Abstract
Arabidopsis displays circadian rhythms in stomatal aperture, stomatal conductance, and CO2 assimilation, each of which peaks around the middle of the day. The rhythmic opening and closing of stomata confers a rhythm in sensitivity and resistance, respectively, to the toxic gas sulfur dioxide. Using this physiological assay as a basis for a mutant screen, we isolated mutants with defects in circadian timing. Here, we characterize one mutant, out of phase 1 (oop1), with the circadian phenotype of altered phase. That is, the timing of the peak (acrophase) of multiple circadian rhythms (leaf movement, CO2 assimilation, and LIGHT-HARVESTING CHLOROPHYLL a/b-BINDING PROTEIN transcription) is early with respect to wild type, although all circadian rhythms retain normal period length. This is the first such mutant to be characterized in Arabidopsis. oop1 also displays a strong photoperception defect in red light characteristic of phytochrome B (phyB) mutants. The oop1 mutation is a nonsense mutation of PHYB that results in a truncated protein of 904 amino acids. The defect in circadian phasing is seen in seedlings entrained by a light-dark cycle but not in seedlings entrained by a temperature cycle. Thus, PHYB contributes light information critical for proper determination of circadian phase.
Circadian rhythms are endogenous rhythms with periods of approximately 24 h. Circadian systems have been extensively described in cyanobacteria, Neurospora crassa, fruitfly (Drosophila melanogaster), mice, and humans; genetic and molecular biological studies have identified a number of the components of circadian systems in these model organisms (Johnson, 2001; Loros and Dunlap, 2001; Reppert and Weaver, 2001; Williams and Sehgal, 2001). One common theme emerging from these studies is that circadian oscillators are composed of two interconnected feedback loops (Glossop et al., 1999; Lee et al., 2000; Shearman et al., 2000; Alabadí et al., 2001; Denault et al., 2001). Although plants have provided many examples of circadian rhythmic outputs, including photoreceptor gene expression, and the study of the photoreceptors of plant circadian input pathways is well advanced (Casal, 2000; Devlin and Kay, 2001), our understanding of the plant circadian system remains incomplete (Harmer et al., 2001; McClung, 2001; McClung et al., 2002). Moreover, with the determination of the complete sequence of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000), it is evident that no obvious Arabidopsis orthologs to most known clock proteins can be found, demonstrating that at least part of the Arabidopsis clock mechanism is novel.
A number of loci have been implicated in the Arabidopsis circadian clock mechanism. At present, the best characterized are TIMING OF CAB 1 EXPRESSION (TOC1), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), and LATE ELONGATED HYPOCOTYL (LHY; Alabadí et al., 2001; Makino et al., 2002; Matsushika et al., 2002). toc1 loss-of-function mutations shorten the period of multiple rhythms, including leaf movement, stomatal conductance (Somers et al., 1998b), and transcription and mRNA accumulation of all clock-regulated genes examined (Millar et al., 1995a; Kreps and Simon, 1997; Strayer et al., 2000). In plants carrying a strong loss-of-function allele of TOC1 (toc1-2), oscillations of LHY and CCA1 mRNA exhibit greatly reduced mRNA abundance, consistent with a role of TOC1 as a positive regulator of CCA1/LHY (Alabadí et al., 2001). CCA1 and LHY encode single Myb domain transcription factors, which, when overexpressed, result in arrhythmicity of multiple clock outputs, including leaf movement and mRNA abundance of all clock-regulated genes tested to date (Schaffer et al., 1998; Wang and Tobin, 1998; Fowler et al., 1999; Matsushika et al., 2002). Critically, overexpression of either LHY or CCA1 results in nonoscillating low-level accumulation of TOC1 mRNA, indicating that CCA1/LHY act as negative regulators of TOC1 expression. The roles of CCA1 and LHY in oscillator function are thought to be at least partially redundant, because loss of CCA1 function shortens the period of mRNA oscillation in at least three clock-controlled genes, but the plants retain rhythmicity (Green and Tobin, 1999).
In all circadian systems, light acts as a powerful resetting signal to induce or repress the expression of clock genes, and photoreceptor mutants show defects in rhythmicity in constant conditions (Devlin and Kay, 2001). In plants, it has recently become clear that photoperception is itself a circadian output because transcription of both CRYPTOCHROME (CRY) genes and four of five PHYTOCHROME (PHY) genes (PHYC is the sole exception) is clock-regulated (Bognár et al., 1999; Tóth et al., 2001). A number of other components of light input pathways have been identified. For example, early flowering 3 (elf3) mutants are conditionally arrhythmic in continuous light and ELF3, which encodes a novel protein (Hicks et al., 2001), plays a role in gating light signals to the clock (Hicks et al., 1996; McWatters et al., 2000; Covington et al., 2001; Liu et al., 2001). Mutational analysis has identified several other components that may participate in light input to the Arabidopsis circadian clock. Loss of function of ZEITLUPE (ZTL, also called ADAGIO1, LOV KELCH PROTEIN 1 [LKP1], and TOC7) lengthens the period of all the rhythms affected by toc1 (Somers et al., 2000). In addition, leaf movement becomes arrhythmic in red light in the adagio1 mutant (Jarillo et al., 2001). Overexpression of the related LKP2 results in arrhythmicity (Schultz et al., 2001). ZTL family members possess known functional domains arranged in a novel fashion: a single PAS/LOV-domain, an F-box, and six Kelch repeats, suggesting a role for light-regulated protein degradation in clock function (Kiyosue and Wada, 2000; Somers et al., 2000; Jarillo et al., 2001). ZTL (ADO1) physically interacts with both PHYB and CRY1 (Jarillo et al., 2001), although a role in photoreceptor degradation has not been established.
We wished to develop an independent and complementary approach to the identification of components of the Arabidopsis circadian system. Circadian rhythms in stomatal aperture and in the responsiveness of guard cells to environmental stimuli have been described in a number of plants (Webb, 1998). In beans, there is circadian control of the underlying biochemical reactions of the Calvin cycle in addition to control of stomatal aperture and gas exchange (Hennessey and Field, 1991). In accordance, we first established that Arabidopsis exhibits circadian rhythms in stomatal opening and CO2 assimilation. We then demonstrated that these rhythms were correlated with a circadian rhythm in resistance and sensitivity to the air pollutant, sulfur dioxide (SO2). We took advantage of this rhythm to screen a population of mutagenized M2 plants for individuals that exhibited damaged leaves in response to SO2 exposure at a time when wild-type plants were resistant. Included among the mutants identified in this screen are several in which the circadian period is altered (either lengthened or shortened from that seen in wild-type plants) or in which circadian phase is disrupted. We show here that one of the mutants isolated from the screen, out of phase 1 (oop1) exhibits a defect in circadian phase but retains wild-type period length. That is, the peak in several rhythms, including leaf movement, CO2 assimilation, and LIGHT-HARVESTING CHLOROPHYLL a/b BINDING PROTEIN (LHCB) transcription, occurs earlier (phase leads) than is seen in wild type. This phenotype has not been previously described in Arabidopsis. oop1 is a new allele of the red-light photoreceptor PHYB gene; oop1 plants accumulate a truncated PHYB lacking most of the C-terminal kinase domain and behave as strong phyB mutants in red light. Surprisingly, the phase phenotype observed in oop1 mutants is found in phyB-9 mutants as well, indicating that it is the loss of PHYB function that confers the phase alteration. A blue-light-dependent enhancement of the hypocotyl phenotype seen in oop1 mutants indicates an alteration of CRY1 signaling, although this is not seen in phyB-9 seedlings and so does not seem to contribute to the phase alteration.
