Abstract
R2R3 Myb genes are widely distributed in the higher plants and comprise one of the largest known families of regulatory proteins. Here, we provide an evolutionary framework that helps explain the origin of the plant-specific R2R3 Myb genes from widely distributed R1R2R3 Myb genes, through a series of well-established steps. To understand the routes of sequence divergence that followed Myb gene duplication, we supplemented the information available on recently duplicated maize (Zea mays) R2R3 Myb genes (C1/Pl1 and P1/P2) by cloning and characterizing ZmMyb-IF35 and ZmMyb-IF25. These two genes correspond to the recently expanded P-to-A group of maize R2R3 Myb genes. Although the origins of C1/Pl1 and ZmMyb-IF35/ZmMyb-IF25 are associated with the segmental allotetraploid origin of the maize genome, other gene duplication events also shaped the P-to-A clade. Our analyses indicate that some recently duplicated Myb gene pairs display substantial differences in the numbers of synonymous substitutions that have accumulated in the conserved MYB domain and the divergent C-terminal regions. Thus, differences in the accumulation of substitutions during evolution can explain in part the rapid divergence of C-terminal regions for these proteins in some cases. Contrary to previous studies, we show that the divergent C termini of these R2R3 MYB proteins are subject to purifying selection. Our results provide an in-depth analysis of the sequence divergence for some recently duplicated R2R3 Myb genes, yielding important information on general patterns of evolution for this large family of plant regulatory genes.
Gene duplications have long been viewed as a main source of evolutionary novelty (Ohno, 1970). The dramatic expansion of the R2R3 Myb family of regulatory genes in the higher plants provides a striking example of how gene amplifications followed by divergence may have impacted plant evolution. Members of this gene family encode proteins characterized by two 50- to 52-residue-long imperfect repeats. Each of these MYB repeats contains three α-helices, with the second and third helices forming a helix-turn-helix structure when bound to DNA (Ogata et al., 1994). Around 125 R2R3 Myb genes are present in the Arabidopsis genome (Reichmann and Ratcliffe, 2000; Stracke et al., 2001), and more than 200 in maize (Zea mays) and related monocots (E.L. Braun and E. Grotewold, unpublished data). This represents a sharp contrast to the small number of genes encoding MYB homologs in the animal and fungal kingdoms (Lipsick, 1996).
Plant R2R3 Myb genes originated from an ancestral gene encoding a three-MYB repeat protein represented in animals today by c-myb and related genes (Lipsick, 1996), and in plants by the small pc-Myb gene family (Braun and Grotewold, 1999a). After the loss of the R1 repeat, an explosive amplification of the R2R3 Myb gene family occurred 250 to 400 million years ago (Mya) (Rabinowicz et al., 1999). R2R3 Myb genes are more or less evenly distributed throughout the plant genome without forming obvious clusters, as found for plant resistance (R) genes, for example (Bergelson et al., 2001).
R2R3 MYB proteins are characterized by the presence of a conserved MYB domain and a longer divergent C-terminal region. Short conserved motifs in the C-terminal region of these proteins have been identified and were used to classify Arabidopsis R2R3 Myb genes into subgroups (Kranz et al., 1998; Stracke et al., 2001). The dramatic divergence of the C-terminal regions does not appear to have a large influence in the regulatory function of the corresponding proteins. For example, related R2R3 Myb genes that act as regulators of anthocyanin biosynthesis in maize, petunia (Petunia hybrida), and Arabidopsis (encoded by C1, An2, and Pap1, respectively) show little or no detectable identity outside their MYB domains. Despite this C-terminal divergence, the maize C1 protein can complement petunia An2 mutants, and vice versa (Quattrocchio et al., 1999, 1993). Similarly, the Arabidopsis GL1 and WER proteins are interchangeable in their ability to regulate root hair or trichome formation, despite sharing only 23% C-terminal identity (Lee and Schiefelbein, 2001). Finally, domain-swapping experiments involving maize P1 and C1 suggest regulatory specificity is largely provided by the MYB domains of these proteins (Grotewold et al., 2000).
