Abstract
Rose (Rosa hybrida) flowers produce and emit a diverse array of volatiles, characteristic to their unique scent. One of the most prominent compounds in the floral volatiles of many rose varieties is the methoxylated phenolic derivative 3,5-dimethoxytoluene (orcinol dimethyl ether). Cell-free extracts derived from developing rose petals displayed O-methyltransferase (OMT) activities toward several phenolic substrates, including 3,5-dihydroxytoluene (orcinol), 3-methoxy,5-hydroxytoluene (orcinol monomethyl ether), 1-methoxy, 2-hydroxy benezene (guaiacol), and eugenol. The activity was most prominent in rose cv Golden Gate, a variety that produces relatively high levels of orcinol dimethyl ether, as compared with rose cv Fragrant Cloud, an otherwise scented variety but which emits almost no orcinol dimethyl ether. Using a functional genomics approach, we have identified and characterized two closely related cDNAs from a rose petal library that each encode a protein capable of methylating the penultimate and immediate precursors (orcinol and orcinol monomethyl ether, respectively) to give the final orcinol dimethyl ether product. The enzymes, designated orcinol OMTs (OOMT1 and OOMT2), are closely related to other plant methyltransferases whose substrates range from isoflavones to phenylpropenes. The peak in the levels of OOMT1 and OOMT2 transcripts in the flowers coincides with peak OMT activity and with the emission of orcinol dimethyl ether.
Roses (Rosa hybrida) are cultivated in nearly all of the countries of the world. They are grown as garden plants, as cut flowers, and as a source of natural fragrances and flavorings (Weiss, 1997). The genus Rosa includes 200 species and more than 18,000 cultivars (Haring, 1986; Gudin, 2000). The damask rose (Rosa damascena) is the most important species used to produce rose water, attar of rose, and essential oils in the perfumery industry. Modern cut-rose varieties are not notable for their scent (Zuker et al., 1998). It could be that because selection in breeding for the cut flower market is geared toward enhancing vase life and modifying color and form, the loss of fragrance may be coincidental (Barletta, 1995).
More than 400 volatile compounds have been identified in the floral scent of various rose cultivars. These compounds can be classified into five major groups based on their functions: hydrocarbons (mostly sesquiterpenes such as β-caryophyllene), alcohols (mostly monoterpenes such as geraniol, nerol, and citronellol, or aromatic such as phenethyl alcohol), esters (mostly acetates such as hexyl-acetate, (Z)-3-hexenyl-acetate, geranyl acetate, and phenethyl acetate), aromatic ethers (mostly 3,5-dimethoxytoluene [orcinol dimethyl ether], benzyl methyl ether, estragole, and methyl-eugenol), and others (including aldehydes such as the monoterpene geranial, the aliphatic chain nonanal, and decanal; the rose oxides; and norisoprenes such as β-ionone) (Flament et al., 1993). The floral scents of most rose cultivars are dominated by representatives of one of these five groups. However, with few exceptions, the phenol ether orcinol dimethyl ether is emitted at some level from the flowers of most rose varieties (Flament et al., 1993).
Most of the research in aroma compounds has historically focused on the chemical elucidation of naturally occurring structures coupled to chemical synthesis to produce the large quantities demanded by the industry (Croteau and Karp, 1991). Although most of the chemical structures of floral scent compounds have been solved, very few studies have focused on their biosynthesis. This situation has recently begun to change with several investigations launched toward understanding the biosynthesis of floral volatiles (Vainstein et al., 2001). For example, it has been shown that in Clarkia breweri, the flowers synthesize scent compounds de novo in the tissues from which these volatiles are emitted, and the emission levels, corresponding enzymes activities, and the level of mRNA are all spatially and temporally correlated (for review, see Dudareva and Pichersky, 2000). In general, the expression of these genes is highest in petals and is restricted to the epidermal cell layer of floral tissues. Enzymes responsible for the biosynthesis of C. breweri scent volatiles and their corresponding genes have been isolated and characterized, including linalool synthase, benzylalcohol acetyltransferase, and two methyltransferases (for review, see Dudareva and Pichersky, 2000; Vainstein et al., 2001). Similar results have been obtained with a methyltransferase catalyzing methyl benzoate formation in snapdragon (Antirrhinum majus) petals (Dudareva et al., 2000).
To date, almost no biosynthetic studies of rose scent have been reported, although the possible contribution of glycosidases to the release of scent volatiles stored in the petals in the form of glycosides has been examined (Oka et al., 1999). To investigate de novo scent biosynthesis in rose flowers, we have begun a project employing genomic approaches (Guterman et al., 2002). The sequences of over 3,000 petal cDNAs have been determined from two rose varieties, cv Fragrant Cloud and cv Golden Gate. Rose cv Fragrant Cloud flowers possess intense scent and red-colored petals (due to anthocyanin pigments) and have a short shelf-life, whereas rose cv Golden Gate flowers have much less noticeable (to the human nose) scent, have yellow-colored petals (due to carotenoid pigments), and are long-lasting (I. Guterman, M. Shalit, M. Menda, D. Piestun, M. Dafny-Yelin, G. Shalev, E. Bar, O. Davydov, M. Ovadis, M. Emanuel, J. Wang, Z. Adam, E. Pichersky, E. Lewinsohn, D. Zamir, A. Vainstein, and D. Weiss, unpublished data). Like most rose varieties, cv Golden Gate flowers emit orcinol dimethyl ether, whereas cv Fragrant Cloud is exceptional in emitting almost undetectable levels of this compound.
