Abstract
1-Deoxy-d-xylulose 5-phosphate reductoisomerase (DXR) catalyzes the first committed step of the 2-C-methyl-d-erythritol 4-phosphate pathway for isoprenoid biosynthesis. In Arabidopsis, DXR is encoded by a single-copy gene. We have cloned a full-length cDNA corresponding to this gene. A comparative analysis of all plant DXR sequences known to date predicted an N-terminal transit peptide for plastids, with a conserved cleavage site, and a conserved proline-rich region at the N terminus of the mature protein, which is not present in the prokaryotic DXR homologs. We demonstrate that Arabidopsis DXR is targeted to plastids and localizes into chloroplasts of leaf cells. The presence of the proline-rich region in the mature Arabidopsis DXR was confirmed by detection with a specific antibody. A proof of the enzymatic function of this protein was obtained by complementation of an Escherichia coli mutant defective in DXR activity. The expression pattern of β-glucuronidase, driven by the DXR promoter in Arabidopsis transgenic plants, together with the tissue distribution of DXR transcript and protein, revealed developmental and environmental regulation of the DXR gene. The expression pattern of the DXR gene parallels that of the Arabidopsis 1-deoxy-d-xylulose 5-phosphate synthase gene, but the former is slightly more restricted. These genes are expressed in most organs of the plant including roots, with higher levels in seedlings and inflorescences. The block of the 2-C-methyl-d-erythritol 4-phosphate pathway in Arabidopsis seedlings with fosmidomycin led to a rapid accumulation of DXR protein, whereas the 1-deoxy-d-xylulose 5-phosphate synthase protein level was not altered. Our results are consistent with the participation of the Arabidopsis DXR gene in the control of the 2-C-methyl-d-erythritol 4-phosphate pathway.
Plants synthesize a large number of isoprenoid compounds that are very diverse in structure and function (Chappell, 1995; McGarvey and Croteau, 1995). Some isoprenoids are essential in all plants. For instance, chlorophylls and carotenoids are required as photosynthetic pigments, ubiquinone and plastoquinone as electron carriers, sterols as structural components of membranes, dolichols as oligosaccharide donors in protein glycosylation, and abscisic acid, brassinosteroids, cytokinins, and gibberellins as growth regulators. In addition, a vast array of specific isoprenoid compounds found in the different plant species are involved in the interaction with other organisms or in the response to environmental challenges. Despite their diversity, all isoprenoids derive from isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), two readily interchangeable five-carbon isomers that can be conceptually viewed as a single isoprenoid building unit. In higher plants, the isoprenoid building unit is formed by two pathways that operate in different subcellular compartments (Eisenreich et al., 1998; Rohmer, 1999; Lichtenthaler, 2000). In the cytosol-endoplasmic reticulum, the two isomers are synthesized by the well-known mevalonate (MVA) pathway. In plastids, IPP and DMAPP are formed by the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway. In the first reaction of this pathway, 1-deoxy-d-xylulose 5-phosphate (DXP) is synthesized from pyruvate and d-glyceraldehyde 3-phosphate. This step is catalyzed by 1-deoxy-d-xylulose 5-phosphate synthase (DXS), which is encoded by the DXS gene. The following reaction, consisting in the conversion of DXP to MEP, is catalyzed by 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR), the product of the DXR gene. Because DXP is a precursor not only of isoprenoids but also of the cofactors thiamine pyrophosphate and pyridoxal phosphate (Julliard and Douce, 1991; Julliard, 1992), the reaction catalyzed by DXR is actually the first committed step of the MEP pathway. Therefore, DXR could play an important role in the control of plastid isoprenoid biosynthesis. In Escherichia coli, three subsequent reactions catalyzed by the products of ygbP, ychB, and ygbB genes, respectively, allow the synthesis of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (Eisenreich et al., 2001), which is converted to (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate by the gcpE gene product (Hecht et al., 2002; Seemann et al., 2002a, 2002b; Wolff et al., 2002). Recent work has shown that the lytB gene product is involved in the final step of the MEP pathway consisting in the conversion of (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate to IPP and DMAPP (Hintz et al., 2001; Rohdich et al., 2002).
The identification of the regulatory steps of the MEP pathway is an issue of major importance in the study of isoprenoid biosynthesis for both theoretical and applied reasons. The fact that two separate but likely coordinated pathways produce the same isoprenoid precursors adds further interest to the research on this issue. So far, the unraveling of the control of the MEP pathway has been focused in DXS and DXR, the first two enzymes identified. Several investigations support that DXS plays a role in the control of plant isoprenoid biosynthesis. A positive correlation was found between the levels of DXS transcript and protein and the accumulation of various plastid isoprenoid products in transgenic Arabidopsis engineered to under- or overexpress DXS (Estévez et al., 2001). A remarkable spatial and temporal correlation was found between the level of DXS transcript and the synthesis of specific isoprenoid products in a variety of systems: lycopene in tomato (Lycopersicon esculentum) fruit during ripening (Lois et al., 2000), apocarotenoids in roots from monocots after colonization by mycorrhizal fungi (Walter et al., 2000), terpenoid indole alkaloids in periwinkle (Catharanthus roseus) cell suspension culture upon hormonal induction (Chahed et al., 2000), and carotenoids in pepper (Capsicum annuum) fruit during chloroplast to chromoplast transition (Bouvier et al., 1998). In addition, 1-deoxy-d-xylulose feeding in mature green tomato fruits induced expression of carotenoid biosynthetic genes and concomitant carotenoid accumulation (Lois et al., 2000).
The participation of DXR in the control of isoprenoid accumulation in plants is also sustained by experimental results. Overexpression of DXR in transgenic peppermint (Mentha piperita) plants led to an increase of essential oil monoterpenes in leaf tissue compared with wild type (Mahmoud and Croteau, 2001). Conversely, partial DXR gene silencing in some of the engineered peppermint plants led to a reduction of essential oil accumulation. In agreement, a positive correlation was found between the accumulation of DXR transcript and apocarotenoids in mycorrhizal roots from monocots (Walter et al., 2000) or terpenoid indole alkaloids in periwinkle cell suspension culture (Veau et al., 2000). In these two systems, both DXS and DXR could have a regulatory role because a parallel increase of DXS and DXR transcripts was observed (Veau et al., 2000; Walter et al., 2000). In contrast to these results, neither DXR transcript nor DXR protein level increased in tomato fruit during ripening, despite the massive carotenoid accumulation, suggesting a non-limiting role for DXR in this system (Rodríguez-Concepción et al., 2001).
So far, all investigations concerning expression of genes encoding plant DXR have been restricted to the analysis of transcript or protein levels in a variety of systems in which the synthesis of specific isoprenoids is induced developmentally or in response to external stimuli (Veau et al., 2000; Walter et al., 2000; Rodríguez-Concepción et al., 2001). The expression pattern of the DXR gene in the whole plant along normal development has not been investigated yet, nor other important related aspects as the subcellular localization of the encoded product. We have chosen Arabidopsis for this research. As a first step, we determined the 5′ end sequence of the Arabidopsis DXR transcript. This allowed the isolation of a cDNA encoding the entire Arabidopsis DXR, the study of the intracellular targeting of the protein, and the construction of DXR-GUS translational fusions. The expression pattern of β-glucuronidase (GUS) driven by the DXR promoter, together with the accumulation profile of DXR transcript and protein, indicates developmental and environmental regulation of the Arabidopsis DXR gene. The DXR protein is targeted to plastids.