The high degree of sequence divergence for the C-terminal regions of many R2R3 Myb proteins, coupled with the apparent absence of functional constraints upon these regions, suggests that they may be diverging at the neutral rate. However, some blocks of C-terminal sequence identity in specific plant R2R3 MYB proteins have persisted over very long evolutionary times. The most compelling example is a block of >50 amino acids with >40% identity found in a moss (Physcomitrella patens) MYB protein and the snapdragon (Antirrhinum majus) MIXTA protein (Kranz et al., 1998). Detailed comparison of sequence divergence in the MYB domains and C-terminal regions of R2R3 MYB proteins might provide insights into these apparent contradictions, and the first attempt to compare the evolution of different regions in MYB proteins found substantial disagreement between trees identified using different regions (Rosinski and Atchley, 1998). This prompted the conclusion that MYB proteins are polyphyletic (Rosinski and Atchley, 1998), a term used for groups based upon shared characters that reflect evolutionary convergence (for review, see Page and Holmes, 1998). The conclusion that Myb genes are polyphyletic must be viewed with caution because incongruence between phylogenetic trees estimated using different regions of the same gene could reflect difficulties with the alignment or analysis rather than genuine differences in tree topology. In fact, it has been demonstrated that random data can show significant incongruence with phylogenetically structured data (Dolphin et al., 2000).
Previous studies have provided a number of specific hypotheses regarding the patterns of evolution for different regions of R2R3 MYB proteins, which differ regarding the rate of evolution and degree of functional constraint upon the C-terminal regions (Braun and Grotewold, 1999a; Stracke et al., 2001). Examining the degree of functional constraint upon proteins using evolutionary comparisons is typically accomplished by calculating the ratio of non-synonymous (KA) to synonymous (KS) substitutions, a value designated ω. If there are no constraints upon the amino acid sequence, estimates of ω will equal one. In contrast, the purifying selection typical of most proteins will result in estimates of ω < 1, whereas the rapid fixation of non-synonymous substitutions that are selectively advantageous will result in estimates of ω > 1 for those sites or regions subject to positive selection. Estimates of ω will exhibit a high variance when they are calculated using data from ancient gene duplications because synonymous sites are expected to saturate rapidly in plants (see Rabinowicz et al., 1999). However, the identification of a group of R2R3 Myb genes that has undergone a recent amplification in the grasses (Braun and Grotewold, 1999b; Rabinowicz et al., 1999) provides us with an excellent opportunity to investigate the patterns of evolution of recently duplicated R2R3 Myb genes.
In this study, we establish an evolutionary framework to explain events that followed the origin of R2R3 Myb genes from widely distributed c-Myb-like genes. To investigate patterns of evolution responsible for the striking divergence that characterizes plant R2R3 Myb genes, we focused upon recently duplicated R2R3 Myb genes. Toward this goal, we characterized ZmMyb-IF35 and ZmMyb-IF25, two maize R2R3 Myb genes that belong to a gene clade that expanded during the evolution of the grasses (Rabinowicz et al., 1999). The physical map positions of these genes were determined and found to be consistent with an origin during the duplication of the maize genome. Comparisons of these and related R2R3 Myb genes provided evidence that MYB domains and C termini are both subject to purifying selection. Furthermore, our analyses indicate substantial heterogeneity in the number of non-synonymous and synonymous substitutions in different regions of recently duplicated R2R3 Myb genes. Together, these studies provide the first insights, to our knowledge, into the possible mechanisms of divergence that accompanied the dramatic expansion of the R2R3 Myb gene family in the plants.