Genomic approaches have proven useful to study genes and enzymes involved in the formation of compounds that contribute to the aroma of plant tissues (Aharoni et al., 2000; Lange et al., 2000; Gang et al., 2001). We have undertaken a similar approach to investigate scent biosynthesis in rose flowers (Guterman et al., 2002). Analysis of the expressed sequence tag (EST) databases of rose cv Golden Gate and cv Fragrant Cloud petals allowed the identification of two cDNAs present in both varieties that are very similar to each other and also exhibit homology to known O-methyltransferases (OMTs), enzymes that catalyze the transfer of a methyl group from S-adenosyl-l-Met (SAM) to an hydroxyl functionality (Ibrahim et al., 1998). Here, we describe the biochemical characterization of the enzymes encoded by these two cDNAs, and we show that they catalyze the last two steps of the formation of orcinol dimethyl ether.
RESULTS
Orcinol Dimethyl Ether in Rose Flower Headspace
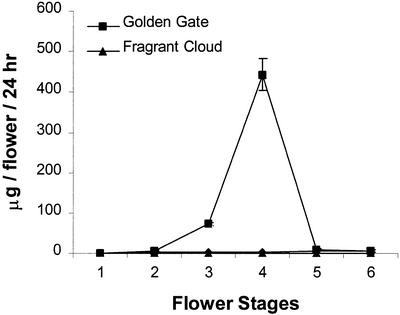
The headspaces of rose cv Fragrant Cloud and cv Golden Gate were assessed for the presence of orcinol dimethyl ether during six stages of flower development (see “Materials and Methods”). The headspace of the rose cv Fragrant Cloud flower is dominated by alcohols, acetate esters, and sesquiterpenes (data not shown), but almost no orcinol dimethyl ether was detected (Fig. 1). In contrast, in rose cv Golden Gate headspace, orcinol dimethyl ether was a major scent component. Its emission sharply increased during flower maturation, with a maximum value at stage 4 of 440 μg flower−1 d−1 (Fig. 1). At this stage of development, orcinol dimethyl ether constituted 55% of the total volatiles present in the rose cv Golden Gate floral bouquet.
Figure 1.
Emission of orcinol dimethyl ether from rose cv Golden Gate and cv Fragrant Cloud flowers as detected by headspace analysis.
Phenolic O-Methyltransferase Activities in Petal Cell-Free Extracts
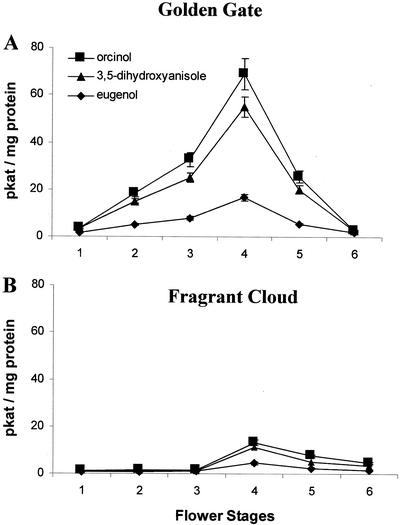
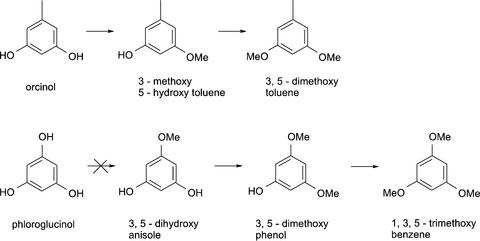
To test for the presence of phenol-sustained OMT activities, cell-free soluble protein extracts were prepared from petals of rose cv Fragrant Cloud and cv Golden Gate flowers at various stages of development. OMT activities determined in these extracts using the substrates 3,5-dihydroxytoluene (orcinol; the first putative hydroxyl-containing precursor for the biosynthesis of 3, 5-dimethoxytoluene), 3,5-dihydroxyanisole (a precursor of 1,3,5-trimethoxybenzene, a volatile emitted by some rose species), and eugenol (the precursor of methyleugenol, which is emitted by rose cv Fragrant Cloud flowers) are shown in Figure 2. Cell-free extracts derived from rose cv Golden Gate petals display higher activities with all three substrates than did similar extracts from rose cv Fragrant Cloud petals, and these activities sharply increased during flower maturation, peaking at stage 4. Calculations (not shown) of the total amount of activity of orcinol methyltransferase in the flower and of the total amount of the methylating activity of the next intermediate, 3-methoxy,5-hydroxytoluene (orcinol monomethyl ether), indicate that more than sufficient levels of methylating activity were present in the flowers to account for the observed emission at peak time from the rose cv Golden Gate flowers.
Figure 2.
OMT activities in crude protein extracts from rose cv Golden Gate (A) and rose cv Fragrant Cloud (B) petals using orcinol, 3,5-dihydroxyanisole, and eugenol.
