Abstract
Dehydrins (DHNs; late embryogenesis abundant D-11) are a family of plant proteins induced in response to abiotic stresses such as drought, low temperature, and salinity or during the late stages of embryogenesis. Spectral and thermal properties of these proteins in purified form suggest that they are “intrinsically unstructured.” However, DHNs contain at least one copy of a consensus 15-amino acid sequence, the “K segment,” which resembles a class A2 amphipathic α-helical, lipid-binding domain found in other proteins such as apolipoproteins and α-synuclein. The presence of the K segment raises the question of whether DHNs bind lipids, bilayers, or phospholipid vesicles. Here, we show that maize (Zea mays) DHN DHN1 can bind to lipid vesicles that contain acidic phospholipids. We also observe that DHN1 binds more favorably to vesicles of smaller diameter than to larger vesicles, and that the association of DHN1 with vesicles results in an apparent increase of α-helicity of the protein. Therefore, DHNs, and presumably somewhat similar plant stress proteins in the late embryogenesis abundant and cold-regulated classes may undergo function-related conformational changes at the water/membrane interface, perhaps related to the stabilization of vesicles or other endomembrane structures under stress conditions.
When plant cells are under environmental stresses such as drought or low temperature, diverse physiological and molecular responses can occur, including alteration of gene expression, changes in metabolism, osmotic adjustment, induction of degradation and repair systems, and elevated expression of late embryogenesis abundant (LEA), chaperone, and mRNA-binding proteins (Ingram and Bartels, 1996; Thomashow, 1999).
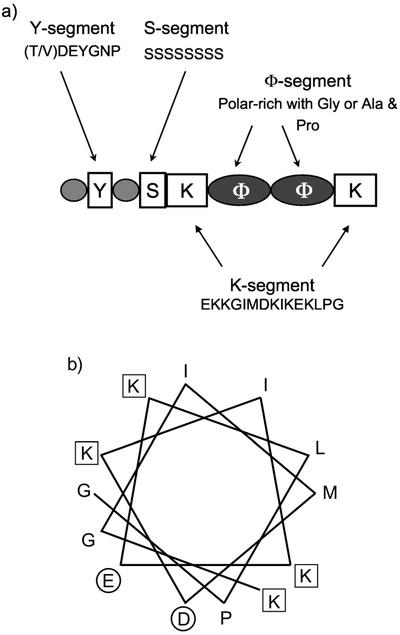
DHNs (LEA D-11 family) are among the most prevalent plant proteins induced during the late stage of embryogenesis or under drought, low temperature, freezing, salinity, or abscisic acid application (Close, 1996). They are characterized by diverse combinations of typical domains (Fig. 1A). The K segment is a Lys-rich 15-amino acid consensus sequence (EKKGIMDKIKEKPLG) that is highly conserved in all plants (Close, 1997). The K segment resembles a lipid-binding class A2 amphipathic α-helical segment found in apolipoproteins and α-synucleins (Fig. 1B; Close, 1997; Davidson et al., 1998). The S-segment is a phosphorylatable Ser-rich tract. The Y segment is an N-terminal conserved sequence. The φ-segment is rich in polar amino acids and either Gly or a combination of Pro and Ala. Each DHN can be subclassified on the basis of these domains (Close, 1997).
Figure 1.
Characteristics of DHN1 protein. A, Typical domains of DHN proteins are described in the text. Here, we show the domain composition of maize (Zea mays) DHN1. B, Helical wheel plot of the predicted amphipathic helix-forming consensus K segment peptide of maize DHN1 (KGIMDKIKEKLPG). Amino acids carrying negatively charged side chains are indicated with a circle, positively charged with a square.
Immunolocalization studies of maize scutellar parenchyma cells have shown that DHN in the cytosol is associated with membrane-rich areas surrounding lipid and protein bodies (Asghar et al., 1994; Egerton-Warburton et al., 1997), and in wheat (Triticum aestivum), an acidic DHN accumulates near the plasma membrane under cold stress (Danyluk et al., 1998). Despite this and other evidence of membrane binding in vivo, DHNs purified from these and numerous other sources are readily soluble in water. However, DHNs appear to lack structure in pure form (Lisse et al., 1996). This apparent lack of structure and a high hydrophilicity of DHNs and other proteins, collectively termed “hydrophilins” (Garay-Arroyo et al., 2000), have been hypothesized to be central to their function. Based on these properties, it has been proposed that “hydrophilins” essentially act as tiny intracellular sponges that help to retain water at low osmotic potential (McCubbin et al., 1985; Garay-Arroyo et al., 2000). Although hydration of purely hydrophilic regions of unstructured DHNs may be important to their function, it is also worth considering the role of membrane binding. An alternative hypothesis is that DHNs are akin to many other “intrinsically unstructured” proteins (Wright and Dyson, 1999) in that their lack of intrinsic structure is relieved when bound to target molecules, and that their function is related to their membrane targets.
SDS has been used as a membrane mimetic for class A2 amphipathic α-helix-forming peptides of apolipoproteins (Rozek et al., 1995), and a 26.5-kD DHN from cowpea (Vigna unguiculata L. Walp.) increased α-helicity in the presence of SDS (Ismail et al., 1999). This gain of structure was hypothesized to be indicative of a propensity of DHNs to associate with phospholipid bilayers. Previous work has shown that the binding of another class A2 α-helix-forming protein, α-synuclein, to phospholipid bilayers requires acidic phospholipids and preferentially small unilamellar vesicles (SUVs; Davidson et al., 1998). A number of similarities between DHNs and α-synuclein include high temperature solubility, a lack of Cys and Trp amino acids, and anomalous migration in SDS-PAGE. To test the hypothesis that DHNs are membrane-binding proteins, here we follow the rationale and methods that have been employed in the study of α-synuclein.