RESULTS
Sequence Analysis of Arabidopsis DXR
The cDNA sequences encoding Arabidopsis DXR reported to date were incomplete at the 5′ end (Lange and Croteau, 1999; Schwender et al., 1999). To determine the 5′ end of the DXR transcript, we performed 5′-RACE using total RNA from 12-d-old Arabidopsis seedlings as a template. Sequencing of four clones derived from the major amplification product showed a single 5′ end that corresponds to the adenine at position +1 of the full-length cDNA sequence deposited in GenBank (accession no. AF148852). This information, together with previous sequence data, allowed the design of PCR primers to amplify a cDNA encoding the entire Arabidopsis DXR. The protein predicted from this cDNA contains 477 amino acid residues and has a molecular mass of 52 kD. The alignment of the cDNA sequence with the corresponding genomic sequence (clone MQB2, accession no. AB009053) revealed the organization of the Arabidopsis DXR gene. This gene maps in chromosome 5 and contains 12 exons and 11 introns extending over a region of 3.2 kb. A databank search showed that no additional sequence homologous to the DXR gene exists in the completely sequenced Arabidopsis genome. In agreement, the pattern of bands obtained in Southern-blot analysis performed under high- and low-stringency conditions, with a 0.84-kb SalI-EcoRV cDNA fragment as a probe, perfectly fits to that predicted from the MQB2 clone (data not shown). It can be concluded that Arabidopsis DXR is encoded by a single-copy gene and that the probe used is DXR specific.
To characterize the sequence of the N-terminal region of Arabidopsis DXR, we aligned this sequence with the equivalent region of the plant DXRs known to date and the E. coli DXR. As shown in Figure 1, the plant enzyme contains an extension of 73 to 80 residues that is not present in the prokaryotic sequence. Data analysis with the ChloroP program (Emanuelsson et al., 1999) predicted a transit peptide for plastids in all plant DXR sequences (Fig. 1). In 11 of the 14 transit peptides, the processing site was predicted at the N terminus of a conserved Cys-Ser-X motif, where X means any of the hydrophobic residues Ala, Val, or Met. The regions at the N- or C-terminal side of the putative processing site have different structural features. At the N-terminal side, the sequence is poorly conserved but enriched in Ser residues, features that are typical of plastid transit peptides (von Heijne et al., 1989). In contrast, the extended region at the C-terminal side (positions 50–80 of Arabidopsis DXR) is highly conserved and particularly rich in Pro residues (Fig. 1). The number of Pro residues in this region ranges from 6 to 8. The consensus motif P(P/Q) PAWPG(R/T) A can be defined in the Pro-rich region of plant DXR (positions 60–68 of the Arabidopsis sequence). The collective sequence analysis suggests that all plant DXRs have a transit peptide for plastids, are processed at a conserved cleavage site, and contain an extended Pro-rich region at the N terminus of the mature protein, which is not present in prokaryotic DXR.
Figure 1.
Multiple sequence alignment of the N-terminal region of plant DXR. The sequence of Arabidopsis DXR was aligned to the other plant DXR sequences known to date, deduced either from complete cDNA clones or expressed sequence tag (EST) entries. Only those EST sequences confirmed by at least two independent entries were considered. Sequence alignment was performed with the ClustalW 1.8 program (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html) and optimized by visual inspection. The origin of the DXR sequence is indicated on the left and the number of amino acid residues on the right. Residues are written in white inside black boxes if they are identical in all plant sequences, in white inside gray boxes if two alternative residues are found in equivalent positions, or in black over white background if a lower conservation is observed. Gaps in the sequence are represented with a dash. The last residue of the transit peptide predicted by the ChloroP program (Emanuelsson et al., 1999) in the different sequences is indicated by an asterisk above the corresponding letter. The putative cleavage site deduced from the collective analysis of all plant DXR sequences is indicated with an arrowhead. The Arabidopsis peptide used for antibody production is marked with a line on the top. The cDNA and EST sequences are accessible at the GenBank with the following accession numbers: Arabidopsis (AF148852), Artemisia annua (AF182287), periwinkle (AF250235), Glycine max (EST 1, BE804032; EST 2, BE211397; and EST 3, BG839054), tomato (AF331705), Lycopersicon hirsutum (EST, AW617386), Medicago truncatula (EST 1, BG456710; and EST 2, BG450566), peppermint (AF116825), Oryza sativa (AF367205), Solanum tuberosum (EST, BE924278), and Zea mays (AJ297566). For reference, the N-terminal sequence of the DXR from E. coli is represented at the bottom.
Functional Analysis and Subcellular Localization of Arabidopsis DXR
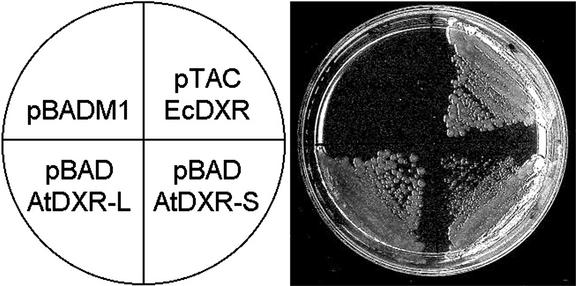
To demonstrate that the isolated Arabidopsis cDNA encodes a functional DXR, we complemented an E. coli lethal mutant defective in the dxr gene (Rodríguez-Concepción et al., 2000). As expected, this mutant can be rescued by expression of plasmid-encoded E. coli DXR (EcDXR, Fig. 2). In addition, the mutant was rescued by expression of either a short derivative of Arabidopsis DXR (AtDXR-S, residues 81–477), which lacked the entire plant-specific N-terminal region, or a longer version of the protein (AtDXR-L, residues 57–477), which lacked the predicted transit peptide and only the first seven residues of the mature protein (Fig. 2). We conclude that the two forms of the Arabidopsis protein have DXR activity. However, AtDXR-L led to a much more vigorous growth than AtDXR-S, as estimated by the colony size (Fig. 2). The same observation was made with dxr mutants generated in two different genetic backgrounds (E. coli strains MC4100 and JC7623). Our results suggest that the N-terminal Pro-rich region of Arabidopsis DXR (residues 57–80) contains elements that are important for activity or stability, at least when expressed in E. coli.
Figure 2.
Complementation of E. coli dxr mutant with Arabidopsis DXR. The E. coli dxr::TET mutant EcAB1–2 (Rodríguez-Concepción et al., 2000) was transformed with a control expression plasmid without DXR insert (pBADM1), a pBADM1 derivative coding for a long form of Arabidopsis DXR (pBAD-AtDXR-L), a pBADM1 derivative coding for a short form of Arabidopsis DXR (pBAD-AtDXR-S), or a expression plasmid coding for the DXR from E. coli (pTAC-EcDXR). Transformants were plated in Luria-Bertani medium containing 6 μg mL−1 tetracycline to select for the dxr::TET mutant, 100 μg mL−1 ampicillin to select for the plasmid, 100 μm isopropyl-β-d-thiogalactoside to induce expression of EcDXR, and 0.02% (w/v) l-Ara to induce expression of AtDXR-S and AtDXR-L. After 17 h at 37°C, the colony size was 2 to 3 mm for the mutant transformed with pTAC-EcDXR or pBAD-AtDXR-L and 0.3 to 0.4 mm for the mutant transformed with pBAD-AtDXR-S.
