Abstract
Neurogenesis relies on the establishment of the proper number and precisely controlled proliferation of neuroblasts, the neuronal precursor cells. A role for the mushroom body defect (mud) gene in both of these aspects of neuroblast behavior, as well as possible roles in other aspects of fruit fly biology, is implied by phenotypes associated with mud mutations. We have localized mud by determining the sequence change in one point mutant, identifying a predicted ORF affected by the mutation, and showing that an appropriate segment of the genome rescues mud mutant phenotypes. An analysis of mud cDNAs and a survey of mud transcripts by Northern blotting indicate that the gene is subject to differential splicing and is expressed primarily during embryogenesis but also, at lower levels, during subsequent developmental stages in a sexually dimorphic manner. The gene is predicted to encode a polypeptide without obvious homologs but with two prominent structural features, a long coiled coil that constitutes the central core of the protein and a carboxyl-terminal transmembrane domain.
Development of the Drosophila central nervous system (CNS) is subdivided into two periods (for an overview, see ref. 1). During embryogenesis, neuroblasts (Nbs) delaminate from the neuroectoderm in a stereotypic pattern and give rise to the neurons that form the basic architecture of the CNS. With the onset of postembryonic development, further Nb proliferation and neuronal differentiation elaborates this pattern to generate a functioning larval CNS, which subsequently is remodeled to serve adult behavior.
Many features of central nervous system development are exemplified in the mushroom bodies (MBs). MBs are paired neuropil structures of the adult midbrain that are the site of a short-term olfactory memory (2) and play important roles in other forms of sensory integration (3). In wild-type flies, the dendrites of the ≈2,500 intrinsic neurons (Kenyon cells, KCs) that make up a single MB form the calyx, a structure that receives input from projection neurons of the antennal lobes. The KC axons extend through the peduncle into a system of lobes. Based on their projection patterns and birth date, three classes of KCs can be distinguished (4). Cell lineage analysis identified four Nbs in each brain hemisphere, each giving rise to all three subtypes of KCs (5). Together with one additional Nb in the antennal lobe, the MB Nbs maintain proliferation throughout development. In contrast, all other Nbs undergo cell cycle arrest at late embryonic stages and only resume mitotic activity several hours after larval hatching (6).
Mutations in the mushroom body defect (mud) gene (7) are associated with an abnormal proliferation pattern of Nbs in embryonic and larval stages and with distorted MBs in adults. In the dorsolateral brain of mud larvae an excess number of MB Nbs is found, whereas in the ventrolateral brain and the ventral nerve cord, Nbs occur in normal numbers but the proliferation period of some of them is extended (8). The excess number of MB Nbs causes a dramatic increase in the number of KCs. Instead of forming a peduncle and lobe system, fibers of these KCs appear to accumulate beneath the cell body layer to form enlarged and misshapen calyces (9). The MB defects of mud mutants become most obvious in the pupal period when, during normal development, many KC fibers are replaced (4, 9). How the proliferation and the axonal outgrowth defects relate to each other is a matter of speculation. Also unknown is whether the hypersensitivity of adult mud flies to general anesthetics like halothane (10, 11) reflects anatomical defects in brain structure or a role for Mud in adult physiology. Finally, a role for Mud in early embryogenesis is suggested by the sterility of mud homozygous females (12). Here, we report the cloning of the gene, its transcription pattern, and its coding potential.
Materials and Methods
Fly Strains and Crosses.
The origin and cytogenetic extent of the deficiencies used in this work have been summarized elsewhere (10). Four alleles of mud have been reported: mud1, mud3, and mud4 were induced in the wild-type Berlin strain by ethyl methanesulfonate (7), and mud2 was isolated after hybrid dysgenesis (13). Stocks of these mutants generally are maintained as balanced lines, from which males can be recovered, albeit with low efficiency. Although homozygous mud2 and mud4 females also can be recovered, in contrast to a previous report (12), homozygous mud1 and mud3females were never observed. Transheterozygous mud1/Df(1)KA9 females can be generated; such heterozygotes are sterile and display a typical mutant MB phenotype. The differential viability of homozygous and hemizygous mud1 suggests that two doses of this allele produce interference with the function of another gene.
Transgenic lines were generated by injecting DNA (purified with a commercial resin) into w1118 embryos. Autosomal inserts were balanced, and males from these lines were crossed to w mud/In(1)FM7, y31d sc8 wa B females. Male offspring that inherited both a mud allele and an insert were phenotyped and used in crosses to generate mud females.
Molecular Biology.
