Abstract
Phloem protein 2 (PP2) is one of the most abundant and enigmatic proteins in the phloem sap. Although thought to be associated with structural P-protein, PP2 is translocated in the assimilate stream where its lectin activity or RNA-binding properties can exert effects over long distances. Analyzing the diversity of these proteins in vascular plants led to the identification of PP2-like genes in species from 17 angiosperm and gymnosperm genera. This wide distribution of PP2 genes in the plant kingdom indicates that they are ancient and common in vascular plants. Their presence in cereals and gymnosperms, both of which lack structural P-protein, also supports a wider role for these proteins. Within this superfamily, PP2 proteins have considerable size polymorphism. This is attributable to variability in the length of the amino terminus that extends from a highly conserved domain. The conserved PP2 domain was identified in the proteins encoded by six genes from several cucurbits, celery (Apium graveolens), and Arabidopsis that are specifically expressed in the sieve element-companion cell complex. The acquisition of additional modular domains in the amino-terminal extensions of other PP2-like proteins could reflect divergence from its phloem function.
In higher vascular plants, sugars and other photoassimilates are transported throughout the plant body in the conducting cells of the phloem tissue. Renewed interest in this tissue arose from evidence accumulating over the past decade that the conducting cells of the phloem also carry numerous structural and informational molecules including a variety of proteins and mRNAs. Furthermore, such macromolecules have been implicated in cell-to-cell trafficking, long-distance transport, and in some cases, are reversibly exchanged between the conducting cells, or sieve elements, and the intimately associated companion cells (for review, see Thompson and Schulz, 1999). The transport of molecular information within the phloem is an obvious mechanism for long distance communication between organs, and indirect evidence supports the hypothesis that events that occur in the phloem tissue can control plant development and physiology (Ruiz-Medrano et al., 2001). However, the mechanisms controlling information flow via the phloem are still largely unknown.
Sieve elements are terminally differentiated cells that undergo exceptional cytoplasmic reorganization to become functional conductive cells capable of long-distance translocation. Mature sieve elements are characteristically devoid of nuclei and ribosomes (Oparka and Turgeon, 1999). Reorganization of the endomembrane system results from degeneration of the tonoplast and dictyosomes and changes in the endoplasmic reticulum (Sjölund and Shih, 1983). Sieve elements are intimately (structurally, developmentally, and functionally) associated with companion cells and are capable of exchanging information via specialized plasmodesmata (for review, see van Bel and Kempers, 1997). Early stages of sieve element differentiation are characterized by the appearance of structurally distinct cytoplasmic proteins, collectively called P-protein. Ultrastructural investigations originally defined P-proteins as idiosyncratic components of the structural architecture of sieve elements (Cronshaw and Esau, 1967). Depending upon the plant species, P-proteins form fibrillar, tubular, or crystalline inclusions whose accumulation and structural state appear to be strictly controlled during differentiation. Elegant work using confocal laser scanning microscopy recently demonstrated a rearrangement of the distribution of P-proteins in the sieve elements after injury or irradiation (Knoblauch and van Bel, 1998, Knoblauch et al., 2001). Plugging of sieve plates to maintain turgor pressure within the sieve tube after injury to a sieve element is the most generally accepted role for these proteins, although other functions in pathogen and pest defense have been proposed (Read and Northcote, 1983).
P-protein, as a structural entity, has been observed in sieve elements of all dicotyledons examined (Evert, 1990) and in the majority of monocotyledons, although conspicuously absent in families such as the Poaceae (Eleftheriou, 1990). The lack of P-protein also appears to be a consistent feature of gymnosperms (Schulz, 1990) and seedless vascular plants. Cucurbits have been used as a model plant for many phloem studies because of their distinctive phloem anatomy and prolific vascular exudation. In Cucurbita spp., two predominant P-proteins, the phloem filament protein or phloem protein 1 (PP1) and the phloem lectin or phloem protein 2 (PP2), have been associated with the structural P-protein filaments (Cronshaw and Sabnis, 1990). In vitro studies have shown PP1 to be the primary structural protein capable of forming P-protein filaments (Kleinig et al., 1975), and PP2, a dimeric poly-GlcNAc-binding lectin, to be covalently linked to the filaments by disulfide bridges (Read and Northcote, 1983). The expression of PP1 and PP2 is developmentally related to defined stages of phloem differentiation (Dannenhoffer et al., 1997). In addition, PP2 has the capacity to interact with mesophyll plasmodesmata to increase the size exclusion limit and traffic cell-to-cell (Balachandran et al., 1997). This property reflects the apparent intercellular movement of PP2 within the sieve element-companion cell complex: PP2 mRNA was detected only in companion cells, although the protein accumulates in the sieve elements (Bostwick et al., 1992; Dannenhoffer et al., 1997). Additional experiments demonstrated that soluble, unpolymerized PP2 subunits translocate within sieve elements from source to sink tissues, and cycle between sieve elements and companion cells (Golecki et al., 1999). Recent in vitro studies have shown that PP2 interacts with a variety of RNAs and could be involved in the long distance movement of viroids (Gomez and Pallas, 2001; Owens et al., 2001).
The presence of translocatable subunits in addition to the structural P-protein polymers offers new functional possibilities for this group of proteins in the plant. Although structural P-protein is widespread among vascular plants, the biochemical and molecular characterization of the P-protein subunits is limited to Cucurbita spp. To further understand the diversity of these proteins and the presence of functionally significant domains, additional PP2 clones were empirically identified and used to anchor database searches of this gene in other angiosperm species. The results show that the phloem lectin is a member of a new family of proteins sharing a signature found in a large number of uncharacterized genes within angiosperms.
RESULTS
Two Forms of the Phloem Lectin Are Found in Cucumis spp.
In the cultivated Cucumis spp., cucumber (Cucumis sativus) and melon (Cucumis melo), two distinct forms of poly-GlcNAc-binding lectins with apparent molecular masses of approximately 26 and 17 kD were identified after affinity chromatography of vascular exudate (Fig. 1). To characterize the genes encoding these two lectins, two strategies were undertaken to isolate from both species their cDNA and the corresponding genomic clones.
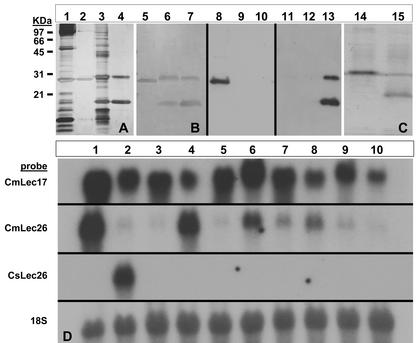
Figure 1.
Characterization of the Cucumis spp. phloem lectins. A through C, Protein analyses of the Cucumis spp. lectins. A, Silver-stained SDS-PAGE of vascular exudate from winter squash (Cbm; lane 1) or melon (Cmm; lane 3) and chitotriose affinity-purified CbmPP2 (lane 2) and Cmm lectins (lane 4). B, Silver-stained SDS-PAGE of chitotriose affinity-purified CbmPP2 (lane 5), cucumber (Cms) lectins (lane 6), and Cmm lectins (lane 7). Immunoblot CbmPP2 (lane 8), Cms lectins (lane 9), and Cmm lectins (lane 10) reacted with anti-CbmPP2 polyclonal antibodies. Immunoblot CbmPP2 (lane 11), Cms lectins (lane 12), and Cmm lectins (lane 13) reacted with anti-Cmm lectin polyclonal antibodies. Chitotriose affinity-purified recombinant CmsLec26 (lane 14) and CmsLec17 (lane 15). Molecular mass markers correspond to A and B. D, RNA-blot analysis of lectin gene expression in 10 Cucumis spp. 1, melon; 2, cucumber; 3, C. africanus; 4, C. callosus; 5, C. dipsaceus; 6, C. heptadactylus; 7, C. meeusei; 8, C. metuliferus; 9, C. myriocarpus; 10, C. sagittatus. RNA blots were sequentially probed with 32P-labeled cDNAs for the CmmLec17, CmmLec26, CmsLec26, and 18S ribosomal genes.
In melon, a full-length cDNA was isolated from a melon cDNA library probed with a partial cDNA generated by reverse transcriptase (RT)-PCR and obtained from the sequence of a putative lectin from cucumber (Toyama et al., 1995). The full-length 751-bp cDNA clone, CmmLec17-1, contains a 465-bp open reading frame (ORF) encoding a putative polypeptide of 154 amino acids with a calculated molecular mass of 17.4 kD (Table I) that fits to the smaller lectin observed in the phloem exudate. In cucumber, a partial cDNA clone, CmsLec17, was generated by RT-PCR and contained a 468-bp ORF encoding a putative polypeptide of similar length (155 amino acids), with a calculated molecular mass of 17.8 kD. CmsLec17 overlapped the uncharacterized partial cDNA clone previously described by Toyama et al. (1995). Cmm and Cms Lec17 proteins deduced from the sequence of these clones shared 75% amino acid identity.
Table I.