RESULTS
Confirmation of SUVs with Leakage of Carboxyfluorescein (CF)
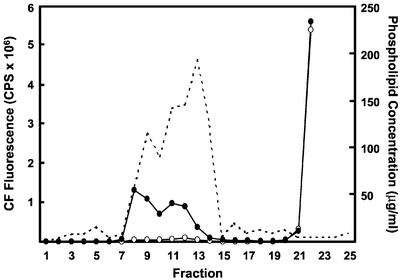
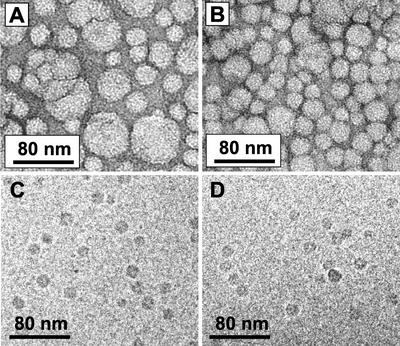
SUVs prepared from brain extract (BE1) phospholipids were prepared and passed over a gel filtration column. The concentration of phospholipid was measured for each fraction. The phospholipid elution profile showed two peaks (Fig. 2), which is typical of mixtures of lipid vesicles prepared by the sonication method. When each fraction was treated with 2% (w/v) Triton X-100, the fractions from both peaks show increased fluorescence, which indicates release of CF because of lysis of liposomes. The first peak (fractions 8 and 9) represents the excluded volume, which contains MLVs. The second peak (fractions 10–14) contains SUVs. The molar ratio of phospholipid to CF is higher in the SUV fractions because the encapsulated volume of SUV is smaller than that of MLV. The existence and size distribution of SUVs in the fractions of the second peak using phosphatidic acid (PA)-derived vesicles were confirmed by negative stain electron microscopy (Fig. 3, A and B).
Figure 2.
Elution profile of BE1-derived SUV. SUV composed of BE1 phospholipids were prepared with 100 mm CF in 20 mm Tris-HCl (pH 7.4), then separated by Superose 6 gel filtration. The fluorescence of CF of each fraction was measured before (white circles) and after (black circles) addition of 2% (w/v) Triton X-100. The concentration of phospholipid was also determined (dashed line). The peak centered on fraction 8 corresponds to multilamellar vesicle (MLV). The peak centered on fraction 12 corresponds to SUV. The left ordinate indicates CF fluorescence (counts per second × 10−6) and right ordinate indicates the concentration of phospholipid.
Figure 3.
Electron microscopic (EM) analysis of liposomes. A, EM image of PA-derived liposomes before separation by gel filtration, negatively stained with uranyl acetate, ×47,000. B, EM image of PA-derived SUV after gel filtration, negatively stained with uranyl acetate, ×47,000. C, Cryo-image of PA-derived SUV before incubation with DHN1, ×47,000. D, Cryo-image of PA-derived SUV after incubation with DHN1, ×47,000.
The Phospholipid-Binding Specificity of DHN1
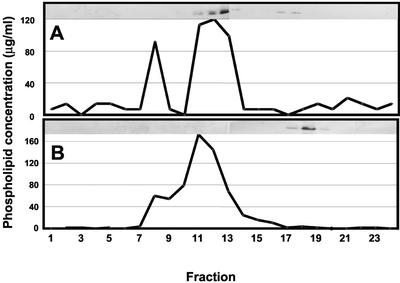
Supernatants containing MLV and SUV derived from BE1 phospholipids were incubated with DHN1 and the mixture was loaded onto a gel filtration column (Fig. 4A). About 75% of the DHN1 that was incubated with BE1 eluted with the SUV, whereas the remainder was found at volumes that are indicative of free protein. DHN1 was not associated with the MLV fraction. With SUV prepared from PC, the most abundant phospholipid in plants (Moreau et al., 1998), all of the protein eluted at a volume corresponding to free protein (Fig. 4B). BE1 is composed of 50% to 55% phosphatidyl-Ser (PS), 10% to 12% phosphatidylinositol (PI), 10% phosphatidylethanolamine (PE), and 23% to 30% unknown phospholipids. Because PS and PI have one net negative charge at physiological pH, BE1 liposomes are very enriched with negative charges when compared with SUV from PC, which is neutral under the conditions that were used.
Figure 4.
Binding of DHN to lipid vesicles. DHN1 was incubated for about 3 h at room temperature with vesicles prepared from either BE1 or soybean (Glycine max Merr.) phosphatidylcholine (PC). The mixtures were then separated by gel filtration as in Figure 2. Because the same precalibrated gel filtration column was used for each sample, actual incubation times before gel filtration chromatography varied from about 3 to 12 h. In preliminary studies with many of these mixtures, we observed no detectable influence of incubation time over the range of 3 to 12 h (data not shown). The phospholipid content of each fraction was measured. Each fraction also was analyzed by western blotting with polyclonal antibody raised against synthetic K segment peptide. The western-blot data are shown in each panel. The ordinate shows concentration of phospholipid and abscissa show fraction number. A, DHN1 with BE1-derived liposomes. B, DHN1 with soybean PC-derived liposomes.
