutant phenotype was attributable to a closely linked mutation. A 4-kb genomic DNA fragment (tACT2g, Fig. 1b) containing the ACT2 gene, including its 5′ promoter region and 3′ polyadenylation sites, was transformed into act2-1/act2-1 plants by vacuum infiltration of Agrobacterium tumenfaciens carrying the insert on a Ti plasmid. Reliable detection of the pear-shaped root hair phenotype of act2-1 mutants permitted a screen for transformants that exhibited normal filamentous root hairs. A population of approximately 4,000 seeds derived from vacuum infiltrated plants was germinated on vertical plates with Suc and no antibiotic selection for the transgene. More than 40 potential transformants displaying wild-type root hairs, as shown in Figure 3a, were identified and transferred to soil. Thus, the phenotypically wild-type plants accounted for more than 1% of the germinated seeds, typical of transformation frequencies we obtained with the vacuum infiltration method (Bariola et al., 1999).
PCR performed on leaf samples confirmed the presence of the complementing transgene (tACT2g) and the absence of a true ACT2 allele in all of the 36 complemented plant lines tested. The independent complementing tACT2g alleles were identified with PCR by pairing a sense primer located outside of the cloned area of ACT2 in the T-DNA (BINTD2369; see “Materials and Methods”) to an antisense primer (ATG30A) located in the first coding exon (Fig. 1b). The presence of a native ACT2 allele was distinguished by pairing a sense primer (1414S) upstream of the 5′ SalI site and the ACT2 promoter with the ATG30A antisense primer. Progeny derived from 29 of the 31 transformants examined exhibited ratios of bulbous to filamentous wild-type root hair phenotypes consistent with segregation of one or two unlinked copies of the tACT2 gene, providing further evidence that the tACT2g gene is responsible for the complementation. Complemented plants had average root hair lengths similar to wild type (Fig. 3d).
Considering the extreme conservation of each plant actin isovariant (Meagher et al., 1999a, 1999b) and recent reports of the functional nonequivalency of plant actins (Kandasamy et al., 2002), we further considered the possibility that only the ACT2 protein could perform the necessary root hair functions for complementation. This was tested by determining whether genes encoding other distant Arabidopsis actin isovariants, ACT7 and ACT1, could rescue the bulbous root hair phenotype in the act2-1 mutant. The ACT7 protein differs in about 6.8% of its amino acid residues from ACT2, and ACT7 promoter-reporter fusions were expressed in transgenic root hairs (McDowell et al., 1996a). Like ACT2, ACT7 encodes a vegetative class actin. A 4-kb genomic DNA fragment containing the entire ACT7 gene (Fig. 1c) was transformed into act2-1/act2-1 plants by vacuum infiltration. From about 500 potential transformants examined growing on vertical Suc plates, five independent seedlings with wild-type root hairs were identified, and two were selected for further study. The root phenotype observed in a segregating population of seedlings derived from one of these complemented lines is shown in Figure 3b. The two complementing plant lines that were examined by PCR contained the tACT7g transgene and were homozygous for the act2-1 allele. Thus, the phenotype of the vegetative actin act2-1 mutation was rescued by overexpression of the ACT7 gene encoding the most distant vegetative actin isovariant, with which ACT2 has not shared a common ancestor for 200 million years (Meagher et al., 1999a). This result suggested that the actin isovariant sequence might not be important, but that the level of actin expression might be critical to root hair development (Fig. 3d) Quantitative data revealed that tACT7g expression restored normal root hair length to mutant plants.
The reproductive actin gene ACT1 encodes the most distant actin isovariant from ACT2 protein in Arabidopsis. Although the ACT1 protein is only slightly more divergent from ACT2 in total amino acid differences (7.2%) than ACT7, ACT1 is a reproductive class actin and contains many more non-synonymous amino acid substitutions relative to ACT2. A construct shown in Figure 1d (tACT1c) containing the cDNA from ACT1 under the control of an ACT2 promoter and terminator (Kandasamy et al., 2001) was transformed into act2-1/act2-1 plants. Approximately 1% of T1 seedlings displayed wild-type root hairs and tACT1c transgene segregated with the normal root hair phenotype as shown in Figure 3c. tACT1c restored root hair length in mutant plants to wild-type levels as shown for two different complemented lines in Figure 3d. Thus, the ACT1 actin isovariant substituted for the loss of ACT2 in root hair development, when the reproductive cDNA was expressed ectopically from the vegetative ACT2 promoter. In contrast to the full complementation of the root hair elongation phenotype, many of the tACT1c transformants of the act2-1 line also exhibited the whole-plant morphological defects and dwarfing of above-ground organs that were seen on wild-type plants expressing ACT1 ectopically (Kandasamy et al., 2002). Ectopic overexpression of ACT1 isovariant in above-ground organs was evidently harmful in both mutant and wild-type backgrounds.