RESULTS
Arabidopsis Displays Circadian Rhythms in Stomatal Aperture and CO2 Assimilation
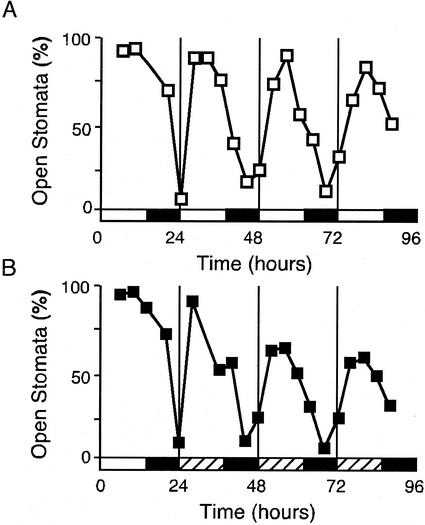
Direct microscopic examination of epidermal peels from rosette leaves of Columbia (Col) plants grown under a 14-h:10-h light-dark cycle for 4 weeks revealed a diurnal oscillation in stomatal opening (Fig. 1A), as has been described in many species (Webb, 1998). The proportion of open stomata is greatest during the middle of the light period and is least late in the dark period. This rhythm persists in continuous conditions (extended dark) and, therefore, is under circadian control (Fig. 1B). This rhythm in stomatal aperture confers a rhythm in stomatal conductance in continuous light (data not shown), as has been described elsewhere (Somers et al., 1998b).
Figure 1.
Circadian rhythm in stomatal aperture in Arabidopsis. A, Epidermal peels were prepared from Col plants grown in a 14-h:10-h light-dark cycle. B, As in A, but plants were grown in a 14-h:10-h light-dark cycle and transferred into extended darkness. Hatched bars indicate subjective day. Each peel was scored by microscopy for the percentage of open stomates. Each point represents the mean of duplicate samples, each of at least 100 stomates.
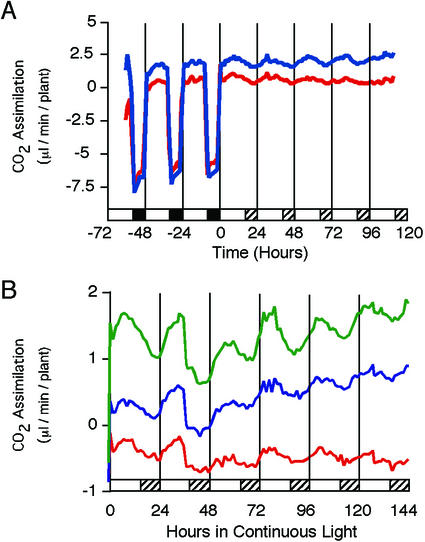
These oscillations in stomatal aperture and stomatal conductance are correlated with a rhythm in CO2 assimilation (Fig. 2). Under entraining light-dark conditions the rate of CO2 assimilation is high throughout the light period and becomes negative as a result of respiration in the dark (Fig. 2A). CO2 assimilation responds immediately to the onset and offset of illumination. We attribute the apparent increase in CO2 fixation occasionally observed before light onset to an artifact of our experimental system in which the lights went on during the sampling interval, yielding an intermediate rate of CO2 assimilation, rather than interpreting this as evidence of dawn anticipation. Upon release into continuous light (Fig. 2B), the rate of CO2 assimilation continues to oscillate, exhibiting a circadian rhythm with a period of 23.59 ± 0.23 h (mean ± se, n = 20).
Figure 2.
The rhythm in stomatal aperture correlates with a rhythm in CO2 fixation. A, Rates of CO2 exchange of representative individual 5- to 6-week-old Col plants grown in a 16-h:8-h light-dark cycle and transferred into continuous light. Hatched bars indicate subjective night. For the first 3 d, plants were maintained under entraining conditions and, therefore, show respiration during the night. B, Rates of CO2 exchange of several representative individual Col plants after transfer into continuous light. Hatched bars indicate subjective night.
Circadian Rhythm in Resistance to SO2
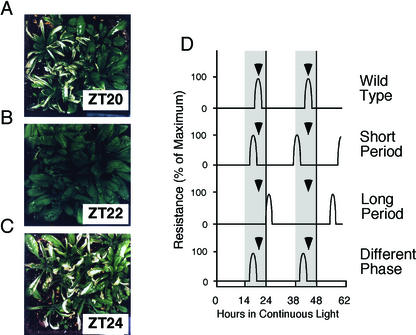
Most gas exchange occurs through stomata, and closed stomata render plants more tolerant to toxic gases (Mansfield and Freer-Smith, 1984). We asked whether the circadian rhythms in stomatal opening and conductance could generate a circadian rhythm in the tolerance of Arabidopsis to SO2. Plants were grown in soil in a 14-h:10-h light-dark cycle for 3 to 4 weeks and transferred into continuous light. Replicate pots of plants were exposed to SO2 at 2-h intervals over 48 h in continuous light. Plants exposed to SO2 when the stomata were open developed pronounced necrotic leaf lesions (Fig. 3, A and C), whereas plants exposed when the stomata were maximally closed exhibited no visible necrosis (Fig. 3B). Plants were resistant only when treated in a narrow temporal window of approximately 1-h duration, 2 h before subjective dawn. Plants treated 2 h earlier, 2 h later, and at all other times were sensitive and developed necrotic lesions. The circadian oscillation in resistance persisted for at least 2 d in continuous light (data not shown). Because the rhythm in CO2 assimilation persists for as long as 6 d in continuous light (Fig. 2B), we expect the rhythm in SO2 resistance to persist after 48 h; however, this has not been experimentally determined.
Figure 3.
Arabidopsis exhibits a rhythm in sensitivity and resistance to SO2. Col plants were grown in a 14-h:10-h light-dark cycle, transferred into continuous light, and then treated with SO2 for 30 min each time, at 2-h intervals. A, Plants gassed 4 h before subjective dawn (ZT20, where ZT refers to Zeitgeber Time, defined as the number of h after the onset of illumination). B, Plants gassed 2 h before subjective dawn (ZT22). C, Plants gassed at subjective dawn (ZT24). D, Diagrammatic representation of the screening strategy for putative clock mutants based on resistance to SO2. Wild-type (WT) plants are resistant to the treatment only approximately 2 h before subjective dawn (ZT22 and ZT46) and, thus, do not develop lesions when treated with SO2 at ZT46. A short-period mutant would already have advanced through the period of resistance and become sensitive because of its open stomata. A long-period mutant would not yet have reached the tolerance period (closed stomata) and would also be sensitive to SO2. Phase mutants would display resistance to SO2 with a WT period, but the timing of the window of resistance is shifted earlier (as shown) or later than that of Col plants. Plants were grown in a 12-h:12-h light-dark cycle and transferred into continuous light at T = 0; the gray area indicates subjective night.
Such a narrow window of resistance to SO2 suggested an easy and sensitive physiological screen to identify mutants with altered circadian properties. Col plants grown in a light-dark cycle and transferred into continuous light would be resistant to a SO2 treatment 2 h before subjective dawn (Fig. 3B). Any mutant unable to control properly its circadian rhythm in stomatal opening would, thus, develop necrotic lesions similar to those shown in Figure 3, A and C. This approach should allow the identification of both short- and long-period mutants in a single screening protocol (Fig. 3D). A short-period mutant would already have advanced through the period of tolerance and become sensitive because of its open stomata. A long-period mutant conversely would not yet have reached the tolerance period (closed stomata) and would also be sensitive to SO2. Moreover, mutants in which the time of day of maximal stomatal closure and, hence, maximal SO2 resistance has been shifted (i.e. circadian phase is altered) would also be expected to develop necrotic lesions.