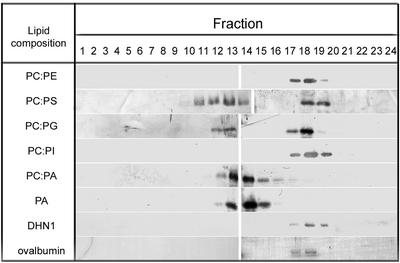
To determine whether binding of DHN1 to BE1-derived SUV results from the abundance of acidic phospholipids, a variety of vesicles prepared from 1:1 mixtures of phospholipids were used (Fig. 5). In a control experiment without vesicles, DHN1 eluted at fraction volumes indicative of free DHN1. Ovalbumin, which is known to have no interaction with SUV, was used as a control protein (Fig. 5). DHN1 migration was unchanged when mixed with SUVs or MLVs composed of PC only or a 1:1 mixture of PC:PE. In contrast, when vesicles were prepared from negatively charged phospholipids (PS, PA, or phosphatidylglycerol [PG]) mixed with PC or PE at 1:1 mass ratio, a considerable amount of DHN bound to SUV. In the case of PI-derived SUV, there was no binding to SUVs. There were also differences in the apparent binding affinity between DHN1 and vesicles derived from anionic phospholipids of various types. For example, in PA-only and 50% PA-containing vesicles, nearly 100% of the DHN1 was associated with the SUV fraction, whereas PG-containing vesicles bound a smaller percentage of DHN1. The results shown in Figure 4A and summarized in Figure 5 also show that DHN1 does not bind to MLVs, which precede SUV fractions.
Figure 5.
Binding specificity of DHN1 to SUVs of different phospholipid compositions. Liposomes prepared from mixtures of neutral and acidic phospholipids were incubated with DHN1, then fractionated and analyzed as described in Figures 1 and 2. Fractions 12 through 14, Vesicle-bound DHN1; fractions 17 through 19, free DHN1. Each fraction was separated on 13% (w/v) SDS-PAGE, transferred to nitrocellulose, and analyzed with anti-K segment antibodies. The two bottom frames show fractions from a sample of DHN1 in the absence of SUV and a sample of ovalbumin in the presence of PA-derived SUV.
Electron microscopy images show that gel filtration increased the uniformity of liposomes by enriching for SUVs (Fig. 3, A versus B), and that the binding of DHN1 to SUV did not have a sufficient structural effect to be detectable by this method (Fig. 3, C versus D). Cryofixation was used (Fig. 3, C and D) to examine a possible structural effect of DHN1 binding to SUV because negative staining could conceivably strip liposomes of protein and thereby invalidate any comparison with and without protein. The liposome size distribution in Figure 3B (negative stain) is approximately 20 to 60 nm in diameter, and in Figure 3C (cryo) approximately 15 nm, but this difference has a trivial explanation. Liposomes that start with the same diameter will appear approximately 1.4 times as large after negative staining as they will after cryofixation. This is because of drying and spreading during negative staining. Also, in cryofixation, the specimen thickness (diameter of liposomes) is dictated by the ice thickness. In thin ice (e.g. 15-nm thickness), 60-nm-diameter liposomes will not be present. The same liposome sample was used for Figure 3, B and C. Bigger liposomes than shown in Figure 3C were seen in thicker ice near the edge of the carbon hole (not shown).
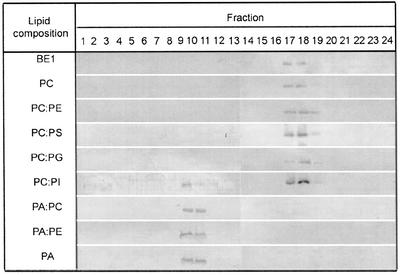
To investigate whether DHN1 can bind to more planar, less curved bilayer surfaces, binding to large unilamellar vesicles (LUVs) was examined. LUVs were prepared from the same battery of phospholipids used for SUVs (Fig. 6). DHN1 did not interact with any LUV except for those containing PA or PI. In the case of both PA:PC and PA:PE, binding to LUV was weaker than to SUV. This suggests that DHN1 has a higher binding affinity to smaller vesicles of anionic phospholipids but will bind to even more planar lipid surfaces if PA, which has a highest charge density of head group among the phospholipids, is present. The binding of DHN1 to PI-derived LUV, but not to PI-derived SUV, suggests that the specific packing properties of PI have an effect on the binding specificity of DHN1 to PI-derived vesicles.
Figure 6.
Specificity of DHN to LUVs of different phospholipid compositions. LUVs prepared from various lipid compositions were incubated with DHN1 and analyzed by western blotting using anti-K segment antibodies, as in Figure 5. Fractions 9 and 10 contain LUVs, whereas fractions 17 through 19 contain free DHN1.