Transcript Levels in act2-1 Mutants
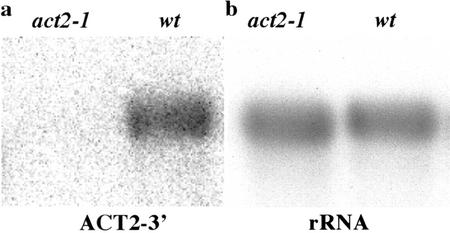
The T-DNA insertion event creating the act2-1 allele separates the ACT2 promoter from its coding region, deletes the first splice acceptor site of the primary transcript, and thus should disrupt any normal ACT2 expression. However, because the downstream ACT2 coding region is intact, there is a formal possibility that transcription from the T-DNA insertion results in expression of a hybrid but functional act2-1 mRNA. Northern analysis was performed on rosette leaves and young inflorescences from mature plants using a double-stranded DNA probe located on the 3′-untranslated region (-UTR) of ACT2 (ACT2 3′-UTR, Fig. 1a). Appreciable levels of ACT2 mRNA were not detected in the act2-1 mutant leaf tissue relative to wild type as shown in Figure 4a or in mutant inflorescence tissue (not shown). After a more extended exposure of the blot shown in Figure 4 or parallel blots, a faint band of RNA was detected in act2-1/act2-1 mature plants. Calculations from densitometer readings on both the northern and RNA dot blots (data not shown) suggest that act2-1 mRNA levels were less than 4% of the wild-type ACT2 message levels. In numerous control experiments, cross-hybridization was not detected between ACT2 DNA and ACT8 DNA (An et al., 1996b) and their respective 3′-UTR probes. These two genes are the most homologous among the Arabidopsis actins but show only 45% sequence homology over their approximately 300-nucleotide 3′-UTR regions, making it unlikely that this weak RNA signal resulted from cross-hybridization between the DNA probe and the ACT8 transcript. Thus, the act2-1 may be a leaky allele. The same blot was rehybridized with a labeled rRNA probe to demonstrate equal loading and transfer to membrane (Fig. 4b). Four repetitions of this experiment failed to detect any higher or more significant levels of act2-1 transcript.
Figure 4.
Steady-state actin RNA levels of the act2-1 mutant. a, Total RNA from 5-week-old act2-1 and wild-type rosette leaves and inflorescences were blotted to a nylon membrane and hybridized with radiolabeled double-stranded DNA from the ACT2-3′-UTR region (see map in Fig. 1a). b, The membrane was reprobed with a radiolabeled 18S ribosomal RNA (rRNA) oligonucleotide (see “Materials and Methods”) to demonstrate equal loading and transfer of RNA to the membrane imprint.
Protein Levels in act2-1 Mutant and Complemented Lines
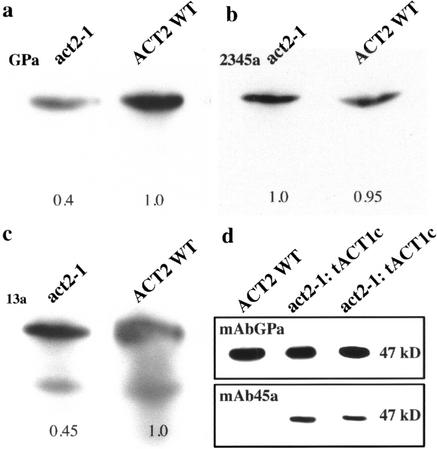
Western analysis was performed on seedlings to assay the reduction in actin protein levels in the act2-1 mutant plants. Membrane imprints of total protein were incubated with several distinct monoclonal antibodies that recognize different subclasses of actin proteins. Monoclonal antibody mAbGPa, which recognizes all plant actins (Kandasamy et al., 1999), was used to examine total actin expression as shown in Figure 5a. This immune reagent clearly shows that total actin protein is reduced more than 2-fold in act2-1/act2-1 plants compared with wild type. Western analysis with the monoclonal antibody mAb13a, which recognizes only subclass 1 and 3 actins (ACT2 and ACT8, and ACT11, respectively [Kandasamy et al., 2001]), also shows a 2-fold decrease of actin protein in independent protein samples (Fig. 5c). The lower band seen in Figure 5c is a common degradation product of actin. Because the ACT11 gene is not significantly expressed in vegetative tissues (Huang et al., 1997), the actin detected should represent only ACT8 and any residual ACT2. This examination of actin protein levels was repeated several times on independent act2-1 plants with similar results. Monoclonal antibody mAb2345a, which recognizes all Arabidopsis actins except the ACT2/8 subclass (ACT7, ACT11, ACT1/3, and ACT4/12 [Kandasamy et al., 2001]), recognizes the low levels of ACT7 in vegetative tissues. This reagent demonstrates approximately equal protein loading and transfer to membranes (Fig. 5b). Thus, the decrease in total actin is specifically attributable to a decrease in the subclass of actins containing ACT2. Furthermore, there was not any reproducible increase in other actins, like the hormone-responsive ACT7 isovariant, to compensate for the loss of the ACT2 isovariant.
Figure 5.