Summary of characterized PP2 genes and proteins
| Gene | Sequencea | Species | Molecular Mass | Amino Acid | ORF | Exon | Intron | Exon | Intron | Exon | GenBank No. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CbmPP2b | g | Winter squash | 24.474 | 219 | 657 | 168 | 97 | 107 | 91 | 381 | L31550 |
| CbpPP2 | c | Pumpkin (Cucurbita pepo) | 24.565 | 219 | 657 | ** | ** | ** | ** | ** | Z22647 |
| CbmosPP2 | c | Cucurbita moschata | 24.49 | 216 | 651 | ** | ** | ** | ** | ** | AF150627 |
| CbaPP2-S1 | c | Cucurbita argyrosperma | 24.491 | 216 | 651 | ** | ** | ** | ** | ** | AF520582 |
| CbaPP2-6 | c | C. argyrosperma | 24.522 | 216 | 651 | ** | ** | ** | ** | ** | L32700 |
| CbaPP2-7 | c | C. argyrosperma | 24.357 | 216 | 651 | ** | ** | ** | ** | ** | L32701 |
| CbdPP2 | c | Cucurbita digitata | 24.379 | 218 | 657 | ** | ** | ** | ** | ** | AF520583 |
| CmsLec26c | c | Cucumber | 25.924 | 225 | 678 | ** | ** | ** | ** | ** | AF520581 |
| CmmLec26 | c | Melon | 26.233 | 226 | 681 | ** | ** | ** | ** | ** | AF517154 |
| CmmLec17-1 | c | Melon | 17.441 | 154 | 465 | ** | ** | ** | ** | ** | AF517156 |
| CmmLec17-3 | c | Melon | 17.429 | 154 | 465 | ** | ** | ** | ** | ** | AF517157 |
| CmsLec17 | c | Cucumber | 17.8 | 155 | 468 | ** | ** | ** | ** | ** | AF517155 |
| CmmLec17 | g | Melon | 17.427 | 154 | 465 | ** | ** | 104 | 187 | 361 | AF520577 |
| CmsLec17-1d | g | Cucumber | 17.497 | 154 | 465 | ** | ** | 104 | 187 | 361 | AF520578 |
| CmsLec17-5 | g | Cucumber | 17.768 | 156 | 468 | ** | ** | 104 | 189 | 364 | AF520579 |
| CmsLec17-7 | g | Cucumber | 17.624 | 155 | 465 | ** | ** | 104 | 188 | 361 | AF520580 |
| AgPP2-1 | c | Celery | 19.849 | 181 | 546 | ** | ** | ** | ** | ** | AY114139 |
| AgPP2-2 | c | Celery | 19.732 | 179 | 540 | ** | ** | ** | ** | ** | AY114140 |
| AtPP2-A1 | c and g | Arabidopsis | 28.100 | 247 | 741 | 246 | 387 | 122 | 238 | 376 | At4g19840 |
| AtPP2-A2 | c and g | Arabidopsis | 17.914 | 155 | 465 | ** | ** | 92 | 91 | 373 | At4g19850 |
Genomic DNA-blot analysis of melon DNA indicated a presence of at least four genes (data not shown), one of which (CmmLec17) was isolated and sequenced. The 1,543-bp genomic clone encoded an ORF composed of two exons that were identical to the CmmLec17-1 cDNA (Table I). In cucumber, four independent genomic clones corresponding to the CmsLec17 cDNA were isolated, three of which were analyzed in detail (CmsLec17-1, CmsLec17-5, and CmsLec17-7; Table I). Intergenic differences in the ORFs of the three genes translated to small differences in the putative proteins, and the CmsLec17-5 gene corresponded to the CmsLec17 cDNA. The intron/exon structure of all three genes was similar to that of the genomic clone isolated from melon (Table I), and the sequence of both introns and exons was highly conserved.
On the basis of the N-terminal amino acid sequence [VEIETEARESLQIQESYGHSLTYILPK] determined from the cucumber 26-kD lectin found in phloem exudate, a nested set of degenerate 5′ primers were designed and used in cucumber to obtain by RT-PCR a 769-bp partial cDNA, CmsLec26. Combining the empirically determined N-terminal amino acid sequence with the sequence deduced from CmsLec26 revealed a 225-amino acid polypeptide with a calculated molecular mass of 25.9 kD. The corresponding melon clone, CmmLec26, was isolated after screening a cDNA library at low stringency with the CmsLec26 probe. The 875-bp CmmLec26 cDNA contained a 681-bp ORF encoding a 226-amino acid polypeptide with a calculated molecular of 26.2 kD. The amino acid sequences of the Cmm and Cms Lec26 proteins were less conserved between the species (65.8% identity) than the Lec17 proteins. The intraspecific conservation between the Lec26 and Lec17 genes was much lower, with only 33.7% and 27.7% identity for melon and cucumber, respectively.
Cucumis spp. Lectins Retain a Chitin-Binding Lectin Activity and Specific Expression in the Phloem
Despite their divergence, Cucumis spp. Lec17 and Lec26 genes are related in their functional characteristics and tissue specificity. Single-step affinity chromatography of recombinant proteins expressed in Escherichia coli, from both the melon CmmLec17 and CmmLec26, revealed that the cloned genes encode chitin-binding lectins (Fig. 1). The most obvious difference between the 17- and 26-kD Cucumis spp. lectins is the absence of 62 amino acids from the N terminus in the 17-kD proteins, suggesting that this region is not essential for the lectin activity. In situ hybridization of digoxygenin-labeled riboprobes generated from CmmLec17 and CmsLec26 showed a similar pattern of companion cell-specific gene expression in the phloem of melon and cucumber stems (Fig. 2).
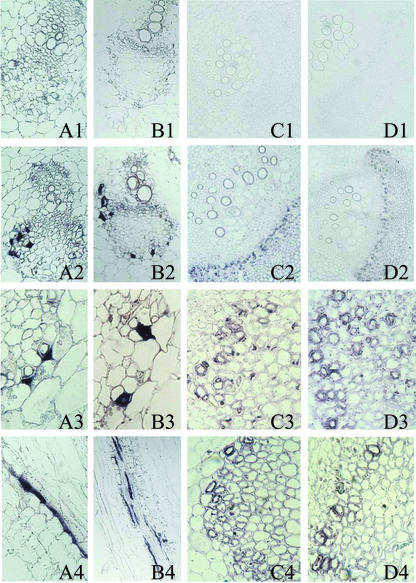
Figure 2.
Phloem localization of PP2 mRNAs in Cucumis spp. and celery. A1 through A4, Localization of mRNA encoding the 17-kD PP2 in the phloem of melon seedling hypocotyls 3 d after germination. A1, Sense riboprobe (40×). A2 through A4, Antisense riboprobes (40×, 100×, and 64×). B1 through B4, Localization of mRNA encoding the 26-kD PP2 in the phloem of cucumber seedling hypocotyls 6 d after germination. B1, Sense riboprobe (26×). B2 through B4, Antisense riboprobes (26×, 100×, and 26×). C1 through C4, Localization of mRNA encoding the AgPP2-1 in the phloem of celery leaf petioles. C1, Sense riboprobe (10×). C2 and C3, Antisense riboprobe (63× and details). C4, Antisense oligonucleotide probe (details 63×). D1, Negative control oligonucleotide probe (10×). D2 through D4, Localization of mRNA encoding the AgPP2-2 in the phloem of celery leaf petioles. D2 and D3, Antisense riboprobe (63× and details). D4, Antisense oligonucleotide probe (details 63×).
Intrageneric and Intergeneric Divergence of the Cucurbitaceae Phloem Lectins
The divergence of the phloem lectins within the Cucumis spp. was even greater when compared with other genera within the Cucurbitaceae family. The 26-kD Cucumis spp. lectins shared only 51.6% (melon) and 47.5% (cucumber) amino acid sequence identity with PP2 of winter squash (Cucurbita maxima; CbmPP2). The identity of the 17-kD Cucumis spp. lectins was further reduced to 44.3% (melon) and 39.7% (cucumber) identity with CbmPP2. This was corroborated by the lack of cross-reactivity of polyclonal antibodies generated against both melon CmmLec17 and CmmLec26 lectins with CbmPP2 from winter squash (Fig. 1). The melon antibodies also failed to cross-react with both the 17- and 26-kD lectins from cucumber. Although the genes encoding the two phloem lectins were not isolated from all Cucumis spp., RNA-blot analysis showed that orthologs of these genes exist in species throughout the genus. Hybridization data using a melon CmmLec17-1 probe (87.5% nucleotide identity between CmmLec17 and CmsLec17 ORF) indicates the orthologous genes encoding the 17-kD lectin are highly conserved among the 10 Cucumis spp. tested (Fig. 1). In contrast, the genes encoding the 26-kD lectins appear to have diverged as evidenced by RNA blots showing considerable variability in the strength of hybridization signals to the melon CmmLec26 probe (Fig. 1). The melon probe strongly hybridized to RNA isolated from four Cucumis spp. (melon, Cucumis callosus, Cucumis heptadactylus, and Cucumis metuliferus) and weakly hybridized to the remaining six species (cucumber, Cucumis africanus, Cucumis dipsaceus, Cucumis meeusei, Cucumis myriocarpus, Cucumis sagittatus) tested. Interestingly, the cucumber probe only hybridized to the cucumber RNA suggesting at least three divergent forms of the gene encoding the 26-kD phloem lectin occur within the genus.