Structural Change in Lipid-Bound DHN1
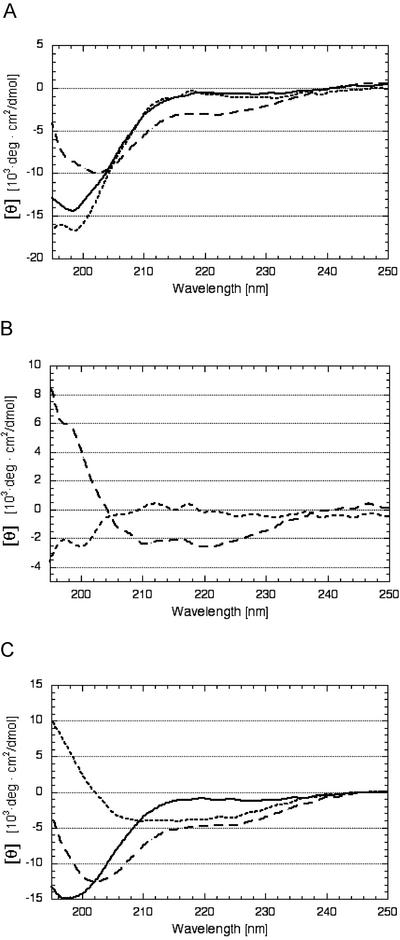
Circular dichroism (CD) analysis was performed to measure structural changes of DHN1 upon binding to vesicles (Fig. 7). The CD spectrum of free DHN1 shows a peak of negative ellipticity at 195 to 200 nm and weak ellipticity around 222 nm, which is characteristic of random coil. Upon incubation with PC-derived vesicles, there was no significant change in the CD spectrum compared with free DHN1 (Fig. 7A). However, incubation of DHN1 with PA-derived vesicles caused a significant shift of the spectrum at 208 to 222 nm (Fig. 7A). The difference spectra (Fig. 7B) indicate a gain of structure when DHN1 binds to PA-derived vesicles, but no gain of structure with PC-derived vesicles. When these spectra were analyzed for evidence of secondary structure, the α-helicity of DHN1 was estimated at 1% in PC-derived SUVs and 9% in PA-derived SUVs. Analysis of the difference spectra (Fig. 7B) by the K2D program (http://www.embl-heidelberg.de/∼andrade/k2d.html) indicated that nearly all of the structure gained by DHN1 upon binding to PA-derived SUV is α-helix. These results with SUV are qualitatively and quantitatively very similar to results obtained by incubating DHN1 with 10 mm SDS (Fig. 7C).
Figure 7.
The effect of PA, PC, and SDS on the secondary structure of DHN1. For incubation with liposomes, the concentration of DHN1 dialyzed against 20 mm phosphate buffer (pH 7.0) was adjusted to 45 μg mL−1 by the addition of buffer alone or liposome in buffer. For incubation with SDS, DHN1 dialyzed against 50 mm NaCl was adjusted to 30 μg mL−1. The CD spectrum was measured at 25°C in a 0.1-cm path length quartz cuvette with eight accumulations, noise reduced, and converted into mean residue molar ellipticity [θ]. DHN1 was incubated with either PC-derived SUVs or PA-derived SUVs for 3 to 6 h at 25°C at a 1:15 mass ratio of DHN1 to phospholipid. A, Buffer- and liposome-corrected spectra of DHN1 alone (solid line), DHN1 in the presence of PC-SUV (dotted line), and DHN1 in the presence of PA-SUVs (dashed line). B, Difference spectra. Buffer- and liposome-corrected DHN1 spectrum in the presence of PA-derived SUVs, minus the spectrum of DHN1 alone (dashed line). Buffer- and liposome-corrected DHN1 spectrum in the presence of PC-derived SUVs, minus the spectrum of DHN1 alone (dotted line). C, DHN1 in 50 mm NaCl (solid line). DHN1 in 50 mm NaCl and 10 mm SDS (dashed line), difference spectrum (spectrum with 10 mm SDS minus spectrum with no SDS; dotted line).
DISCUSSION
Immunohistochemical studies have suggested that cytoplasmic maize DHN1 is concentrated near endomembrane systems within maize scutellar parenchyma cells (Asghar et al., 1994; Egerton-Warburton et al., 1997). Yet, this and other DHNs (and LEAs and CORs in general) can be found abundantly in soluble protein extracts (Ceccardi et al., 1994; Lisse et al., 1996; Ismail et al., 1999). It may seem hard to reconcile evidence for membrane association with solubility properties of DHNs. However, DHNs are retained on hydrophobic interaction matrices (Ceccardi et al., 1994) and, despite their apparently extreme hydrophilicity on hydropathy plots, contain one or more putative lipid-binding class A2 amphipathic α-helices. DHNs also have a basic pI, which is typical of peripheral membrane proteins, and there are many examples of water-soluble peripheral membrane proteins (Sankaram and Marsh, 1993). Together, the facts are not inconsistent with DHNs binding to membrane surfaces.
The affinity of DHN1 for vesicles containing anionic phospholipids is intriguing. There are numerous examples of lipid- or membrane-binding proteins that require acidic phospholipids to bind, in some cases to regulate the biological activity or structure of the proteins. CTP:phosphocholine cytidyltransferase, a key regulatory enzyme in the PC biosynthetic pathway in animals, is active only when bound to acidic phospholipid bilayers, and the membrane-binding region is predicted to form amphipathic α-helices (Wang and Kent, 1995). The Escherichia coli SecA protein, a principal component of the protein export machinery, binds and inserts into acidic phospholipid membranes in parallel with a conformational change to an active form (Ulbrandt et al., 1992). The initiator protein of chromosome replication in E. coli, DnaA, requires acidic membrane phospholipids for displacement of ADP from inactive ADP-DnaA, which is followed by ATP binding and activation of DnaA as a replication initiator (Sekimizu and Kornberg, 1993). During the activation of protein kinase C by Ca2+, a membrane microdomain enriched in PS forms, which then induces membrane association and activation of protein kinase C (Orr et al., 1992). Human Src protein binds to membranes via electrostatic interactions of its basic amino terminus with acidic phospholipids and hydrophobic insertion of a myristoyl group (Sigal et al., 1994).