Actin protein expression of the act2-1 mutant. a through c, Membrane imprints of total protein from act2-1/act2-1 and ACT2 wild-type seedlings resolved by SDS-PAGE were incubated with three distinct monoclonal antibodies that recognize different subsets of actin proteins. Relative signal strengths for these antibodies are shown below each band. a, Monoclonal antibody mAbGPa is a general antibody that recognizes all plant actins. b, Monoclonal antibody mAb2345a recognizes all Arabidopsis actins (ACT1, -3, -4, -7, -11, and -12) except those in subclass 1 (ACT2 and ACT8). c, Monoclonal antibody mAb13a recognizes only ACT2, -8, and -11 of subclasses 1 and 3. d, Western-blot analysis of late pollen actin protein expression in leaves of the act2-1/act2-1 mutant complemented with tACT1c. Duplicate membranes were reacted with mAbGPa (top) and mAb45a (bottom). Monoclonal antibody mAb45a reacts with the late pollen actins including ACT1, which are not expressed in vegetative tissue of wild-type plants. Lane 1, Wild-type (WT) control. Lanes 2 and 3, Two mutant independent plant lines in which the act2-1 mutation is complemented with tACT1c construct expressing ACT1.
Western-blot analysis revealed that act2-1 mutant plants complemented with tACT7g or tACT1c transgenes and showing the wild-type root hair morphology expressed higher levels of total actin protein in vegetative organs than their mutant parents. For example, when protein extracts from wild-type plants and mutants complemented with tACT1c are reacted with the general actin antibody, mAbGPa, almost similar levels of total actin were detected in all plants (Fig. 5d, top panel). When duplicates of these filters were reacted with monoclonal antibody mAb45a, which reacts only with late-pollen-specific actins including ACT1, ACT1 protein is found at significant levels in the complemented lines (Fig. 5d, act2-1:tACT1c) but not in the wild-type plants (Fig. 5d, WT). Similar levels of total actin were seen in the tACT7g complemented lines (not shown) and in several repetitions of these experiments.
The Phenotypes of an act2-1 + act7-1 Double Mutant
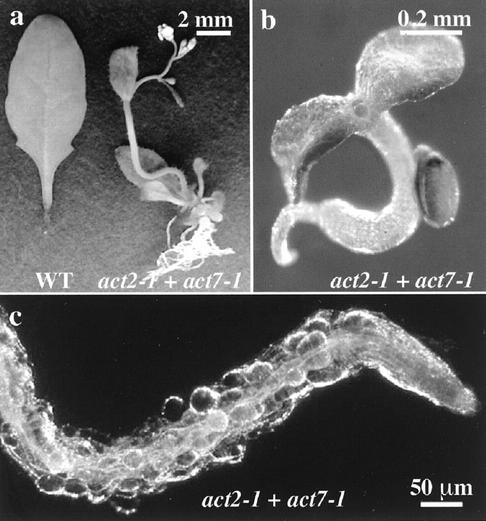
Considering that the ACT2 gene encodes the most highly expressed Arabidopsis actin in nearly all mature vegetative tissues, it was surprising that after its disruption, only defects in root hairs were observed. This suggested that the remaining actins were sufficiently redundant in function and expression patterns that they contributed to the nearly normal development of act2-1 mutant plants. To asses the need for other vegetative actins, double mutant plants homozygous for both act2-1 and act7-1 alleles were generated from crosses of plants containing the individual mutant alleles. The act7-1 allele is a T-DNA insertion with undetectable levels of ACT7 protein in seedlings (McKinney et al., 1995; Gilliland et al., 2002). Thus, the act2-1 + act7-1 double mutant plant is effectively null for two of the three vegetative actin genes, leaving ACT8 as the only functional vegetative actin gene (An et al., 1996b). The double mutant has severe abnormalities in nearly every aspect of development, as shown in Figure 6. When grown on a Suc-containing medium, the seedlings and plants are severely dwarfed (Fig. 6a, right) but are able to grow and produce functional reproductive structures and a few viable seeds by self-fertilization. The leaves are small and curled under and have high densities of phenotypically normal trichomes. A wild-type leaf from the same aged plant (Fig. 6a, left) is shown for comparison. Only a small number of flowers are produced, and siliques never fully elongate compared with wild-type plants, but otherwise the double mutant flowers appear fully normal. The root system is highly branched (Fig. 6a). Compared with elongated wild-type root epidermal cells, the surface cells of the double mutant roots appear rounded, swollen, and even bulbous (Fig. 6c). The root epidermal cells are not arranged in regular files and almost always lack root hairs. The plants are poorly anchored into the solid growth medium.
Figure 6.
Phenotypes of the act2-1 + act7-1 double mutant. a, A 5-week-old double mutant grown on Murashige and Skoog medium with 1% (w/v) Suc. The plant has started to bolt and is shown next to a rosette leaf from a wild-type plant of the same age. The crooked stem is a result of prolonged growth on a petri plate and not an abnormality of the double mutant (bar = 2 mm). b, Eleven-day-old double mutant arrested in development after germinating on Murashige and Skoog medium lacking exogenous sugar; bar = 0.2 mm. c, Dark field image of root from act2-1 act7-1 double mutant grown on Suc showing bulging surface cells and absence of root hairs.