Two PP2 Genes Are Expressed in the Phloem of Celery (Apium graveolens)
In the phloem of celery, one of the most abundant mRNA species, as revealed by a relative high frequency of expressed sequence tags (ESTs) in several celery phloem cDNA libraries (F. Vilaine and S. Dinant, unpublished data), corresponded to a gene encoding a PP2-like protein. The full-length cDNA, AgPP2-1, isolated from one of these cDNA libraries, was 818 bp and contained a 546-bp ORF encoding a 181-amino acid polypeptide with a calculated molecular mass of 19.8 kD. The deduced protein encoded by AgPP2-1 shares 34.1% amino acid identity with winter squash CbmPP2 that is similar to the 39.7% identity between CbmPP2 and the cucumber CmsLec17 protein. Another related celery cDNA corresponding to a second PP2 gene, AgPP2-2, was identified in the celery phloem cDNA libraries from a less abundant EST. The 796-bp sequence contained an 540-bp ORF encoding a 179-amino acid deduced protein with a calculated molecular mass of 19.7 kD that shared 58.9% amino acid identity with AgPP2-1. In the petiole of celery, transcript profiling with cDNA macroarrays (F. Vilaine and S. Dinant, unpublished data) showed that AgPP2-1 was expressed at a high level in the phloem tissue, but not in xylem and storage parenchyma. AgPP2-2 was also expressed in the phloem, although at a lower level.
In situ hybridization of digoxygenin-labeled riboprobes generated from AgPP2-1 and AgPP2-2 showed a similar pattern of expression in the petioles of newly expanding or mature leaves of celery that was restricted to the phloem (Fig. 2). Examination of the phloem tissue at higher magnification showed strong signal in the companion cell-sieve element complex in the phloem of petioles (Fig. 2). These results were further confirmed by in situ hybridization of digoxygenin-labeled oligonucleotide probes using gene-specific oligonucleotides designed in the 5′ most variable region of their coding region, to rule out the possibility of cross-hybridization (Fig. 2). Although the expression of AgPP2-1 appeared to be strictly confined to these cells, a faint signal was sometimes detected in parenchyma cells with the AgPP2-2 oligonucleotide probe (data not shown).
Comparison of Cucurbit and Celery PP2 Proteins Revealed a Conserved PP2 Signature
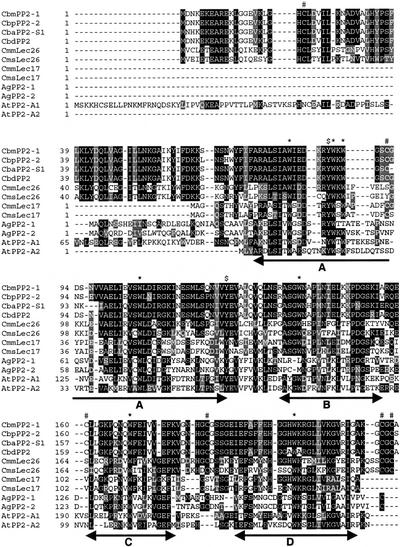
Apiales, which include Apiacae, are unrelated to Cucurbitales and belong to highly divergent subgroups, asterids and rosids, respectively. However, alignment of cucurbit and celery sequences showed a succession of four conserved motifs (A, B, C, and D) common to sequences in the two subgroups (Fig. 3). This PP2 domain signature was also found in sequences of other species such as Arabidopsis (Fig. 3) and is characterized by a high frequency of charged residues and seven conserved Trp residues (CbmPP2 Trp-80, -87, -89, -105, -138, -168, and -199). Two Tyr residues appeared to be also very well conserved (CbmPP2 Tyr-86 and -128). Although reported to play an important functional role in dimerization of the PP2 subunits and oxidative cross-linking to the phloem filaments (Read and Northcote, 1983), Cys residues were poorly conserved in both their number and location within the different PP2 proteins (Fig. 3). This PP2 signature was further used to anchor database searches for PP2-like genes in other species.
Figure 3.
Amino acid sequence alignment of celery and cucurbit PP2 proteins. The alignment was performed using the ClustalW method (v1.7; Thompson et al., 1994) available in the GCG package. White letters on black are identical amino acid residues; white on dark gray are strongly similar; and black on light gray are weakly similar. The conserved motifs A through D of the PP2 domain are underlined. Gaps were introduced to produce the alignment. *, Conserved Trp residues. #, Conserved Cys residues. $, Conserved Tyr residues.
Identification of 30 PP2-Like Genes in Arabidopsis
Thirty unique sequences encoding a PP2-like domain were identified in database searches of the Arabidopsis genome (Table II), only one of which had been previously verified as a transcribed gene (AtPP2-B9; Farràs et al., 2001). The PP2-like genes are distributed on each of the five chromosomes, and several genes appeared to result from local gene duplication. The most extreme duplication of PP2-like genes appears to have occurred on chromosome 2 where a cluster of 10 tandem repeats (AtPP2-B1 to AtPP2-B10) was identified within a 30-kb region.
Table II.
Summary of PP2-like genes identified in the Arabidopsis genome
| Name | Genea | Amino Acidb | Molecular Massc | pId | EST or cDNAe |
|---|---|---|---|---|---|
| AtPP2-A1 | At4g19840 | 246 | 28.1 | 9.81 | AV557741; AV527977; AI994461; AV553593; Z26078; AI995445;T41813; T88033; R65550; H37543; AV565967; AI099762; H36105; R30274; AV557705; F15502; T44114 |
| AtPP2-A2 | At4g19850 | 154 | 17.9 | 6.08 | AV567627; AV518792; AV558075; AV522456; AV563053; AV567198; AI998961; AV537530; AV520283 |
| AtPP2-A3 | At2g26820 | 463 | 52.2 | 6.94 | – |
| AtPP2-A4 | At1g33920 | 165 | 19.1 | 4.98 | – |
| AtPP2-A5 | At1g65390 | 431* | 50.2 | 5.89 | AI996860 |
| AtPP2-A6 | At5g45080 | 392 | 44.1 | 9.01 | – |
| AtPP2-A7 | At5g45090 | 297 | 33.2 | 5.00 | – |
| AtPP2-A8 | At5g45070 | 354 | 40.1 | 5.85 | AV551981; AV541701; AI998857T42739; AA395151 |
| AtPP2-A9 | At1g31200 | 180 | 20.3 | 10.25 | AV543510; AV558142; BE522427; BE039440; AV518209; AA597395 |
| AtPP2-A10 | At1g10150 | 184 | 20.9 | 9.94 | AA651176 |
| AtPP2-A11 | At1g63090 | 289 | 32.5 | 7.80 | F13737; AI998874; AV536500 |
| AtPP2-A12 | At1g12710 | 291 | 33.2 | 7.81 | AV550312; AV552118 |
| AtPP2-A13 | AT3g61060 | 254 | 29.1 | 9.03 | AV553381; AV544847; BE039382; AV526557, AV527225; AV553919; R90540; F14157 |
| AtPP2-A14 | At5g52120 | 291 | 33.6 | 8.93 | AI997403 |
| AtPP2-A15 | At3g53000 | 300 | 34.2 | 6.59 | – |
| AtPP2-B1 | At2g02230 | 317 | 34.7 | 5.05 | AI995319 |
| AtPP2-B2 | At2g02250 | 305 | 34.4 | 8.11 | – |
| AtPP2-B3 | At2g02270 | 265 | 30.5 | 6.14 | – |
| AtPP2-B4 | At2g02280 | 144 | 16.5 | 9.8 | – |
| AtPP2-B5 | At2g02300 | 284 | 32.0 | 8.9 | – |
| AtPP2-B6 | At2g02310 | 307 | 34.5 | 5.37 | – |
| AtPP2-B7 | At2g02320 | 306 | 35.5 | 7.80 | – |
| AtPP2-B8 | At2g02340 | 305 | 35.2 | 8.48 | – |
| AtPP2-B9(SKIP3) | At2g02350 | 182* | 20.7 | 8.62 | AV556684; AF263379 |
| AtPP2-B10 | At2g02360 | 272 | 31.3 | 8.41 | – |
| AtPP2-B11 | At1g80110 | 264 | 29.7 | 5.78 | BE522361 |
| AtPP2-B12 | At5g24560 | 253 | 29.4 | 4.64 | – |
| AtPP2-B13 | At1g56240 | 284 | 31.9 | 6.88 | BE662898 |
| AtPP2-B14 | At1g56250 | 283 | 31.8 | 5.86 | – |
| AtPP2-B15 | At1g09155 | 288 | 32.5 | 6.99 | BE524163 |
Gene accession nos.
No. of predicted amino acids annotated in TAIR; *, no. of amino acids not annotated.
Predicted calculated Mr.
Predicted pI.
GenBank accession nos. for EST or cDNA (in italics).
The presence of one or more ESTs or cDNAs corresponding to the Arabidopsis PP2-like genes indicate that at least 15 of the 30 genes are actively transcribed during normal Arabidopsis growth and development (Table II). Two tandem genes, AtPP2-A1 and -A2 were of particular interest because they showed the highest amino acid identities with CbmPP2 and CmmLec26 (37%–40%) as well as AgPP2-1 and AgPP2-2 (33%–38%). The predicted intron/exon junctions were correctly annotated for AtPP2-A1 and showed that three exons (246, 122, and 376 bp) separated by two introns (387 and 238 bp) encoded a 246-amino acid protein with a calculated molecular mass of 28.1 kD (Table I). The cDNA sequences for AtPP2-A2 revealed that the predicted gene sequence with its continuous ORF was incorrectly annotated. The cDNA sequences aligned with two exons (92 and 373 bp) encoding a 155-amino acid protein with a calculated molecular mass of 17.9 kD separated by a single 91-bp intron.