Clones containing genomic DNA from the 12E region on the X chromosome used in this work were as follows: cos24 was from the NotBamNot-CoSpeR library of an isogenized y;cn bw sp strain (14); pZGRC was made by cloning an 18-kb SnaBI subfragment of cos24 into the StuI site of the pCaSpeR4 transformation vector; cos80G4 was from the European Drosophila Genome Project library of an Oregon-R strain (15); pJMD1 was generated by cloning a 16-kb XhoI subfragment of cos80G4 into the pW8 transformation vector. Expressed sequence tag clones LD24364 and LD31911 were identified by partial sequences deposited in the Berkeley Drosophila Genome Project database (16) and were obtained from Research Genetics, Huntsville, AL. Additional cDNAs were identified on filters containing spotted cDNA clones, obtained from RZPD, the Resource Center of the German Human Genome Project (17); these were screened with genomic and cDNA probes. The ID number for cDNA clone RZ15 is ICRFp520J1117Q4. A cDNA fragment, AP-C, from the putative 5′ end of the gene was amplified from a 4–8 h embryonic cDNA library (kindly provided by N. Brown, University of Cambridge, Cambridge, U.K.) by standard PCR procedures using oligonucleotides 5′-GCTGGAGTATACGGACCGCATTCG-3′ and 5′-CATTGCTCGTTCGGAGCAGTTGTG-3′. Sequencing of cDNA clones, genomic clones, and PCR products was done by using the DyeDeoxy Terminator Cycle Sequencing kit (Perkin–Elmer) or the PRISM dRhodamine Terminator kit (Applied Biosystems).
For Northern analysis of embryonic or larval transcripts, 5–10 μg of poly(A)+ RNA (prepared with an Amersham Pharmacia mRNA Purification Kit) was separated on 0.9% agarose-formaldehyde gels and blotted onto nylon membranes. The blots were hybridized with 32P-labeled DNA probes (prepared with an Amersham Pharmacia MegaPrime Kit); they were stripped and reprobed up to six times. A probe for the rp49 gene was used to verify the loading and integrity of the different RNA samples. The position of the mud gene probes is diagrammed in Fig. 1. Probe A was the AP-C fragment described above. Probe B was a NotI–SalI fragment derived from cDNA clone RZ3, a 5′ truncated version of RZ15. Probe C, encompassing exon 7, was amplified from a cloned genomic fragment using the oligonucleotides 5′-TAAATGGCATGGTCGCCGGCGA-3′ and 5′-CGGGCGGCACGCATTTTTAAAGGA-3′. Probe D was a PstI–BglII fragment of cDNA clone LD31911. Probe E was a BamHI–BglI fragment of cDNA clone RZ2, a 5′ truncated version of LD31911. Northern analysis of adult transcripts followed a similar protocol but used the following antisense oligonucleotides that were 32P-labeled with terminal transferase: GZ44, 5′-CCACTGGAGTAGGACCTTGCGCCAGCTGCGCGTGTCCAT-3′; GZ45, 5′-CTTTGTTGAGAACTTTAATGAGCTCGTCCTTCTCCATTTGCTGCTCC-3′; GZ51, 5′-CTCTCATTCAACTTTGAATGCACGGTATGCGATACAGTGTTG-3′.
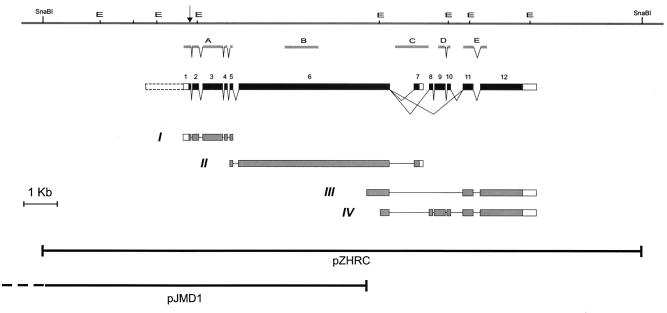
Figure 1.
A physical and transcription map of the mud gene. The horizontal line at the top represents the genomic DNA; the centromere is to the right. Restriction sites for EcoRI (E) and SnaBI enzymes are indicated. The position of the codon that is changed in mud4 is indicated by an arrow. Below the genomic DNA is diagrammed the exon-intron structure of the mud transcription unit, as derived from the analysis of various cDNAs isolated by PCR (line I, clone AP-C) or from libraries (line II, clone RZ15; line III, clone LD24364; line IV, clone LD31911). Filled boxes represent potential coding regions and open boxes correspond to untranslated sequences. The dashed open box indicates a noncoding upstream portion for the mud transcription unit that is predicted by fgenesh, but corresponding cDNAs have not yet been found. For each splice variant, only the longest isolated cDNA is diagrammed. Complete sequences for these clones have been deposited, lines I and II as GenBank accession no. AF209068 and lines III and IV as GenBank accession no. AF174134. The sequences show that two versions of exon 12 are used: exon 12a (for cDNA III) having its 5′ end 12 nt upstream of the 5′ end of exon 12b (for cDNAIV). Below the cDNAs are shown the inserts of two partially overlapping genomic constructs that respectively succeed and fail to rescue mud phenotypes: pZHRC and pJMD1 (which extends 4.4 kb distally, as indicated by the dashed line). Between the genomic and exon-intron maps are shown the positions of probes A-E, used for Northern blot analyses.