RESULTS AND DISCUSSION
An Evolutionary Framework for Plant R2R3 MYB Proteins
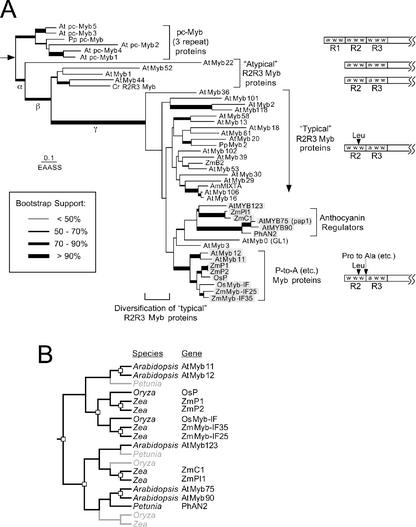
Plant MYB proteins are encoded by a very large and diverse gene family (Braun and Grotewold, 1999a; Rabinowicz et al., 1999; Stracke et al., 2001), with most plant Myb homologs characterized by two MYB repeats and the insertion of a single amino acid (Leu) in the first (R2) repeat, relative to animal Myb homologs (Fig. 1A). To provide a framework to examine plant Myb gene evolution, we estimated the phylogeny of a large number of Arabidopsis and selected monocot Myb proteins representing all major groups. Two data sets were used, a small alignment (45 sequences) and a large alignment (106 sequences; see supplemental data at www.plantphysiol.org). The topology of the trees obtained using these data sets, or by including additional plant Myb proteins from our own collection or available from public databases, were essentially the same with none of the well-supported clade shown in Figure 1A rearranged (not shown). Analyses of both alignments revealed two distinct types of plant Myb homologous that lack the Leu insertion in the R2 repeat: (a) the three repeat pc-Myb proteins (Braun and Grotewold, 1999a), also described as 3Rmyb (Kranz et al., 2000); and (b) a subset of MYB proteins designated the “atypical” R2R3 MYB proteins (Braun et al., 2001). Surprisingly, some of these atypical R2R3 MYB proteins (e.g. AtMyb22) have a Trp residue in the first helix of R3, like the pc-Myb proteins but unlike the majority of plant MYB proteins (Braun and Grotewold, 1999a).
Figure 1.
Evolutionary relationships among plant MYB domain proteins. A, An estimate of phylogeny obtained using weighted neighbor joining of ML distance estimates obtained using the WAG+ Γ model of sequence evolution (parameters are provided in “Materials and Methods”). Sequences are from Arabidopsis (At), Chlamydomonas reinhardtii (Cr), rice (Oryza sativa; Os), P. patens (Pp), and maize (Zm). An arrow indicates the position of the root discussed in the text. The support for each of the branches is indicated by the thickness of the lines. Estimates of phylogeny obtained using both alignments were identical, with the exception of the C. reinhardtii R2R3 MYB, which shifted to a position outside of a clade containing AtMyb1, AtMyb44, and AtMyb55 in analyses of the large alignment. Branch lengths are proportional to the expected number of amino acid substitutions per site under the WAG+ Γ model. The specific molecular changes that occurred during the evolution each of the major phylogenetic groups are indicted by the structures of the MYB domains on the right of the tree. Sequences included in the figure were selected to sample the diversity of Mybs based upon C-terminal motifs. B, Pattern of gene duplications for sequence duplication/divergence of the groups of R2R3 Myb genes discussed in this study. The pattern of gene duplications is shown as a reconciled tree, showing genes that have been inferred but not identified in specific lineages in light gray text. These genes have either been lost during evolution or have not been sampled in the relevant lineages.
The root of the plant Myb phylogeny is likely to be between the pc-Myb proteins and the R2R3 MYB proteins (indicated with an arrow in Fig. 1A). Three repeat MYB homologs are broadly distributed in animals, plants, and slime molds (Lipsick, 1996; Braun and Grotewold, 1999a), whereas typical R2R3 MYB proteins are limited to the green plant lineage (Braun et al., 2001; Stracke et al., 2001). Restricting our consideration to plant MYB proteins, a three-step model for the origin for typical R2R3 MYB proteins is suggested (Fig. 1A). This model would involve loss of the R1 repeat (branch α), mutation of the Trp residue in the first helix of R3 to a Phe (branch β), and insertion of the Leu residue in R2 (branch γ). Loss of the first repeat (branch α) occurred before divergence of land plants and chlorophyte algae because an atypical R2R3 Myb is present in C. reinhardtii (Fig. 1A). The Leu insertion and diversification of the typical R2R3 MYB proteins occurred before the divergence of mosses and angiosperms because a typical R2R3 Myb (PpMyb2) is present in the moss P. patens (Leech et al., 1993).