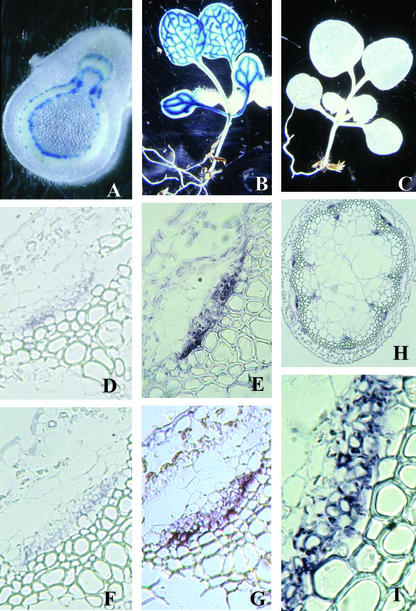
Tissue-specific expression for the tandem AtPP2 genes was first evaluated using histochemical localization of promoter-β-glucuronidase (GUS) fusions in transgenic Arabidopsis and tobacco (Nicotiana tabacum) plants. Approximately 1 kb of 5′-flanking sequence from AtPP2-A1 was sufficient to direct high levels of GUS activity in the vascular tissue of Arabidopsis and in the phloem of the bicollateral vascular bundles of tobacco stems (Fig. 4). A similar pattern of vascular expression was observed in transgenic Arabidopsis with 1 kb of 5′-flanking sequence from the AtPP2-A2 gene, although detected at very low levels (Fig. 4). Phloem-localized expression of AtPP2-A1 and AtPP2-A2 was confirmed by in situ hybridization experiments. Digoxigenin-labeled antisense oligonucleotide-specific probes for both genes hybridized to mRNA in the phloem tissue of Arabidopsis floral stem sections (Fig. 4). The hybridization signal was restricted to the companion cell-sieve element complex and was mostly associated with companion cells. In a few cases, a faint signal was also observed with AtPP2-A1 and AtPP2-A2 probes in a few small cells of the xylem that was not detected with sense oligonucleotide probes.
Figure 4.
Tissue and cellular specificity of AtPP2-A1 and AtPP2-A2 gene expression. A through C, Histochemical localization in transgenic tobacco and Arabidopsis plants of AtPP2-A1 and AtPP2-A2 promoter:GUS fusions. A, GUS expression directed by approximately 1 kb of 5′-flanking region from AtPP2-A1 in a cross-section of a transgenic tobacco stem. B, GUS expression directed by approximately 1 kb of 5′-flanking region from AtPP2-A1 in an Arabidopsis seedling. C, GUS expression directed by approximately 1 kb of 5′-flanking region from AtPP2-A2 in an Arabidopsis seedling. D through I, Localization of mRNA encoding the AtPP2-A1 and AtPP2-A2 in the phloem of Arabidopsis floral stem by in situ hybridization, using oligonucleotide probes. E, H, and I, Localization of mRNA encoding AtPP2-A1 (E, 63×; H, 5×; I, details 63×). G, Localization of mRNA encoding AtPP2-A2 (63×). D and F, Sense oligonucleotide probe (negative control) for AtPP2-A1 (D; 63×) and AtPP2-A2 (F; 63×).
Identification of Widely Conserved Domains
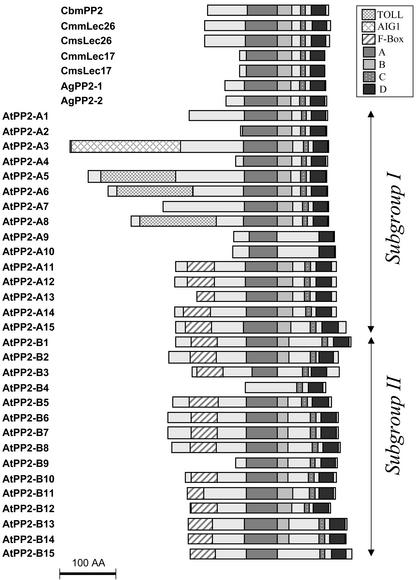
Comparison of the 30 Arabidopsis predicted proteins with CbmPP2 using the GAP program showed a range of amino acid identity extending up to 41.0% identity. Proteins with molecular masses corresponding to the two subgroups of cucurbit phloem lectins (17 and 24–26 kD) and the celery AgPP2 proteins (19–20 kD) were also present in Arabidopsis. In addition, proteins with much higher theoretical molecular masses such as AtPP2-A8 (50.2 kD) and AtPP2-A7 (52.2 kD) were identified (Table II). The Arabidopsis PP2-like proteins separate into subgroups I and II based on the conservation of the conserved motifs of the PP2 domain and the presence of N-terminal extensions. Figure 5 shows a schematic diagram of the Arabidopsis PP2-like proteins. AtPP2-like proteins were named after their phylogenetic relationship with the PP2 type member CbmPP2 and relatedness into the two subgroups. The PP2 domain is 120 to 150 amino acids in length and in all cases is located in the central and C-terminal regions of the protein. The A, C, and D motifs of the PP2 domain are highly conserved among most proteins, whereas the B motif is less conserved and precedes a divergent region that is not found in cucurbit phloem lectins. The length of this inter-domain region is variable and explains most of the length heterogeneity detected in the central and C-terminal region. Besides size polymorphism, the Arabidopsis PP2-like proteins showed variations in their electric charge, as revealed by their predicted pI (Table II).
Figure 5.
Schematic diagram of conserved domains in PP2 and PP2-like proteins. The presence or absence of each of the four motifs (A–D) of the PP2 domain in cucurbit, celery, or Arabidopsis proteins is shown. Also shown are the predicted AIG1, F-Boxes, and TOLL domains located in the N-terminal extensions of the Arabidopsis proteins. The Arabidopsis proteins have been grouped (I and II) according to the overall domain structure. Bar = 100 amino acids.
Analysis of Arabidopsis PP2-like proteins with the domain search programs available at the Swiss Institute for Experimental Cancer Research Bioinformatics (Epalinges, Switzerland) allowed the identification of three additional domains in the N-terminal region of some of the predicted proteins (Fig. 5). Three proteins in subgroup I, AtPP2-A5, AtPP2-A6, and AtPP2-A8, present a Toll/Il-1R (TIR) domain consisting of 125 residues in the N terminus of the proteins. The TIR domain of the protein encoded by the N resistance gene from Nicotiana glutinosa (Whitham et al., 1994) matched with significant conservation (35%–39% identity, E value = 5e−19) to these three AtPP2 proteins. In the same subgroup, the 175 N-terminal residues of the AtPP2-A3 protein shows high conservation (51.3% identity, E value = 1e−46) with the N-terminal domain of the protein encoded by the Arabidopsis defense gene AIG1 (Reuber and Ausubel, 1996) and other AIG1-like genes in Arabidopsis and other plant species (data not shown). The third motif, a typical F-Box of approximately 40 residues, was located in the N-terminal extension of most proteins in the second subgroup and in a few proteins in subgroup one (Fig. 5).
Phylogenetic Analyses Revealed Ancient Duplication of PP2-Like Genes
PP2-like genes were found in eight dicot species including members of the families Solanaceae, Fabaceae, Malvaceae, and Aizoaceae and in four monocot species in the Poaceae (Table III) with significant probability (E value > 1e−18). The presence of several putative paralogs within multiple species, including the monocot rice and the dicots tomato and soybean, suggests that multigene PP2 families are common in angiosperms (Table III). Furthermore, several related EST sequences associated with at least three different genes were found in the gymnosperm Pinus taeda, and an EST showing a low, although significant score, was found in the moss Physcomitrella patens. Although most of these PP2-like sequences were deduced from ESTs, and therefore partial, when available the sequences readily aligned with the conserved PP2 domain. The A, C, and D motifs of the PP2 domain appeared to be very well conserved throughout these species, whereas the B motif was less conserved. The length of the central and C-terminal region was conserved throughout these sequences, and most of the length variation occurred in the N-terminal region of the predicted proteins. Six of the seven conserved Trp residues initially identified were well conserved among proteins (CbmPP2 Trp-80, -87, -89, -105, -168, and -199) and the two conserved Tyr residues (CbmPP2 Tyr-86 and -128). Although several Cys residues were found throughout these proteins, their position was not conserved.
Table III.