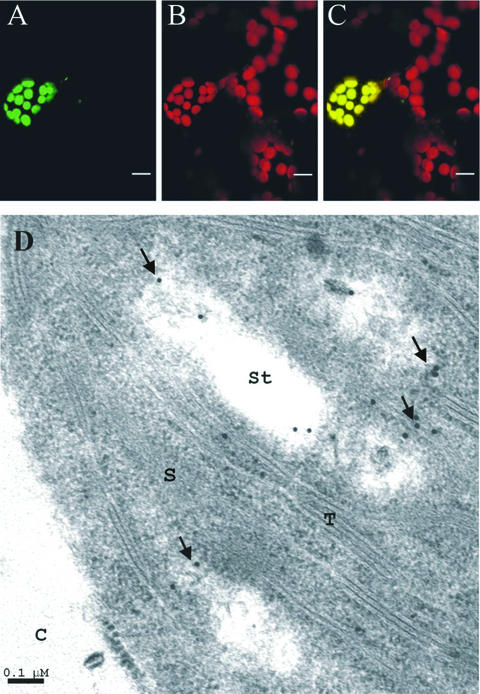
The intracellular targeting of Arabidopsis DXR was studied by transient expression of a construct encoding the entire DXR protein fused to the N terminus of an optimized version of the soluble green fluorescent protein (GFP). Leaves from 15-d-old light-grown Arabidopsis seedlings were microbombarded with this construct. The DXR-GFP fusion protein accumulated in the chloroplasts of leaf cells as shown by its colocalization with chlorophyll autofluorescence (Fig. 3, A–C). In a second approach, we analyzed the distribution of DXR in cells from Arabidopsis leaves, using a polyclonal antibody (Ab-AtDXR1) raised against peptide EAPRQSWDGPK, which corresponds to the N-terminal extended region of Arabidopsis DXR (Fig. 1, residues 71–81). Immunogold particles were found in chloroplasts (Fig. 3D) and rarely outside this organelle. These observations, which to our knowledge are the first experimental evidence of plastid localization of plant DXR, are in agreement with the proposed role of this protein in the synthesis of plastid-derived isoprenoids.
Figure 3.
Targeting and subcellular localization of plant DXR. Leaves of 15-d-old Arabidopsis seedlings were microbombarded with a construct encoding Arabidopsis DXR fused to the N terminus of GFP. Cells expressing the fusion protein were studied by laser confocal microscopy. The images show green fluorescence of DXR-GFP (A), red autofluorescence of chlorophyll (B), and the superimposed green and red fluorescence (C). Bars in A through C indicate 10 μm. Arabidopsis DXR was immunolocalized in leaves of 15-d-old seedlings with the Ab-AtDXR1 polyclonal antibody. The electron micrograph (D) shows localization of 15-nm gold particles in chloroplast. C, Cytoplasm; S, stroma; St, starch granule; T, thylakoid. Bar in D indicates 0.1 μm.
Expression Analysis of the Arabidopsis DXR Gene
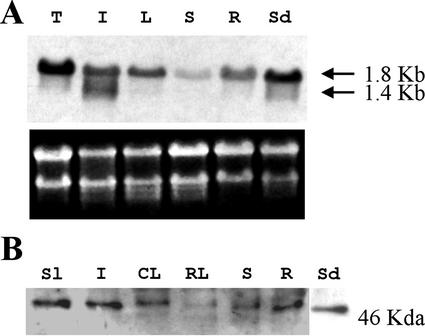
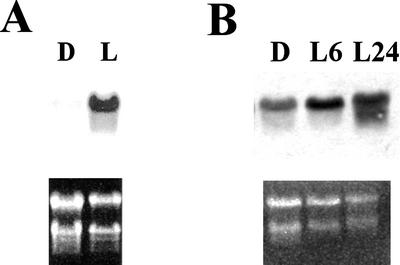
As a first step to evaluate the importance of the DXR gene in the control of the MEP pathway, we analyzed the distribution of the DXR transcript and protein in Arabidopsis, studied the accumulation of these molecules in response to particular conditions and characterized the expression pattern of a chimeric construct containing the GUS reporter gene under control of the DXR promoter. Northern-blot analysis of total RNA from different tissues revealed a transcript of about 1.8 kb (Fig. 4A), in agreement with the size of full-length DXR cDNA. This transcript is present in all tissues analyzed, but accumulates at higher levels in seedlings and inflorescences (Fig. 4A). Lower transcript levels were observed in leaves of adult plants and roots of 15-d-old seedlings. The lowest transcript level was detected in stems. The DXR transcript is also present at a high level in the light-grown cell suspension line T87 derived from Arabidopsis leaves (Axelos et al., 1992; Fig. 4A). A good correlation in tissue distribution was found between the 1.8-kb transcript and a 46-kD protein detected by western blot with the Ab-AtDXR1 antibody (Fig. 4B). The DXR protein accumulates at higher levels in 15-d-old seedlings, inflorescences, and fruits, and at lower levels in cauline leaves and stems. The protein is barely detectable in rosette leaves of adult plants. The apparent molecular mass of the immunodetected DXR is consistent with processing at the conserved site described above. The detection of this protein by western blot confirms that the N-terminal Pro-rich sequence is present in the mature form of the Arabidopsis DXR. In addition to the 1.8-kb message, a transcript of about 1.4 kb was detected with the DXR-specific probe in inflorescences (Fig. 4A). The level of the 1.8-kb transcript was much higher in seedlings grown in the light than in seedlings grown in the dark (Fig. 5A). The DXR transcript rapidly accumulated upon de-etiolation (Fig. 5B), indicating that the increase in the transcript level was a direct response to light and not an indirect effect of the changes in the morphogenetic pattern.
Figure 4.
Distribution of DXR transcripts and protein in Arabidopsis plants. A, Fifteen micrograms of total RNA from Arabidopsis tissues was analyzed by northern blot as described in “Materials and Methods.” Exposure time was 48 h. Ethidium bromide staining of the gel before transfer is also shown. B, Crude extracts (30 μg of protein) from Arabidopsis tissues were analyzed by western blot as described in “Materials and Methods.” DXR protein was detected with the Ab-AtDXR1 polyclonal antibody. Developing time was 1 min. CL, Cauline leaves, Sl, siliques; I, inflorescences; L, rosette and cauline leaves; R, roots of 15-d-old seedlings grown either in Murashige and Skoog plates exposed to light (A) or in soil (B); RL, rosette leaves; S, stems from adult plant; Sd, 15-d-old seedlings grown in 16-h light/8-h dark regime; T, Arabidopsis cell suspension line T87.
Figure 5.
Accumulation of Arabidopsis DXR transcript induced by light. Samples were analyzed by northern blot as described in “Materials and Methods.” A, Twenty micrograms of total RNA from light-grown (L) and dark-grown (D) 12-d-old seedlings. B, Ten micrograms of total RNA from dark-grown 15-d-old seedlings (D) or 14-d-old dark-grown seedlings exposed to light for 6 h (L6) or 24 h (L24). Exposure time was 40 h for the filter of A and 72 h for the filter of B.