Isolation and Sequence Characterization of Rose OMT cDNAs
EST databases from rose petals of cv Fragrant Cloud and cv Golden Gate have been constructed (I. Guterman, M. Shalit, M. Menda, D. Piestun, M. Dafny-Yelin, G. Shalev, E. Bar, O. Davydov, M. Ovadis, M. Emanuel, J. Wang, Z. Adam, E. Pichersky, E. Lewinsohn, D. Zamir, A. Vainstein, and D. Weiss, unpublished data). A search in these databases for potential OMTs revealed 33 cDNAs (2.5% of total) similar to known OMT sequences in the rose cv Fragrant Cloud database and 30 cDNAs (3% of total) in the rose cv Golden Gate EST database. Two types of cDNAs, designated OOMT1 (for orcinol OMTs, see below) and OOMT2 were identified. Thirteen cDNAs from rose cv Golden Gate and 22 cDNAs from rose cv Fragrant Cloud represented OOMT1, and OOMT2 was represented by 17 rose cv Golden Gate cDNAs and 11 rose cv Fragrant Cloud cDNAs. OOMT1 and OOMT2 are 97% identical to each other in their coding regions and 94% identical in their 3′ non-coding regions, and they encode proteins of 367 and 366 amino acids, respectively, that are 96% identical to each other, differing from each other at 13 positions (Fig. 3). The 13 differences include one deletion/addition (Tyr-60 in OOMT1 is lacking in OOMT2) and 12 mostly conservative substitutions. Only one difference (Asn-138 in OOMT1 versus Thr-137 in OOMT2) occurs at the substrate binding site, based on three-dimensional structural modeling (Gang et al., 2002; data not shown).
Figure 3.
Alignments of the two rose OOMT sequences with two related OMTs: eugenol OMT (EOMT; Gang et al., 2002) from basil (Ocimum basilicum) and isoflavone OMT (IOMT; He and Dixon, 1996) from alfalfa (Medicago sativa). Differences between OOMT1 and OOMT2 are highlighted.
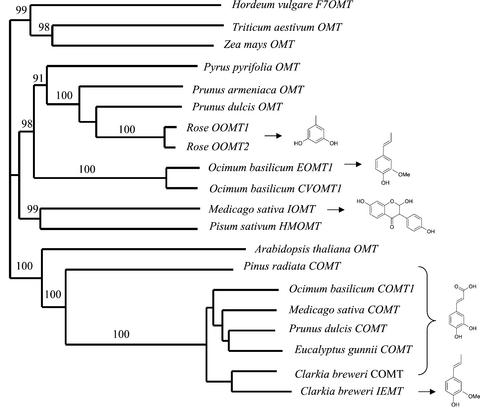
OOMT1 and OOMT2 proteins were also very similar (>70%) to other published OMT sequences (Figs. 3 and 4), including alfalfa IOMT (He and Dixon, 1996) and sweet basil EOMT and chavicol OMT (Gang et al., 2002), and in particular, to an OMT from a related Rosae species, almond (Prunus dulcis, Rosaceae), that was shown to be specifically expressed in the flowers, but whose substrate has not been determined (Suelves and Puigdomenech, 1998).
Figure 4.
A neighbor-joining tree based on degree of sequence similarity between rose OOMTs and other related OMTs. The accession numbers of OOMT1 and OOMT2 are AF502433 and AF502434, respectively. The sources of the other OMTs are cited by Gang et al. (2002) and Wang and Pichersky (1999). The substrates of selected enzymes (see text) are shown.
Enzymatic Activity of Recombinant OOMT1 and OOMT2
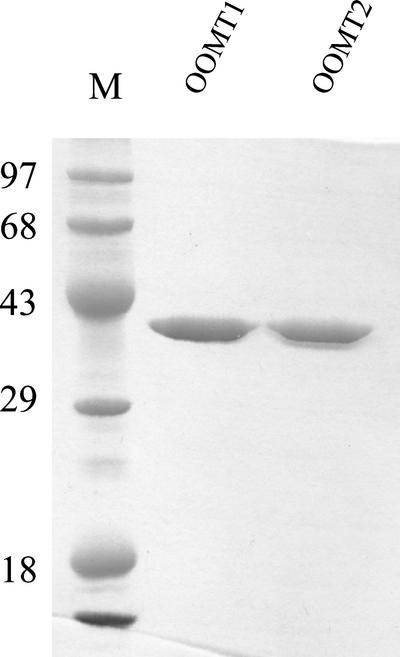
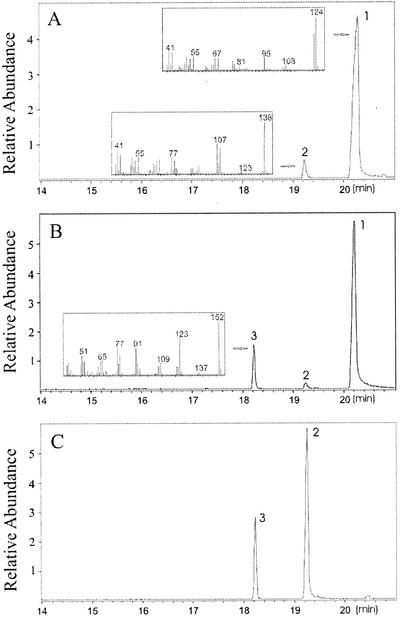
The coding regions of OOMT1 and OOMT2 were transferred to the expression vector pET(11a) (Studier et al., 1990) for functional analysis. The OMT proteins were purified to homogeneity (Fig. 5) using affinity chromatography as described by Wang et al. (1997). Purified enzymes for each recombinant protein and from cell-free extracts of stage 4 petals were used to evaluate their ability to catalyze the SAM-dependent O-methylation of a large number of potential substrates (Table I) and to determine their general catalytic properties and kinetic parameters with their preferred substrates (Table II). Both enzymes were able to methylate both intermediates in the biosynthesis of orcinol dimethyl ether, 3,5-dihydroxytoluene (orcinol), and orcinol monomethyl ether (Figs. 6 and 7). OOMT1 and OOMT2 had Km values for these substrates in the 13 to 48 μm range, similar to values found for other OMTs with their primary substrates (Ibrahim et al., 1998). However, the catalytic efficiency of OOMT1 with orcinol was twice that of OOMT2, and the catalytic efficiency of OOMT2 with orcinol monomethyl ether was about 5-fold higher than that of OOMT1 (Table II). Consistent with these results, when OOMT1 was incubated with orcinol, the product obtained was the monomethylated orcinol monomethyl ether (Fig. 7A), but when OOMT2 was incubated with orcinol, most of the product obtained was the dimethylated orcinol dimethyl ether (Fig. 7B), suggesting that the monomethylated intermediate could be efficiently methylated by OOMT2 before accumulating.