M2 populations of ethyl methanesulfonate-mutagenized Col plants grown in a 14-h:10-h light-dark cycle and transferred into continuous light were treated with SO2 2 h before subjective dawn on the 2nd d after transfer, when wild-type plants are resistant to SO2. Plants that developed necrotic lesions were allowed to regrow from undamaged young leaves and to set seeds. The screen involved 6,500 ethyl methanesulfonate-mutagenized plants from 13 independent M2 pools. Approximately 100 putative mutants were isolated that developed necrotic lesions when wild type did not. Of these, approximately one-third developed necrotic lesions without exposure to SO2 and so were not studied further.
The CO2 assimilation patterns of the remaining 65 mutants were analyzed to identify those with altered circadian rhythm properties. Twelve mutants with altered period lengths and/or altered phase relationships were found and fell into the three expected classes (long period, short period, and altered phase) of mutants. The period mutants have been designated circadian timing defective. The remaining mutants that exhibited wild-type period lengths in CO2 assimilation rhythm but in which the phase of the peak of CO2 assimilation was either earlier (phase leading) or later (phase lagging) than in wild type have been designated out of phase (oop). Because no mutants with this phenotype (leading circadian phase but wild-type period) have yet been described in Arabidopsis, one of these, oop1, was chosen for further characterization.
Characterization of oop1, a Circadian Phase Mutant of Arabidopsis
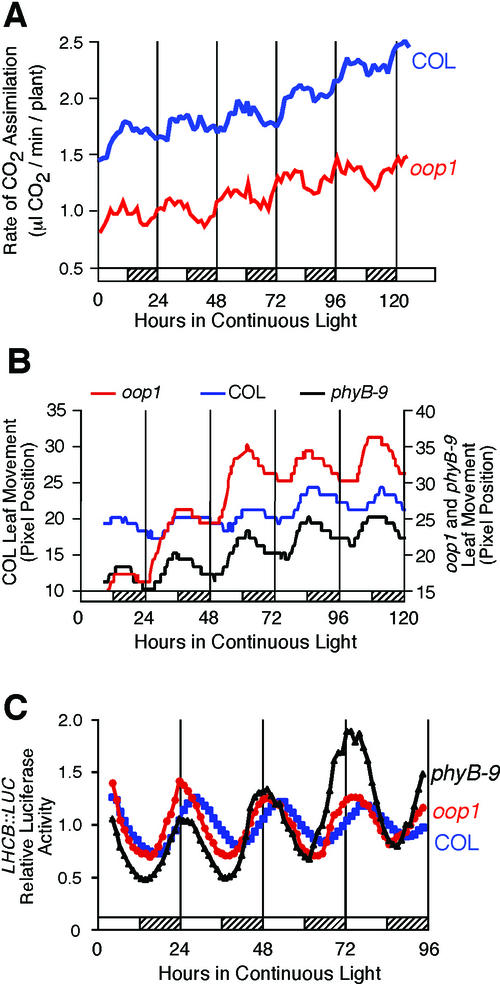
We compared rhythmicity of oop1 with wild-type Col plants in terms of both period and phase of the peak (acrophase; Fig. 4; Table I). To facilitate comparison of phase among rhythms with slightly different periods, phase values were normalized to the period length of the rhythmic trace and are recorded in circadian time (CT = phase/period × 24 h). The period of the CO2 assimilation rhythm in oop1 was similar to that of the wild-type Col; in contrast, the acrophase (peak) of the rhythm occurred 3.4 h earlier than in Col (Fig. 4A; Table I). This difference in phase of CO2 assimilation was significant (Student's two-tailed heteroscedastic t test, P = 0.0036).
Figure 4.
The oop1 mutation causes an altered phase in circadian rhythms. A, Net CO2 fixation levels in oop1 and Col. oop1 and Col plants were grown for 4 weeks in a 12-h:12-h light-dark cycle and transferred into continuous light. CO2 assimilation levels were recorded for 6 d. B, Cotyledon movement in Col and oop1 seedlings. oop1 and Col seedlings were grown for 4 to 5 d in a 12-h:12-h light-dark cycle and transferred to 24-well cloning plates, one seedling per well, in continuous light. Cotyledon movement was recorded for 7 d. C, LHCB::LUC transcription. Col, oop1, and phyB-9 seedlings homozygous for the LHCB::LUC transgene were grown in a 12-h:12-h light-dark cycle and transferred into continuous light at T = 0. Luciferase activity was recorded from each group of seedlings after transfer into continuous light and temperature conditions and is presented as the average of multiple seedlings (see Table I) for four complete circadian cycles. Hatched bars indicate subjective night. Col, Blue squares; oop1, red circles; and phyB-9, black triangles.
Table I.
Alterations in circadian phase in oop1 and phyB-9 mutants
| Rhythm | Genotype | Period | Phasea |
|---|---|---|---|
| h | CT h | ||
| CO2 | Col | 22.20 ± 0.52 | 13.58 ± 1.50 (n = 5) |
| Assimilation | oop1 | 22.80 ± 0.36 (P = 0.0667)b | 10.15 ± 1.15 (n = 5; P = 0.0036) |
| Leaf | Col | 24.24 ± 0.90 | 17.32 ± 1.54 (n = 13) |
| Movement | oop1 | 24.30 ± 0.54 (P = 0.8007) | 13.69 ± 1.62 (n = 24; P < 0.0001) |
| phyB-9 | 23.72 ± 1.02 (P = 0.2213) | 15.15 ± 3.39 (n = 9; P = 0.0548) | |
| LHCB::LUC | Col | 24.53 ± 0.88 | 3.81 ± 2.15 (n = 7) |
| oop1 | 24.22 ± 0.80 (P = 0.4104) | 1.93 ± 1.91 (n = 17; P = 0.0459) | |
| phyB-9 | 24.23 ± 0.74 (P = 0.3830) | 1.32 ± 2.14 (n = 21; P = 0.0131) |
Values are mean ± sd.
Phase values are normalized to the period length and are given in circadian time (CT = phase/period × 24 h).
P value from Student's two-tailed heteroscedastic t test comparing the mutant value with Col is given in parentheses. The oop1 and phyB-9 values were not significantly different within any treatment.
Leaf movement allows us to look at an unrelated clock-controlled output. Although the periods of the rhythms in Col and oop1 were not significantly different, the acrophase of the cotyledon movement in oop1 occurred 3.6 h earlier than in Col (Fig. 4B; Table I), consistent with the results seen for CO2 assimilation. Again, the difference in circadian phase between Col and oop1 was statistically significant (Student's two-tailed heteroscedastic t test, P < 0.0001).
We also determined the effect of the oop1 mutation on transcription of a luciferase transgene driven by the LHCB 1*1 (CAB2) promoter (LHCB::LUC). Luciferase activity was recorded over 4 d in continuous light after 7 d of entrainment in a 12-h:12-h light-dark (Fig. 4C; Table I). Again, the period of the rhythm in LHCB transcription was not statistically different in oop1 versus Col seedlings. However, the phase of the rhythm in LHCB transcription was 1.9 h earlier, and this difference was statistically significant (Student's two-tailed heteroscedastic t test, P = 0.0459).