In this work, we have shown that DHN1 binds liposomes that contain a range of acidic phospholipids. Preferential binding of DHN1 to SUVs versus LUVs of the same lipid composition may be because of distinctive packing properties of lipids in the vesicle surface. LUVs have a relatively planar bilayer with the acyl chains generally buried within a hydrophobic environment. In comparison, the more highly curved surface of SUV has more lipid packing defects, through which penetration of segments of peripheral or integral proteins can be initiated (Berkhout et al., 1987; Riley et al., 1997; Johnson et al., 1998). DHN1 might be adsorbed to the membrane surface or it may penetrate somewhat into the hydrophobic inner core. In addition to the nature of the head group of lipids, their acyl chain composition (e.g. di-oleoyl versus palmitoyl-oleoyl) and degree of saturation could also affect the intrinsic curvature of the membrane, and also potentially the interaction of the membrane with DHN1. Certainly, there is a major influence of lipid acyl chain saturation on the interaction of synucleins with liposomes (Perrin et al., 2001). Synucleins bind and form multimers upon exposure to vesicles containing certain polyunsaturated fatty acid acyl groups, including arachidonoyl and docosahexaenoyl, even when the head group is neutral. The possible influence of acyl chain composition on the binding of DHN1 to liposomes remains to be explored.
In any case, the binding of DHN1 to SUV is associated with an apparent gain of secondary structure such that there is a transition to about 9% α-helical structure, similar to DHN1 in the presence of 10 mm SDS (Fig. 7). Because there are two K segments in the DHN1 protein, and because these portions of the protein comprise only about 15% of the total protein length (26 amino acids in a total of 168) and are the only portions of the protein predicted to form α-helices, these results are consistent with the K segments being about 58% α-helical under the conditions used. Alternatively, one K segment may be 100% α-helical and the other may be relatively unstructured under the conditions used.
There are many examples of the induction of α-helical or β-sheet structures that accompany the binding of proteins to membranes (Ladokhin and White, 1999), notably the adoption of α-helical structure by the binding of α-synuclein to liposomes containing anionic phospholipids (Davidson et al., 1998). The binding of DHN1 to PI-derived LUVs but not to PI-derived SUVs is intriguing in that it may help define the specificity of the interaction in more detail. To get more detailed information of DHN protein structure in the presence of SDS and liposomes, multidimensional NMR studies are under way. CD analyses similar to those shown using SUV in Figure 7 indicate that DHN1 gains structure also in association with LUV (data not shown).
One or more copies of the 15-amino acid consensus K segment, which resembles the lipid-binding class A2 amphipathic α-helical peptides (Epand et al., 1995), are present in all DHN proteins. Class A2 amphipathic helices are thought to stabilize the membrane, protecting it from fusion and lysis by decreasing the monoloayer negative curvature strain (Epand et al., 1995; Steponkus et al., 1998). As does a 26.5-kD cowpea DHN (Ismail at al., 1999), maize DHN1 also assumes increased α-helical structure in the presence of 10 mm SDS (Fig. 7C). The increase of α-helicity of maize DHN1 when bound to lipid vesicles in vitro suggests that DHN1 takes on α-helical structure also when associated with vesicles in vivo and that the K segment is involved in membrane binding.
It is interesting to consider the apparent preference of DHN1 for PA-containing vesicles. Specific physical attributes of PA may underlie the apparent preference of DHN1 for this particular acidic phospholipid. PA-derived vesicles undergo bilayer-to-hexagonal phase transitions at acidic pH and in the presence of high concentrations of Ca2+ (Cullis et al., 1986). PA is a minor lipid in plant cells, representing only about 1% to 2% of the total phospholipids, but PA levels typically increase with activation of phospholipase D activity in response to a number of signals or physical stress, including drought stress (Moreau et al., 1998; Frank et al., 2000; Munnik, 2001). For example, in green algae (Chlamydomonas moewvsii Gerloff) and cultured tomato (Lycopersicon esculentum Mill.) and alfalfa (Medicago sativa) cells, PA increased rapidly and persisted for a few hours when cells were challenged with NaCl (Munnik et al., 2000). Also, when barley aleurone protoplasts were treated with the plant water stress signal molecule abscisic acid, both phospholipase D activity and PA levels were transiently elevated for 10 to 15 min (Ritchie and Gilroy, 1998). The case has been made recently that PA acts as a second messenger in a wide variety of stress responses, including osmotic stress and others (Munnik, 2001). Also, one could imagine that PA-enriched lipid domains might form within membranes, and, as a consequence, DHN1 might then associate with these PA-rich domains. The abundance of PA also increases the net negative charge of the membrane, which may generally affect protein-membrane electrostatic interactions (Wang, 2000).
We hypothesize that DHN1 stabilizes membranes either by reducing the negative curvature strain of PA-enriched monolayers, possibly to inhibit transition to the hexagonal II phase, or by altering the membrane interfacial charge density to decrease the facilitated fusion of negatively charged vesicles. The Cor15am protein from Arabidopsis, which lowers occurrence of freeze-induced lamellar to hexagonal phase II transitions (Steponkus et al., 1998), also binds to PA-derived SUVs (data not shown). Further studies with liposomes will help develop a more complete understanding of the mode of action of DHN1 and a number of other stress-related proteins that adopt amphipathic α-helices at the bilayer/sol interface.
MATERIALS AND METHODS
Phospholipids and Other Reagents
PC (soybean [Glycine max Merr.], +99%), dioleyl PA, 1-palmitoyl 2-oleyl PE, bovine liver PI (+99%), 1-palmitoyl 2-oleyl PG, PS (soybean, +98%) and brain phospholipid extract type 1 (BE1; Folch fraction I from bovine brain) were purchased from Sigma (St. Louis). All other reagents were analytical or reagent grade.