The PP2-like gene family in plant species
| Species | Family | Gene | Accession No.a |
|---|---|---|---|
| Celery | Apiaceae | AgPP2 | Table I |
| Arabidopsis | Brassicaceae | AtPP2 | Table II |
| Cicer arietinum | Fabaceae | CaPP2 | AJ271666 |
| C. argyrosperma | Cucurbitaceae | CbaPP2 | Table I |
| C. digitata | Cucurbitaceae | CbdPP2 | Table I |
| C. moschata | Cucurbitaceae | CbmosPP2 | Table I |
| Winter squash | Cucurbitaceae | CbmPP2 | Table I |
| Pumpkin | Cucurbitaceae | CbpPP2 | Table I |
| Melon | Cucurbitaceae | CmmPP2 | Table I |
| Cucumber | Cucurbitaceae | CmsPP2 | Table I |
| Cotton (Gossypium hirsutum) | Malvaceae | GhPP2 | AI731302 |
| Soybean (Glycine max) | Fabaceae | GmPP2-1 | AJ010265 |
| GmPP2-2 | AW620778, AW203522 | ||
| GmPP2-3 | AW309707 | ||
| GmPP2-4 | BE658530, AW310374 (+) | ||
| Common ice plant (Mesembryanthemum crystallinum) | Aizoaceae | McPP2 | BE035638 |
| Medicago truncatula | Fabaceae | MtPP2-1 | AW584333 |
| MtPP2-2 | BG647197 | ||
| Tobacco | Solanaceae | Nt Nictaba | AF389848 |
| Solanum lycopersicum | Solanaceae | SlPP2-1 | AW617857, AW036335 |
| SlPP2-2 | AW616959 | ||
| SlPP2-3 | AW621391 | ||
| SlPP2-4 | AI488236 | ||
| Potato (Solanum tuberosum) | Solanaceae | StPP2 | BG5999036 |
| Barley (Hordeum vulgare) | Poaceae | HvPP2-1 | AW982836 |
| HvPP2-2 | BE216413 | ||
| Rice (Oryza sativa) | Poaceae | OsPP2-1 | AQ794590 |
| OsPP2-2 | AU058265, AU092429 (+) | ||
| OsPP2-3 | AAK38315 (g) | ||
| Sorghum (Sorghum bicolor) | Poaceae | SbPP2 | AW745611, AW285727 |
| Winter rye (Secale cereale) | Poaceae | ScPP2 | BE438497 |
| Pinus taeada | Pinaceae | PtPP2-1 | BE643948, BF049852 |
| PtPP2-2 | BG317827 | ||
| PtPP2-3 | AW064927 | ||
| Physcomitrella patens | Funariaceae | PpPP2 | BI436506 |
(+), Additional ESTs identified matching the same gene; (g), genomic clone.
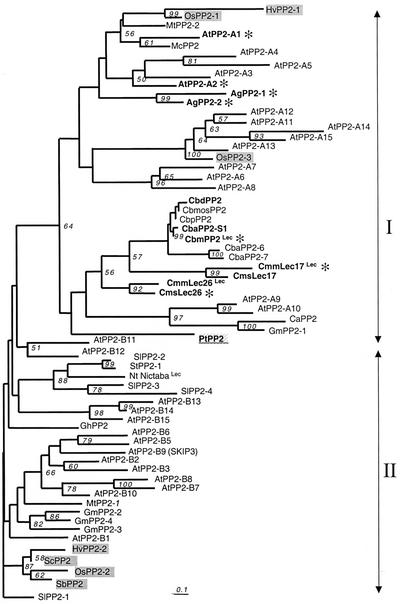
A neighbor-joining tree was constructed by using the aligned conserved domains and determined that the PP2 protein sequences fall into two distinct groups (Fig. 6). The two main branches correspond to the two AtPP2 subgroups. This reflects a polymorphism and evolution of the B motif, characterized in the second group by insertions of additional amino acid residues and loss of an otherwise well-conserved Trp residue (CbmPP2 Trp-138). In both subgroups, the divergence remained however limited, as estimated by the short branch length. A few proteins, including the Arabidopsis AtPP2-A9 and AtPP2-A10 or AtPP2-A14 and AtPP2-A15, showed faster rates of substitutions that are likely to be associated with an even more rapid evolution of the B domain.
Figure 6.
Phylogenetic relationship of PP2-like proteins. Amino acid sequences of the PP2 conserved domain (motifs A–D) were aligned using ClustalW (Thompson et al., 1994). After corrections for an high rate of substitution using the Kimura 2-parameters distance option included in ClustalW, a neighbor-joining tree (Saitu and Nei, 1987) depicting relationships among the PP2-like proteins was constructed by using the aligned amino acid sequences described in Table III. Two sequences, PpPP2 and AtPP2-B4, that showed only partial alignments were not included in the tree. The protein sequence deduced from a P. taeda EST (PtPP2) was included in the phylogenetic analysis represented as an unrooted tree. All branches are drawn to scale as indicated by the scale bar (bar = substitution/site rate of 0.1%), and their length indicates the level of divergence among sequences. Only percentages of bootstrap values supported by more than 50% of the 1,000 replicates are indicated above nodes. These sequences can be assembled into two main groups (arrows). For each protein, the prefix indicates the species from which it is derived. GenBank accession numbers and acronyms are detailed in Table III. Proteins from Arabidopsis, cucurbits, and celery that were more extensively described in this work are indicated in bold letters. Monocot proteins are indicated on a gray background. The gymnosperm protein PtPP2 is underlined on a gray background. Lec, Proteins for which GlcNAc-binding activity was demonstrated. A gray asterisk (*) indicates proteins corresponding to genes expressed in the companion cell-sieve element complex.
The grouping of proteins in either of the two branches did not clearly correlate to known properties of the phloem lectin. Chitin-binding lectin activity, demonstrated for the cucurbit proteins CmLec26, CmLec17, and CbmPP2 and the tobacco nictaba lectin (Nt nictaba; Chen et al., 2002), was found in both subgroups. In contrast, phloem-specific expression that was demonstrated for seven PP2 genes (CbmPP2, CmsLec17, CmmLec26, AgPP2-1, AgPP2-2, AtPP2-A1, and AtPP2-A2) appeared to be associated with one subgroup (Fig. 6). Interestingly, the presence of an F-Box at the N-terminal region position (Fig. 5) appeared in proteins found in the two branches. Thus, the acquisition of this domain was probably very ancient. Moreover, several species, including the monocots rice and barley and the dicots soybean and M. truncatula, showed paralogs in the two main branches, again revealing a very ancient duplication of PP2-like genes.
DISCUSSION
P-protein is a distinctive feature in the sieve elements of most vascular angiosperms and as a structural entity is widespread among disparate taxa. The phloem lectin in Cucurbita spp. immunolocalizes to P-protein filaments where it is thought to cross-link via disulfide bonds to polymers of phloem filament protein (PP1; Read and Northcote, 1983; Smith et al., 1987). However, the evidence for interactions between PP1 and PP2 in P-protein filament formation is ambiguous. Purified PP2 does not form filaments in vitro (Kleinig et al., 1975), and the presence of Cys residues is not a strictly conserved feature among PP2 proteins (Thompson, 1999). PP2 monomers had a high affinity for PP1 in non-covalent protein-binding assays (G.A. Thompson and A.M. Clark, unpublished data), raising the possibility that the in vivo associations are non-covalent and transient. PP2 is translocated in sieve elements (Golecki et al., 1999), and two independent reports have recently implicated a role for a PP2-like protein in the long-distance transport of viroids in cucumber plants (Gomez and Pallas, 2001; Owens et al., 2001). Thus, it appears that the phloem lectin has functions beyond being a putative component of the structural phloem filaments.
PP2 genes, initially described in winter squash (CbmPP2), belong to small multigene families that are highly conserved among species within the genus Cucurbita (Bostwick et al., 1994). However, the lack of nucleic acid cross-hybridization (Golecki et al., 1999) and polyclonal antibody cross-reactivity (Read and Northcote, 1983; Fig. 1) between species in different genera indicates that PP2 genes have diverged within the Cucurbitaceae, probably in part by ancient duplications. This was confirmed by the identification of several genes in Cucumis spp. encoding two distinct forms of the phloem lectin, 17 and 26 kD, that shared only approximately 40% to 50% amino acid identity with CbmPP2. The difference in the size and sequence of the two forms of the phloem lectin reflects the divergence of the PP2 genes while maintaining cell-specific gene expression, overall domain structure, and lectin activity. Furthermore, differences in antibody cross-reactivity showed that the divergence of PP2 extended to the two subgenera Cucumis (cucumber) and Melo (melon and other Cucumis spp.) described by Jobst et al. (1998). RNA hybridization studies provided additional evidence that the genes encoding the 26-kD lectin have diverged both between and within the subgenera to a greater extent than the genes encoding the 17-kD lectin. In parallel studies, two PP2 genes were identified in an unrelated plant family, the Apiaceae. These genes, AgPP2-1 and AgPP2-2, isolated from celery, were expressed in the sieve element-companion cell complex of the phloem in petioles of mature or immature leaves.
On the basis of alignments with the signature PP2 domain from Cucurbitaceae and Apiaceae proteins, extensive molecular and phylogenetic analyses identified PP2 genes found in 21 species belonging to eight dicot as well as monocot families. Within the completely sequenced Arabidopsis genome, PP2-like genes belong to a large multigene family constituted of 30 PP2-like members. Such large multigene families are not unusual in Arabidopsis, reflecting large-scale ancestral duplications (Arabidopsis Genome Initiative, 2000). Several PP2-like genes were found in tandem repeats or as clusters in the Arabidopsis genome, revealing the more recent acquisition by local duplication of some genes. However, the phylogenetic relationships between the angiosperm PP2-like genes do not appear to be hierarchical. In five species for which multiple paralogs were identified, such as Arabidopsis and rice, genes from one species are more closely related to genes of other species than to other genes within the species. This indicates that the duplications giving rise to the different subgroups of PP2-like genes were ancient and must have predated the monocot-dicot split. Although such duplications frequently generate the acquisition of new functions, no evidence supports the speculation that PP2 diversification was associated to the acquisition of distinct functions or to a distinct pattern of expression for the group of genes that are the most diverged.