Previous analyses suggest that many gene duplications in the family encoding typical R2R3 MYB proteins occurred early in the history of land plants (Rabinowicz et al., 1999). This model of rapid diversification early in the history of land plants is consistent with the relatively short and poorly supported (by the bootstrap and posterior probabilities in Bayesian analyses) branches at the base of the typical R2R3 MYB group. Establishing the mechanisms by which the plant R2R3 Myb gene family expanded and diverged is complicated by the ancient nature of these duplications. However, several recently duplicated groups of R2R3 Myb genes have been identified in maize (Braun and Grotewold, 1999b), and the analysis of members of one of these groups provides clues on the possible mechanisms of evolutionary divergence after gene duplication of R2R3 Myb genes.
ZmMyb-IF25 and ZmMyb-IF35 Belong to a Group of Recently Duplicated R2R3 Myb Genes
We previously have analyzed more than 80 sequences of MybBRH and these studies provided evidence that several groups of maize R2R3 Myb genes amplified within the past 50 million years (Braun and Grotewold, 1999b; Rabinowicz et al., 1999). One of these groups corresponds to the P-to-A clade, united by the change of Pro-63 to Ala (Fig. 1A) in the hinge region between the R2 and R3 MYB repeats (Rabinowicz et al., 1999). The analysis of the MybBRH sequences suggested two types of genes within the P-to-A clade: (a) those diverging at a relatively low rate (e.g. P1, a regulator of 3-deoxy flavonoid biosynthesis; Grotewold et al., 1994); and (b) those diverging more rapidly (e.g. ZmMyb-IF25 and ZmMyb-IF35; Rabinowicz et al., 1999).
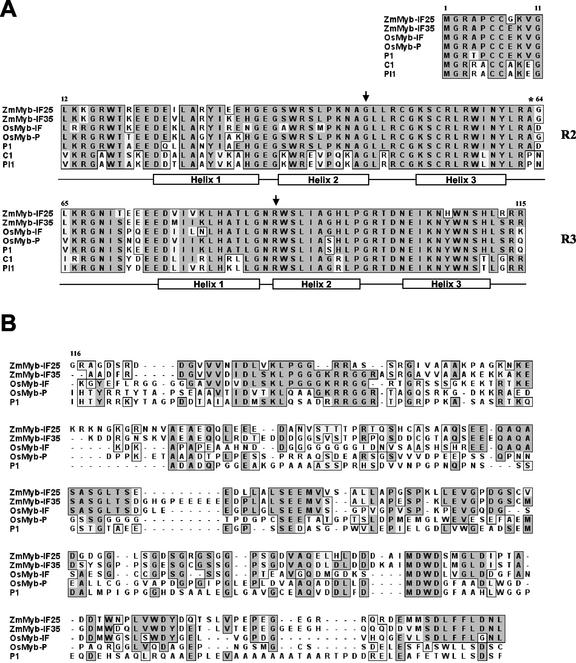
To better understand the patterns of evolution of these recently duplicated genes, we cloned ZmMyb-IF35 (accession no. AF521880) and ZmMyb-IF25 (accession no. AF521881; see “Materials and Methods”). A full-length cDNA clone for ZmMyb-IF25 was obtained from a yeast (Saccharomyces cerevisiae) one-hybrid screen, using the high-affinity P-binding sites present in the A1 gene (Grotewold et al., 1994), in an effort to identify additional proteins that recognize this binding site (M.-G. Kim and E. Grotewold, unpublished data). The molecular analysis of the genomic and cDNA clones showed that the intron-exon structure of ZmMyb-IF35 and ZmMyb-IF25 (Fig. 2A) was identical to that of the maize P1, C1, and Pl1 genes (Paz-Ares et al., 1987; Grotewold et al., 1991; Cone et al., 1993). Like in P1 (Grotewold et al., 1991), the second intron of ZmMyb-IF35 and ZmMyb-IF25 is unusually long, more than 2 kb for ZmMyb-IF35 and over 6 kb for ZmMyb-IF25.
Figure 2.