The phenotype of the double mutant is drastically different when grown without a supplemental sugar source (Fig. 6b). The seeds germinate, but seedlings arrest in growth at the appearance of the first leaves. Primary roots are much thinner than the hypocotyls, lack root hairs, and do not grow into the solid medium. The cotyledons look normal but do not extend on petioles and turn chlorotic after 2 weeks. The first true leaves are just visible at the base of the cotyledons and usually remain green well after bleaching of the cotyledons. There are few visible trichomes on the true leaves. Double mutants germinated on plates supplemented with mannitol or higher concentrations of agar were less drastic and exhibited phenotypes closely resembling those germinated on Suc. This effect may indicate that the phenotype is modulated by osmolarity similar to the effect on act2-1 single mutants.
DISCUSSION
ACT2-Specific Functions in Root Hair Elongation
The disruption of a highly expressed vegetative actin gene, ACT2, resulted in an extreme root hair phenotype. We observed no phenotypes in any above-ground organs or cells including leaves, trichomes, stems, and flowers. Although the above-ground plant structures were unaffected, the process of root hair elongation is compromised. It has been shown with both genetic (Schiefelbein and Somerville, 1990; Parker et al., 2000) and biochemical studies (Bibikova et al., 1999; Miller et al., 1999) that root hair growth can be separated into three distinct phases: determination of the trichoblast cell to form a root hair, initiation of the hair as a bulge, and tip elongation. Loss of ACT2 in the act2-1 mutant does not affect root hair number or positioning, a finding that agrees well with existing data that new actin polymerization is not needed to determine which cells initiate root hair development (Miller et al., 1999; Baluska et al., 2000). However, both visualization of actin filaments and chemical inhibition of actin has suggested a strong role for actin in root hair elongation. Actin filament bundles are seen running the length of the root hair during root hair elongation and thin out to undetectable levels in the apical tip (Miller et al., 1999). The actin perturbing drug, cytochalasin D, stops elongation of root hairs but does not prevent initiation of root hair bulges (Miller et al., 1999). It is interesting to note here that the role of actin in tip growth has also been established in organisms from several kingdoms. For example, fruitfly (Drosophila melanogaster) bristle elongation (Hopmann et al., 1996; Tilney et al., 2000) and hyphal tip morphogenesis in Saprolegnia ferax and Neurospora crassa (Heath et al., 2000) are also dependent on the actin cytoskeleton.
The phenotypes of the act2-1 mutant and complementation data with the wild-type gene not only corroborates the requirement of actin for root hair elongation, but identifies a specific actin gene required for this function: ACT2. Our analysis of protein levels with subclass-specific monoclonal antibodies indicate that the mutant plants have significantly less total actin than wild-type plants (Fig. 5). Evidence is presented demonstrating that other actin isovariants can substitute for the loss of ACT2 and restore normal root hair elongation to the act2-1 mutant: An exogenous genomic copy of the ACT7 gene and ectopic expression of the ACT1 coding region under the regulatory regions of ACT2 both suppressed the short and bulbous root hair phenotypes. These data argue that individual actin proteins are, at least in part, functionally redundant for root hair elongation and suggest that the normal expression pattern of ACT2 may be the primary factor responsible for its dominance in the root hair elongation process. The predominantly pollen-specific ACT1 might substitute for ACT2 function, despite the hundreds of millions of years separating these protein sequences, because the parallel functions of pollen tube and root hair elongation by tip growth may have conserved similar functional domains on these actin proteins. Kumar et al. (1997) report a similar example of actin functional redundancy in mice, where the coding region of one actin isovariant expressed under an appropriate promoter can partially rescue a defect caused by the loss of a different actin isovariant. Nevertheless, although other actin proteins, ACT8 and ACT7, are found in trichoblasts (An et al., 1996b; McDowell et al., 1996a), they appear to be less effective at performing the task of root hair elongation. A mutation in ACT7, which is strongly expressed in root hairs, does not affect root-hair elongation (Gilliland et al., 2002), further indicating that Arabidopsis specifically uses ACT2 protein as the main actin responsible for root hair elongation.
These data showing functional redundancy of plant actin for root hair elongation are in sharp contrast to the results from ectopically expressing ACT1 protein in the rest of the plant organs and tissues (Kandasamy et al., 2002). Ectopic ACT1 protein expression from the ACT2 promoter and terminator regulatory regions disrupted leaf, shoot, flower, and silique development, and resulted in severe dwarf phenotypes. Even the epidermal cells of leaves and hypocotyls were dwarfed when ecotopically expressing ACT1. Overexpression of ACT2 itself did not give these dwarf phenotypes. Ectopic expression of tACT1c in the act2-1 mutant background produced the same above-ground phenotypes (not shown). There appear to be common functional elements required for root hair elongation that are shared among the diverse ACT1 and ACT2 isovariants and not compatible with many other aspects of above-ground plant development.
Bulbous Root Hair Phenotype and Cell Wall Defects
The bulging of the root hair base in the act2-1 mutants is more difficult to explain than shortened root hairs partly because this root hair phenotype is far less commonly reported than defects in elongation. The mutation root hair development 1-1, which maps to a different location than any actin gene (McKinney and Meagher, 1998; Parker et al., 2000), causes swelling of the root hair base with root hair length varying greatly depending on the gelling agent used in the medium (Schiefelbein and Somerville, 1990; Favery et al., 2001). Similarly, the pronounced bulbous root hair phenotype of the act2-1 mutant was observed only when the Phytagel gelling agent was used instead of Phytagar and the phenotype required the presence of an osmoticum in the medium. This sensitivity of the phenotype to growth substrate suggests that part of the selection against the act2-1 mutant may be intermittent (Brookfield, 1997; Nowak et al., 1997) and dependent upon environmental conditions such as contact with the substrate or soil.