These rhythms are measured at different stages of development: Leaf movement and LHCB transcription were recorded on 5- to 12-d-old seedlings, whereas CO2 assimilation was measured on 4- to 5-week-old plants. Thus, the oop1 mutation affects multiple rhythmic outputs at different developmental stages. Moreover, the oop1 mutation alters the phase of rhythms that display distinct circadian phases. The period of the rhythm in CO2 assimilation was shorter than that for leaf movement or LHCB transcription. We suspect that differences in the growth conditions, possibly including the light conditions, as well as the age of the plants for each assay, may account for the variation in period lengths for the measured rhythms (Table I; for details, see “Materials and Methods”).
oop1 Is Primarily Impaired in Red-Light Photoperception
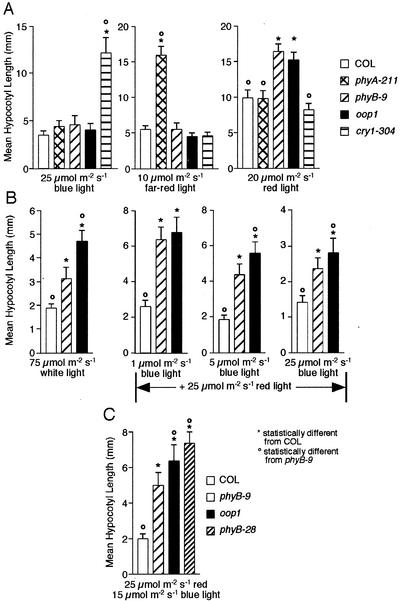
Initial observations of the oop1 mutant suggested that its hypocotyl was longer than that of wild-type seedlings when grown under continuous white light. Mutant seedlings were, thus, grown under different light qualities (100 μmol m−2 s−1 white, 20 μmol m−2 s−1 red, 25 μmol m−2 s−1 blue, and 10 μmol m−2 s−1 far-red) to better define the response of oop1 seedlings to light. Hypocotyls of oop1 seedlings are much longer than Col under white and red light but not under blue or far red light (Fig. 5, A and B), indicating that oop1 is primarily impaired in red-light photoperception through PHYB. The extent of the hypocotyl elongation phenotype seen in oop1 under red light is similar to that of phyB-9, a null allele of PHYB in the Col background (Reed et al., 1993), suggesting that the oop1 mutation eliminates PHYB-dependent signaling. In white light, the oop1 hypocotyl phenotype was inherited as a single recessive Mendelian locus. As with phyB loss of function alleles, in red light, the hypocotyl length of seedlings heterozygous for oop1 is intermediate between wild-type and oop1 homozygotes (data not shown), suggesting semidominant or incompletely recessive inheritance of the oop1 mutation.
Figure 5.
The oop1 mutant is primarily impaired in red-light perception. A, oop1 was grown in continuous blue, far-red, or red light for 5 d before hypocotyl length was measured with NIH Image v1.62. The known photoreceptor mutants phyA-211, phyB-9, and cry1–304 were also used as controls for loss of PHYA-, PHYB-, or CRY1-mediated light perception, respectively. Hypocotyl length (mean ± sd) is given for each genotype and treatment. B, Hypocotyl phenotype of oop1 and phyB-9 in response to combinations of red and blue lights. Col, oop1, and phyB-9 seedlings were grown as described in A, under 25 μmol m−2 s−1 red light combined with 1, 5, or 25 μmol m−2 s−1 blue light. C, Enhancement of hypocotyl phenotype in oop1 and phyB-28. Col, oop1, phyB-9, and phyB-28 seedlings were grown as described in A, under a combination of 25 μmol m−2 s−1 red light and 15 μmol m−2 s−1 blue light. *, Hypocotyl length is significantly different (Student's two-tailed heteroscedastic t test, P < 0.001) from Col; °, hypocotyl length is significantly different (P < 0.001) from phyB-9.
The oop1 Locus Is PHYB
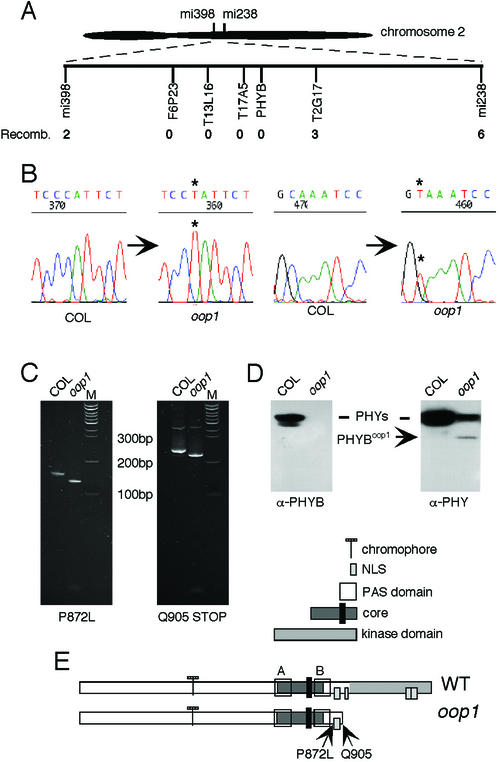
Cloning the mutated gene responsible for the phase defect seen in several rhythms was the next step in trying to understand the molecular determinants of phase regulation in Arabidopsis. For low-resolution mapping, 41 lines homozygous for the oop1 mutation were selected from the F2 progeny of a cross of oop1 to Landsberg erecta, based on their long hypocotyl in red light. Using CAPS, RFLP, and simple sequence length polymorphism markers, the oop1 mutation was located centromere proximal, on the bottom arm of chromosome 2 (Fig. 6A). A large region, from 29 to 36 cM, was identified in which no recombinant chromosomes could be detected. Of particular interest to us was the fact that PHYB lies at 34.46 cM within this region. We determined the genomic sequence of PHYB in the oop1 background. As shown in Figure 6B, two mutations were found on two overlapping PCR products, a missense mutation (P872L), and a nonsense mutation, introducing a stop codon after amino acid 904 (Q905X). These mutations were confirmed by sequencing independent PCR amplification products from independent DNA preparations and with the creation of mutation-specific dCAPS markers (Fig. 6C).
Figure 6.
oop1 is a new allele of PHYB. A, Map position of the oop1 locus. The number of recombinant chromosomes among a population of 41 plants homozygous for the oop1 mutation is indicated for each marker. mi398 and mi238 are RFLP markers at 29.27 and 39.02 cM, respectively. B, Mutations in the PHYBoop1 gene. Sequencing results from Col and oop1 are shown. The positions of the mutations are highlighted by asterisks. C, dCAPS analysis of PHYB in oop1. Two dCAPS markers were developed, one specific to each mutation. The PCR products were amplified from Col and oop1 DNA and digested with StyI for the P872L mutation and BsrGI for the Q905X mutation. The restriction digests were run on a 10% (w/v) acrylamide gel and stained with ethidium bromide. Left, dCAPS results obtained for the P872L mutation. Right, Q905X mutation. M indicates the 100-bp Plus ladder (MBI Fermentas, Hanover, MD). D, Western-blot analysis of oop1 plants. Left, The membrane was probed with a PHYB-specific monoclonal antibody that recognizes an epitope in the carboxy terminus, which is not retained in oop1. This antibody fails to detect full-length PHYB protein in oop1. Right, The membrane was probed with a monoclonal antibody raised against a conserved epitope in the central region of the molecule, which recognizes all phytochromes; the oop1 sample shows a smaller protein species of the Mr predicted for a PHYB protein comprising the first 904 amino acids only. E, Maps of PHYB and PHYBoop1. The functional domains of PHYB are indicated, as well as the position of the two mutations in oop1. The Q905X mutation introduces a premature stop codon at amino acid 904 and causes the loss of the His-kinase related domain and three of the four putative nuclear localization sequences.
PHYBoop1 is predicted to encode a truncated protein of an apparent molecular mass of approximately 95 kD. This truncation would eliminate the C terminus of phytochrome, which is the region with most divergence among PHYs. The PHYB-specific monoclonal antibody B6B3 (Hirschfeld et al., 1998) fails to detect any PHYB protein in oop1 (Fig. 6D), presumably because the epitope to this antibody is C-terminal to the oop1 truncation. However, a second monoclonal antibody (3B5), which recognizes all five Arabidopsis PHYs (Hirschfeld et al., 1998), detects in oop1 homozygotes both a reduced quantity of full-length PHYs and a peptide of smaller Mr than a full-length PHY protein (Fig. 6D). This protein has the predicted approximately 95-kD size for a truncated PHYBoop1 protein lacking the last 268 amino acids. The large C-terminal truncation may reduce protein stability and contribute to the low levels of the PHYB photoreceptor in oop1. We, therefore, conclude that oop1 is a new allele of the PHYB gene encoding the red-light photoreceptor PHYB and that the oop1 mutant accumulates low levels of a truncated PHYB lacking most of the kinase domain (Fig. 6E).