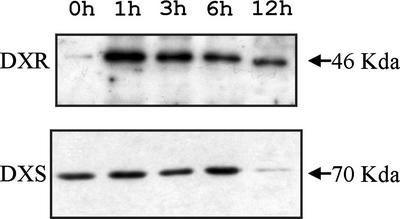
To determine whether the DXR protein level was altered in response to a block of the MEP pathway, we treated 7-d-old Arabidopsis seedlings with fosmidomycin, a bleaching herbicide that specifically inhibits DXR (Kuzuyama et al., 1998; Zeidler et al., 1998). The level of the DXS protein was also determined for comparison. As shown in Figure 6, fosmidomycin treatment caused a sharp increase in DXR protein, whereas the amount of DXS did not change significantly. The maximal level of DXR was observed 1 h after treatment. The progressive decrease of DXR and DXS proteins, observed after 3, 6, and 12 h, might be due to cytotoxicity of the inhibitor (Fig. 6).
Figure 6.
Accumulation of Arabidopsis DXR protein induced by fosmidomycin. One hundred micromolar fosmidomycin was added to liquid cultures containing 7-d-old Arabidopsis seedlings grown under continuous light with agitation. Samples were collected just before treatment or at different times after treatment and analyzed by western blot. Arabidopsis DXR protein was detected with the Ab-AtDXR1 antibody. Arabidopsis DXS protein was detected with a specific polyclonal antibody (Estévez et al., 2000). Collection times are indicated at the top.
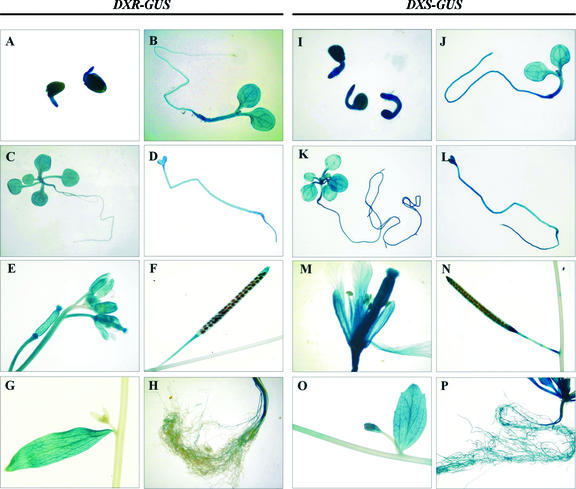
The expression pattern of the DXR gene was further studied with transgenic plants carrying a translational fusion between a 1.3-kb fragment of DXR and the entire coding sequence of the GUS reporter gene. The DXR fragment included 1.2 kb of the 5′-flanking region, the entire 5′-transcribed untranslated region (UTR), and the first 6 bp of the DXR coding sequence. Histological analysis of GUS activity was performed in plants from 6 independent lines of generation T2. All the lines showed the same GUS expression pattern with little variation in the intensity of staining. High expression of GUS was observed right after germination (Fig. 7A). In the first stages of development in the light, GUS activity was high in the hypocotyl and cotyledons, and less intense in the root with a progressive decline toward the tip (Fig. 7B). High expression was observed in true leaves emerging in subsequent stages of development (Fig. 7C). In etiolated seedlings, GUS staining distributed quite uniformly (Fig. 7D). In adult plants, the highest level of GUS expression was observed in inflorescences (Fig. 7E). GUS staining was higher in the gynoecium than in sepals and petals. Upon maturation, the staining of the silique was progressively restricted to the basal and distal ends (Fig. 7F). An intense staining was observed in cauline leaves, in sharp contrast with the adjacent stem (Fig. 7G). Only those parts of the stem proximal to the inflorescences showed blue staining (Fig. 7E). GUS activity was also detected in roots of adult plants (Fig. 7H).
Figure 7.
Histochemical analysis of GUS activity in transgenic Arabidopsis plants expressing the GUS gene under control of the DXR (A–H) or the DXS (I–P) promoter. A and I, Germinating seeds imbibed for 24 h in Murashige and Skoog medium; B and J, 6-d-old light-grown seedlings; C and K, 15-d-old light-grown seedlings; D and L, 9-d-old dark-grown seedlings; E and M, flowers; F and N, mature siliques; G and O, cauline leaf with emerging axillary inflorescence; H and P, roots of adult plant.
The expression pattern of DXR-GUS transgenic plants was compared with that of transgenic plants carrying the GUS reporter gene under control of the Arabidopsis DXS promoter. The chimeric DXS-GUS construct included 1.7 kb of the DXS 5′-flanking region, the entire 5′-UTR, and 12 bp of the DXS coding sequence. As previously reported (Estévez et al., 2000), the Arabidopsis DXS gene is mainly expressed in developing photosynthetic and non-photosynthetic tissues, but also to some extent in most tissues of the plant. Whereas our results confirm the published data, we observe, in addition, previously unreported high GUS expression driven by the DXS promoter in etiolated seedlings (Fig. 7L), petals and the whole gynoecium (Fig. 7M), cauline leaves (Fig. 7O), and roots of the adult plant (Fig. 7P). Therefore, the GUS expression pattern of the DXS-GUS transgenic plants (Fig. 7, I–P) closely parallels that of the DXR-GUS transgenic plants described above (Fig. 7, A–H). In many instances, however, the staining pattern of DXR-GUS transgenic plants appears to be either more restricted or less intense. This is more evident in the stems and roots of adult plants and in emerging axillary inflorescences (Fig. 7, G, H, O, and P). In emerging inflorescences, the onset of DXS expression clearly precedes that of DXR (Fig. 7, compare G with O).
DISCUSSION
Two fundamental aspects of the Arabidopsis DXR gene have been addressed in the present work: the analysis of the expression of this gene and the subcellular localization of the encoded protein. The identification of the 5′ end of the Arabidopsis DXR transcript opened the way to the isolation of a full-length cDNA and the construction of a DXR-GUS chimeric gene appropriate for the analysis of the DXR expression pattern in transgenic plants. The protein predicted from the Arabidopsis cDNA has an N-terminal extension of 80 amino acid residues that is not present in the prokaryotic DXR homologs. A comparative analysis of this part of the protein in all plant DXR sequences known to date uncovered two regions with clearly different structural features: an N-terminal region with the features of a plastid transit peptide and a highly conserved region particularly rich in Pro residues (Fig. 1). The two regions join at a putative cleavage site located at the N terminus of the consensus motif Cys-Ser-(Ala/Met/Val). The sequence preceding this motif is not strictly conserved in plant DXR (Fig. 1), but it always contains preferred residues of the sequence around the cleavage site of known plastid transit peptides (Emanuelsson et al., 1999). Therefore, although a different processing site was originally predicted with the ChloroP program in three of the 14 sequences (Fig. 1), the collective analysis suggests that all plant DXRs are processed at the conserved cleavage motif. In agreement with the sequence analysis, Arabidopsis DXR was shown to be targeted to plastids by transient expression of a DXR-GFP fusion protein in Arabidopsis cells and to localize into chloroplasts of Arabidopsis leaf cells by immunoelectron microscopy. The consensus P(P/Q) PAWPG(R/T) A is conserved in the Pro-rich region of all plant DXR sequences (Fig. 1) and may be used as a signature for this protein. The presence of the Pro-rich region in the mature Arabidopsis DXR was supported by immunodetection with a specific polyclonal antibody in western blot and electron microscopy. A well-known function of protein domains rich in Pro residues is to mediate protein-protein interactions (Kay and Williamson, 2000). The Pro-rich region of plant DXR might be required for oligomerization of the protein. However, this seems unlikely because no extended Pro-rich region is present in the DXR protein from E. coli that was purified as a homotetramer (Takahashi et al., 1998). It is tempting to speculate that the Pro-rich region of plant DXR might be involved in specific interactions with regulatory proteins or other enzymes of the MEP pathway.