Extrapolation of the phenotypes in yeast actin mutants and proposed actin functions in plants suggest a compelling explanation for the bulge observed at the base of the act2-1 mutant root hairs. Both cell wall deposition and transport of secretory vesicles are likely functions for actin in expanding yeast cells (Novick and Botstein, 1985; Gabriel and Kopecka, 1995) and plants (Kobayashi et al., 1987; Foissner and Wasteneys, 1997; Boevink et al., 1998). Although root-hair determination and initiation remain intact, there may still be a stimulus for tip growth. But the loss of actin may result in inadequate transportation of vesicles carrying the necessary building blocks and enzymes to the growing tip and the accumulation of these vesicles at the base of the root hair. Mislocalization and/or accumulation of cell wall-loosening components at the base of the root hair may allow the cell wall to expand and bulge. The effect of altering medium osmolarity on the phenotype of the root hairs is particularly striking, because yeast actin mutants also have osmotic sensitivity. Increased osmotic strength of the medium significantly lowered the permissive temperature for the yeast phenotype (Novick and Botstein, 1985). This finding suggests a parallel function for plant actin in osmoregulation, because in our study the bulbous root hair phenotype is conditionally dependent on a high-osmotic-strength medium.
The Essential Role for Actin in Plant Development
The gross morphological phenotypes of the act2-1 + act7-1 double mutant give us further insight into the functions of the actin in plants. Although six other functional actin genes remain, there is only one fully functioning vegetative actin gene, ACT8. The double act2-1 + act7-1 mutant has severe phenotypes in practically all aspects of vegetative plant growth, underscoring the diverse functions of these two actins in the plant cytoskeleton and linked developmental processes. In fact, the plants are not viable without a metabolizable sugar source. The severity of the phenotype of the double mutant is not simply an additive combination of the single mutants, which would indicate the action of independent genes. Rather, the sever phenotype suggests a synergistic relationship between the two actin genes. This synergistic relationship argues against stringent sorting of isovariants into specific filaments. If each isovariant was sorted into specific filaments, the removal of one isovariant would eliminate specific groups of filaments responsible for a subset of functions. The resultant double mutant would then be expected to have an additive combination of the two phenotypes of the single mutants, because both subsets of filaments and their dependent functions are disrupted. A microfilament system using a common pool of actin monomers in heteropolymers would alternately be only mildly disturbed by the loss of a single actin gene product, because other isovariants are present and all classes of filaments could still be assembled. A quantitative loss of actin protein would affect polymerization rates of the whole microfilament system, and thereby of most systems dependent on actin, and would result in the severe, synergistic phenotype seen in the double mutant. Future cell biological research with actin isovariant-specific antibodies or in vitro biochemical analysis of purified actin isovariants can test this model for isovariant sorting.
The dwarfed size, high density of trichomes on leaves, and rounded shape of surface root cells in the double mutant may suggest that a shortage of actin prevents some cells from expanding or elongating properly (Staiger and Cande, 1991; Meagher et al., 1999b). The swollen nature of the root surface cells may indicate a defect in the cell wall, an idea further supported by the observation that the double mutant root was permeated more rapidly by the fixative osmium tetroxide than a wild-type root (data not shown). The lack of root hairs on the double mutant further underscores the requirement of actin for root hair elongation, and the extensive lateral root formation may be an attempt to compensate for the decrease of surface area vital for nutrient uptake. The presence of normal and fertile reproductive structures in double mutant plants grown on Suc is consistent with the normally low expression or absence of both ACT7 and ACT2 in reproductive structures and with the strong expression of the five reproductive actin genes that are still functional.
The root-hair-specific phenotype of the act2-1 single mutant contrasts with the broad systemic phenotypes of the act2-1 + act7-1 double mutant. The enhanced fitness of plants maintaining the ACT2 allele instead of the act2-1 allele (Asmussen et al., 1998; Gilliland et al., 1998) implies ACT2 plays an essential and distinct role in plant development. The decrease in root hair length found on the act2-1 mutant plants demonstrates the role of ACT2 in root hair development and probably explains the loss of fitness when act2-1 plants were grown at high density on soil and competing for nutrients. When the root-hair-specific act2-1 phenotype is compared with the extensive systemic phenotypes of the double mutants, it appears that many of the roles of ACT2 can be seen as partially redundant with other actins. The functions lost in the double mutant must normally be performed jointly and redundantly with the other actin isovariants present in vegetative tissues (e.g. ACT7 and ACT8). The severity and range of defects in the double mutant demonstrate that both the universal and essential roles of actin in vegetative cells can use multiple isovariants with partially overlapping functions.