The single-strand conformation polymorphism scan for mud mutations used RNase A-treated genomic DNA that had been cut with one of several arbitrarily chosen restriction enzymes of four-base specificity. About 6 μg of this material was loaded on each lane of a 20% GeneAmp Detection gel (Perkin–Elmer) and electrophoresed overnight at 4°C. After electrotransfer to a nylon membrane, the DNA was probed with 4- to 10-kb genomic fragments that had been labeled with the Random Primers Labeling Kit (GIBCO/BRL); hybridization was at 68°C in QuikHyb Hybridization Solution (Stratagene).
Predictions concerning the mud gene were obtained with the Wisconsin Sequence Package of the Genetics Computer Group (peptidestructure for α-helix content and seg for globular domains) or at the following electronic sites: http://genomic.sanger.ac.uk/ for exon/intron boundaries (fgenesh); http://www.ch.embnet.org/software/COILS_form.html for coiled coil (coils); and http://genome.cbs.dtu.dk/services/ for transmembrane (tmhmm).
Results and Discussion
Genomic Localization of mud.
Complementation studies involving mud alleles and various deficiencies previously showed that the gene is uncovered by Df(1)KA9 (7) but not by Df(1)g-l and Df(1)RK5 (K. F. Fischbach and M.H., unpublished observations). This placed the gene in the 12E region of the X chromosome, in the vicinity of the recently cloned yolkless (yl) gene (18). The previously established positions of the proximal breakpoint of Df(1)g-l and the distal breakpoint of Df(1)KA9 (18) indicated that mud extended in the centromere-proximal direction from yl. To delimit a boundary for mud in this direction, we investigated the distal breakpoint of Df(1)RK5. We obtained a set of overlapping cosmid clones covering the yl-proximal region from C. Schonbaum (University of Chicago) and used them to establish a dense restriction map (see Figs. 5 and 6, which are published as supplemental material on the PNAS web site, www.pnas.org). Comparison of restriction patterns of Df(1)RK5 with the Π2 line from which this deficiency was derived (19) revealed that several alterations were clustered in the region of the genome corresponding to the middle of cosmid 80G4 (15), as reported (20). If Df(1)RK5 ends in the region of these alterations (see Figs. 5 and 6), the mud gene would be confined to the 60 kb proximal to yl. Study of other deficiencies did little to improve this mapping. Df(1)NP5, which complements mud4, was made the same way as Df(1)RK5 and is reported (20) to have the same alterations in cosmid 80G4. Three deficiencies that fail to complement mud mutations—Df(1)RK3, Df(1)CO1, and Df(1)CO2 (19, 21)—appear to delete all genetic material from the 60-kb window (data not shown), a result consistent with the assignment of mud to this window but one that fails to refine the map.
An obvious candidate for the mud gene was not apparent within the 60-kb window. Northern blots of RNA extracted from embryos, larvae, and adults detect more than 15 distinguishable mRNAs (unpublished observations). Genomic analysis of the mud alleles did not detect a physical alteration in the window, not even mud2, which was generated in a dysgenic cross (13). Moreover, a screen of 800 X-linked P element lines turned up only two within the 60-kb window, neither of which disrupted mud. The initial clue to finding the gene came from a single-strand conformation polymorphism (SSCP) scan of the window for sequence alterations in the mutant lines. As described in Materials and Methods, we used large fragments from the cosmid walk to probe restriction digests of genomic DNA from mud1, mud3, and mud4, the three alleles that had been derived from the same parental strain. Alterations in the SSCP pattern were detected for one mutant (mud4) with an 8-kb fragment from the centromere-proximal end of the window (see Figs. 5 and 6). Probing the same digests with shorter fragments localized the alteration to a region of ≈1 kb. The corresponding region of genomic DNA was amplified by PCR and sequenced. This revealed a single nucleotide change in the mud4 mutant by comparison with the parental line and the other mud alleles.