The loss and acquisition of modular domains in PP2-like proteins could allow heterogeneity in their physiological properties. The overall structure of the conserved domain with its four motifs was much more variable in Arabidopsis PP2 than the cucurbit and celery. All AtPP2 proteins share the central A motif and the carboxy-terminal D motif, whereas 25 of the proteins have all four motifs of the conserved PP2 domain. Like the previously characterized PP2 proteins, size polymorphism of the N-terminal region of the proteins was found to occur among the AtPP2 proteins. However, an important variation from the previous PP2 proteins was the acquisition in more than one-half of AtPP2 proteins of additional modular domains within the N-terminal region. A TIR domain, reported in plants to be involved in the initial interaction with specific ligands that activates intracellular signaling cascades in response to pathogens (Van der Biezen and Jones, 1998) was identified in three AtPP2 proteins. A yet unassigned domain found in the N terminus of the AIG1 protein (Reuber and Ausubel, 1996) of a number of other Arabidopsis and plant proteins was also identified in one AtPP2 protein. This domain was also found in the N-terminal region of the proteins encoded by the imap-like genes that are associated to the immune response in mammals (Stamm et al., 2002). F-boxes, typically involved in targeting proteins to the E3 ubiquitinylation degradation pathway, were also identified in 18 of the predicted AtPP2 proteins, in addition to one (AtPP2-B9) identified in a screen for SKP1 interacting proteins (Farràs et al., 2001). In plants, many physiological processes such as hormone and defense responses, light signaling, circadian rhythms, and pattern formation use F-box function to direct negative regulators to the ubiquitin-mediated degradation pathway (Callis and Viestra, 2000; del Pozo and Estelle, 2000). Both the TIR domain and F-box are relatively common elements in Arabidopsis proteins (Arabidopsis Genome Initiative, 2000) and proteins with such motifs have been shown to participate in protein-protein interactions involved in various responses. The acquisition of such N-terminal extensions was found in other PP2-like genes from other species, suggesting that some PP2-like proteins participate in processes requiring protein-protein interactions. In addition to size polymorphism, variations in the pI of these proteins were observed that could be associated to many properties, such as changes in subcellular localization, polymerization, or conformation.
PP2-like genes appear to have evolved and specialized in parallel with the onset of the vascular tissues. In situ hybridization experiments in hypocotyls of winter squash seedlings established that CbmPP2 mRNA accumulates in both immature and differentiated sieve element-companion cell complexes and was tightly linked to vascular differentiation (Dannenhoffer et al., 1997). Cucumis spp., Arabidopsis, and celery all have PP2 genes that are specifically expressed in the phloem, and their pattern of expression in the sieve element-companion cell complex appears to parallel that of CbmPP2 in winter squash. The conservation of this pattern of gene expression in the highly specialized cells of the phloem in multiple unrelated taxa suggests that these proteins play a widespread role in this vascular tissue. Interestingly, the expression of one PP2 gene, CmsLec17, is down-regulated by cytokinins (Toyama et al., 1995). Cytokinins are transported in the phloem (Hoad, 1995) and in combination with auxin, are involved in vascular differentiation (Aloni, 1995).
Genes encoding PP2-like proteins were also identified in Pinus taeada and several cereals, which was surprising, because structural P-protein is not a characteristic feature in the sieve elements of gymnosperms or cereals. Chitin-binding lectins of approximately 25 kD were previously reported in sieve elements of Pinus sabiniana (Schulz et al., 1989). The polypeptide was immunolocalized in the proteinaceous crystal inclusions of sieve element plastids and was related serologically to peptides found in three other Pineaceae species. This suggests that PP2 arose early in the evolution of land plants, and that the gene duplications giving rise to the different subgroups could have also predated the angiosperm/gymnosperm split. A PP2-like gene was also found in the nonvascular plant, P. patens, possibly revealing an ancestral form of the protein. Although a trend toward the differentiation of conducting tissues is not exceptional in the Bryophyta, the organization of the leptoids found in a few species is far from that of the vascular tissues found in Spermatophyta.
The presence of a PP2-like gene in a nonvascular plant raises the possibility that PP2-like proteins have physiological properties that are not exclusively related to structural P-protein or vascular-specific functions. Some of the Arabidopsis PP2-like genes are expressed in other tissues (C. Kusiak and S. Dinant, unpublished data) or other cell types (Y. Zhu and G.A. Thompson, unpublished data). The synthesis of a PP2-like agglutinin was recently shown to be induced by jasmonate methyl ester in the cytoplasm of tobacco leaf mesophyll cells but not minor veins (Chen et al., 2002). The diversification of PP2-like genes suggests that PP2 proteins play important basic functions in higher plants with distinct physiological roles in vivo. Because more than a dozen PP2-like genes are specifically expressed in sieve element-companion cell complexes in three different angiosperms taxons, it is tempting to speculate that the functions associated with PP2 proteins favored an important step in the development and function of the vascular system. Moreover, the observation that many of these proteins have acquired additional domains generally associated to protein-protein interactions or signal transduction pathways, as well as the recent observations of the long distance translocation of CbmPP2 in sieve elements, indicate roles in intracellular and/or intercellular signaling.
MATERIALS AND METHODS
Isolation of cDNAs Encoding the 26-kD Lectin from Cucumis spp.
The 26-kD phloem lectin was isolated from vascular exudate collected from the severed stems of cucumber (Cucumis sativus cv Straight 8) plants. The exudate was diluted 1:4 in exudate extraction buffer (1 m Tris-HCl, pH 8.2, 5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 20 mm dithiothreitol, and 3.75 mm NaN3) and exudate proteins were modified with 60 mm iodoacetic acid for 2 h in the dark at 0°C. The proteins were precipitated with (NH4)2SO4 at 67% of saturation, resuspended, and desalted in a PD-10 column (Pharmacia AB, Uppsala) according to the manufacturer's instructions. Total exudate proteins (approximately 16 μg) were separated in a 15% (w/v) SDS-PAGE mini-gel, then transferred onto 0.2-μm polyvinylidene difluoride membrane (Immobilon-P, Millipore, Bedford, MA) by electroblotting (10 mm CAPS, pH 11, and 10% [v/v] methanol) and stained with Coomassie Blue R250. The predominant approximately 26-kD protein band was cut from the membrane, and the N-terminal protein sequence was determined on an ABI 477A pulsed-liquid protein sequencer (Applied Biosystems, Foster City, CA) using Edman chemistry and HPLC C18 reverse phase PTH column (Laboratory for Protein Sequence Analysis, Arizona Research Laboratories, Tucson). A nested set of two degenerate oligonucleotide primers, 5′-CCGCTCGAGAATGGTNGARATHGARAC-3′ and 5′-CCGCTCGAGCARATHCARGARAGYTAYGG-3′, were designed from the N-terminal amino acid sequence of the 26-kD protein and overlapped the nine N-terminal amino acids. Total RNA (1.5 μg) extracted from cucumber cv Straight 8 seedling hypocotyls 6 d after germination was reverse transcribed using an oligo(dT) primer according to the Superscript II (Invitrogen, Carlsbad, CA) manufacturer's instructions. The single-stranded cDNA template was PCR amplified (94°C, 5 min; 40 cycles of 94°C, 1 min; 50°C, 1 min; 72°C, 2 min; and 72°C, 10 min) using Lec26-N5′ and oligo(dT) primers. The double-stranded cDNA template was re-amplified as above using the Lec26-I5′ and oligo(dT) primers. The second round PCR product was agarose gel-purified, cloned into the BluescriptII vector, sequenced, and designated as CmsLec26 (cucumber lectin 26). CmsLec26 was used to probe a melon (Cucumis melo cv AR 5) hypocotyl cDNA library (Lambda-Zap II, Stratagene, La Jolla, CA) from which CmmLec26 was isolated.
Isolation of cDNAs Encoding the 17-kD Lectin from Cucumis spp.
A gene-specific 5′ primer (5′-CACGATATCGGCAGGCCAAAGCACAC-3′) was designed from the 5′ partial cDNA sequence of the putative phloem lectin CRR80 (GenBank accession no. D63388; Toyama et al., 1995). This primer was used in combination with an oligo(dT) primer for RT-PCR (methods described above) of total RNA isolated from cucumber cv Straight 8 or melon cv PMR5. The amplicons were cloned into the BluescriptII vector, sequenced, and designated CmmLec17. The 5′ ends of the melon cv PMR5 cDNAs were obtained by 5′-RACE from the primer 5′-GTGTGCTTTGGC-CTGC-3′. CmsLec17 was used to probe a melon cv AR 5 hypocotyl cDNA library (Stratagene Lambda-Zap II) from which CmmLec-17-1 and CmmLec17-3 were isolated.
Isolation of Genomic Clones Encoding the 17-kD Lectins from Cucumis spp.
Genomic DNA purified from leaves of either cucumber cv Straight 8 or melon cv AR 5 (Saghai-Maroof et al., 1984) was digested to completion with EcoRI. Size-specific genomic libraries were constructed from DNA fragments of ±1 kb of the respective positions of positive signals in genomic DNA blots that were separated in 0.8% (w/v) TAE agarose gel, purified, and inserted into the EcoRI site of the cloning vector Lambda Zap II (Stratagene). The bacteriophage were packaged with Gigapack II Gold (Stratagene) packaging extract and incubated with the Escherichia coli host XL1-Blue MRF′. For isolation of genomic clones, approximately 1.6 × 104 phage were transferred to nitrocellulose filters as described by Sambrook et al. (1989). Genomic clones were identified by hybridization with their respective cDNAs radiolabeled with [32P]dATP by random priming labeling (DECAprime kit, Ambion, Austin, TX). Hybridizations and washes were as described by Sambrook et al. (1989). Positive plaques were identified by autoradiography.