Figure 5.
SDS-PAGE analysis of affinity-purified OOMT1 and OOMT2. The size of the protein markers in lane M is shown (in kilodaltons) on the left.
Table I.
Relative activity of crude extracts from petals of rose cv Golden Gate and cv Fragrant Cloud and of purified OOMT1 and OOMT2 gene products with selected substrates
| Substrate Tested and Substitution Pattern | Crude Extracts
|
Gene Products
|
||
|---|---|---|---|---|
| cv GoldenGate | cv Fragrant Cloud | OOMT1 | OOMT2 | |
| meta-Dihydroxy | ||||
| Orcinol | 100 | 100 | 100 | 100 |
| Resorcinol (1,3-dihydroxybenzene) | 62 | 94 | 49 | 35 |
| Phloroglucinol (1,3,5-trihydroxybenzene) | 21 | 73 | 6 | 0 |
| 3,5-Dihydroxyanisole (1-methoxy,3,5-dihydroxybenzene) | 81 | 80 | 80 | 73 |
| meta-Methoxy-hydroxy | ||||
| Orcinol monomethyl ether | 73 | 47 | 80 | 193 |
| 3,5-Dimethoxyphenol (1,3-dimethoxy,5-hydroxybenzene) | 61 | 69 | 56 | 79 |
| 3-Methoxyphenol (1-methoxy,3-hydroxybenzene) | 41 | 61 | 25 | 86 |
| ortho-Dihydroxy | ||||
| Catechol (1,2-dihydroxybenzene) | 28 | 61 | 10 | 30 |
| Protocatechuic aldehyde | 19 | 45 | 1 | 0 |
| Pyrogallol (1,2,3-trihydroxybenzene) | 24 | 52 | 7 | 14 |
| Caffeic acid | 3 | 3 | 0 | 0 |
| para-Hydroxy | ||||
| 4-Methoxyphenol (1-methoxy,4-hydroxybenzene) | 13 | 0 | 2 | 17 |
| Coumaric acid | 0 | 0 | 0 | 0 |
| ortho-Methoxy-hydroxy | ||||
| Guaiacol (1-methoxy,2-hydroxybenzene) | 98 | 99 | 38 | 145 |
| O-Cresol (2-hydroxytoluene) | 35 | 42 | 27 | 55 |
| Eugenol | 26 | 41 | 3 | 16 |
| Ferulic acid | 0 | 0 | 0 | 0 |
For both extracts and purified enzymes, activity with orcinol was set arbitrarily as 100%. All substrates were tested at a 1 mm concentration.
Table II.
Kinetic parameters of OOMT1 and OOMT2
| Substrate | Km | Kcat | Kcat/Km |
|---|---|---|---|
| μm | s−1 × 10−3 | nm−1 s−1 | |
| OOMT1 | |||
| Orcinol | 13 | 4.47 | 0.34 |
| Orcinol monomethyl ether | 10 | 1.79 | 0.2 |
| 3,5-Dihydroxyanisole | 27 | 4.41 | 0.16 |
| 3,5-Dimethoxyphenol | 15 | 2.84 | 0.19 |
| Guaiacol | 23 | 8.62 | 0.37 |
| SAM | 10 | ||
| OOMT2 | |||
| Orcinol | 48 | 9.52 | 0.19 |
| Orcinol monomethyl ether | 10 | 9.62 | 0.96 |
| 3,5-Dihydroxyanisole | 41 | 9.71 | 0.24 |
| 3,5-Dimethoxyphenol | 55 | 5.35 | 0.1 |
| Guaiacol | 18 | 39.2 | 2.81 |
| SAM | 10 |
Figure 6.
The reactions catalyzed by rose OOMT1 and OOMT2. SAM is the methyl donor in all these methylation reactions. Both OOMT1 and OOMT2 show very little activity with phloroglucinol.
Figure 7.
Gas chromatography-mass spectrometry (GC-MS) analysis of products produced in the reactions catalyzed by OOMT1 and OOMT2. Total ion chromatograms are shown. A, A reaction using orcinol as the substrate with OOMT1 as the enzyme. B, A reaction using orcinol as the substrate with OOMT2 as the enzyme. C, A reaction using orcinol monomethyl ether as the substrate with OOMT2 as the enzyme (similar results were obtained with OOMT1). Peak 1, orcinol; peak 2, orcinol monomethyl ether; and peak 3, orcinol dimethyl ether. Mass spectra of the peaks, shown here, matched the mass spectra of the corresponding authentic standards. Reactions were carried out in a total volume of 1 mL with substrate concentration of 2 mm and a total of 1.5 μg of purified protein and were allowed to proceed for 120 min.
Because some rose species such as Rosa chinensis (but not rose cv Golden Gate or cv Fragrant Cloud) also emit 1,3,5-trimethoxybenzene (N. Watanabe, personal communication), we tested the cell-free extracts and OOMT1 and OOMT2 gene products with the three intermediates in the biosynthesis of this compound (Tables I and II; Fig. 6). Interestingly, both recombinant enzymes could catalyze the last two reactions in the conversion of phloroglucinol to 1,3,5-trimethoxybenzene, but had little activity with phloroglucinol itself (because this reaction did not proceed, Km values for phloroglucinol could not be determined).