Altered Phase in LHCB::LUC Transcription Is a General Property of phyB Mutants
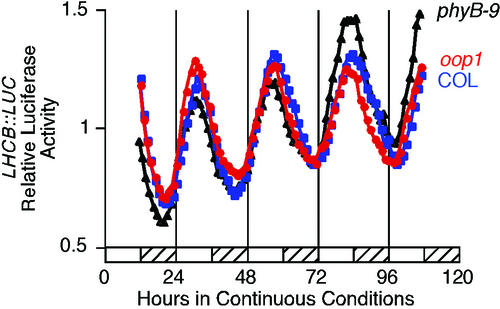
The isolation of a new phyB allele that confers altered phasing of several circadian rhythms was unexpected. The role of PHYB has been well documented for the establishment of the proper period length in red light and in entrainment of the circadian clock. In intermediate and high-fluence red light, loss of PHYB function lengthens the period, whereas PHYB overexpression shortens the period (Somers et al., 1998a; Devlin and Kay, 2000, 2001). Little has been reported on the effect of phyB mutations on circadian rhythms under white light. In white light, overexpression of PHYB does not shorten nor does loss of PHYB function lengthen the period for LHCB transcription (D.E. Somers, personal communication), consistent with our observations that neither oop1 nor phyB-9 confer lengthened period in white light. Blue-light signaling through PHYA, CRY1, and CRY2 also is important in the establishment of period length (Somers et al., 1998a; Devlin and Kay, 2000, 2001), and we suspect that blue-light signaling is sufficient to establish wild-type period length in oop1 under white light. We have considered two explanations of the leading (early) phase observed in oop1. The truncated PHYB protein accumulating in oop1 could display dominant negative interference with another signal transduction pathway and, together with the loss of PHYB activity, cause the leading phase. As an alternative, the phase alteration could be a general feature of phyB mutants. In accordance, we crossed a LHCB::LUC transcriptional fusion into the oop1 and phyB-9 mutant backgrounds. Luciferase activity was recorded over 4 d in continuous light after 7 d of entrainment in a 12-h:12-h light-dark cycle. F3 seedlings homozygous for oop1 LHCB::LUC and phyB-9 LHCB::LUC show the same leading phase when compared with seedlings of their respective wild types (Fig. 4C; Table I). This is consistent with the oop1 mutant phenotype resulting from the mutation of the PHYB locus as opposed to resulting from a mutation in a second locus tightly linked to PHYB. Moreover, because phyB-9 is a complete loss-of-function allele (Reed et al., 1993), it is the loss of PHYB function in white light that is responsible for the leading phase of LHCB::LUC transcription and of leaf movement (Table I). Importantly, this phase alteration is not seen in oop1 or phyB-9 seedlings entrained by a 12-h:12-h temperature cycle of 12°C: 22°C (Fig. 7; Col phase = 3.30 ± 3.27 CT h [n = 9], oop1 phase = 3.14 ± 2.75 CT h [n = 19], and phyB-9 phase = 2.83 ± 2.46 CT h [n = 24]). This shows that the phase alteration is solely due to the loss of photoperception through PHYB and does not indicate a defect in the clock oscillator, which retains a wild-type period.
Figure 7.
Phase alteration of LHCB::LUC transcription in oop1 is not seen after entrainment by temperature cycles. Col and oop1 seedlings homozygous for the LHCB::LUC transgene were grown for 7 d under entraining conditions consisting of 12 h at 22°C followed by 12 h at 12°C. Luciferase activity was recorded from each group of seedlings after transfer into continuous light and temperature conditions and is presented as the average of multiple seedlings. Col, Blue squares; oop1, red circles; and phyB-9, black triangles. The hatched bars represent the subjective cold (12°C) period of the day.
PHYBoop1 Affects Blue-Light Photoperception
When grown in white light, oop1 seedlings displayed a longer hypocotyl than phyB-9 seedlings (Fig. 5B), raising the possibility that oop1 may affect more than PHYB signaling in white light. We grew the seedlings under a combination of red or far-red light (25 μmol m−2 s−1) plus blue light at 1, 5, or 15 μmol m−2 s−1. Although no differences between oop1 and phyB-9 could be seen in combined far-red plus blue lights (data not shown), hypocotyls of oop1 are significantly longer than those of Col, cry1-304, and phyB-9 when seedlings are grown in mixtures of red and blue lights. This effect was observed at 5 and 25 μmol m−2 s−1 blue light, but not at 1 μmol m−2 s−1 blue light (Fig. 5B).
Because the enhancement of the hypocotyl phenotype is seen only when red light and fluence rates of blue light 5 μmol m−2 s−1 and higher are combined, we conclude that oop1 causes a red-light-dependent alteration of CRY1 signaling. This does not reflect changes in CRY1 protein levels, which are unaltered in oop1 plants (data not shown). Although CRY2 has been shown to interact with PHYB in vivo (Más et al., 2000), CRY1 is the primary blue-light photoreceptor at high-fluence rates, whereas CRY2 and PHYA are both important in the perception of blue light at low-fluence rates (Lin, 2000). A similar enhancement of hypocotyl elongation was seen in the phyB-28 mutant (Fig. 5C), which lacks most of the kinase domain due to a frame shift at amino acid 991 that results in the premature termination of the protein at amino acid 995 (Krall and Reed, 2000). The phenotype in combined red plus blue lights was even more pronounced than in oop1, presumably because phyB-28 accumulates more truncated PHYB than oop1 (Krall and Reed, 2000). Because oop1 and phyB-28 were isolated from two independent screens and show the same phenotype with respect to hypocotyl elongation in combined lights, we believe this phenotype to be due to an interference with another signaling pathway, possibly originating from CRY1, rather than to the presence of a second, linked mutation in the oop1 background. Consistent with the oop1 mutation interfering with blue-light signaling through CRY1, oop1 plants grown in short day conditions display elongated internodes before the transition to reproductive meristem has occurred (data not shown). This phenotype is similar to, although not as extreme as, that seen in phyB cry1 double mutants grown in the same conditions (Casal and Mazzella, 1998).
DISCUSSION
The oop1 phenotype in which the phases but not the periods of multiple rhythms are affected in white light is unusual and may allow insight into the mechanisms of phase determination. oop1 is a new allele of the red-light photoreceptor PHYB gene. This represents the first report of PHYB as a regulator of circadian phase, which adds to its known involvement in controlling the period of the clock in red light (Somers et al., 1998a; Devlin and Kay, 2000, 2001). oop1 plants accumulate a truncated form of PHYB that lacks the C-terminal Ser/Thr kinase domain (Yeh and Lagarias, 1998), but that retains the two PAS domains, which mediate protein-protein interactions (Lindebro et al., 1995; Ni et al., 1998). Elongation of hypocotyls in red light shows that oop1 is a putative null allele of PHYB in red light, which suggests that this truncated PHYB protein is not active. The characterization of phyB-9, a known null PHYB allele (Reed et al., 1993), shows that loss of PHYB function affects the phase of LHCB::LUC transcription. The similar phase alteration caused by these two phyB mutations suggests that the truncated photoreceptor accumulating in oop1 is not responsible for the circadian phenotype but rather, it is the loss of PHYB signaling that is primarily responsible for changing the phase.