Purification of DHN1 from Maize (Zea mays) Kernels
Purification of maize DHN1 (Mr = 17,070 and ε278 = 7,000 m−1 cm−1) from inbred W22 was performed essentially as described previously (Ceccardi et al., 1994). In brief, maize kernels were ground and extracted with 25 mm MES buffer (pH 6.0), 20 mm NaCl, and 1 mm phenylmethylsulfonyl fluoride. The delipidated extract was heated, then the soluble supernatant was dialyzed against 25 mm MES (pH 6.0) and 20 mm NaCl and loaded onto a Source 15S column and eluted with a gradient of 20 to 340 mm NaCl. DHN1 fractions were pooled and dialyzed against 50 mm phosphate buffer (pH 7.5) and 1.4 m ammonium sulfate, loaded onto a phenyl superose HR 10/10 FPLC column, and eluted with a gradient of 1.4 to 0 m ammonium sulfate. After this step of hydrophobic interaction chromatography (HIC), the protein was approximately 95% DHN1, and we refer to this grade of purity as HIC grade. Further purification by anion-exchange chromatography was accomplished by loading onto a Source 15 Q column equilibrated with 20 mm Tris (pH 8.8) and eluting with a gradient of 0 to 1.0 m NaCl. This resulted in four fractions of >99% DHN1, each representing a different state of phosphorylation. Before incubation with SUVs or LUVs, fractions containing DHN1 were pooled and dialyzed either against 40 mm Tris (pH 7.4; for gel filtration) or 20 mm potassium phosphate buffer (pH 7.0; for CD measurements), and then concentrated using a filter (Amicon Ultra-YM3, Amicon, Bedford, MA).
Preparation of SUVs and LUVs
Lipids were dissolved in chloroform and dried under a stream of nitrogen gas. SUVs of each lipid composition were generated by a sonication method (Barenholz et al., 1977). In brief, 10 mg of dried lipid was suspended in 1 mL of 40 mm Tris (pH 7.4; for gel filtration) or 20 mm potassium phosphate buffer (pH 7.0; for CD). Lipid dispersions were sonicated using a sonifier (Sonifier 450, Branson Ultrasonics, Danbury, CT). Sonication was carried out in an ice-filled bucket for 20 to 30 min with 1- to 2-min pulse intervals followed by 1-min cooling periods. SUVs and MLVs were isolated by ultracentrifugation at 100,000g in a TLA 100.2 rotor (Beckman Coulter, Fullerton, CA) for 1 h at 25°C. The upper portion of the supernatant contained SUVs and MLVs. MLVs are larger vesicles than SUVs and are composed of stacked bilayers with only about 10% of the lipid exposed in the outermost layer.
LUVs of various lipid compositions were prepared by an extrusion method (Hope et al., 1985). In brief, lipid mixtures were resuspended in 40 mm Tris buffer (pH 7.4) to generate large MLVs, which were then introduced into an extruder (Lipex Biomembranes, Vancouver). The large MLVs were passed through polycarbonate filters of 100-nm pore size 10 times under pressure of 100 to 500 p.s.i. This method produced unilamellar vesicles with hydrodynamic diameters of 130 ± 30 nm. The sizes of SUVs and LUVs were confirmed using negatively stained electron microscopy.
Binding of DHN1 to SUVs and LUVs
To investigate the interaction of DHN1 with lipid vesicles of different surface curvatures, the SUV and LUV dispersions in 40 mm Tris-HCl (pH 7.4) were incubated with DHN1 essentially as described previously for α-synuclein (Davidson et al., 1998). In brief, the vesicles and DHN1 at a 15:1 mass ratio of phospholipid to DHN1 were incubated in buffer for 3 to 12 h at 25°C. The incubation mixture was loaded onto a calibrated Superose 6 (10 mm × 30 cm) gel filtration column (Amersham Pharmacia Biotech, Piscataway, NJ) and eluted at 0.5 mL min−1 with 40 mm Tris-HCl (pH 7.4) to separate unreacted lipid, liposome with DHN1, and free DHN1 protein. The results presented in this manuscript represent highly reproducible results of numerous replicates.
Protein Concentration Determination
DHN1 concentration in pure solution (after anion exchange) was determined using a molar extinction coefficient of 7,000 m−1 cm−1 at 278 nm calculated on the basis of the amino acid sequence of DHN1 (Gill and von Hippel, 1989) and verified by the bicinchoninic acid method (Smith et al., 1985). HIC-grade DHN1 concentration was determined by the bicinchoninic acid method with a correction for the estimated 5% impurity.
Concentration of Phospholipids
Phospholipids in each fraction were measured by an ashing procedure, in which the phosphorus contents were determined by color development at 820 nm because of the formation of an ammonium phosphomolybdate complex (Ames and Dubins, 1960).
Immunoblotting
To determine the interactions of DHN1 with SUVs or LUVs, every fraction from the gel filtration column was collected, and an aliquot separated on 13% (w/v) SDS-PAGE and analyzed by western blotting with polyclonal anti-dehydrin antibody directed against the conserved K segment peptide of DHNs, as described previously (Close et al., 1993).