Affinity Purification, Immunoblot, and Recombinant Protein Analyses of Cucumis spp. Lectins
Vascular exudate collected from severed stems of melon or cucumber was diluted 1:4 in exudate extraction buffer. The lectins were affinity purified by adsorption to ovomucoid-acryl beads and eluted with 1 mm chitotriose as described by Bostwick et al. (1992). After SDS-PAGE, the gels were either silver-stained or the proteins were transferred from the gel to Immobilon-P membrane (Millipore) by electroblotting. Polyclonal antibodies against the combined affinity-purified melon 17- and 26-kD lectins were raised in New Zealand White rabbits. Blots were incubated overnight with either of the melon lectin (1:20,000 [v/v]) polyclonal antibodies. Alkaline phosphatase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) was diluted 1:10,000 for the secondary antibody reaction. The immunoblotting procedure and the detection with the chemiluminescent substrate were adapted from the Western-Light kit protocol (Tropix, Bedford, MA). The complete ORFs encoding the 17- and 26-kD proteins were PCR amplified from the corresponding cDNAs (CmmLec17 and CmmLec26) and ligated in-frame into the protein expression vector pRSETB (Invitrogen). Each construction was sequenced to confirm the maintenance of the reading frame, and fusion protein was synthesized according to the manufacturer's instructions. The recombinant proteins were purified from the E. coli lysate by ovomucoid affinity chromatography and elution with chitotriose.
Cucumis spp. RNA Isolation and Blot Analysis
Total RNA was extracted from hypocotyls of individual Cucumis spp. (melon, cucumber, Cucumis africanus, Cucumis callosus, Cucumis dipsaceus, Cucumis heptadactylus, Cucumis meeusei, Cucumis metuliferus, Cucumis myriocarpus, and Cucumis sagittatus) following the method of Gustincich et al. (1991) as modified by Clark et al. (1997). Ten micrograms of total RNA was electrophoresed in a 1.2% (w/v) agarose-glyoxal gel. RNA was transferred onto MagnaGraph membrane and sequentially probed with [32P]DNA probes generated by random priming the cDNAs CmcLec17, CmcLec26, or CscLec26. Hybridization and wash conditions were as described by the membrane manufacturer. Hybridization with 18S rDNA probes verified equal loading of total RNA.
Localization of Cucumis spp. Lectin mRNAs by in Situ Hybridization
Melon and cucumber seedling hypocotyl tissues were fixed at room temperature in 2% (v/v) glutaraldehyde/50 mm KPO4, pH 7.0, dehydrated in ethanol and tertiary butyl alcohol, and embedded in paraffin. Paraffin blocks were sectioned at 7 to 10 μm. Digoxigenin-labeled sense and antisense riboprobes were synthesized by in vitro transcription using CmmLec17 and CmsLec26 templates. Probe preparation and hybridization protocols were previously described by Bostwick et al. (1992).
Isolation of cDNAs Encoding Two 20-kD Proteins in Celery
EST determined from cDNA libraries (F. Vilaine and S. Dinant, unpublished data) constructed from phloem strands isolated from the petioles of newly expanding or mature leaves of celery (Apium graveolens var Dulce cv Vert d' Elne) were searched for PP2-like sequences. Four EST were found and corresponded to four cDNA clones from a library constructed in Lambda ZAP II (Stratagene). These sequences fell into two contigs: PA171, PA213, and PA516 corresponded to a first gene AgPP2-1; and PA554 corresponded to a second gene AgPP2-2, which shared 75% nucleotide identity. These clones were sequenced at least twice on both strands. Additional AgPP2-1 cDNA clones were obtained from a cDNA library enriched in phloem expressed sequences, generated by a suppressive subtractive hybridization kit (PCR Select cDNA Subtraction Kit, BD Biosciences Clontech, Palo Alto, CA) performed following the manufacturer's instructions (F. Vilaine and S. Dinant, unpublished data).
Localization of Celery and Arabidopsis PP2 mRNAs by in Situ Hybridization
For celery, the petioles of fully expanded leaves were used for in situ hybridization. For Arabidopsis (ecotype Wassilewskija), the stems of 8- to 12-week-old plants were used. Plants were fixed in 4% (v/v) formaldehyde in phosphate-buffered saline under vacuum twice for 20 min each, and left in fixative overnight. After fixation, tissues were washed, dehydrated, and embedded in paraffin, essentially as described by Jackson (1991). Paraffin sections (8- to 10-μL thick) were cut and attached to precoated glass slides (DAKO, Buckinghamshire, UK). Sense and antisense riboprobes were synthesized from AgPP2-1 and AgPP2-2 cDNA templates using digoxigenin (DIG-UTP, Roche Diagnostics, Indianapolis) according to the manufacturer's instructions. Gene-specific oligonucleotide probes were labeled using the DIG oligonucleotide tailing kit (Roche Diagnostics) according to the manufacturer's instruction. Sense and antisense oligonucleotides designed for the detection of AgPP2-1 transcripts were 5′-TCGATCGATACACAAACATTT-3′ and 5′-AAATCATCCCTGGTTACCA-3′. Sense and antisense oligonucleotides designed for the detection of AgPP2-2 transcripts were 5′-CAAAGCTCGTTAAGCAAGGA-3′ and 5′-AGGTGATGTTGGCAAGTGG-3′. Sense and antisense oligonucleotides designed for the detection of AtPP2-A1 were 5′-ACCAAGACTCGAAATACTTGATC-3′ and 5′- GGACTTGGTTTACCGAAAAAGAG-3′. Sense and antisense oligonucleotides designed for the detection of AtPP2-A2 were 5′-ATGAGGGTAAAGCGAAGAAAAAC-3′ and 5′- GAATGTGAACCTGTTAGAAAGG-3′. In situ hybridization with oligonucleotide probes was performed according to the manufacturer's instructions, at a hybridization temperature of 60°C. Immunodetection of the DIG-labeled probes was performed using an anti-DIG Fab fragments conjugated to alkaline phosphatase and visualized using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate substrates.
Expression of AtPP2 Promoter-GUS Constructs in Transgenic Plants
A DNA fragment from the 5′-flanking sequence of either AtPP2-A (887 bp) or AtPP2-B (1,000 bp) extending from, but not including, the first nucleotide 5′ of the translation initiating Met codon was ligated 5′ of the uidA gene in the binary vector pGPTV-BAR (Becker et al., 1992). Constructs were inserted into the Agrobacterium tumefaciens strain EHA105, and Arabidopsis ecotype Columbia was transformed using the floral dip procedure (Clough and Bent, 1998). Transgenic plants were selected with BASTA (glufosinate). Tobacco (Nicotiana tabacum cv W38) leaf discs were infected with A. tumefaciens, and BASTA (glufosinate)-resistant shoots were selected after 3 to 4 weeks in culture. Transgenic tobacco plants were rooted and grown in a greenhouse for 4 weeks before analysis of GUS expression. Transgenic Arabidopsis or tobacco plants were screened for GUS activity using the histochemical techniques described by Jefferson et al. (1987). GUS-positive Arabidopsis plants were allowed to self-pollinate and selected to the T2 generation. Transformation was confirmed by genomic DNA-blot analysis, and only plants with a single T-DNA insert were analyzed.
Identification of PP2-Like Proteins and Domain Analysis
The conserved domains of CbmPP2-1 and AgPP2-1 were used to perform BLAST (Altschul et al., 1997) similarity searches at The Arabidopsis Information Resource (http://www.Arabidopsis.org/cgi-bin/Blast) or at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Both BLASTP and TBLASTN searches were done. The sequences showing homology to the PP2-like domain were obtained from the Arabidopsis Database or from the National Center for Biotechnology Information. The amino acid predicted sequences were analyzed using the Swiss Institute for Experimental Cancer Research Bioinformatics (http://hits.isb-sib.ch/cgi-bin/PFSCAN), which identified putative domains within the sequences (PROSITE profiles and pfam collection of hidden Markov). Location of the genes on the Arabidopsis genome was determined using the Arabidopsis Sequence Map Overview of The Arabidopsis Information Resource (http://www.Arabidopsis.org/cgi-bin/maps/schom). Multiple alignments were visualized using BOXSHADE (v3.21; http://www.ch.embnet.org/software/BOX_form.html).
Alignment and Phylogenetic Reconstruction
The degree of identity and similarity among pairs of sequences was calculated using the GAP program available on GCG (Genetics Computer Group, Madison, WI) using default parameters. Sequences were aligned by Pileup available on the GCG package. Phylogenetic reconstruction was performed using the neighbor-joining method (Saitu and Nei, 1987) based on corrected distances for a high rate of substitution using the program ClustalW (v1.81; Thompson et al., 1994) available on GCG. The phylogenetic tree was generated using default parameters, except for correction for the substitution rate using the Kimura distances. Bootstrapping was performed using 1,000 replicates. The phylogenetic tree was displayed using the Tree View software (Page, 1996).