At least three possible mechanisms may explain the phase alteration seen in oop1 and phyB-9. PHYB signaling could affect light input to the clock. As an alternative, PHYB signaling could directly affect levels of a critical clock component. Finally, PHYB signaling could affect a component of an output pathway acting downstream of the oscillator. Because the period length is unaffected by the oop1 or phyB-9 mutations, we conclude that oscillator function is not affected, and it is unlikely that an oscillator component is the direct target of PHYB signaling. However, our data are inadequate to distinguish between a defect in light input to the clock and light modulation of an output component. It is often difficult to distinguish between input and output pathways based solely on an altered clock parameter like period or phase (Roenneberg and Merrow, 1998; Foster and Lucas, 1999; Merrow et al., 1999). As noted above, the expression of Arabidopsis PHY and CRY photoreceptor genes oscillates, indicating that the ability to perceive light (a clock input) is modulated by the clock and, hence, is a clock output (Bognár et al., 1999; Tóth et al., 2001). However, we do note that because the phase of multiple rhythms (SO2 resistance, CO2 assimilation, leaf movement, and LHCB::LUC transcription) is affected in oop1 and phyB-9 mutants, if PHYB affects clock output, then the defect must occur at an output component that is common to all of these output pathways, clock-proximal to any branch points that distinguish these output pathways.
Both light and temperature signaling can entrain the circadian clock (Somers et al., 1998a; Devlin and Kay, 2000, 2001). The altered phase in the oop1 mutant is only evident in response to an entraining light-dark cycle and is not detected after entrainment to temperature cycles. Temperature signaling, which is not affected in oop1 plants, apparently compensates for the impairment in red-light signaling and is sufficient for the establishment of proper phase in both oop1 and phyB-9 plants after entrainment by temperature cycles. However, in the temperature cycles used in Figure 7, the seedlings are grown under continuous light and one might expect that the light signaling defect of oop1 and phyB-9 should be evident as a phase-angle alteration. Nonetheless, under these conditions the phase of LHCB transcription is identical in all genotypes. This argues that the oop1 (phyB) defect does not affect parametric entrainment under continuous light but, rather, that PHYB is required to establish the wild-type phase relationship (phase angle) during discrete entrainment to light-dark cycles.
Loss of PHYB signaling would be expected to attenuate light input to the clock. This would reduce the acute induction of a light-induced clock component at dawn (lights on) or, conversely, delay the light-regulated degradation of some critical clock component. In either case, the timing of the acrophase after dawn would be affected. One known target of PHYB signaling is PHY INTERACTING FACTOR 3 (PIF3), a bHLH-PAS transcription factor that binds to G-box motifs (Ni et al., 1998). Upon illumination with red light, PHYB relocalizes from the cytoplasm into the nucleus (Kircher et al., 1999; Yamaguchi et al., 1999) where it binds directly to PIF3 and modulates transcriptional activity (Ni et al., 1999; Martínez-García et al., 2000). Transcription of two known clock components, CCA1 and LHY, is activated by PHYB through PIF3 bound to G-boxes in their promoter regions, and in PIF3 antisense lines, the induction of both CCA1 and LHY is attenuated (Martínez-García et al., 2000). We suggest that the loss of PHYB-mediated induction of CCA1 and LHY at dawn might confer the phase defect seen in oop1 and phyB-9 plants. The mutant phase leads (precedes) the phase seen in wild-type plants, which suggests that the PHYB/PIF3 target should function as a negative clock element. This is consistent with the demonstrated role of CCA1 as a negative regulator of TOC1 transcription (Alabadí et al., 2001). Thus, we suggest that PHYB and PIF3 may constitute an important phase-determination pathway providing input to either the Arabidopsis clock or to a downstream output component. This pathway need not be exclusive to PIF3 signaling and may include one or more of the many PIF3 relatives found in Arabidopsis. PHYA also provides light input through PIF3 to induce CCA1 expression (Tepperman et al., 2001). CCA1 and LHY expression is still detectable to levels close to wild type in oop1 and phyB-9 (data not shown), suggesting that induction of these putative clock components is not exclusively mediated by PHYB. It is interesting to note that both PHYB and PIF3 contain PAS domains, as do the ZTL/LKP2/FKF1 proteins that are implicated in light-regulated protein degradation in the Arabidopsis circadian system (Kiyosue and Wada, 2000; Nelson et al., 2000; Somers et al., 2000; Schultz et al., 2001). The PAS domain features prominently in other clock-associated light input pathways including that of N. crassa, where the WHITE COLLAR proteins and VIVID (VVD) contain PAS domains (Heintzen et al., 2001; Loros and Dunlap, 2001; Merrow et al., 2001).
Phase mutants have been described in several other clock systems. In fruitfly, two phase-angle (psi-2 and psi-3) mutants dramatically alter the phase of the eclosion rhythm, although the role of these genes in the establishment of phase remains unknown (Jackson, 1983). In N. crassa, mutation of the vvd locus alters the phase of the rhythm in FREQUENCY expression (Heintzen et al., 2001). Deletion of a calcium/calmodulin-dependent protein kinase, camk-1,confers both a lagging phase and slight period lengthening in N. crassa (Yang et al., 2001). In cyanobacteria, inactivation of cikA, which encodes a bacteriophytochrome, alters circadian phase but also shortens the period length (Schmitz et al., 2000). Thus, PHYB is analogous to VVD and CikA in playing a role in the determination of both period and phase.
Although the circadian phase defect seen in the oop1 mutant probably reflects simple loss of PHYB signaling, the observed hypocotyl phenotypes of oop1 and other phyB mutants suggests interactions with other light-signaling pathways. Through its PAS domains, PHYBoop1 may be able to interact with at least some PHYB-interacting factors, but would be unable to transduce the light signal to them, because it lacks all of the kinase domain. The enhancement of hypocotyl elongation seen in red plus higher intensities of blue light suggests that PHYBoop1 and PHYBphyB-28 interact with a CRY1 signaling pathway, possibly through the titration of some common interacting protein. Because PHYBoop1 lacks the three nuclear localization sequences shown to be sufficient for nuclear translocation of a GUS::PHYB898–1172 fusion (Sakamoto and Nagatani, 1996), we predict that these potential interactions are cytosolic. PIF3 is constitutively nuclear (Ni et al., 1998) and is, therefore, unlikely to be a partner for this aberrant PHYBoop1 interaction. ELF3 affects light input to the circadian clock (McWatters et al., 2000) and light-mediated inhibition of hypocotyl elongation in both red and blue wavelengths (Zagotta et al., 1996). In addition, ELF3 interacts directly with PHYB (Liu et al., 2001). However, ELF3 is a nuclear protein (Liu et al., 2001), so it is unlikely that PHYBoop1 titrates ELF3. PHY KINASE SUBSTRATE 1 (PKS1) is cytosolic (Fankhauser et al., 1999), but PKS1 overexpressing or antisense lines show no modified sensitivity to blue light, indicating that PKS1 is unlikely to be the relevant target of PHYBoop1. PKS2 (Munich Information Center for Protein Sequences no. At1g14280), a protein 55% identical to PKS1, is predicted to be cytosolic and might represent such in interactor. As an alternative, PHYBoop1 could physically interact with CRY1. Labeling studies showed a red-light-dependent, far-red-light reversible phosphorylation of CRY1, which could indicate that PHYB may interact with CRY1 in vivo (Ahmad et al., 1998).
It has been clear for some time that PHYB plays multiple important roles in light regulation of multiple developmental processes. The characterization of oop1, a new allele with a truncation of the carboxy terminus of PHYB, has revealed two previously unknown roles of PHYB: a potential interaction with CRY1-mediated blue-light regulation of hypocotyl elongation, and an important contribution to white-light-mediated phase determination of the circadian clock.