Sequence comparisons of recently duplicated R2R3 Myb genes. A, Amino acid sequence alignment in the MYB domain. Comparison of amino acid sequences of the predicted proteins encoded by ZmMyb-IF25, ZmMyb-IF35, OsMyb-IF, OsMyb-P, P1, C1, and Pl1. Dark-shaded boxes indicate identical amino acids and lighter shaded boxes indicate similar amino acids. The positions of the three helices forming the R2 and R3 MYB repeats are shown with the clear boxes and the MYB domain interrupted by two introns are indicated with arrows. The change from Pro to Ala that defines the MybPtoA clade is marked with a star. B, Amino acid sequence alignment for the C termini. Comparison of the predicted C-terminal regions encoded by MybPtoA clade members ZmMyb-IF25, ZmMyb-IF35, OsMyb-IF, OsMyb-P, and P1. Dashes indicate gaps introduced to reflect insertions and deletions during evolution.
We mapped the ZmMyb-IF35 and ZmMyb-IF25 loci using polymorphisms in the 5′ sequence. Primers designed from the 5′ region of ZmMyb-IF35 detected a direct size polymorphism between B73 and Mo17, the parents of the intermated B73/Mo17 (IBM) mapping population (Lee et al., 2002). Genotypes were determined for the 94 core individuals of the IBM population and ZmMyb-IF35 was placed against a framework map of 219 loci. Primers designed from the 5′ region of ZmMyb-IF25 detected a direct size polymorphism between T218 and GT119, the parents of a quantitative trait loci mapping population also used to map a large number of the maize SSR loci (Sharopova et al., 2002). Genotypes were determined for the 93 F2 individuals from this population and ZmMyb-IF25 was mapped against a framework map of 96 loci. The IBM and T218 X GT119 maps are available at MaizeDB (http://www.agron.missouri.edu/maps.html). ZmMyb-IF35 mapped to chromosome 3, bin 3.04 and ZmMyb-IF25 mapped to chromosome 8, bin 8.03–8.04. These map positions correspond to duplicated regions of the maize genome, based upon comparative mapping analyses (Helentjartis et al., 1988; Gaut and Doebley, 1997). Thus, similar to the maize regulators of anthocyanin biosynthesis C1 and Pl1, ZmMyb-IF35 and ZmMyb-IF25 appear to have diverged during the reversion of the segmental allotetraploid ancestor of maize to disomic inheritance, an event that has been estimated to have occurred approximately 11 Mya (Gaut and Doebley, 1997).
Patterns of Evolution of Recently Duplicated R2R3 Myb Genes
The origin of the P-to-A clade precedes the divergence of monocots and eudicots. Evidence for this is provided by the presence of two Arabidopsis sequences (AtMyb11 and AtMyb12) with a similar change of Pro-63 to Ser, AtMyb111 with a change of Pro-63 to Arg, and the cotton (Gossypium hirsutum) GhMyb-J that also has a Pro-63 to Ala substitution (Loguercio et al., 1999). Molecular clock estimates indicated that the expansion of this group to yield a large number of maize paralogs was an event unique to the grasses (Rabinowicz et al., 1999), a clade of plants that diversified within the past 55 million years. Consistent with this hypothesis, AtMyb11, AtMyb12, P1, ZmMyb-IF35, and ZmMyb-IF25 form a well-supported clade, with AtMyb11 and AtMyb12 basal to a monophyletic group of monocot P-to-A R2R3 Myb sequences (Fig. 1, A and B). However, two rice genes encoding P-to-A Mybs (accession nos. BAB64029 and AAL84631) provided evidence that the ZmMyb-IF35 and ZmMyb-IF25 genes diverged from the P lineage, characterized by the recently duplicated P1 and P2 genes (Zhang et al., 2000).
The evidence for orthology of the rice P-to-A sequences and the maize P1, ZmMyb-IF35, and ZmMyb-IF25 genes was strengthened by finding substantial C-terminal identity between these genes (Fig. 2B), prompting us to call the rice genes OsMyb-P and OsMyb-IF. The genomic sequence of OsMyb-P (accession no. AF474141) and OsMyb-IF (accession no. AP002873) shows the presence of relatively long (>4 kb) introns in the same position as the second intron in P1. The OsMyb-IF gene lacks an intron in a position homologous to the first intron in P1 (Fig. 2A), whereas other grass P-to-A Myb genes have short introns in this position.