Both OOMT1 and OOMT2 also showed activity with several other phenolic compounds but not with phenylpropanoids. As has been shown with other OMTs (Gang et al., 2002), it appears that both of the rose OMT enzymes can efficiently accept phenolic substrates that are somewhat smaller than the natural substrates (guaiacol is the best example), but not larger molecules such as the phenylpropanoids, probably because they cannot efficiently fit in the active site. However, it appears that OOMT1 is a bit more active toward meta-dihydroxy compounds, whereas OOMT2 is more active toward ortho- and meta-methoxy-hydroxy compounds (Table I). The cell-free petal extracts of the two varieties displayed broader substrate specificity, efficiently methylating pyrogallol, catechol, eugenol, protocatechuic aldehyde, and phloroglucinol (Table I).
The OMT gene products of OOMT1 and OOMT2 eluted from a calibrated gel filtration column as approximately 80-kD proteins. Because the calculated molecular weights of the OOMT1 and OOMT2 subunits are 41,278 and 41,226, respectively, this result suggests that they exist as homodimers in solution, similar to what has been found for other OMTs (Ibrahim et al., 1998). The recombinant proteins possessed a narrow pH optimum from 7.5 to 8. Increasing concentrations of salt (NaCl or KCl) reduced enzymatic activities. Optimum temperature for activity was determined to be 30°C to 37°C.
RNA Gel-Blot Analysis of OOMT1 and OOMT2 mRNA
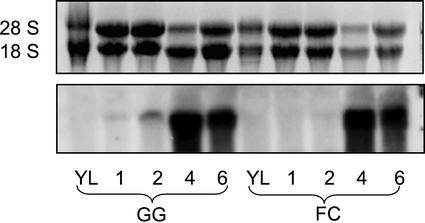
We examined the levels of OOMT transcripts from rose petals of different developmental stages as well as from leaves (Fig. 8). No OOMT transcripts were detected in leaves of either variety. In petals of both rose cv Golden Gate and cv Fragrant Cloud, transcript levels were below detection in the buds and were increasing from stage 2 to a peak at stage 4. Overall, OOMT transcript levels seemed to be similar in both rose cv Golden Gate and cv Fragrant Cloud. It should be pointed out that because of the high similarity in the sequences of the mRNA of OOMT1 and OOMT2 in the coding and non-translated regions, the probe used could not distinguish between the two types of transcripts.
Figure 8.
Northern-blot analysis of RNA samples derived form rose tissues. Top, Ethidium bromide staining of the gel. Bottom, Autoradiography of the gel blot hybridized with a 32P-labeled OOMT1 cDNA probe as described in “Materials and Methods.” YL, Young leaves; 1, 2, 4, and 6, Flowers at stages 1, 2, 4, and 6, respectively. GG, Rose cv Golden Gate. FC, Rose cv Fragrant Cloud.
DISCUSSION
Orcinol dimethyl ether is one of the most common volatiles of rose flowers, and it is known to contribute up to 60% of the total volatiles in some rose varieties (Flament et al., 1993). Similar to other rose volatiles, the emission of orcinol dimethyl ether displays a circadian rhythm pattern (Helsper et al., 1998). In addition to roses, this compound has so far been identified in very few other plants: In the floral headspace of Narcissus tazetta, a native of central Europe and the Mediterranean region, orcinol dimethyl ether constitutes up to one-third of the total (Mookherjee et al., 1989); and in the floral headspace of Acnistus arborescens, the so-called “Maria Mole” of the Solanaceae from the southern Atlantic rain forest (Kaiser, 2000).
In roses, orcinol dimethyl ether is emitted by most varieties, and it is absent only from some varieties whose floral scent is rich in alcohols, such as the classic Rosa gallica and damask rose (Flament et al., 1993). Of the two cultivars we examined, rose cv Golden Gate emits relatively high levels of orcinol dimethyl ether, whereas rose cv Fragrant Cloud, a variety with prominent scent, nonetheless emits almost undetectable levels of this compound. Interestingly, both varieties contain similar steady-state levels of OOMT1 and OOMT2 transcripts in the petals (Fig. 8). Because the levels of OOMT enzymatic activity are 5-fold lower in rose cv Fragrant Cloud than in rose cv Golden Gate (Fig. 2), the involvement of posttranscriptional processes in modulating OOMT enzymatic activity levels is indicated. However, lowers levels of OOMT activity by themselves do not fully explain the almost complete lack of orcinol dimethyl ether emission from rose cv Fragrant Cloud. Additional causes may lie, at least in part, in earlier steps in the pathway, which might be blocked in rose cv Fragrant Cloud petals, or in the efficient shunting of the substrates into glycoside or other non-volatile derivatives, which would not be detected in our analytical conditions but would still decrease the concentration of the free substrate available for the OOMT enzymes. It is not clear why cv Fragrant Cloud flowers express the OOMT enzymes at all, because they barely emit any orcinol dimethyl ether. Nevertheless, high levels of scent-producing enzymes without the concomitant emission of the product has been observed before (Dudareva and Pichersky, 2000).