Computer predictions with the fgenesh program (22), trained to recognize the structure of Drosophila genes, suggested that several small exons were present in the ≈1-kb region and that the change in the mud4 strain created a stop codon in a predicted translated region of one of them. To explore the extent of the candidate mud gene, we determined additional DNA sequence from an appropriately positioned cosmid, 80G4. In the end, more than 16 kb of sequence (GenBank accession no. AF174134) was needed to satisfy fgenesh and other computerized predictions that a full-length gene had been covered. Many (but not all) of these predictions assigned the first exon to the one that was altered in mud4 and the last exon to a segment nearly 10 kb away (Fig. 1). A cosmid clone from a library designed for genomic rescue (14) was identified by restriction mapping as nearly coextensive with cos80G4 (see Figs. 5 and 6) and thus expected to contain the putative mud ORF. Three independent autosomal insertions of the ≈38-kb genomic segment of this construct, cos24, were recovered and crossed into mud1, mud3, and mud4 strains. As judged by appearance of brain sections (23), two of the three transgenic lines rescued the MB anatomy (calyx, peduncle, lobe system) of all male flies analyzed (see Fig. 2 for an example). To quantify the degree of rescue, the calyx volume was determined by planimetric measurements of serial frontal sections. The MB of mud mutants shows a high degree of variability: in individual flies, the increase in calyx volume ranges from 2-fold to 10-fold and is accompanied by a correspondingly severe defect in the peduncle and lobe system. Nevertheless, compared with the calyx volume (μm3 ± SEM) of wild-type males (20,200 ± 465, n = 20), the calyx volume of mud1(108,000 ± 23,800, n = 20) and mud4 (143,000 ± 17,200, n = 20) males is clearly increased. Moreover, a single copy of the cos24 construct is sufficient to restore the wild-type calyx size and shape in mud1(20,700 ± 780, n = 20) and mud4 (22,200 ± 1,183, n = 10) males. In addition, a copy of the cos24 transgene produced rescue for: the lethality of homozygous mud1 females, which proved to be fertile; the sterility of homozygous mud4 and transheterozygous mud1/Df(1)KA9 females; and the MB anatomy in mud1 and mud4 females. A single copy of cos24 also reduced the hypersensitivity of a mud mutant to halothane. Specifically, the concentration of halothane required to reduce performance by 50% in a behavioral test (10) was 0.20 vol/vol% (95% confidence limits = 0.17–0.23) for transheterozygous mud4/Df(1)KA9 females carrying a control transgene but 0.39 vol/vol% (95% confidence limits = 0.34–0.48) when such females carried a cos24 transgene.
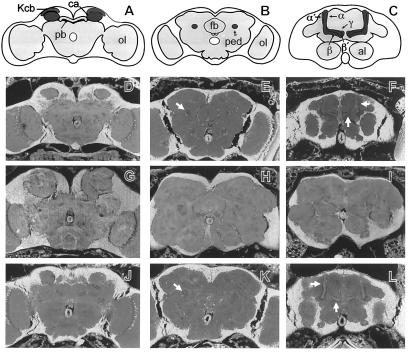
Figure 2.
Rescue of the brain anatomy phenotype of mud by a genomic fragment. (A-C) Schematic frontal sections of wild-type Drosophila heads at the level of the calyces (A), the peduncle (B), and the lobe system (C). The overall neuorophil structure is outlined in gray, the MBs are highlighted in dark gray. The KC bodies (Kcb) of the MBs are located in the dorsal-posterior cortex. KC dendrites and extrinsic input fibers constitute the calyx (ca). The KC fibers form the peduncle (ped), which projects anterior-ventrally where it divides into the dorsally projecting α and α′ lobes and the medially projecting β, β′, and γ lobes. pb: protocerebral bridge; fb: fan-shaped body; al: antennal lobes; ol: optic lobes. (D–L) Frontal sections (7 μm) of paraffin-embedded heads (23) examined with a fluorescence microscope: wild type (D–F), mud1 (G–I), and mud1 flies that carry a single copy of cos24 (J–L). Note that the KC fibers of mud1 do not form a peduncle (H) or lobe system (I) but instead form a greatly enlarged and misshaped calyx region (G). In these flies, distortions of brain anatomy outside the MBs may be secondary to the enlargement of the calyces and/or reflect prolonged proliferation of other central brain Nbs (8). When mud1 flies bear a cos24 transgene, the organization of the entire MB is restored to a wild-type appearance, including calyces (D and J), peduncles (arrows in E and K), and lobe system (arrows in F and L).
Further evidence that the candidate ORF is indeed the mud gene came from transformation with a subclone derived from cos24, pZHRC. Although this construct leaves only a few kb upstream and downstream of the predicted ORF (Fig. 1), one copy of this construct is sufficient (in one of two independent insertions) to rescue the MB phenotype in mud1 and mud4 males. The degree of rescue was similar to that with the cos24 transgene: in males that carry a single autosomal copy of pZHRC, the calyx volume (μm3 ± SEM, n = 20) for mud1 and mud4 was, respectively, 22,200 ± 877 and 22,000 ± 474. It must be noted that in some cases the calyces of the rescued lines still appeared to be slightly misshaped. Similarly, although pZHRC rescued the sterility and MB phenotype in homozygous mud4 females and the sterility of transheterozygous mud1/Df(1)KA9 females, homozygous and fertile mud1 females were recovered only at low frequency (data not shown). This suggests that the region flanking mud in pZHRC is insufficient to achieve full expression of the gene. In contrast to the successful rescues with cos24 and pZHRC, consistent failure to achieve any rescue of mud phenotypes was observed with a genomic fragment that encompasses only part of the predicted mud ORF (pJMD1, Fig. 1) or with various genomic fragments from the 60-kb window that do not overlap with the predicted mud ORF (J.M., A.P., and T.R., unpublished observations).
Expression of the mud Gene.