Two transcripts of about 1.8 and 1.4 kb were detected in northern-blot analysis. The 1.8-kb transcript corresponds to the cloned cDNA and is widely distributed in the plant. In contrast, the 1.4-kb transcript is only found in inflorescences. We conclude that the longer transcript corresponds to the immunodetected DXR because the apparent molecular mass of this protein fits to the size of the predicted mature protein and the two molecules have a parallel distribution in the plant. Most likely, the shorter transcript also derives from the DXR gene because the probe used for RNA detection was gene specific. The cloning of this transcript is being pursued at present to confirm its identity and determine the sequence of the putative encoded product. The 1.8-kb transcript accumulates at high levels in the Arabidopsis cell suspension line T87. Previous work in the same system showed expression of the two Arabidopsis genes coding for 3-hydroxy-3-methylglutaryl CoA reductase, a key regulatory enzyme of the MVA pathway (Enjuto et al., 1995; Lumbreras et al., 1995). These data indicate that the two pathways for IPP and DMAPP biosynthesis operate simultaneously in Arabidopsis tissue culture cells.
The expression pattern of the GUS reporter gene under control of the DXR promoter was fully consistent with the distribution of the 1.8-kb transcript detected in northern-blot analysis. This suggests that the DXR-GUS chimeric construct contains all the cis elements required for spatial and temporal control of expression and that the levels of DXR transcript in different parts of the plant reflect the transcriptional activity of the promoter. The expression of the DXR gene closely parallels that of the DXS gene. Both genes are mainly expressed in seedlings and inflorescences, but also at significant levels in most tissues of the plant. The high expression of DXS and DXR in young photosynthetic tissues and the induction of these genes by light are in agreement with their role in chlorophyll and carotenoid biosynthesis. The expression of DXS and DXR in inflorescences is consistent with the production of a high variety of still uncharacterized isoprenoids in this part of the Arabidopsis plant (Tholl et al., 2001). We observed high expression of the DXS-GUS construct in the whole gynoecium, unnoticed in a previous report (Estévez et al., 2000). This apparent discrepancy might be due to the presence of a longer DXS promoter region in our construct (2.0 versus 1.4 kb used in the preceding work). Interestingly, inflorescences are also a major site for expression of all genes of the MVA pathway identified to date in Arabidopsis (Lluch et al., 2000). Therefore, both the MEP and the MVA pathways likely contribute to the production of specialized isoprenoids in Arabidopsis reproductive organs. Some of the isoprenoid products derived from the MEP pathway in Arabidopsis might be essential for flower development because flower primordia of the cla1-1 mutant or dark-grown wild-type plants never mature into normal flowers (Mandel et al., 1996). It was reported previously that DXS and DXR are expressed in roots of cereals upon colonization by mycorrhizal fungi (Walter et al., 2000). We observed expression of Arabidopsis DXS and DXR in the roots of seedlings and adult plants. Because Arabidopsis does not form mycorrhiza (Harrison, 1997), our results suggest that other non-photosynthetic processes occurring in roots might likewise require specific isoprenoid products synthesized by the MEP pathway.
The expression of Arabidopsis DXR is modulated throughout development, in contrast to the constitutive DXR expression observed in tomato fruit during ripening (Rodríguez-Concepción et al., 2001). Therefore, the apparent non-limiting nature of DXR in tomato fruit isoprenoid biosynthesis might be particular of this system. The slightly more restricted expression pattern of DXR versus DXS in Arabidopsis might be related to the dual role of DXS gene product in isoprenoid and cofactor biosynthesis, but also to a hypothetical regulatory role of DXR in plastid isoprenoid biosynthesis. In emerging inflorescences, for instance, DXS starts to be expressed earlier than DXR, suggesting that DXR instead of DXS might be limiting for the onset of isoprenoid biosynthesis. Our experiments with fosmidomycin clearly suggest a metabolic control of DXR at the protein level. This is consistent with the hypothesis that Arabidopsis DXR might have a regulatory role. On the other hand, overexpression of DXS in transgenic Arabidopsis led to an increased accumulation of diverse isoprenoid products, supporting a regulatory role of this gene in plastid isoprenoid biosynthesis (Estévez et al., 2001). Taken together, the present data suggest that, in plants, several enzymes of the MEP pathway may contribute to the control of this metabolic route.
MATERIALS AND METHODS
Plant Culture and Treatments
All biological material used in this work derived from Arabidopsis plants of the ecotype Columbia. Axenic cultures were prepared by surface-sterilizing seeds in 0.27% (w/v) Bayrochlore (Bayrol Gmb, Planegg, Germany) dissolved in ethanol. Sterile seeds were germinated on petri dishes containing solid Murashige and Skoog medium (ICN Biochemicals Division, Aurora, OH) supplemented with 0.5 g L−1 MES (pH 5.7). Unless otherwise stated, plants were grown under 16-h light/8-h dark illumination regime at 22°C to 24°C on a 1:1:1 (v/v) perlite:vermiculite:sphagnum mixture irrigated with mineral nutrients. Roots were obtained either from adult plants or 3-week-old seedlings grown on filter paper layered onto Murashige and Skoog medium. For treatment with fosmidomycin, seedlings were grown in liquid culture containing Murashige and Skoog medium with 0.1% (w/v) agar under continuous light and agitation. Fosmidomycin (Molecular Probes, Eugene, OR) was dissolved at 0.1 m in 10 mm Tris-HCl (pH 8.5) and added to the culture at a final concentration of 100 μm. Arabidopsis cell suspension line T87 was cultured as described (Axelos et al., 1992).
Cloning of Arabidopsis DXR cDNA
As a first step for the isolation of a full-length Arabidopsis cDNA, we determined the 5′ end of the Arabidopsis DXR transcript. RACE was carried out with 5′-RACE System (version 2.0, Life Technologies/Gibco-BRL, Cleveland) following the instructions of the supplier. The first strand of the cDNA was synthesized using total RNA from 12-d-old Arabidopsis seedlings as a template and the oligonucleotide DXR-GSP1 (Table I) as specific downstream primer. The 3′ end region of the single-stranded cDNA was amplified by two nested PCR reactions. In the first PCR, the specific downstream primer was the oligonucleotide DXR-GSP2 and the upstream primer was the oligonucleotide 5′-RACE-AAP supplied in the kit. In the second PCR, the specific downstream primer was the oligonucleotide DXR-GSP3 and the upstream primer was the oligonucleotide AUAP supplied in the kit. The major amplification product of the second PCR was purified by agarose-gel electrophoresis, cloned, and sequenced.