The native species R. chinensis, which served as an ancestral species in the breeding of the modern tea roses, has rich emission of 1,3,5-trimethoxybenzene (N. Watanabe, personal communication), and it also possesses several OMTs capable of synthesizing 1,3,5-trimethoxybenzene as well as dimethoxytoluene (Scalliet et al., 2002). Both rose cv Golden Gate and cv Fragrant Cloud, like most other cultivated roses, did not emit detectable levels of 1,3,5-trimethoxybenzene under our experimental conditions. Nonetheless, our results indicate that both OOMT1 and OOMT2 can catalyze the last two putative steps in the synthesis of 1,3,5-trimethoxybenzene, but they cannot efficiently methylate phloroglucinol (Table II; Fig. 6). Because petal cell-free extracts from these varieties contain such an activity (Table I), we hypothesize that an additional, yet unidentified, OMT expressed in these rose varieties, capable of methylating phloroglucinol ought to be present. However, the phloroglucinol-methylating activity found in the petals is apparently not sufficient to bring about the synthesis of 1,3,5-trimethoxybenzene, possibly because the substrate itself may be lacking or unavailable to the enzymes. The biosynthetic pathways leading to both phloroglucinol and orcinol are not fully known, but these compounds are probably derived from phenylpropanoid acids by a combination of β-oxidation and decarbonylation or decarboxylation reactions (Croteau and Karp, 1991; Laempe et al., 2001).
The finding that these cultivated varieties contain the enzymes that can methylate the intermediates in trihydroxybenzene biosynthetic (even though the complete pathway may not be operative) may simply be due to inheritance and segregation. Rose cv Fragrant Cloud and cv Golden Gate are “tea hybrids,” and they have inherited genes from both of their R. gallica and R. chinensis ancestors. In subsequent breeding, some of the genes in this pathway may have been lost by segregation.
Petal extracts of rose cv Golden Gate and especially of rose cv Fragrant Cloud also displayed a broad array of methylating activities with substrates such as eugenol, pyrogallol, catechol, and protocatechuic aldehyde, substrates that are not methylated by OOMT1 and OOMT2 (Table I). This might also indicate the expression of an additional, yet unknown OMT gene or genes.
The substrate specificities of OOMT1 and OOMT2 are similar, but not identical. Their high sequence identity to each other in coding (97%) and non-coding (94%) regions suggests that they may be encoded by homeologous loci in the tetraploid rose genome. OOMT1 and OOMT2 belong to the family of OMTs that includes chavicol OMT, EOMT, and IOMT, but they have a substrate that is smaller. Another closely related sequence encodes a putative OMT in almond flowers. This gene has previously been postulated to be involved in flavonoid metabolism, but it is possible that it is involved in scent emission from almond flowers, because almond flowers emit methoxylated phenolic compounds, and display orcinol-dependent OMT activity (N. Lavid and E. Lewinsohn, unpublished data). Sequence alignments show that among closely related OMT sequences, there is a wide diversity of substrates (Fig. 4), and, therefore, such information by itself appears to be insufficient to predict substrate specificity with any accuracy (but see Schröder et al., 2002). The range of plant specialized compounds produced by these OMTs remain to be experimentally determined.
MATERIALS AND METHODS
Plant Materials
Flowers of Rosa hybrida cv Fragrant Cloud and cv Golden Gate were harvested from plants grown in a greenhouse in Newe-Ya'ar, Israel. Flower development was divided into six stages: At stage 1, flower buds are closed and petals are green. At stage 2, petals start to emerge from the sepals and their color changes to red (rose cv Fragrant Cloud) or yellow (rose cv Golden Gate). Stages 3 and 4 are characterized by petal elongation and further accumulation of pigments. At stage 5, petals unroll, reaching full size at stage 6.
Headspace of Volatiles
Intact individual rose flowers, still attached to the bush, were enclosed in a 1-L glass container with the appropriate openings, and headspace was trapped, eluted, and concentrated using a method modified from Raguso and Pichersky (1995), using a Porapak Q 80/100 polydivinylbenzene filter (Waters, Milford, MA) for 24 h. Volatiles were eluted using 10 mL of HPLC grade hexane containing 10 μg mL−1 ethylmyristate as an internal standard and evaporated to 0.5 mL. One microliter of each sample was analyzed by GC-MS.
GC-MS Analysis
The volatile compounds collected from the headspace were analyzed on a HP-GCD apparatus equipped with an HP-5 (30-m × 0.25-mm) fused-silica capillary column. Helium (1 mL min−1) was used as a carrier gas. The injector temperature was 250°C, set for splitless injection. The oven was set to 50°C for 1 min, and then the temperature was increased to 200°C at a rate of 4°C min−1. The detector temperature was 280°C. Mass range was recorded from 45 to 450 m/z, with electron energy of 70 eV. Identification of the main components was done by comparison of mass spectra and retention time data with those of authentic samples and supplemented with a Wiley GC-MS library. The quantitative analyses were determined using isobutylbenzene as an internal standard (Lewinsohn et al., 2001; Shalit et al., 2001).
Chemicals and Radiochemicals
All chemicals were purchased from Sigma (St. Louis), unless otherwise noted. Orcinol dimethyl ether was a generous gift from Prof. N. Watanabe (Department of Applied Biological Chemistry, Shizuoka University, Japan). Orcinol monomethyl ether was synthesized according to Henrich and Nachtigall (1903). The purified product was characterized by thin-layer chromatography on silica gel plate, GC-MS, and NMR. Proton NMR data were in agreement with published data (Kakiuchi et al., 1991). S-[3H-methyl]adenosyl-l-Met (specific activity 15 Ci mmol−1) and S-[14C-methyl]adenosyl-l Met (specific activity 55 mCi mmol−1) were from Amersham (Buckinghamshire, UK).