The genomic sequence described above was used to identify cDNAs in the Berkeley Drosophila Genome Project database of expressed sequence tags (16) and on cDNA array filters (17). Representative clones were fully sequenced; these clones fall into three different classes, the longest members of which are diagrammed in lines II, III, and IV in Fig. 1. These clones have in common sequences corresponding to an exon from the middle of the gene but have divergent 3′ ends, indicating the existence of alternative splicing. The clones appear to be less than full length: their 5′ ends fall far short of the computer-predicted start of the gene and none contains a complete ORF preceded by in-frame stop codons. A cDNA fragment from the putative beginning of the gene could be amplified from an embryonic cDNA library (kindly provided by N. Brown) using a primer predicted to be upstream of the translated portion of the mud gene and a primer complementary to the 5′ end of the cDNA diagrammed in Fig. 1, line II. Sequence analysis verified that this cDNA fragment (diagrammed in Fig. 1, line I) includes the putative start codon, preceded by stop codons in all three frames; it also includes the codon altered in mud4. Together, these cDNAs define 12 exons, 11 of which were predicted by fgenesh.
Northern blots of poly(A)+ RNA have been hybridized with probes corresponding to various exons. The observed patterns (Fig. 3) are complex but several points are clear. First, expression in embryos (lanes 3) is generally much higher than expression in larvae (lanes 1 and 2) and adults (not shown). Preliminary study indicates that mud RNA is most abundant in midembryogenesis, 5–9 h after egg laying (data not shown). Second, the detected transcripts are large, typically 6–9 kb, suggesting that all isoforms include the 4.5 kb of exon 6. Third, as expected if the 3′ exons are incorporated into alternatively spliced mud messages, probes for these exons detect distinct transcripts. Of particular interest is the preferential use of exon 7 in male larvae, hinting at a special role for an unusual form of Mud protein (see below). Fourth, at least in embryo RNA, multiple species of different sizes are detected by virtually every probe. It must be pointed out that, for a given probe, the relative intensity of these species often varies between different preparations of embryo RNA. Differential degradation is an unlikely explanation for this variability because the integrity of each RNA sample was verified by hybridization with a probe corresponding to the ubiquitously expressed rp49 gene (data not shown). Instead, we suspect that the observed variation in mud transcripts reflects differential expression of various isoforms during the stages of embryogenesis, coupled with varying representation of these stages in material collected for extraction. Importantly, sequential hybridization of the same blot of embryonic RNA with probes for exons 1–5 (probe A) and exon 6 (probe B) detected an identical set of bands (data not shown), indicating that the 5′ portion of mud is expressed as a unit, at least in embryos. Moreover, in poly(A)+ RNA extracted from adult flies an apparently identical set of large transcripts is detected by probes (oligonucleotides GZ44, GZ45, and GZ51) that are antisense to portions of exons 1, 3, and 6 (data not shown). This suggests that exons 1–6 also are expressed as a unit in adults. Because this unit includes the position of the mud4 mutation, which converts the sixth codon of exon 1 to a stop signal, mud4 is a presumptive null allele. However, we cannot rule out the possibility that sometimes exon 1 is skipped or, despite that presence of a fair match to the consensus for a translational start in this exon, that alternative translational starts are used.
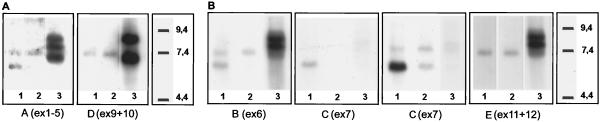
Figure 3.
Northern analysis of mud transcripts. Each panel presents a single blot hybridized sequentially with the indicated probes, which are described in Fig. 1. Samples 1 and 2 are RNA isolated, respectively, from male and female third instar larvae; sample 3 is RNA isolated from unstaged embryos. Different embryonic and larval RNA preparations were used in A and B. Note that the smallest larval transcript (detected only by probes A, B, and C) is predominantly expressed in males. Two exposures are shown of the blot hybridized with probe C; the one on the right has been overexposed to illustrate that the small message is expressed in female larvae, albeit at very low levels.
Taken together, the results with cDNAs and Northern blots indicate that mud is expressed at various times in the life of a fly and that the gene is subject to a complex program of differential splicing. This suggests there are different functions for the gene at different times and places.
The Coding Potential of the mud Gene.
Each of the mud gene transcripts can be considered to encode a product with three domains: a central core flanked by amino- and carboxyl-terminal domains. The core and amino-terminal domains are encoded by exons 1–6 and are thus likely to be common to all isoforms. On the other hand, the carboxyl terminus should vary between isoforms, being encoded by different combinations of exons 7–12.