Table I.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| DXR-34 | 5′-CAAGAGTAGTAGTGCGGTTCTCTGG-3′ |
| DXR-E2 | 5′-CAGTTTGGCTTGTTCGGATCACAG-3′ |
| DXR-end | 5′-ACGAATTCATTATGCATGAACTGGCCTAGCACC-3′ |
| DXR-GSP1 | 5′-ATTCGAACCAGCAGCTAGAG-3′ |
| DXR-GSP2 | 5′-CCAGTAGATCCAACGATAGAG-3′ |
| DXR-GSP3 | 5′-GGCCATGCTGGAGGAGGTTG-3′ |
| DXR-SalIA | 5′-TTCATGTCGACACTGGCCTAGCACCAGAA-3′ |
| Mut1CLA1 | 5′-GAAGGAAAAGCAAATCTAGAAGAAGCCATTGG-3′ |
| 5′-MVKPI | 5′-GGCATATGGTGAAACCCATCTCTATCGTTGGATC-3′ |
| 5′-MQQQ | 5′-TGCATATGCAACAACAACCTCCTCCAGC-3′ |
| pBAD-mut1 | 5′-CTGAGAGTGCACCATCTGCGGTGTGAAATACC-3′ |
| PRI1 | 5′-GTGAGCGGCTCGAAGCAAACCGGA-3′ |
| PRIBASE-XbaI | 5′-GTTTCTAGACATCATCAGAGTCTTTTAAAAATCG-3′ |
| T7-21 | 5′-GTAATACGACTCACTATAGGG-3′ |
The full-length Arabidopsis DXR cDNA was amplified from a cDNA library from the cell suspension line T87 by two consecutive PCR reactions. The reaction mixture of the first PCR contained 4 × 105 plaque-forming units of the library, 0.5 μm of the primers DXR-34 and DXR-E2, 1.25 units of Pfu DNA polymerase (Stratagene, La Jolla, CA), and reaction buffer supplied with the enzyme, in a final volume of 25 μL. After a hot start, the reaction mixture was incubated for 35 cycles consisting of 30 s at 94°C, 40 s at 55°C, and 6.5 min at 72°C, followed by a final step of 15 min at 72°C. An aliquot (0.5 μL) of the resulting sample was used as a template for the second PCR, which was performed in the same conditions as the first, except that the final volume of the reaction mixture was 50 μL and the number of cycles 15. The amplification product was purified by agarose-gel electrophoresis, cloned into plasmid pBluescript SK+, and sequenced. The plasmid containing this cDNA was designated pDXR-At.
Complementation of Escherichia coli dxr Mutant
A modified version of plasmid pBAD-GFPuv (CLONTECH Laboratories, Palo Alto, CA) was generated by removing the NdeI site located at position 4,926 by site-directed mutagenesis (Kunkel et al., 1987) with the oligonucleotide pBAD-mut1 as mutagenic primer. The resulting plasmid was designated pBADM1. Arabidopsis DXR cDNA was amplified by PCR with primers 5′-MQQQ and DXR-end to obtain a cDNA fragment coding for a nearly complete mature Arabidopsis DXR (AtDXR-L, positions 57–477 of the protein sequence) or primers 5′-MVKPI and DXR-end to obtain a cDNA fragment coding for a shorter protein (AtDXR-S, positions 81–477). The cDNA fragments were digested with NdeI and EcoRI restriction enzymes and cloned into plasmid pBADM1, previously digested with the same restriction enzymes, to generate plasmids pBAD-AtDXR-L and pBAD-AtDXR-S. In these plasmids, the cloned cDNAs are under control of the pBAD promoter, which can be induced with l-Ara. The E. coli dxr::TET mutant EcAB1–2 (Rodríguez-Concepción et al., 2000) was complemented by transformation with pBAD-AtDXR-L or pBAD-AtDXR-S plasmid.
Isolation and Analysis of Nucleic Acids
Genomic DNA from 12-d-old Arabidopsis seedlings was prepared as described (Ausubel et al., 1987). For Southern-blot analysis, aliquots containing 10 μg of DNA were digested with PstI, ClaI, EcoRV, or EcoRI restriction enzyme, size fractionated by electrophoresis in 0.8% (w/v) agarose gels, and transferred to Hybond C nitrocellulose membranes (Amersham, Buckinghamshire, UK). Hybridization was for 18 h at either 65°C (high stringency) or 58°C (low stringency) in 0.7 m sodium chloride, 40 mm sodium phosphate (pH 7.6), 4 mm EDTA, 0.1% (w/v) SDS, 0.2% (w/v) polyvinylpyrrolidone, 0.2% (w/v) Ficoll, 9% (w/v) dextran sulfate, and 200 μg mL−1 denatured salmon sperm DNA. The probe used was a 32P-labeled 840-bp SalI-EcoRV fragment excised from the EST clone 120E8T7. High stringency washes were performed at 65°C twice in 1× SSC, 0.5% (w/v) SDS, and twice in 0.2× SSC and 0.5% (w/v) SDS. Low-stringency washes were performed at 58°C twice in 2× SSC and 0.5% (w/v) SDS.
Total RNA from Arabidopsis tissues or cells was isolated as described (Dean et al., 1985). For northern-blot analysis, the RNA samples were fractionated by electrophoresis in a 1% (w/v) agarose gel containing 2.2 m formaldehyde and transferred to Neutral Nylon membrane (Schleicher & Schuell, Keene, NH). Hybridization with the 840-bp probe indicated above was performed for 18 h at 68°C in ExpressHyb hybridization solution (CLONTECH Laboratories). High stringency washes were performed at 68°C twice in 2× SSC and 0.1% (w/v) SDS, and twice in 0.1× SSC and 0.1% (w/v) SDS.
Immunological Methods
The polyclonal antibody Ab-AtDXR1 (Sigma, Cambridge, UK) was raised in a rabbit injected with peptide EAPRQSWDGPK, corresponding to positions 71 through 81 of Arabidopsis DXR. For western-blot analysis, crude protein extracts from Arabidopsis tissues were obtained by harvesting into liquid nitrogen and grinding in ice-cold homogenization buffer (0.1 m Tricine [pH 7.2], 30% [w/v] Suc, 1% [w/v] Ficoll 400, 1 mm EDTA, 1 mm MgCl2, and 10 mm KCl). Protein concentration was determined using a protein reagent (Bio-Rad Laboratories, Hercules, CA) according to the dye-binding procedure (Bradford, 1976). Proteins were subjected to SDS-PAGE on 10% (w/v) polyacrylamide gels (Laemmli, 1970) and either stained with Coomassie Blue or transferred to a Hybond P membrane (Amersham). Arabidopsis DXR was detected with the Ab-AtDXR1 serum (dilution 1:200 [v/v]) as a primary antibody and anti-rabbit immunoglobulin horseradish peroxidase-conjugate (Amersham, dilution 1:10,000 [v/v]) as a secondary antibody. Arabidopsis DXS was detected with the anti-GST-CLA1 ascites fluid (Estévez et al., 2000; 1:500 [v/v]) as a primary antibody and anti-mouse immunoglobulin horseradish peroxidase-conjugate (Amersham, 1:10,000) as a secondary antibody. Chemiluminescent detection was carried out with the ECL+plus system (Amersham), following the recommendations of the supplier. For electron microscopy analysis, Arabidopsis leaf cross-sections were fixed, embedded, and immunolabeled as described (Araus et al., 1993) with minor modifications. The fixation buffer contained 4% (v/v) paraformaldehyde, 0.1% (v/v) glutaraldehyde, and 0.1 m cacodylate (pH 7.4). Nonspecific antibody sticking was blocked by incubating for 30 min in Tris-buffered saline buffer containing 10 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 1% (w/v) bovine serum albumin. The Ab-AtDXR1 antibody was used at 1:200 (v/v) dilution and the anti-rabbit-gold antibody (15 nm, British BioCell International, Cardiff, UK) at 1:25 (v/v). Specimens were observed in a Hitachi 600 electron microscope (Hitachi, Tokyo).