MATERIALS AND METHODS
Plant Growth and Genotypes
Plants were sown on standard soil mixture (Pro-mix “BX,” Premier, Rivière-du-Loup, Canada) and watered with Arabidopsis nutrient solution (Somerville and Ogren, 1982). Plants were germinated and grown under fluorescent white light (model TL-741, Philips, Somerset, NJ). The wild-type ecotype was Col-2 (Col, ABRC no. CS907, Ohio State University, Columbus). Several null alleles of photoperception mutants were used in this study. phyA-211, phyB-9, and cry1-304 are null alleles of PHYA, PHYB, and CRY1, respectively, and all are in the Col ecotype. The cry1-304 seeds were a gift of C. Lin (University of California, Los Angeles).
Determination of Stomatal Opening, Stomatal Conductance, and Measurement of CO2 Assimilation
Epidermal peels were prepared from Col plants grown for 3 weeks under approximately 100 μmol m−2 s−1 in a 14-h:10-h light-dark cycle or grown in a light-dark cycle and transferred into extended darkness. Stomatal opening was scored by microscopic inspection. For each time point, at least 10 fresh peels were made and at least 10 stomata per peel (at least 100 stomata total) were scored as either open or closed. Each point in Figure 1 represents the mean of duplicate samples. Stomatal conductance was measured with a photosynthesis system (CI-301PS, CID, Vancouver, WA).
For the measurement of CO2 assimilation, plants were grown under 100 μmol m−2 s−1 white light in a 12-h:12-h light-dark cycle (Fig. 4A) or in a 16-h:8-h light-dark cycle (Fig. 2) at 20°C to 22°C in soil in 50-mL conical tubes from which the tips had been removed to permit bottom watering. The soil was initially wetted with a nutrient solution (Somerville and Ogren, 1982) and plants were subsequently bottom watered. After approximately 1 week of growth, seedlings were thinned to one per tube. After 3 to 4 weeks, individual plants in their conical tubes were transferred to 100-mL beakers containing approximately 50 mL of deionized water. The upper surface of the beakers was sealed with laboratory film (Parafilm, American National Can, Greenwich, CT) to reduce evaporation. The beakers and the plants were then placed individually into 1-quart Mason jars that were individually attached to Micro Oxymax Respirometers (Columbus Instruments, Columbus, OH). The plants were further entrained in 12-h:12-h light-dark cycle (Fig. 4A) or in 16-h:8-h light-dark cycle (Fig. 2) for another 2 d before being transferred into continuous white light (approximately 100 μmol m−2 s−1). CO2 concentration in the sample chambers was determined by infrared gas analysis at approximately 1-h intervals. Because the system is closed, after each sampling cycle, the air in the sample chamber was replaced with compressed air (Northeast Airgas, White River Junction, VT) of constant composition (350 μL L−1 CO2).
SO2 Sensitivity/Resistance Assay
For determination of sensitivity to SO2, 4-week-old Col plants were grown in a 14-h:10-h light-dark cycle, transferred to continuous light for 2 d, and then treated with SO2 at 2-h intervals over two complete circadian cycles. Time is given in Zeitgeber Time (ZT; zeitgeber is German for time giver), where ZT0 corresponds to lights on; for example, ZT20 corresponds to 20 h from the onset of illumination. The plants were gassed in a black Plexiglas box with an approximate volume of 130 L. SO2 (445 μL L−1, Merriam-Graves, White River Junction, VT) was added to the box at a flow rate of 5.63 L min−1 for 2 min (total of 11.26 L) to yield a final SO2 concentration of 38.5 μL L−1; plants were left under these conditions for 15 min. The box was then flushed with compressed air at approximately 8 L min−1 for 15 min. Plants were removed from the Plexiglas box, returned to the light-dark cycle, and scored for the presence of lesions (water-soaked, wilted areas) 2 h after treatment and again 2 d after treatment, when the lesions dried and turned white.
Leaf Movement Assay
Assessment of rhythmicity in leaf movement was carried out as described (Millar et al., 1995a, 1995b; Hicks et al., 1996). Seeds were surface-sterilized by the vapor-phase method (Clough and Bent, 1998) and then plated on Murashige and Skoog salts + 2% Suc (MS2S). Seedlings were grown in white light for 4 to 5 d in a 12-h:12-h light-dark cycle on MS2S plates and then transferred to 24-well cloning plates (Greiner Labortechnik, Frickenhausen, Germany), one seedling per well. The plates were transferred to continuous white light (30–40 μmol m−2 s−1), and leaf movement was recorded every 20 min over 7 d by Panasonic CCTV cameras (model WV-BP120, Matsushita Communications Industrial, Laguna, Philippines). Post-run analysis was performed using the Kujata software program (Millar et al., 1995a, 1995b), and traces were analyzed by fast Fourier transform-nonlinear least squares (Plautz et al., 1997; Zhong et al., 1997).
Hypocotyl-Length Characterization
Seeds were surface-sterilized by the vapor-phase method (Clough and Bent, 1998) and then plated along the diagonal of MS2S petri plates. The plates were stratified for 3 d at 4°C and released in white light (approximately 100 μmol m−2 s−1) for 12 h to induce germination. The plates were then transferred into the dark for 12 h before being released into the appropriate light conditions. For monochromatic light treatments, plates were placed in an E-30LED chamber (Percival, Boone, IA) in combination with red Plexiglas filters (Rohm and Haas no. 2423, Cadillac Plastics, Manchester, NH). For combined light studies, the filters were omitted because both blue and red light were provided. After 5 to 6 d of growth, the plates were scanned in Adobe Photoshop (Adobe Systems, Mountain View, CA), and hypocotyl length was determined by using NIH Image 1.62.
Genetic Mapping of oop1 and Sequencing of PHYBoop1
The map position of oop1 was determined by the analysis of 41 oop1 × Landsberg erecta F3 lines homozygous at the oop1 locus, by a combination of CAPS (Konieczny and Ausubel, 1993), RFLP, and simple sequence length polymorphism (Bell and Ecker, 1994) markers. The PHYBoop1 genomic sequence was determined from three overlapping PCR products on both strands.
Luciferase Imaging of Gene Expression
A transcriptional fusion of the LHCB1*1 promoter and luciferase (LHCB::LUC) was introduced into Col, oop1, and phyB-9 plants by crossing and selection on MS2S plates containing 50 μg/mL kanamycin (Sigma Chemicals, St. Louis). Seven-day-old seedlings grown under approximately 100 μmol m−2 s−1 white light and entrained in a 12-h light:12-h dark photoperiod were transferred into 96-well plates, each well containing 150 μL of MS2S and 30 μL of luciferin (2.5 mg/mL; Biosynth, Staad, Switzerland). The plates were covered with sealing tape and entrained for an additional 2 d before being moved into continuous white light (15–25 μmol m−2 s−1) on a Packard TopCount Luminometer. The data were analyzed by fast Fourier transform-nonlinear least squares.
Protein Extraction and Immunoblotting
Proteins were extracted from 8-d-old white light-grown seedlings, ammonium sulfate fractionated, separated on 6% SDS-polyacrylamide gels, and immunoblotted as described by Aukerman et al. (1997). Monoclonal antibodies used were: B6-B3 (PHYB-specific) and 3B5 (universal) as described by Hirschfeld et al. (1998).
ACKNOWLEDGMENTS
We thank Chentao Lin (University of California, Los Angeles), Steve Kay (Scripps Research Institute, La Jolla, CA), Jason Reed (University of North Carolina, Chapel Hill), David Somers (Ohio State University, Columbus), and the Arabidopsis Biological Resource Center (Ohio State University) for Arabidopsis lines and for constructs and clones. We thank Mary Lou Guerinot (Dartmouth College, Hanover, NH) for comments on the manuscript. We also thank Jay Dunlap (Dartmouth Medical School, Hanover, NH) for suggesting the experiment described in Figure 7.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN–9808801 to R.A.S. and grant nos. MCB–9723482 and MCB–0091008 to C.R.M.) and by the American Cancer Society (institutional grant to the Norris Cotton Cancer Center at Dartmouth College).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.003418.