The central core is defined by a long region (encoded by part of exon 3 and all of exons 4–6) of high α-helical potential. Moreover, as shown in Fig. 4, the α-helices of the central region are predicted (24) to form a long stretch of coiled–coil structure, albeit interrupted by segments calculated to have low coiled–coil potential. The multicoil program (25) agrees largely with the assignment of Fig. 4 and further predicts that the intertwined helices of the central core are likely to be two-stranded rather than three-stranded (not shown). Thus, for more than 1,600 aa, a dimeric coiled coil is the predominant feature of the mud gene product. Because the structure is built from α-helices that have a regular repeat pattern of hydrophobic and hydrophilic residues (24, 25), all coiled–coil proteins would be expected to display some sequence similarity. Correspondingly, blast searches (26) with the mud gene product yield many highly significant “hits” on proteins with long coiled coils, including myosins, kinesins, plectins, and golgins. In these proteins, the coiled–coil portion, which often is interrupted to introduce points of flexibility into an otherwise rigid structure (27), is exploited for several different ends. One use for the structure is the formation of either head-to-head or head-to-tail homodimers (28). In other cases, it appears that at least a part of the structure is used to make specific contacts with different molecules (29–31). In this context it should be pointed out that, within the region predicted to form the coiled coil of Mud, there is a sequence of 42 aa that is repeated, almost perfectly, 10 times (Fig. 4). It is attractive to imagine that this feature subserves a specific function, perhaps as a repeated interaction site. Indeed, almost every one of the repeats has a good match to a motif found in several actin binding proteins, a motif associated with the binding of the modulator phosphatidylinositol 4,5-bisphosphate (32).
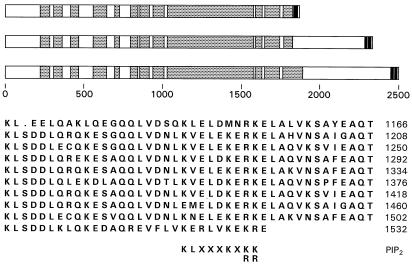
Figure 4.
Features of the product of the mud gene. The three proteins that are deduced to arise from translation of alternatively spliced mud transcripts are diagrammed. All three isoforms include exons 1–6 (which end, respectively, at protein residues 13, 71, 266, 302, 338, and 1824). In successive diagrams, this common segment is extended to incorporate the segments encoded by the isoform-specific exons shown in lines II, III and IV of Fig. 1. Regions of these gene products that are calculated to have a >60% propensity to form a coiled coil (24) are indicated by shading. Filled boxes mark those portions of each Mud isoform that are predicted (34) to have a >80% propensity to span a membrane bilayer. Below the isoform diagrams is the peptide sequence (one-letter code) of a segment of the central core, arrayed to emphasize its tandemly repeated nature. To the right of each line is the number of the last residue shown. Beneath the array is aligned the consensus motif associated with binding of phosphatidylinositol 4,5-bisphosphate (32).
Of course, a potential role for the coiled–coil structure is to provide physical separation between functional domains of a protein. As expected from this surmise, virtually all of the long coiled–coil proteins do have at least one globular domain attached to an end of the coiled coil. Such domains often have an identified catalytic activity or specific affinity for a partner: examples include the motor domains of kinesins, the actin binding domains of plectin and related cytoskeletal cross-linkers, and the microtubule binding domains of cytoplasmic linker protein (CLIP) family members. However, none of the flanking domains of Mud match any of these or any other well-characterized domain. The ≈225 aa of the amino terminus, encoded by exons 1 and 2 plus part of exon 3, is predicted (33) to contain a globular domain. However blast searches failed to identify a highly significant match either to proteins of known function or to translated DNA sequence databases. Concerning the carboxyl terminus of Mud, of the three splice isoforms that we have identified (Fig. 1), the shortest adds 47 aa to the end of the coiled coil. In the next larger splice isoform, a noncoiled-coil segment of 500 aa is abutted to the core (Fig. 4). Finally, the longest isoform extends the coiled–coil domain by ≈70 aa (Fig. 4) and to it adds a segment of ≈600 aa. Both the 500- and 600-aa termini are predicted (33) to be at least partly globular. Nevertheless, like the amino terminus, none of the carboxyl-terminal domains has an obvious match in databases of either well-studied or anonymous proteins.