Microbombardment Assays
A cDNA fragment containing the Arabidopsis DXR coding sequence was amplified by PCR using plasmid pDXR-At as template and oligonucleotides DXR-SalIA and T7–21 as primers. The amplification product was cloned in pGEM-T plasmid (Promega, Madison, WI) and subsequently transferred as a 1.5-kb SalI-SalI fragment to pGFP-MRC plasmid (Rodríguez-Concepción et al., 1999) previously digested with XhoI and SalI. The resulting construct encodes the entire Arabidopsis DXR fused to the N terminus of GFP. Expression of this construct is under control of the CaMV 35S promoter. Leaves of 15-d-old Arabidopsis seedlings were microbombarded with plasmid DNA-coated tungsten particles and examined by confocal laser scanning microscopy as described (Lois et al., 2000).
Generation and Analysis of Arabidopsis Transgenic Plants
A 1.3-kb fragment of the DXR 5′-flanking region was amplified by PCR using Arabidopsis genomic DNA as template and the oligonucleotides PRIBASE-XbaI and PRI-1 as primers. The amplification product was cloned in pGEM-T vector (Promega) and sequenced. The insert was recovered as an XbaI-SphI fragment and transferred to plasmid pBI221 (CLONTECH Laboratories), to substitute for the CaMV 35S promoter. The resulting plasmid was named pLBI2PRI1.2. The DXR-GUS chimeric gene of this plasmid contains 1,158 bp of DXR 5′-flanking untranscribed region, the entire DXR 5′-UTR (85 bp), and the first 6 bp of the DXR coding sequence, cloned in phase with the coding region of the E. coli uidA reporter gene. This chimeric gene was recovered as an EcoRI-HindIII fragment and ligated to the equivalent sites of plasmid pBI121 (CLONTECH Laboratories) to produce plasmid pLBI1PRI1.1. A BglII-BglII 2.0-kb fragment corresponding to the 5′-flanking region of the Arabidopsis DXS gene was recovered from plasmid 1C10AP64 and cloned in the BamHI restriction site of plasmid pBluescript SK− (Stratagene). A XbaI site was introduced 12 bp downstream of the ATG translation start site of the genomic fragment by site-directed mutagenesis (Kunkel et al., 1987), using the oligonucleotide Mut1CLA1 as mutagenic primer. The DXS promoter region was recovered as a 2.0-kb HindIII-XbaI fragment and ligated to the equivalent sites of plasmid pBI121, to substitute for the CaMV 35S promoter. The resulting plasmid was designated pLBI1PS2. The DXS-GUS chimeric gene of this plasmid contains 1,738 bp of the DXS 5′-flanking untranscribed region, the entire DXS 5′-UTR (202 bp), and the first 12 bp of the DXS coding sequence, fused in frame to the uidA reporter gene. Arabidopsis transgenic plants carrying either the DXR-GUS or the DXS-GUS chimeric gene were generated and analyzed as described (Cunillera et al., 2000). The staining time in the histochemical analysis of GUS activity was 14 to 18 h for samples obtained from DXR-GUS transgenic plants and 6 to 18 h for samples from DXS-GUS plants.
ACKNOWLEDGMENTS
We thank Dr. Helmut Hilbert (QIAGEN GmbH, Hilden, Germany) for the genomic clone 1C10AP64, Oscar Besumbes (Universitat de Barcelona) for the plasmid pTAC-EcDXR, and Dr. Patricia León (Universidad Nacional Autónoma de México, Cuernavaca) for the anti-GST-CLA1 polyclonal antibody. The EST clone 120E8T7 was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). We thank Dr. Nuria Cortadellas, Dr. Palmira Ros, and Dr. Almudena García (Serveis Cientificotècnics, Universitat de Barcelona) for their expert technical assistance in immunoelectron microscopy. We also thank the staff from the Serveis de Camps Experimentals and Serveis Cientificotècnics (Universitat de Barcelona) for their help in plant culture and DNA sequencing. We thank Pablo Leivar and Susanna Sauret (Universitat de Barcelona) for critical reading of the manuscript.
Footnotes
This work was supported by the Spanish Ministerio de Educación y Cultura (grant no. BIO2000–0334), by the Generalitat de Catalunya (grant no. CIRIT 1999SGR–00032), and by the Agencia Española de Cooperación Internacional (predoctoral MUTIS fellowship to I.A.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003798.
LITERATURE CITED
- Araus JL, Bort J, Brown RH, Bassett CL, Cortadellas N. Immunocytochemical localization of phosphoenolpyruvte carboxylase and photosynthetic gas-exchange characteristics in ears of Triticum durum Desf. Planta. 1993;191:507–514. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley and Sons, Inc.; 1987. [Google Scholar]
- Axelos M, Curei C, Mazzolini L, Bardet C, Lescure B. A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem. 1992;30:123–128. [Google Scholar]
- Bouvier F, d'Harlingue A, Suire C, Backhaus RA, Camara B. Dedicated roles of plastid transketolases during the early onset of isoprenoid biogenesis in pepper fruits. Plant Physiol. 1998;117:1423–1431. doi: 10.1104/pp.117.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chahed K, Oudin A, Guivarc'h N, Hamdi S, Chénieux J-H, Rideau M, Clastre M. 1-Deoxy-d-xylulose 5-phosphate synthase from periwinkle: cDNA identification and induced gene expression in terpenoid indole alkaloid-producing cells. Plant Physiol Biochem. 2000;38:559–566. [Google Scholar]
- Chappell J. Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:521–547. [Google Scholar]
- Cunillera N, Boronat A, Ferrer A. Spatial and temporal patterns of GUS expression directed by 5′ regions of the Arabidopsis thaliana farnesyl diphosphate genes FPS1 and FPS2. Plant Mol Biol. 2000;44:747–758. doi: 10.1023/a:1026588708849. [DOI] [PubMed] [Google Scholar]
- Dean C, van den Elzen P, Tamaki S, Dunsmuir P, Bedbrook J. Differential expression of eight genes of the petunia ribulose bisphosphate carboxylase small subunit multi-gene family. EMBO J. 1985;4:3055–3061. doi: 10.1002/j.1460-2075.1985.tb04045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W, Rohdich F, Bacher A. Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci. 2001;6:78–84. doi: 10.1016/s1360-1385(00)01812-4. [DOI] [PubMed] [Google Scholar]
- Eisenreich W, Schwarz M, Cartayrade A, Arigoni D, Zenk MH, Bacher A. The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem Biol. 1998;5:R221–R233. doi: 10.1016/s1074-5521(98)90002-3. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuto M, Lumbreras V, Marín C, Boronat A. Expression of the Arabidopsis HMG2 gene, encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase, is restricted to meristematic and floral tissues. Plant Cell. 1995;7:517–527. doi: 10.1105/tpc.7.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez JM, Cantero A, Reindl A, Reichler S, León P. 1-Deoxy-d-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem. 2001;276:22901–22909. doi: 10.1074/jbc.M100854200. [DOI] [PubMed] [Google Scholar]
- Estévez JM, Cantero A, Romero C, Kawaide H, Jiménez LF, Kuzuyama T, Seto H, Kamiya Y, León P. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-d-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 2000;124:95–103. doi: 10.1104/pp.124.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ. The arbuscular mycorrhizal symbiosis: an underground association. Trends Plant Sci. 1997;2:54–60. [Google Scholar]
- Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F. Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc Natl Acad Sci USA. 2002;98:14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintz M, Reichenberg A, Altincicek B, Bahr U, Gschwind RM, Kollas AK, Beck E, Wiesner J, Eberl M, Jomaa H. Identification of (E)-4-hydroxy-3-methyl-but-2-enyl pyrophosphate as a major activator for human γδ T cells in Escherichia coli. FEBS Lett. 2001;509:317–322. doi: 10.1016/s0014-5793(01)03191-x. [DOI] [PubMed] [Google Scholar]
- Julliard JH. Biosynthesis of the pyridoxal ring (vitamin B6) in higher plant chloroplasts and its relationship with the biosynthesis of the thiazole ring (vitamin B1) CR Acad Sci Ser III. 1992;314:285–290. [Google Scholar]
- Julliard JH, Douce R. Biosynthesis of the thiazole moiety of thiamin (vitamin B1) in higher plant chloroplasts. Proc Natl Acad Sci USA. 1991;88:2042–2045. doi: 10.1073/pnas.88.6.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay BK, Williamson MP. The importance of being a proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kuzuyama T, Shimizu T, Takahashi S, Seto H. Fosmidomycin, a specific inhibitor of 1-deoxy-d-xylulose 5-phosphate reductoisomerase in the nonmevalonate pathway for terpenoid biosynthesis. Tetrahedron Lett. 1998;39:7913–7916. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lange BM, Croteau R. Isoprenoid biosynthesis via a mevalonate-independent pathway in plants: cloning and heterologous expression of 1-deoxy-d-xylulose-5-phosphate reductoisomerase from peppermint. Arch Biochem Biophys. 1999;365:170–174. doi: 10.1006/abbi.1999.1168. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Non-mevalonate isoprenoid biosynthesis: enzymes, genes and inhibitors. Biochem Soc Trans. 2000;28:785–789. [PubMed] [Google Scholar]
- Lois LM, Rodríguez-Concepción M, Gallego F, Campos N, Boronat A. Carotenoid biosynthesis during tomato fruit development: regulatory role of 1-deoxy-d-xylulose 5-phosphate synthase. Plant J. 2000;22:503–513. doi: 10.1046/j.1365-313x.2000.00764.x. [DOI] [PubMed] [Google Scholar]
- Lumbreras V, Campos N, Boronat A. The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates a mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with and extended N-terminal region. Plant J. 1995;8:541–549. doi: 10.1046/j.1365-313x.1995.8040541.x. [DOI] [PubMed] [Google Scholar]
- Lluch MA, Masferrer A, Arro M, Boronat A, Ferrer A. Molecular cloning and expression analysis of the mevalonate kinase gene from Arabidopsis thaliana. Plant Mol Biol. 2000;42:365–376. doi: 10.1023/a:1006325630792. [DOI] [PubMed] [Google Scholar]
- Mahmoud SS, Croteau RB. Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc Natl Acad Sci USA. 2001;98:8915–8920. doi: 10.1073/pnas.141237298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, León P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996;9:649–658. doi: 10.1046/j.1365-313x.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- McGarvey DJ, Croteau R. Terpenoid metabolism. Plant Cell. 1995;7:1015–1026. doi: 10.1105/tpc.7.7.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Ahumada I, Diez-Juez E, Sauret-Güeto S, Lois LM, Gallego F, Carretero-Paulet L, Campos N, Boronat A. 1-Deoxy-d-xylulose 5-phosphate reductoisomerase and plastid isoprenoid biosynthesis during tomato fruit ripening. Plant J. 2001;27:213–222. doi: 10.1046/j.1365-313x.2001.01089.x. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Campos N, Lois LM, Maldonado C, Hoeffler JF, Grosdemange-Billiard C, Rohmer M, Boronat A. Genetic evidence of branching in the isoprenoid pathway for the production of isopentenyl diphosphate and dimethylallyl diphosphate in Escherichia coli. FEBS Lett. 2000;473:328–332. doi: 10.1016/s0014-5793(00)01552-0. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Yalovsky S, Zik M, Fromm H, Gruissem W. The prenylation status of a novel plant calmodulin directs plasma membrane or nuclear localization of the protein. EMBO J. 1999;18:1996–2007. doi: 10.1093/emboj/18.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohdich F, Hecht S, Gärtner K, Adam P, Krieger C, Amslinger S, Arigoni D, Bacher A, Eisenreich W. Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc Natl Acad Sci USA. 2002;99:1158–1163. doi: 10.1073/pnas.032658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Prod Rep. 1999;16:565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]
- Schwender J, Müller C, Zeidler J, Lichtenthaler HK. Cloning and heterologous expression of a cDNA encoding 1-deoxy-d-xylulose-5-phosphate reductoisomerase of Arabidopsis thaliana. FEBS Lett. 1999;455:140–144. doi: 10.1016/s0014-5793(99)00849-2. [DOI] [PubMed] [Google Scholar]
- Seemann M, Campos N, Rodríguez-Concepción M, Hoeffler JF, Grosdemange-Billiard C, Boronat A, Rohmer M. Isoprenoid biosynthesis via the methylerythritol phosphate pathway: accumulation of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate in a gcpE deficient mutant of Escherichia coli. Tetrahedron Lett. 2002a;43:775–778. [Google Scholar]
- Seemann M, Campos N, Rodríguez-Concepción M, Ibáñez E, Duvold T, Tritsch D, Boronat A, Rohmer M. Isoprenoid biosynthesis in Escherichia coli via the methylerythritol phosphate pathway: enzymatic conversion of methylerythritol cyclodiphosphate into a phosphorylated derivative of (E)-2-methylbut-2-ene-1,4-diol. Tetrahedron Lett. 2002b;43:1413–1415. [Google Scholar]
- Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci USA. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D, Ohnuma S, Farooq A, Gershenzon J. Arabidopsis thaliana: a model system for the analysis of terpenoid secondary metabolism? In: Knöss W, editor. 5th European Symposium on Plant Isoprenoids. University of Bonn. 2001. [Google Scholar]
- Veau B, M C, Oudin A, Chénieux J-C, Rideau M, Clastre M. Cloning and expression of cDNAs encoding two enzymes of the MEP pathway in Catharanthus roseus. Biochim Biophys Acta. 2000;1517:159–163. doi: 10.1016/s0167-4781(00)00240-2. [DOI] [PubMed] [Google Scholar]
- von Heijne G, Stepphuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Walter MH, Fester T, Strack D. Arbuscular mycorrhizal fungi induce the non-mevalonate methylerythritol phosphate pathway of isoprenoid biosynthesis correlated with accumulation of the “yellow pigment” and other apocarotenoids. Plant J. 2000;21:571–578. doi: 10.1046/j.1365-313x.2000.00708.x. [DOI] [PubMed] [Google Scholar]
- Wolff M, Seemann M, Grosdemange-Billiard C, Tritsch D, Campos N, Rodríguez-Concepción M, Boronat A, Rohmer M. Isoprenoid biosynthesis via the methylerythritol phosphate pathway. (E)-4-Hydroxy-3-methyl but-2-enyl diphosphate: chemical synthesis and formation from methylerythritol cyclodiphosphate by a cell-free system from Escherichia coli. Tetrahedron Lett. 2002;43:2555–2559. [Google Scholar]
- Zeidler J, Schwender J, Müller C, Wiesner J, Weidemeyer C, Beck E, Jomaa H, Lichtenthaler K. Inhibition of the non-mevalonate 1-deoxy-d-xylulose-5-phosphate pathway of plant isoprenoid biosynthesis by fosmidomycin. Z Naturforsch. 1998;53:980–986. [Google Scholar]