The sister group to the P-to-A clade includes several anthocyanin regulators (e.g. the products of the maize C1 and the petunia An2 genes). Interestingly, we found limited support for a clade containing AtMyb123 and C1/Pl1 from maize, but excluding AtMyb75 (Pap1; Borevitz et al., 2000) and An2 from petunia. Based upon the exchangeability of C1 and An2 (Quattrocchio et al., 1993, 1999) and clustering in previous phylogenetic analyses (Braun and Grotewold, 1999b; Rabinowicz et al., 1999), AtMyb75, petunia An2, and maize C1 had been thought to be orthologous. However, C1, Pl1, and AtMyb123 have two short boxes of identity in the C-terminal region. One of these boxes is a nine-amino acid signature previously reported (Stracke et al., 2001) and the second is a 15-amino acid signature starting with amino acid 172 in C1 (E.L. Braun, unpublished data). These conserved motifs in the divergent C-terminal regions of these proteins, together with our phylogenetic analyses (Fig. 1), provide additional evidence that C1 and AtMyb123 are orthologs. Interestingly, AtMyb123 (TT2) was characterized to be a regulator of proanthocyanidin accumulation in developing seed (Nesi et al., 2001). It is likely that C1 and An2 are paralogs related by a duplication event before the divergence of monocots and eudicots, although they have clearly retained similar functions (Quattrocchio et al., 1999).
Taken together, these findings provide evidence for a model in which: (a) the origin of the P-to-A clade precedes the divergence of monocots and eudicots; (b) this group of R2R3 Myb genes underwent a recent expansion in the grasses; and (c) this expansion involved genome duplication (e.g. ZmMyb-IF35 and ZmMyb-IF25), tandem gene duplication (e.g. P1 and P2; Zhang et al., 2000), and a more ancient duplication (e.g. the P1/P2 and ZmMyb-IF35/ZmMyb-IF25 lineages). The study of the divergence patterns of R2R3 Myb genes that duplicated recently by diverse mechanisms provides a unique opportunity to understand the general evolutionary patterns of plant R2R3 Myb genes.
Divergence of the Conserved MYB Domains and Variable C-Terminal Regions
A general characteristic of plant R2R3 Myb genes is the high conservation of the MYB domains coupled with the dramatic divergence of C-terminal regions. Does this divergence difference reflect functional constraints upon the MYB domain with the C-terminal regions evolving at the neutral rate? Or are MYB domains and C-terminal regions both subject to purifying selection? To investigate the constraints upon the MYB and C-terminal regions of recently duplicated R2R3 Myb genes, the numbers of KS and KA substitutions were estimated for pairs of recently duplicated R2R3 Myb genes (Table I). As expected, separate estimates of ω (KA/KS) from the MYB and C-terminal regions indicated more constraints upon MYB domains than upon C-terminal regions. However, estimates of ω for C-terminal regions were lower than expected under the neutral model (Table I), suggesting the existence of moderate purifying selection for these regions.
Table I.