LITERATURE CITED
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Bognár LK, Hall A, Ádám É, Thain SC, Nagy F, Millar AJ. The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proc Natl Acad Sci USA. 1999;96:14652–14657. doi: 10.1073/pnas.96.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Photochem Photobiol. 2000;71:1–11. doi: 10.1562/0031-8655(2000)071<0001:pcppii>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Mazzella MA. Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB, and hy4 simple, double, and triple mutants in Arabidopsis. Plant Physiol. 1998;118:19–25. doi: 10.1104/pp.118.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Covington MF, Panda S, Liu XL, Strayer CA, Wagner DR, Kay SA. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell. 2001;13:1305–1316. doi: 10.1105/tpc.13.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denault DL, Loros JJ, Dunlap JC. WC-2 mediates WC-1-FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J. 2001;20:109–117. doi: 10.1093/emboj/20.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12:2499–2510. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Kay SA. Circadian photoperception. Annu Rev Physiol. 2001;63:677–694. doi: 10.1146/annurev.physiol.63.1.677. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Yeh K-C, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Foster RG, Lucas RJ. Clocks, criteria and critical genes. Nat Genet. 1999;22:217–219. doi: 10.1038/10270. [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glossop NRJ, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- Green RM, Tobin EM. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc Natl Acad Sci USA. 1999;96:4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Panda S, Kay SA. Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol. 2001;17:215–254. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Loros JJ, Dunlap JC. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell. 2001;104:453–464. doi: 10.1016/s0092-8674(01)00232-x. [DOI] [PubMed] [Google Scholar]
- Hennessey TL, Field CB. Oscillations in carbon assimilation and stomatal conductance under constant conditions. Plant Physiol. 1991;96:831–836. doi: 10.1104/pp.96.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR. EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell. 2001;13:1281–1292. doi: 10.1105/tpc.13.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA. Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA. Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics. 1998;149:523–535. doi: 10.1093/genetics/149.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson FR. The isolation of biological rhythm mutations on the autosomes of Drosophila melanogaster. J Neurogenet. 1983;1:3–15. doi: 10.3109/01677068309107068. [DOI] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Tang R-H, Yang H-Q, Alonso JM, Ecker JR, Cashmore AR. An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature. 2001;410:487–490. doi: 10.1038/35068589. [DOI] [PubMed] [Google Scholar]
- Johnson CH. Endogenous timekeepers in photosynthetic organisms. Annu Rev Physiol. 2001;63:695–728. doi: 10.1146/annurev.physiol.63.1.695. [DOI] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schafer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Wada M. LKP1 (LOV kelch protein 1): a factor involved in the regulation of flowering time in Arabidopsis. Plant J. 2000;23:807–815. doi: 10.1046/j.1365-313x.2000.00850.x. [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Krall L, Reed J. The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc Natl Acad Sci USA. 2000;97:8169–8174. doi: 10.1073/pnas.140520097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Simon AE. Environmental and genetic effects on circadian clock-regulated gene-expression in Arabidopsis thaliana. Plant Cell. 1997;9:297–304. doi: 10.1105/tpc.9.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Loros JJ, Dunlap JC. Interconnected feedback loops in the Neurospora circadian system. Science. 2000;289:107–110. doi: 10.1126/science.289.5476.107. [DOI] [PubMed] [Google Scholar]
- Lin C. Plant blue-light receptors. Trends Plant Sci. 2000;5:337–342. doi: 10.1016/s1360-1385(00)01687-3. [DOI] [PubMed] [Google Scholar]
- Lindebro MC, Poellinger L, Whitelaw ML. Protein-protein interaction via PAS domains: role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13:1293–1304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros JJ, Dunlap JC. Genetic and molecular analysis of circadian rhythms in Neurospora. Annu Rev Physiol. 2001;63:757–794. doi: 10.1146/annurev.physiol.63.1.757. [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 2002;43:58–69. doi: 10.1093/pcp/pcf005. [DOI] [PubMed] [Google Scholar]
- Mansfield TA, Freer-Smith PH. The role of stomata in resistance mechanisms. In: Koziol MJ, Whatley FR, editors. Gaseous Air Pollutants and Plant Metabolism. London: Butterworths; 1984. pp. 131–146. [Google Scholar]
- Martínez-García JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Más P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408:207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: II. Characterization with CCA1-overexpressing plants. Plant Cell Physiol. 2002;43:118–122. doi: 10.1093/pcp/pcf006. [DOI] [PubMed] [Google Scholar]
- McClung CR. Circadian rhythms in plants. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:139–162. doi: 10.1146/annurev.arplant.52.1.139. [DOI] [PubMed] [Google Scholar]
- McClung CR, Salomé PA, Michael TP. The Arabidopsis circadian system. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. pp. 1–25.http://www.aspb.org/publications/arabidopsis/ . DOI 10.1199/tab.0044 http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature. 2000;408:716–720. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- Merrow M, Brunner M, Roenneberg T. Assignment of circadian function for the Neurospora clock gene frequency. Nature. 1999;399:584–586. doi: 10.1038/21190. [DOI] [PubMed] [Google Scholar]
- Merrow M, Franchi L, Dragovic Z, Görl M, Johnson J, Brunner M, Macino G, Roenneberg T. Circadian regulation of the light input pathway in Neurospora crassa. EMBO J. 2001;20:307–315. doi: 10.1093/emboj/20.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua N-H, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995a;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua N-H, Kay SA. The regulation of circadian period by phototransduction pathways in Arabidopsis. Science. 1995b;267:1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell. 2000;101:331–340. doi: 10.1016/s0092-8674(00)80842-9. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photo-induced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, Merrow M. Molecular circadian oscillators: an alternative hypothesis. J Biol Rhythms. 1998;13:167–179. doi: 10.1177/074873098129000011. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G. LATE ELONGATED HYPOCOTYL, an Arabidopsis gene encoding a MYB transcription factor, regulates circadian rhythmicity and photoperiodic responses. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell. 2001;13:2659–2670. doi: 10.1105/tpc.010332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GTJ, Hastings MH et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin P, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998a;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AAR, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998b;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. Isolation of photorespiration mutants of Arabidopsis. In: Edelman M, Hallick RB, Chua NH, editors. Methods in Chloroplast Molecular Biology. New York: Elsevier; 1982. pp. 129–139. [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang H-S, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R, Kevei É, Hall A, Millar AJ, Nagy F, Kozma-Bognár L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Webb AAR. Stomatal rhythms. In: Lumsden PJ, Millar AJ, editors. Biological Rhythms and Photoperiodism in Plants. Oxford: BIOS Scientific Publishers, Ltd.; 1998. pp. 69–79. [Google Scholar]
- Williams JA, Sehgal A. Molecular components of the circadian system in Drosophila. Annu Rev Physiol. 2001;63:729–755. doi: 10.1146/annurev.physiol.63.1.729. [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Cheng P, Zhi G, Liu Y. Identification of a calcium/calmodulin-dependent protein kinase that phosphorylates the Neurospora circadian clock protein FREQUENCY. J Biol Chem. 2001;276:41064–41072. doi: 10.1074/jbc.M106905200. [DOI] [PubMed] [Google Scholar]
- Yeh K-C, Lagarias JC. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 1996;10:691–702. doi: 10.1046/j.1365-313x.1996.10040691.x. [DOI] [PubMed] [Google Scholar]
- Zhong HH, Resnick AS, Straume M, McClung CR. Effects of synergistic signaling by phytochrome A and cryptochrome 1 on circadian clock-regulated catalase expression. Plant Cell. 1997;9:947–955. doi: 10.1105/tpc.9.6.947. [DOI] [PMC free article] [PubMed] [Google Scholar]