Preparation of Crude Cell-Free Extracts from Petals
Fresh rose flowers were weighed and frozen by liquid nitrogen in a chilled mortar. Cell-free extracts were prepared as follows, all stages at 4°C: The tissues (approximately 1 g) were ground with a pestle in the presence of approximately 0.5 g of polyvinylpoly-pyrrolidone to adsorb phenolic materials and extraction buffer A (50 mm BisTris propane, pH 7.5, containing 10% [v/v] glycerol, 5 mm Na2S2O5, 10 mm NaCl, 1 mm EDTA, 14 mm 2-mercaptoethanol, and 1% [w/v] polyvinylpyrrolidone-10) was added at 10 times the fresh weight. The slurry was mixed and then centrifuged at 20,000g for 10 min. The supernatant was used for enzymatic assays (Lewinsohn et al., 2000).
Cloning of Rose OMTs
OOMT1 and OOMT2 were identified in the EST databases by homology search (BLAST) with other OMTs. Both clones were recloned into the T7-dependent expression vector pET(11a) by PCR with the appropriate oligonucleotides as previously described (Wang and Pichersky, 1999).
Expression of OOMT1 and OOMT2 in Escherichia coli and Enzyme Purification
Individual bacterial colonies (E. coli strain BL21(DES) pLysS) from freshly streaked plates were grown in liquid culture, and induction, harvesting, and protein purification by affinity chromatography were as previously described (Wang et al., 1997; Wang and Pichersky, 1999).
Enzyme Activity Assays, Product Identification, and Determination of Kinetic Parameters and Native Molecular Mass
Procedures were as previously described (Wang et al., 1997; Wang and Pichersky, 1999; Gang et al., 2002).
RNA Purification and RNA Gel-Blot Analysis
Total RNA was extracted from petals and leaves as previously described (Manning, 1991). RNA samples (10 μg) were fractionated in a 1% (w/v) agarose gel containing formaldehyde and blotted into HyBond N+ membranes (Amersham). The blots were hybridized in a solution containing 0.263 m Na2PO4, 7% (w/v) SDS, 1 mm EDTA, and 1% (w/v) bovine serum albumin at 60°C with 32P-labeled OOMT1 cDNA probe (Redprime, Amersham). The membranes were washed twice in 2× SSC and 0.1% (w/v) SDS at 60°C for 20 min each and exposed to x-ray film (Fuji Photo Film, Tokyo).
ACKNOWLEDGMENT
We thank Dr. N. Watanabe for his generous gift of chemicals.
Footnotes
This work was supported by the Israeli Ministry of Science, Culture and Sports (grant no. 1410–2–00 to E.L., D.Z., Z.A., A.V., and D.W.), by a Binational Agricultural Research and Development Fund scholarship (to E.P.), by the National Science Foundation (grant no. MCB–9974463 to E.P.), by the United States-Israel Binational Science Foundation and the Israel Science Foundation (to S.S.), and by a Deutscher Akademischer Austauschdienst fellowship (Gemeinsames Hochschulprogramm III von Bund und Ländern; to T.B.). This is publication no. 113/2002 of the Agricultural Research Organization (Bet Dagan, Israel).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.005330.
LITERATURE CITED
- Aharoni A, Keizer LC, Bouwmeester HJ, Sun Z, Alvarez-Huerta M, Verhoeven HA, Blaas J, van Houwelingen AM, De Vos RC, van der Voet H et al. Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell. 2000;12:613–616. doi: 10.1105/tpc.12.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta A. Scent makes a comeback. Floraculture. 1995;5:23–25. [Google Scholar]
- Croteau R, Karp F. Origin of natural odorants. In: Muller P, Lamparsky D, editors. Perfumes: Art, Science and Technology. New York: Elsevier Applied Science; 1991. pp. 101–126. [Google Scholar]
- Dudareva N, Murfitt LM, Mann CJ, Gorenstein N, Kolosova N, Kish CM, Bonham C, Wood K. Developmental regulation of methyl benzoate biosynthesis and emission in snapdragon flowers. Plant Cell. 2000;12:949–961. doi: 10.1105/tpc.12.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E. Biochemical and molecular genetic aspects of floral scents. Plant Physiol. 2000;122:627–633. doi: 10.1104/pp.122.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flament I, Debonneville C, Furrer A. Volatile constituents of roses: characterization of cultivars based on the headspace analysis of living flower emissions. In: Teranishi R, Buttery RG, Sugisawa H, editors. Bioactive Volatile Compounds from Plants. Washington, DC: American Chemical Society; 1993. pp. 269–281. [Google Scholar]
- Gang D, Lavid N, Zubieta C, Cheng F, Beuerle T, Lewinsohn E, Noel JP, Pichersky E. Characterization of phenylpropene O-methyltransferases from sweet basil: facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell. 2002;14:505–519. doi: 10.1105/tpc.010327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gang D, Wang J, Dudareva N, Hee Nam K, Simon JE, Lewinsohn E, Pichersky E. An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol. 2001;125:539–555. doi: 10.1104/pp.125.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudin S. Rose: genetics and breeding. Plant Breed. 2000;17:159–189. [Google Scholar]
- Guterman I, Dafny-Yelin M, Shalit M, Emanuel M, Shaham M, Piestun D, Zuker A, Ovadis M, Lavy M, Lavid N et al. An integrated genomic approach to discovering fragrance-related genes in rose petals. Flowering Newslett. 2002;32:31–37. [Google Scholar]