Despite lacking an identifiable homolog, the carboxyl termini of Mud all are characterized by the same structural feature, a transmembrane (TM) domain that is calculated (34) to occur at the very end of the protein. For the shortest isoform, a single TM segment is predicted to precede the last 15 aa of the protein (Fig. 4). For both longer isoforms, a pair of TM segments, separated by only 2 aa, are predicted to precede the final 6 aa of the protein (Fig. 4). TM segments that occur very close to the carboxyl terminus have been identified in several proteins, although most of these have only a single TM segment (35). Like Mud, these proteins are predicted not to have a signal sequence at their amino terminus, so they are not expected to use the conventional pathway for export across or incorporation into lipid bilayers. Nevertheless, proteins with a terminal TM domain have been found to be associated with membranes and, accordingly, this feature has been described as a carboxyl-terminal membrane anchor (36). As exemplified by Bax, synaptobrevin, and LAP2, proteins with this feature serve a variety of functions and can be associated with mitochondrial, vesicular, and nuclear membranes (35, 37, 38). It is therefore difficult to predict how Mud protein might use this feature but, together with the extensive coiled coil, it forms a kind of signature for the protein. A survey of proteins with long coiled–coil domains has identified a few with a predicted carboxyl-terminal anchor but none of them match all of the features of Mud. For example, the interaptin protein of Dictyostelium discoideum (39) has an actin binding domain at the amino terminus that is followed in turn by a segment that is predicted to form ≈1,200 aa of discontinuous coiled coil, a short globular domain, and a single TM span that ends 8 aa from the carboxyl terminus. Despite the structural similarity to a large portion of interaptin, Mud lacks a conventional actin binding domain. Similarly, the Golgi protein giantin (40) is believed to have a very long stretch of interrupted coiled coil followed by a single TM domain. However, there is little similarity between the globular portions of Mud and nonhelical segments of giantin.
Concluding Remarks.
Of the large collection of viable structural brain mutants of Drosophila, few have been characterized at the molecular level. Three of these are involved in the proliferation pattern of optic lobe Nbs. Mutations in the anachronism (ana) gene, which encodes a glycoprotein secreted by glia, cause a massive disorganization of optic lobe structures caused by precocious entry of quiescent postembryonic central brain and optic lobe Nbs into S phase (41). Conversely, failure to reactivate quiescent Nbs results from mutations in no optic lobes (nol), a gene that also may encode a secreted glycoprotein (K. Koizumi, personal communication). A marked reduction in optic lobe volume but no gross alterations in optic lobe architecture is observed in minibrain (mnb) mutants. The mnb gene product is a serine/threonine protein kinase that is required for the correct spatial arrangement of optic lobe Nbs, which seems to be critical for normal proliferation (42). In mud, the Nb proliferation phenotype is restricted to certain Nbs of the ventral ganglion and the ventrolateral brain (8). For these, intact Mud protein is required for timely cell-cycle arrest. On the other hand, the MB defect in mud mutant flies is probably a consequence of both the excess number of MB Nbs and the misrouting of KC fibers. This defect has its counterpart in mutants of mushroom bodies tiny (mbt) encoding a p21-activated kinase. In mbt mutant flies, the number of MB Nbs can be reduced but no misrouting of KC fibers is observed (43). An interesting possibility is that during development of the wild-type brain, a single Nb delaminates from the procephalic neuroectoderm and undergoes two equal divisions to give rise to the four MB Nbs, which only then adopt stem cell behavior. According to this view, the number of equal divisions would be increased in mutant mud and decreased in mutant mbt flies. Clonal expansion of groups of Nbs may be a crucial step in the establishment of some cerebral subsystems. The molecular characterization of the mud gene should not only help to determine how the diverse phenotypes observed in mutant flies are causally linked but also to define the features involved in the establishing the correct number of Nbs.
Supplementary Material
Acknowledgments
We thank Robert Scott and Gertrud Gramlich for technical assistance and Eike Kibler for help with the isolation and characterization of clones from the vicinity of mud. We are particularly indebted to Chris Schonbaum for providing information and material from the genomic walk of the region, and we thank Kevin Mitchell for sharing unpublished results. We gratefully acknowledge Gunnar von Heijne, Jürgen Knoblich, and especially Linda Restifo for comments on this manuscript. T.R. was supported by funds from the Deutsche Forschungsgemeinschaft (Ra561/2–4), and A.P. was supported by funds from the Spanish Ministry of Education and Science and from the Basque government.