SUV Analysis Using CF
Determination of the release of a water-soluble marker from the liposome is a useful way to identify and confirm the intact liposome. CF, a hydrophilic fluorescent reagent, was used for this purpose. In brief, SUV composed of bovine brain extract phospholipids (BE1; Sigma; Folch fraction I; approximately 10% PI and approximately 50% PS) were prepared in a solution of 0.1 m CF in 20 mm Tris-HCl buffer (pH 7.4) by the sonication procedure described previously (Weinstein et al., 1981; Sun et al., 1996). The suspension was passed through a Superose 6 gel filtration column to separate liposome-enclosed CF from free CF. By addition of Triton X-100, SUVs were lysed and the CF contained within the SUVs was released into the solution. The fluorescence of each fraction was measured with and without addition of 2% (w/v) Triton X-100 using a Flurolog 112A fluorescence spectrometer (SPEX Inc., Edison, NJ) with excitation at 460 nm and emission at 550 nm. The slit widths for excitation and emission were 0.5 mm.
Electron Microscopy of SUVs
For cryo electron microscopy, SUVs in column buffer were applied to freshly glow-discharged holey carbon grids and after blotting excess liquids, the grids were plunged into liquid ethane cooled by liquid nitrogen. For negative staining, SUVs were diluted in column buffer and applied to 300 mesh copper grids covered with continuous carbon film. SUVs were stained with 2% (w/v) uranyl acetate. Grids were examined in a Philips CM300 transmission electron microscope (Analytical Electron Microscopy Facility, University of California, Riverside) equipped with a Gatan cryo setup and CCD camera.
CD Measurement and Analysis
The overall secondary structures of DHN1 in liposome-free and -bound states were analyzed using CD spectroscopy. The protein sample was dialyzed against 20 mm potassium phosphate buffer (pH 7.0) and concentration was adjusted to 45 μg mL−1 by addition of buffer or liposome in buffer. Spectra were measured at 25°C in a 0.1-cm path length quartz cuvette using a J-715 spectropolarimeter (Jasco, Easton, MD). The DHN1 spectra were corrected against buffer and liposome. The content of α-helicity was estimated using the learning neural network program K2D (Andrade et al., 1993).
ACKNOWLEDGMENTS
The authors thank Eugene Nothnagel (University of California, Riverside) for advice on the phosphorous assay and use of the fluorescence spectrometer, Anthony Huang (University of California, Riverside) for use of a sonicator, David Johnson (University of California, Riverside) for advice on fluorescence spectroscopy, Carl Frieden (Washington University School of Medicine, St. Louis) for use of his CD spectrometer during the sabbatical leave of T.J.C., and Larry Vickery for access to the CD spectrometer at University of California (Irvine).
Footnotes
This work was supported by the National Science Foundation (grant no. IBN 92–05269), by the University of California (Biotechnology Research and Education grant no. 97–15), and by the California Agricultural Experiment Station Funds (Hatch grant no. 5306–H).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.011171.
LITERATURE CITED
- Ames BN, Dubin DT. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J Biol Chem. 1960;235:769–775. [PubMed] [Google Scholar]
- Andrade MA, Chacon P, Merelo JJ, Moran F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- Asghar R, Fenton RD, DeMason DA, Close TJ. Nuclear and cytoplasmic localization of maize embryo and aleurone dehydrin. Protoplasma. 1994;177:87–94. [Google Scholar]
- Barenholz Y, Gibbes D, Litman BJ, Goll J, Thompson TE, Carlson RD. A simple method for the preparation of homogenous phospholipid vesicles. Biochemistry. 1977;16:2806–2810. doi: 10.1021/bi00631a035. [DOI] [PubMed] [Google Scholar]
- Berkhout TA, Rietveld A, de Kruijff B. Preferential lipid association and mode of penetration of apocytochrome c in mixed model membranes as monitored by tryptophanyl fluorescence quenching using brominated phospholipids. Biochim Biophys Acta. 1987;897:1–4. doi: 10.1016/0005-2736(87)90308-7. [DOI] [PubMed] [Google Scholar]
- Ceccardi TL, Meyer NC, Close TJ. Purification of a maize dehydrin. Protein Expr Purific. 1994;5:266–269. doi: 10.1006/prep.1994.1040. [DOI] [PubMed] [Google Scholar]
- Close TJ. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant. 1996;97:795–803. [Google Scholar]
- Close TJ. Dehydrins: a commonalty in the response of plants in dehydration and low temperature. Physiol Plant. 1997;100:291–296. [Google Scholar]
- Close TJ, Fenton RD, Moonan F. A view of plant dehydrins using antibodies specific to the carboxy terminal peptide. Plant Mol Biol. 1993;23:279–286. doi: 10.1007/BF00029004. [DOI] [PubMed] [Google Scholar]
- Cullis PR, Hope MJ, Tilcock CPS. Lipid polymorphism and the roles of lipids in membranes. Chem Phys Lipids. 1986;40:127–144. doi: 10.1016/0009-3084(86)90067-8. [DOI] [PubMed] [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell. 1998;10:623–638. doi: 10.1105/tpc.10.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- Egerton-Warburton LM, Balsamo RA, Close TJ. Temporal accumulation and ultrastructural localization of dehydrins in Zea mays L. Physiol Plant. 1997;101:545–555. [Google Scholar]
- Epand RM, Shai Y, Segrest JP, Anantharamaiah GM. Mechanisms for the modulation of membrane bilayer properties by amphipathic helical peptides. Biopolymers. 1995;37:319–338. doi: 10.1002/bip.360370504. [DOI] [PubMed] [Google Scholar]