- Haring PA. Modern Roses. Shreveport, LA: American Rose Society; 1986. [Google Scholar]
- He XZ, Dixon RA. Affinity chromatography, substrate/product specificity and amino acid sequence analysis of an isoflavone O-methyltransferase from alfalfa (Medicago sativa L.) Arch Biochem Biophys. 1996;336:121–129. doi: 10.1006/abbi.1996.0539. [DOI] [PubMed] [Google Scholar]
- Helsper JPF, Davies JA, Bouwmeester HJ, Krol AF, van Kampen MH. Circadian rhythmicity in emission of volatile compounds by flowers of Rosa hybrida L. cv. Honesty Planta. 1998;207:88–95. [Google Scholar]
- Henrich F, Nachtigall G. Über die Einwirkung von Salpetersäure auf den Monomethylather des Orcins. Chem Ber. 1903;36:889–895. [Google Scholar]
- Ibrahim RK, Bruneau A, Bantignies B. Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol Biol. 1998;36:1–10. doi: 10.1023/a:1005939803300. [DOI] [PubMed] [Google Scholar]
- Kaiser R. Scents from rain forests. Chimia. 2000;54:346–363. [Google Scholar]
- Kakiuchi K, Ue M, Yamaguchi B, Nishimoto A, Tobe Y. Photochemical lumiketone-type rearrangement of 3-methoxyphenol promoted by AlBr3. Bull Chem Soc Jpn. 1991;64:3468–3470. [Google Scholar]
- Laempe D, Jahn M, Breese K, Schägger H, Fuchs G. Anaerobic metabolism of 3-hydroxybenzoate by the denitrifying bacterium Thauera aromatica. J Bacteriol. 2001;183:968–979. doi: 10.1128/JB.183.3.968-979.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R. Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Natl Acad Sci USA. 2000;97:2934–2939. doi: 10.1073/pnas.97.6.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn E, Schalechet F, Wilkinson J, Matsui K, Tadmor Y, Kyoung-Hee N, Amar O, Lastochkin E, Larkov O, Ravid U et al. Enhanced levels of the aroma and flavor compound S-linalool by metabolic engineering of the terpenoid pathway in tomato fruits. Plant Physiol. 2001;127:1256–1265. [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn E, Ziv-Raz I, Dudai N, Tadmor Y, Lastochkin E, Larkov O, Chaimovitsh D, Ravid U, Putievsky E, Pichersky E et al. Biosynthesis of estragole and methyl-eugenol in sweet basil (Ocimum basilicum L): developmental and chemotypic association of allylphenol O-methyltransferase activities. Plant Sci. 2000;160:27–35. doi: 10.1016/s0168-9452(00)00357-5. [DOI] [PubMed] [Google Scholar]
- Manning K. Isolation of nucleic-acids from plants by differential solvent precipitation. Anal Biochem. 1991;195:45–50. doi: 10.1016/0003-2697(91)90292-2. [DOI] [PubMed] [Google Scholar]
- Mookherjee BD, Trenkle RW, Wilson RA. Live vs. dead: Part II. A comparative analysis of the headspace volatiles of some important fragrance and flavor raw materials. J Ess Oil Res. 1989;2:85–90. [Google Scholar]
- Oka N, Ohishi H, Hatano T, Hornberger M, Sakata K, Watanabe N. Aroma evolution during flower opening in Rosa damascena Mill. Z Naturforsch Sect C Biosci. 1999;54:889–895. [Google Scholar]
- Raguso RA, Pichersky E. Floral volatiles from Clarkia breweri and C. concinna (Onagraceae): recent evolution of floral scent and moth pollination. Plant Syst Evol. 1995;194:55–67. [Google Scholar]
- Scalliet G, Journot N, Jullien F, Baudino S, Magnard JL, Channeliere S, Vergne P, Dumas C, Bendahmane M, Cock JM, Hugueney P (2002) Biosynthesis of the major scent components 3,5-dimethoxytoluene and 1,3,5-trimethoxybenzene by novel rose O-methyltransferases. FEBS Lett (in press) [DOI] [PubMed]
- Schröder G, Wehinger E, Schröder J. Predicting the substrates of cloned plant O-methyltransferases. Phytochemistry. 2002;59:1–8. doi: 10.1016/s0031-9422(01)00421-6. [DOI] [PubMed] [Google Scholar]
- Shalit M, Katzir N, Tadmor Y, Larkov O, Burger Y, Schalechet F, Lastochkin E, Ravid U, Amar O, Edelstein M et al. Acetyl CoA: alcohol acetyl transferase activity and aroma formation in ripening melon fruits. J Agric Food Chem. 2001;49:794–799. doi: 10.1021/jf001075p. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Suelves M, Puigdomenech P. Specific mRNA accumulation of a gene coding for an O-methyltransferase in almond (Prunus amygdalus, Batsch) flower tissues. Plant Sci. 1998;134:79–88. [Google Scholar]
- Vainstein A, Lewinsohn E, Pichersky E, Weiss D. Floral fragrance: new inroads into an old commodity. Plant Physiol. 2001;127:1383–1389. [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dudareva N, Bhakta S, Raguso RA, Pichersky E. Floral scent production in Clarkia breweri (Onagraceae): II. Localization and developmental modulation of the enzyme SAM (iso) eugenol O-methyltransferase and phenylpropanoid emission. Plant Physiol. 1997;114:213–221. doi: 10.1104/pp.114.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pichersky E. Identification of specific residues involved in substrate discrimination in two plant O-methyltransferases. Arch Biochem Biophys. 1999;368:172–180. doi: 10.1006/abbi.1999.1304. [DOI] [PubMed] [Google Scholar]
- Weiss EA. Rosaceae. Wallingford, Oxon, UK: CAB International; 1997. Essential oil crops; pp. 393–416. [Google Scholar]
- Zuker A, Tzfira T, Vainstein A. Cut-flower improvement using genetic engineering. Biotech Adv. 1998;16:33–79. doi: 10.1016/s0734-9750(97)00063-3. [DOI] [PubMed] [Google Scholar]