ACKNOWLEDGMENTS
We thank Dr. Hervé Philippe for his help and advice in performing the phylogenetic studies. We thank Dr. Yves Chupeau for his constant interest in this work, and Megan Skaggs, Dr. Bettina Golecki, and Dr. Patrick Moran for their technical contributions.
Footnotes
This work was supported in part by the Association Franco Israèlienne pour la Recherche Scientifique et Technologique and by the National Science Foundation Integrative Plant Biology Program (grant no. IBN–9727626).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.013086.
LITERATURE CITED
- Aloni R. The induction of vascular tissues by auxin and cytokinins. In: Davies PJ, editor. Plant Hormones: Physiology, Biochemistry, and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 531–546. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Balachandran S, Xiang Y, Schobert C, Thompson GA, Lucas WJ. Phloem sap proteins from Cucurbita maxima and Ricinus communishave the capacity to traffic cell to cell through plasmodesmata. Proc Natl Acad Sci USA. 1997;94:14150–14155. doi: 10.1073/pnas.94.25.14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Bostwick DE, Dannenhoffer JM, Skaggs MI, Lister RM, Larkins BA, Thompson GA. Pumpkin phloem lectin genes are specifically expressed in companion cells. Plant Cell. 1992;4:1539–1548. doi: 10.1105/tpc.4.12.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostwick DE, Skaggs MI, Thompson GA. Organization and characterization of Cucurbitaphloem lectin genes. Plant Mol Biol. 1994;26:887–897. doi: 10.1007/BF00028856. [DOI] [PubMed] [Google Scholar]
- Callis J, Viestra RD. Protein degradation in signalling. Curr Opin Plant Biol. 2000;3:381–386. doi: 10.1016/s1369-5266(00)00100-x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Peumans WJ, Hause WJ, Bras J, Kumar M, Proost P, Barre A, Rougé P, Van Damme EJM (2002) Jasmonate methyl ester induces the synthesis of a cytoplasmic/nuclear chitooligosaccharide-binding lectin in tobacco leaves. FASEB J express article 101096/fj01–0598fje [DOI] [PubMed]
- Clark AM, Jacobsen KR, Bostwick DE, Dannenhoffer JM, Skaggs MI, Thompson GA. Molecular characterization of a phloem-specific gene encoding the filament protein, phloem protein 1 (PP1) from Cucurbita maxima. Plant J. 1997;12:49–61. doi: 10.1046/j.1365-313x.1997.12010049.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cronshaw J, Esau K. Tubular and fibrillar components of mature and differentiating sieve elements. J Cell Biol. 1967;34:801–815. doi: 10.1083/jcb.34.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw J, Sabnis DD. Phloem proteins. In: Behnke HD, Sjölund RD, editors. Sieve Elements. Berlin: Springer; 1990. pp. 257–283. [Google Scholar]
- Dannenhoffer JM, Schulz A, Skaggs MI, Bostwick DE, Thompson GA. Expression of the phloem lectin is developmentally linked to vascular differentiation in cucurbits. Planta. 1997;201:405–414. [Google Scholar]
- del Pozo JC, Estelle M. F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol Biol. 2000;44:123–128. doi: 10.1023/a:1006413007456. [DOI] [PubMed] [Google Scholar]
- Eleftheriou EP. Monocotyledons. In: Behnke HD, Sjölund RD, editors. Sieve Elements. Berlin: Springer; 1990. pp. 139–159. [Google Scholar]
- Evert RJ. Dicotyledons. In: Behnke HD, Sjölund RD, editors. Sieve Elements. Berlin: Springer; 1990. pp. 103–137. [Google Scholar]
- Farràs R, Ferrando A, Jàsik J, Kleinow T, Ökrész L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C. SKP1-SnRK protein kinase interactions mediated proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 2001;20:2742–2756. doi: 10.1093/emboj/20.11.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golecki B, Schulz A, Thompson GA. Translocation of structural P proteins in the phloem. Plant Cell. 1999;11:127–140. doi: 10.1105/tpc.11.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez G, Pallas V. Identification of an in vitro ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber plants. Mol Plant-Microbe Interact. 2001;14:910–913. doi: 10.1094/MPMI.2001.14.7.910. [DOI] [PubMed] [Google Scholar]
- Gustincich S, Manfioletti G, Del Sal G, Schneider C, Carninci P. A fast method for high-quality genomic DNA extraction from whole human blood. Biotechniques. 1991;11:298–302. [PubMed] [Google Scholar]
- Hoad GV. Transport of hormones in the phloem of higher plants. Plant Growth Regul. 1995;16:173–182. [Google Scholar]
- Jackson DP. In situhybridization in plants. In: Bowles DJ, Gurr SJ, McPhereson M, editors. Molecular Plant Pathology: A Practical Approach. Oxford: Oxford University Press; 1991. pp. 163–174. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobst J, King K, Hemleben V. Molecular evolution of the internal transcribed spacers (ITS1 and ITS2) and phylogenetic relationship among species of the family Cucurbitaceae. Mol Phylogenet Evol. 1998;9:204–219. doi: 10.1006/mpev.1997.0465. [DOI] [PubMed] [Google Scholar]
- Kleinig H, Thones J, Dorr I, Kollmann R. Filament formation in vitro of sieve tube protein from Cucurbita maxima and Cucurbita pepo. Planta. 1975;127:163–170. doi: 10.1007/BF00388377. [DOI] [PubMed] [Google Scholar]
- Knoblauch M, Peters WS, Ehlers K, van Bel AJE. Reversible calcium-regulated stopcocks in legume sieve tubes. Plant Cell. 2001;13:1221–1230. doi: 10.1105/tpc.13.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch M, van Bel AJE. Sieve tubes in action. Plant Cell. 1998;10:35–50. [Google Scholar]
- Oparka KJ, Turgeon R. Sieve elements and companion cells: traffic control centers of the phloem. Plant Cell. 1999;11:739–750. doi: 10.1105/tpc.11.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens RA, Blackburn M, Ding B. Possible involvement of the phloem lectin in long-distance viroid movement. Mol Plant-Microbe Interact. 2001;14:905–909. doi: 10.1094/MPMI.2001.14.7.905. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic tree on personal computers. Computer applications. Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Read SM, Northcote DH. Chemical and immunological similarities between the phloem proteins of three genera of the Cucurbitaceae. Planta. 1983;158:119–127. doi: 10.1007/BF00397704. [DOI] [PubMed] [Google Scholar]
- Reuber TL, Ausubel FM. Isolation of Arabidopsisgenes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell. 1996;8:241–249. doi: 10.1105/tpc.8.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cazares B, Lucas WJ. The phloem as a conduit for inter-organ communication. Curr Opin Plant Biol. 2001;4:202–209. doi: 10.1016/s1369-5266(00)00162-x. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal spacer-length polymorphism in barley Mendelian inheritance chromosomal location and population dynamics. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitu N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schulz A. Conifers. In: Behnke HD, Sjölund RD, editors. Sieve Elements. Berlin: Springer; 1990. pp. 63–88. [Google Scholar]
- Schulz A, Alosi MC, Sabnis DD, Park RB. A phloem-specific lectin-like protein is located in pine sieve-element plastids by immunocytochemistry. Planta. 1989;179:506–515. doi: 10.1007/BF00397590. [DOI] [PubMed] [Google Scholar]
- Sjölund RD, Shih CY. Freeze-fracture analysis of phloem structure in plant tissue cultures I: the sieve element reticulum. J Ultrastruct Res. 1983;82:111–121. doi: 10.1016/s0022-5320(83)90101-6. [DOI] [PubMed] [Google Scholar]
- Smith LM, Sabnis DD, Johnson RPC. Immunocytochemical localisation of phloem lectin from Cucurbita maximausing peroxidase and colloidal-gold labels. Planta. 1987;170:461–470. doi: 10.1007/BF00402980. [DOI] [PubMed] [Google Scholar]
- Stamm O, Krücken J, Schmitt-Wrede H-P, Benten WPM, Wunderlich F. Human ortholog to mouse gene imap38encoding an ER-localizable G-protein belongs to a gene family clustered on chromosome 7q32-36. Gene. 2002;282:159–167. doi: 10.1016/s0378-1119(01)00837-x. [DOI] [PubMed] [Google Scholar]
- Thompson GA. P-protein trafficking through plasmodesmata. In: van Bel AJE, Van Kesteren WJP, editors. Plasmodesmata: Nanochannels with Megatasks. Berlin: Springer-Verlag; 1999. pp. 296–313. [Google Scholar]
- Thompson GA, Schulz A. Macromolecular trafficking in the phloem. Trends Plant Sci. 1999;4:354–360. doi: 10.1016/s1360-1385(99)01463-6. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama T, Teramoto H, Takeba G, Hideo T. Cytokinin induces a rapid decrease in the levels of mRNAs for catalase, 3-hydroxy-3-methylglutaryl CoA reductase, lectin and other unidentified proteins in etiolated cotyledons of cucumber. Plant Cell Physiol. 1995;36:1349–1359. [PubMed] [Google Scholar]
- van Bel AJE, Kempers R. The pore/plasmodesm unit: key element in the interplay between sieve element and companion cell. Prog Bot. 1997;58:278–291. [Google Scholar]
- Van der Biezen EA, Jones JDG. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]