MATERIALS AND METHODS
Growth Conditions and Media
A homozygous act2-1 line that was free of other T-DNA insertions (McKinney et al., 1995) was backcrossed to the wild-type Wassilewskija ecotype. The offspring of this backcross were used for all analyses. All plants were grown at 22°C at 16-h days/8-h nights. Plants exhibiting bulbous root hairs were grown on vertical plates containing one-half strength Murashige and Skoog salts, 0.3% (w/v) Phytagel (Sigma-Aldrich), and 1% (w/v) Suc. This medium was adjusted to 1.0% to 3.0% (w/v) Suc to assess changes in the phenotype. Liquid culture medium contained one-quarter Murashige and Skoog salts and 0.5% (w/v) Suc. In some experiments the Phytagel gelling agent was replaced with Phytagar (Invitrogen). Mature plants used for northern analysis were grown on soil under artificial light. Nine-day-old seedlings used for western analysis were grown in liquid culture.
Immunocytochemistry
Root hair samples were prepared for F-actin labeling using the rapid freeze fixation and freeze substitution methods described previously (Kandasamy et al., 2002). The general actin monoclonal antibody mAbGPa, which reacts with all the Arabidopsis actin isovariants, was used for immunocytochemistry at 5 μg mL−1 concentration.
PCR Primers and Methods
The following primers were used to distinguish the various ACT2 alleles (arrows in Fig. 1). Pairing AAc2-I485S with a primer found in the right border of the T-DNA (RB16843S) will amplify the act2-1 allele, whereas pairing it with an antisense actin primer (AAc2-ATG30A) located in the first coding exon at the insertion site will preferentially amplify the wild-type ACT2 allele (Gilliland et al., 1998). Pairing a sense primer located outside of the cloned area of ACT2, in the T-DNA (BINTD2369, ACTGGAAAGCGGGCAGT- GAGCGCAACGCAAT, or BINTD2683, AAGGAGCGGGCGCCATTCAGGCTGCGCAAC-TGT) or the Arabidopsis genome (AAc2-1414S, TATAAGTGACGAGGACACCAACAAAC-TATT), to the common antisense primer ATG30A specifically amplifies only the complementing (tACT2) allele and native ACT2 alleles, respectively, using PCR. Leaf or cotyledon tissue DNA was prepared as described by Gilliland et al. (1998) and used as template in PCR for genotype determination.
Complementation of the act2-1 allele: A 5.5-kb HindIII genomic clone of ACT2 (An et al., 1996b) was digested with SalI and yielded a 4-kb fragment containing the full ACT2 gene including 750 bp of sequence upstream of the TATA box. This product was cloned into the SalI site of pBluescript and subcloned into the plant transformation vector pBIN19. The construct was electroporated into Escherichia coli using Gene Pulser (Bio-Rad, Hercules, CA) according to manufacturer's instructions. Plasmids containing the genomic fragments were retrieved and isolated using QIAprep Spin Miniprep kit (Qiagen USA, Valencia, CA) and directly transformed into Agrobacterium tumenfaciens strain C58C1. The configuration of the insertions was verified by both restriction digests and PCR. The constructs were then transformed by vacuum infiltration into 5-week-old act2-1/act2-1 plants as described by Bariola et al. (1999). Seeds were collected about 3 weeks after vacuum infiltration and surface sterilized. The seeds were imbibed at 4°C for 2 d and then placed in horizontal lines at approximately 4 per cm on vertical plates containing 1% (w/v) Suc and 0.3% (w/v) Phytagel. The plants were screened under a dissecting scope (12–25×) for elongated root hairs after 2 weeks of growth. Any plants containing long wild-type root hairs were gently removed and transplanted to moist soil. Approximately 20 to 35 progeny from each of the potential tACT2 transformants were plated vertically on Phytagel medium and scored for root hair phenotype. The tACT7 and tACT1c constructs were prepared as described by Gilliland et al. (2002) and Kandasamy et al. (2002), respectively, and vacuum infiltrated into act2-1/act2-1 plants as above.
RNA Preparation and Northern Analysis
RNA was prepared as by Logemann et al. (1987), and 30 μg was resolved on a 1% (w/v) agarose, 16% (v/v) formaldehyde gel and transferred onto a Biotrans (+) nylon membrane by chromatography. A PCR product using primers AAc2-376S (An et al., 1996b) and AAc2-3′A2 (ACTAAAACGCAAAACGAAAGCGGTT; Fig. 1) was gel purified and labeled with [α-32P]dATP by Klenow (Feinberg and Vogelstein, 1983). The probe was purified through a Sepharose 6B column. Blots were hybridized for 48 h as described by An et al. (1996a) and washed twice for 15 min with 6× SSC and 0.5% (w/v) SDS, once for 10 min with 2× SSC and 0.2% (w/v) SDS, and once for 5 min with 1× SSC and 0.1% (w/v) SDS. The blots were exposed to X-film (Eastman Kodak, Rochester, NY) for 4 to 16 d. A densitometer (Molecular Dynamics, Sunnyvale, CA) was used to compare hybridization strength. An rRNA probe was hybridized to the previously used blot to demonstrate equal loading. The 26-nucleotide antisense rRNA oligo complementary to the 18S subunit sequence starting at nucleotide 1,627 (Eckenrode et al., 1985) was radiolabeled as described by Tanzer and Meagher (1995). Prehybridization was performed with Blotto mix of 0.25% (w/v) dry milk, 2× SSC, and 0.2% (w/v) SDS. The blot was hybridized at 48°C and washed three times with a solution of 2× SSC and 0.5% (w/v) SDS. Blot was exposed to X-film for 10 min.