Synonymous (KS) and non-synonymous (KA) distances between the pairs of recently duplicated R2R3 Myb genes examined for this study
| Pair Comparison | KS (Mean/Myb/C Term) | KA (Mean/Myb/C Term) | ω (Mean/Myb/C Term) |
|---|---|---|---|
| C1/Pl1 | 0.2515/0.1239/0.2819 | 0.0707/0.0236/0.1354 | 0.2811/0.1907/0.4803 |
| AtMyb75/AtMyb90 | 0.4368/0.4970/0.3776 | 0.1254/0.0443/0.2558 | 0.2871/0.0891/0.6775 |
| ZmMyb-IF35/ZmMyb-IF25 | 0.4871/0.4708/0.5212 | 0.1136/0.0305/0.1654 | 0.2331/0.0647/0.3174 |
| AtMyb11/AtMyb12 | 1.4303/0.6542/1.3962 | 0.2748/0.0563/0.4383 | 0.1921/0.0861/0.3139 |
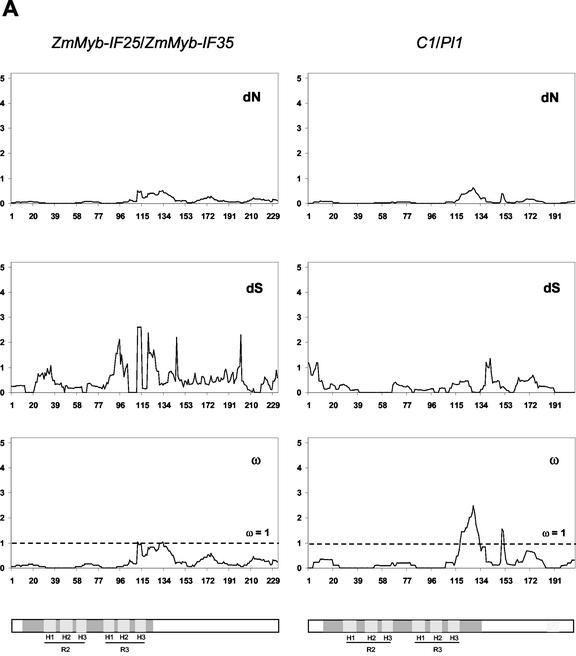
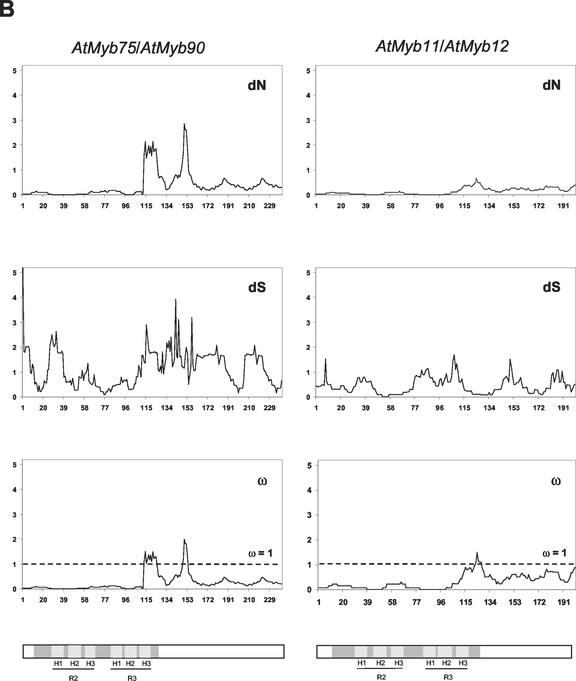
As a heuristic to examine which regions of these recently duplicated pairs of R2R3 Myb genes had accumulated more non-synonymous substitutions, we conducted a “sliding window” analysis (Fig. 3). These analyses provided evidence for functional constraints upon the MYB domain, consistent with the key role of MYB domains in DNA binding (for review, see Lipsick, 1996) and protein-protein interactions (Grotewold et al., 2000). However, the C-terminal regions of all pairs examined also show regions exhibiting very limited non-synonymous divergence (Fig. 3).
Figure 3.
Sliding window analysis of duplicated pairs of R2R3 Myb genes. Numbers of synonymous and non-synonymous substitutions in 15 codon windows were estimated using the YN00 model of sequence evolution (constraining nuisance parameters as described in “Materials and Methods”). Bars in gray indicate the three α-helices in MYB repeat 1 (H1, 22–33 amino acids; H2, 36–45 amino acids; and H3, 51–60 amino acids) and MYB repeat 2 (H1, 75–86 amino acids; H2, 88–97 amino acids; and H3, 102–111 amino acids), where the second and third helices form a helix-turn-helix (HTH) structure when bound to DNA. The TAD in ZmMyb-C1/ZmMyb-Pl1 is shown in light gray.