Abbreviations
- KC
Kenyon cell
- mud
mushroom body defect gene
- MB
mushroom body
- Nb
neuroblast
- TM
transmembrane
Footnotes
References
- 1.Truman J W, Taylor B J, Awad T A. In: The Development of Drosophila melanogaster. Bate M, Martinez Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1245–1275. [Google Scholar]
- 2.Zars T, Fischer M, Schulz R, Heisenberg M. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- 3.Heisenberg M. Learn Mem. 1998;5:1–10. [PMC free article] [PubMed] [Google Scholar]
- 4.Lee T, Lee A, Luo L. Development (Cambridge, UK) 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 5.Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. Development (Cambridge, UK) 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- 6.Ito K, Hotta Y. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- 7.Heisenberg M. In: Development and Neurobiology of Drosophila. Siddiqi O, Babu P, Hall L M, Hall J C, editors. New York: Plenum; 1980. pp. 373–390. [Google Scholar]
- 8.Prokop A, Technau G M. Dev Biol. 1994;161:321–337. doi: 10.1006/dbio.1994.1034. [DOI] [PubMed] [Google Scholar]
- 9.Technau G, Heisenberg M. Nature (London) 1982;295:405–407. doi: 10.1038/295405a0. [DOI] [PubMed] [Google Scholar]
- 10.Guan Z, Scott R L, Nash H A. J Neurogenet. 2000;14:25–42. doi: 10.3109/01677060009083475. [DOI] [PubMed] [Google Scholar]
- 11.Walcourt A, Nash H A. J Neurobiol. 2000;42:69–78. [PubMed] [Google Scholar]
- 12.de Belle J S, Heisenberg M. Proc Natl Acad Sci USA. 1996;93:9875–9880. doi: 10.1073/pnas.93.18.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischbach K F, Boschert U, Barleben F, Houbé B, Rau T. J Neurogenet. 1987;4:126–128. [Google Scholar]
- 14.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 15.Sidén-Kiamos I, Saunders R D C, Spanos L, Majerus T, Treanar J, Savakis C, Louis C, Glover D M, Ashburner M, Kafatos F C. Nucleic Acids Res. 1990;18:6261–6270. doi: 10.1093/nar/18.21.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.FlyBase Consortium. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/26.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vente A, Korn B, Zehetner G, Poustka A, Lehrach H. Nat Genet. 1999;22:22. doi: 10.1038/8734. [DOI] [PubMed] [Google Scholar]
- 18.Schonbaum C P, Lee S, Mahowald A P. Proc Natl Acad Sci USA. 1995;92:1485–1489. doi: 10.1073/pnas.92.5.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drysdale R, Warmke J, Kreber R, Ganetzky B. Genetics. 1991;127:497–505. doi: 10.1093/genetics/127.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell K J, Doyle J L, Serafini T, Kennedy T E, Tessier-Lavigne M, Goodman C S, Dickson B J. Neuron. 1996;17:203–215. doi: 10.1016/s0896-6273(00)80153-1. [DOI] [PubMed] [Google Scholar]
- 21.Oh C E, McMahon R, Benzer S, Tanouye M A. J Neurosci. 1994;14:3166–3179. doi: 10.1523/JNEUROSCI.14-05-03166.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solovyev V, Salamov A. Ismb. 1997;5:294–302. [PubMed] [Google Scholar]
- 23.Heisenberg M, Böhl K. Z Naturforsch. 1979;34:143–147. [Google Scholar]
- 24.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 25.Wolf E, Kim P S, Berger B. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown J H, Cohen C, Parry D A D. Proteins Struct Funct Genet. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 28.Lupas A. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 29.Vitale G, Rybin V, Christofordis S, Thornqvist P-O, McCaffrey M, Stenmark H, Zerial M. EMBO J. 1998;17:1941–1951. doi: 10.1093/emboj/17.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Chang M C Y, Zylka D, Turley S, Harrison R, Turley E A. J Biol Chem. 1998;273:11342–11348. doi: 10.1074/jbc.273.18.11342. [DOI] [PubMed] [Google Scholar]
- 31.Fong K S K, de Couet H G. Genomics. 1999;58:146–157. doi: 10.1006/geno.1999.5817. [DOI] [PubMed] [Google Scholar]
- 32.Stock A, Steinmetz M O, Janmey P A, Aebi U, Gerisch G, Kammerer R A, Weber I, Faix J. EMBO J. 1999;18:5274–5284. doi: 10.1093/emboj/18.19.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wootton J C, Federhen S. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 34.Sonnhammer E L L, von Heijne G, Krogh A. Ismb. 1998;6:175–182. [PubMed] [Google Scholar]
- 35.Whiteley P, Grahn E, Kutay U, Rapoport T A, von Heijne G. J Biol Chem. 1996;271:7583–7586. doi: 10.1074/jbc.271.13.7583. [DOI] [PubMed] [Google Scholar]
- 36.Kutay U, Hartmann E, Rapoport T A. Trends Cell Biol. 1993;7:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 37.Nechushtan A, Smith C L, Hsu Y-T, Youle R J. EMBO J. 1999;18:2330–2341. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furukawa K, Pante N, Aebi U, Gerace L. EMBO J. 1995;14:1626–1636. doi: 10.1002/j.1460-2075.1995.tb07151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivera F, Kuspa A, Brokamp R, Matzner M, Noegel A A. J Cell Biol. 1998;142:735–750. doi: 10.1083/jcb.142.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linstedt A D, Foguet M, Renz M, Seelig H P, Glick B S, Hauri H P. Proc Natl Acad Sci USA. 1995;92:5102–5105. doi: 10.1073/pnas.92.11.5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebens A J, Garren H, Cheyette B N, Zipursky S L. Cell. 1993;74:15–27. doi: 10.1016/0092-8674(93)90291-w. [DOI] [PubMed] [Google Scholar]
- 42.Tejedor F, Zhu X R, Kaltenbach E, Ackermann A, Baumann A, Canal I, Heisenberg M, Fischbach K F, Pongs O. Neuron. 1995;14:287–301. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- 43.Melzig J, Rein K H, Schafer U, Pfister H, Jäckle H, Heisenberg M, Raabe T. Curr Biol. 1998;8:1223–1226. doi: 10.1016/s0960-9822(07)00514-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.