Protein Preparation and Western Analysis
Plant extracts were prepared in actin stabilization buffer as described by Kandasamy et al. (1999). Protein samples were separated on a 12% (w/v) SDS-PAGE gel, and western was performed as described by Kandasamy et al. (1999), but washed five times before incubation with the secondary antibody. Western blots in Figure 5 were exposed to Hyperfilm (Amersham Biosciences AB, Uppsala) for 20 to 60 s. A Molecular Dynamics densitometer was used to compare the signals.
Generation of Double Mutants
The pollen of an ACT7/act7-1 plant was crossed to an emasculated act2-1/act2-1 parent, the progeny were screened by PCR for the presence of the act7-1 allele, and these F1 plants were allowed to self-pollinate. F2 progeny displaying the dwarf phenotypes were checked by PCR for presence of both mutant alleles and the absence of both wild-type alleles.
ACKNOWLEDGMENTS
We thank Libby McKinney for helpful suggestions during a number of experiments and Gay Gragson for editing the manuscript.
Footnotes
This work was supported by the National Institutes of Health (Training grant no. 2T32–GM 07103–27 to the Genetics Department and grant no. GM 36397–14 to L.U.G.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.014068.
LITERATURE CITED
- An Y-Q, Huang S, McDowell JM, McKinney EC, Meagher RB. Conserved expression of the Arabidopsis ACT1 and ACT3 actin subclass in organ primordia and mature pollen. Plant Cell. 1996a;8:15–30. doi: 10.1105/tpc.8.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y-Q, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB. Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 1996b;10:107–121. doi: 10.1046/j.1365-313x.1996.10010107.x. [DOI] [PubMed] [Google Scholar]
- Asmussen MA, Gilliland LU, Meagher RB. Detection of deleterious genotypes in multigenerational studies: II. Theoretical and experimental dynamics with selfing and selection. Genetics. 1998;149:727–737. doi: 10.1093/genetics/149.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F, Salaj J, Mathur J, Braun M, Jasper F, Samaj J, Chua NH, Barlow PW, Volkmann D. Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev Biol. 2000;227:618–632. doi: 10.1006/dbio.2000.9908. [DOI] [PubMed] [Google Scholar]
- Bariola PA, MacIntosh GC, Green PJ. Regulation of S-like ribonuclease levels in Arabidopsis: Antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiol. 1999;119:331–342. doi: 10.1104/pp.119.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova TN, Blancaflor EB, Gilroy S. Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 1999;17:657–665. doi: 10.1046/j.1365-313x.1999.00415.x. [DOI] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;15:441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- Brookfield JFY. Genetic redundancy. In: Hall JC, Friedmann T, Dunlap JC, Giannelli F, editors. Advances in Genetics. Vol. 36. San Diego: Academic Press; 1997. pp. 137–155. [DOI] [PubMed] [Google Scholar]
- Eckenrode VK, Arnold J, Meagher RB. Comparison of the nucleotide sequence of soybean 18S rRNA with the sequence of other small subunit rRNAs. J Mol Evol. 1985;21:259–269. doi: 10.1007/BF02102358. [DOI] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 2001;15:79–89. doi: 10.1101/gad.188801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Foissner I, Wasteneys GO. A cytochalasin-sensitive actin filament meshwork is a prerequisite for local wound wall deposition in Nitella internodal cells. Protoplasma. 1997;200:17–30. [Google Scholar]
- Fyrberg EA, Fyrberg CC, Biggs JR, Saville D, Beall CJ, Ketchum A. Functional nonequivalence of Drosophila actin isoforms. Biochem Genet. 1998;36:271–287. doi: 10.1023/a:1018785127079. [DOI] [PubMed] [Google Scholar]
- Gabriel M, Kopecka M. Disruption of the actin cytoskeleton in budding yeast results in formation of an aberrant cell wall. Microbiology. 1995;141:891–899. doi: 10.1099/13500872-141-4-891. [DOI] [PubMed] [Google Scholar]
- Gilliland LU, McKinney EC, Asmussen MA, Meagher RB. Detection of deleterious genotypes in multigenerational studies: I. Disruptions in individual Arabidopsis actin genes. Genetics. 1998;149:717–725. doi: 10.1093/genetics/149.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland LU, Pawloski L, Kandasamy MK, Meagher RB (2002) The Arabidopsis actin isovariant, ACT7, plays a vital role in germination and root growth. Plant J (in press) [DOI] [PubMed]
- Heath IB, Gupta G, Bai S. Plasma membrane-adjacent actin filaments, but not microtubules, are essential for both polarization and hyphal tip morphogenesis in Saprolegnia ferax and Neurospora crassa. Fungal Genet Biol. 2000;30:45–62. doi: 10.1006/fgbi.2000.1203. [DOI] [PubMed] [Google Scholar]