- Frank W, Munnik T, Kerkmann K, Salamini F, Bartels D. Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. Plant Cell. 2000;12:111–123. doi: 10.1105/tpc.12.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem. 2000;275:5668–5674. doi: 10.1074/jbc.275.8.5668. [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Hope MJ, Bally MB, Webb G, Cullis PR. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim Biophys Acta. 1985;812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Ismail AM, Hall AE, Close TJ. Purification and partial characterization of a dehydrin involved in chilling tolerance during seedling emergence of cowpea. Plant Physiol. 1999;120:237–244. doi: 10.1104/pp.120.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Rao NM, Hei SW, Cornell RB. Conformation and lipid binding properties of four peptides derived from the membrane-binding domain of CTP:phosphocholine cytidylyltransferase. Biochemistry. 1998;37:9509–9519. doi: 10.1021/bi980340l. [DOI] [PubMed] [Google Scholar]
- Ladokhin AS, White SH. Folding of amphipathic α-helices on membranes: energetics of helix formation by melittin. J Mol Biol. 1999;285:1363–1369. doi: 10.1006/jmbi.1998.2346. [DOI] [PubMed] [Google Scholar]
- Lisse T, Bartels D, Kalbitzer HR, Jaenicke R. The recombinant dehydrin-like desiccation stress protein from the resurrection plant Craterostigma plantagineum displays no defined three-dimensional structure in its native state. J Biol Chem. 1996;377:555–567. doi: 10.1515/bchm3.1996.377.9.555. [DOI] [PubMed] [Google Scholar]
- McCubbin WD, Kay CM, Lane BG. Hydrodynamic and optical properties of the wheat germ Em protein. Can J Biochem Cell Biol. 1985;63:803–811. [Google Scholar]
- Moreau P, Bessoule JJ, Mongrand S, Testet E, Vincent P, Cassagne C. Lipid trafficking in plant cells. Prog Lipid Res. 1998;37:371–391. doi: 10.1016/s0163-7827(98)00016-2. [DOI] [PubMed] [Google Scholar]
- Munnik T. Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci. 2001;6:227–233. doi: 10.1016/s1360-1385(01)01918-5. [DOI] [PubMed] [Google Scholar]
- Munnik T, Meijer HJG, ter Riet B, Hirt H, Frank W, Bartels D, Musgrave A. Hyperosmotic stress stimulates phospholipase D activity and elevates the level of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 2000;22:147–154. doi: 10.1046/j.1365-313x.2000.00725.x. [DOI] [PubMed] [Google Scholar]
- Orr JF, Keranen LM, Newton AC. Reversible exposure of the pseudosubstrate domain of protein kinase C by phosphatidylserine and diacylglycerol. J Biol Chem. 1992;267:15263–15266. [PubMed] [Google Scholar]
- Perrin RJ, Woods WS, Clayton DF, George JM. Exposure to long chain polyunsaturated fatty acids triggers rapid multimerization of synucleins. J Biol Chem. 2001;276:41958–41962. doi: 10.1074/jbc.M105022200. [DOI] [PubMed] [Google Scholar]
- Riley ML, Wallance BA, Flitsch S, Booth PJ. Slow α helix formation during folding of a membrane protein. Biochemistry. 1997;36:192–196. doi: 10.1021/bi962199r. [DOI] [PubMed] [Google Scholar]
- Ritchie S, Gilroy S. Abscisic acid signal transduction in the barley aleurone is mediated by phospholipase D activity. Proc Natl Acad Sci USA. 1998;95:2697–2702. doi: 10.1073/pnas.95.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozek A, Buchko GW, Cushley RJ. Conformation of two peptides corresponding to human apolipoprotein C-I residues 7–24 ad 35–53 in the presence of sodium dodecyl sulfate by CD and NMR spectroscopy. Biochemistry. 1995;34:7401–7408. [PubMed] [Google Scholar]
- Sankaram MB, Marsh D. Protein-lipid interactions with peripheral membrane proteins. In: Watt A, editor. Protein-Lipid Interactions. 25. New Comprehensive Biochemistry. New York: Elsevier; 1993. pp. 127–162. [Google Scholar]
- Sekimizu K, Kornberg A. Cardiolipin activation of DnaA protein, the initiation protein of replication in Escherichia coli. J Biol Chem. 1993;263:7131–7135. [PubMed] [Google Scholar]
- Sigal CT, Zhou W, Buser CA, Mclaughlin S, Resh MD. Amino-terminal basic residues of Src mediate membrane binding through electrostatic interaction with acidic phospholipids. Proc Natl Acad Sci USA. 1994;91:12253–12257. doi: 10.1073/pnas.91.25.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Steponkus PL, Uemera M, Joseph RA, Gilmour SJ, Thomashow MF. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1998;95:14570–14575. doi: 10.1073/pnas.95.24.14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun WQ, Leopold AC, Crowe LM, Crowe JH. Stability of liposomes in sugar glasses. Biophys J. 1996;70:1769–1776. doi: 10.1016/S0006-3495(96)79740-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Ulbrandt ND, London E, Oliver DB. Deep penetration of a portion of the Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J Biol Chem. 1992;267:15184–15192. [PubMed] [Google Scholar]
- Wang X. Multiple forms of phospholipase D in plants: the gene family, catalytic and regulatory properties, and cellular functions. Prog Lipid Res. 2000;39:109–149. doi: 10.1016/s0163-7827(00)00002-3. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kent C. Identification of an inhibitory domain of CTP:phosphocholine cytidyltransferase. J Biol Chem. 1995;270:18948–18952. doi: 10.1074/jbc.270.32.18948. [DOI] [PubMed] [Google Scholar]
- Weinstein JN, Kluser RD, Innerarity T, Ralston E, Blumenthal R. Phase transition release, a new approach to the interaction of proteins with lipid vesicles. Biochim Biophys Acta. 1981;647:270–284. doi: 10.1016/0005-2736(81)90255-8. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson JH. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]