- Hightower RC, Meagher RB. The molecular evolution of actin. Genetics. 1986;114:315–332. doi: 10.1093/genetics/114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann R, Cooper JA, Miller KG. Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. J Cell Biol. 1996;133:1293–1305. doi: 10.1083/jcb.133.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, An Y-Q, McDowell JM, McKinney EC, Meagher RB. The Arabidopsis ACT11 actin gene is strongly expressed in tissues of the emerging inflorescence, pollen and developing ovules. Plant Mol Biol. 1997;33:125–139. doi: 10.1023/a:1005741514764. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, Gilliland L, McKinney E, Meagher RB. One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell. 2001;13:1541–1554. doi: 10.1105/TPC.010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney E, Meagher RB. The late pollen specific actins in angiosperms. Plant J. 1999;18:681–691. doi: 10.1046/j.1365-313x.1999.00487.x. [DOI] [PubMed] [Google Scholar]
- Kandasamy MK, McKinney EC, Meagher RB. Functional non-equivalency of actin isovariants in Arabidopsis. Mol Biol Cell. 2002;13:251–261. doi: 10.1091/mbc.01-07-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Fukuda H, Shibaoka H. Reorganization of actin filaments associated with the differentiation of tracheary elements in Zinnia mesophyll cells. Protoplasma. 1987;138:69–71. [Google Scholar]
- Kumar A, Crawford K, Close L, Madison M, Lorenz J, Doetschman T, Pawlowski S, Duffy J, Neumann J, Robbins J et al. Rescue of cardiac alpha-actin-deficient mice by enteric smooth muscle gamma-actin. Proc Natl Acad Sci USA. 1997;94:4406–4411. doi: 10.1073/pnas.94.9.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. The problem of genetic diversity. Harvey Lect. 1974;70:1–20. [PubMed] [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method of isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Mascarenhas JP. Molecular mechanisms of pollen tube growth and differentiation. Plant Cell. 1993;5:1303–1314. doi: 10.1105/tpc.5.10.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell J, An Y-Q, McKinney EC, Huang S, Meagher RB. The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol. 1996a;111:699–711. doi: 10.1104/pp.111.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Huang S, McKinney EC, An Y-Q, Meagher RB. Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics. 1996b;142:587–602. doi: 10.1093/genetics/142.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EC, Ali N, Traut A, Feldmann KA, Belostotsky DA, McDowell JM, Meagher RB. Sequence-based identification of T-DNA insertion mutations in Arabidopsis: actin mutants act2-1 and act4-1. Plant J. 1995;8:613–622. doi: 10.1046/j.1365-313x.1995.8040613.x. [DOI] [PubMed] [Google Scholar]
- McKinney EC, Meagher RB. Members of the Arabidopsis actin gene family are widely dispersed in the genome. Genetics. 1998;149:663–675. doi: 10.1093/genetics/149.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher RB, McKinney EC, Kandasamy MK. Isovariant dynamics expand and buffer the responses of complex systems: the diverse plant actin gene family. Plant Cell. 1999a;11:995–1006. doi: 10.1105/tpc.11.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher RB, McKinney EC, Vitale AV. The evolution of new structures: clues from plant cytoskeletal genes. Trends Genet. 1999b;15:278–284. doi: 10.1016/s0168-9525(99)01759-x. [DOI] [PubMed] [Google Scholar]
- Meagher RB, McLean BG. Diversity of plant actins. Cell Motil Cytoskeleton. 1990;16:164–166. doi: 10.1002/cm.970170403. [DOI] [PubMed] [Google Scholar]
- Meagher RB, Williamson RE. The plant cytoskeleton. In: Meyerowitz E, Somerville C, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 1049–1084. [Google Scholar]
- Miller DD, de Ruijter NCA, Bisseling T, Emons AMC. The role of actin in root hair morphogenesis: studies with lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug. Plant J. 1999;17:141–154. [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Nowak MA, Boerlijist C, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- Parker JS, Cavell AC, Dolan L, Roberts K, Grierson CS. Genetic interactions during root hair morphogenesis in Arabidopsis. Plant Cell. 2000;12:1961–1974. doi: 10.1105/tpc.12.10.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C. Genetic control of root hair development in Arabidopsis thaliana. Plant Cell. 1990;2:235–243. doi: 10.1105/tpc.2.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger CJ, Cande WZ. Microfilament distribution in maize meiotic mutants correlates with microtubule organization. Plant Cell. 1991;3:637–644. doi: 10.1105/tpc.3.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer MM, Meagher RB. Degradation of the soybean ribulose-1,5-bisphosphate carboxylase small-subunit mRNA, SRS4, initiates with endonucleolytic cleavage. Mol Cell Biol. 1995;15:6641–6652. doi: 10.1128/mcb.15.12.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Connelly PS, Vranich KA, Shaw MK, Guild GM. Actin filaments and microtubules play different roles during bristle elongation in Drosophila. J Cell Sci. 2000;113:1255–1265. doi: 10.1242/jcs.113.7.1255. [DOI] [PubMed] [Google Scholar]