Estimates of KS were substantially higher for the segments of C1/Pl1 and AtMyb11/AtMyb12 that encode the C-terminal regions of these gene products than for the segments encoding the MYB domains of these proteins (Table I; Fig. 3). The divergent C-terminal regions of AtMyb75/AtMyb90 have accumulated slightly fewer synonymous differences than the highly similar MYB domains. However, the difficulty in estimating numbers of synonymous substitutions suggests that it is reasonable to conclude that any differences in KS for the MYB domain and the C terminal of both ZmMyb-IF35/ZmMyb-IF25 and AtMyb75/AtMyb90 are extremely modest.
These findings provide strong evidence that the C-terminal regions of all pairs of R2R3 Myb genes studied here are subject to moderate purifying selection and confirm the fact that MYB domains are subject to strong purifying selection. The existence of moderate purifying selection upon C-terminal regions is surprising because the R2R3 Myb genes examined here are members of clades for which experiments have demonstrated limited functional roles for the C-terminal domains (Goff et al., 1991; Sainz et al., 1997; Grotewold et al., 2000). However, based on the evolutionary analyses presented here, we propose that the C termini of these proteins are likely to play a more important role in the function of R2R3 MYB transcription factors than previously anticipated.
These data also provide evidence that KA and KS values exhibit a positive intragenic correlation in some genes, extending the results of studies using mammalian sequences (Alvarez-Valin et al., 1998; Smith and Hurst, 1999) to the flowering plants. Clearly, these results challenge the use of KS values to provide a time scale for evolution, and question the biological basis for the observed intragenic KA-KS correlation.
Understanding the Divergence of C-Terminal Regions between R2R3 Myb Genes
Although sliding window analyses did show some regions with ω = 1, consistent with specific regions evolving under a neutral model, there were no regions where estimates of ω greatly exceeded values consistent with the neutral or purifying selection models of evolution. These observations suggest that recently duplicated R2R3 Myb genes related to P1 and C1 in maize have not diverged because of the action of positive selection, suggested as a possible model for the evolution of specific MADS box transcription factors in species of the genera Argyroxiphium, Dubautia, and Wilkesia, a group known as the Hawaiian silverswords (Barrier et al., 2001). Thus, our results suggest that the observed non-synonymous acceleration does not represent a general feature of transcription factor genes in polyploids, because this study did not show evidence for accelerated accumulation of non-synonymous substitutions in maize, an ancient polyploid (Gaut and Doebley, 1997).
The C-terminal regions of R2R3 MYB proteins also accumulate a number of insertion and deletion changes (e.g. Fig. 2), making it difficult to align these regions for proteins that diverged more than 100 Mya. The difficulty in aligning these regions could be largely responsible for the observed incongruence in the evolution rate between different regions of R2R3 MYB proteins. This model cannot be excluded by the results of this study, unlike the neutral model that was excluded by the values of ω <1 for R2R3 MYB protein C-terminal regions that we observed. The notion that much of the incongruence between the MYB domain and C-terminal regions reflects difficulty in alignment represents an excellent alternative to the polyphyly model of Rosinski and Atchley (1998), and it is consistent with the observation that many R2R3 MYB clades identified by analyses of the MYB domain share C-terminal motifs. It remains possible that domain shuffling have contributed to the divergence of specific Myb genes. In fact, some alleles of the P1 gene have a putative Cys-containing metal binding domain at their C terminus that could have arisen by domain shuffling (Chopra et al., 1996). However, there is no evidence that this pattern of evolution is more common than divergence at different rates coupled with substantial numbers of insertion-deletion mutations.
A surprising aspect of the current study is the fact that regions of R2R3 Myb genes that encode the divergent C-terminal regions often exhibit higher KS values than the regions encoding the MYB domains (see the comparisons of C1/Pl1 and AtMyb11/AtMyb12; Table I). The observation in this study is difficult to reconcile with neutral models, which postulate that different regions of the genome show different rates of mutation. Likewise, we believe that models that invoke selection for specific mRNA structures (e.g., Eyre-Walker and Bulmer, 1993) represent an unlikely explanation for the observations in this study. The GC-content of genes should have a strong impact upon RNA structure, so it is likely that selection on synonymous sites because of mRNA structure would be stronger and more consistent in the GC-rich genes of grasses. However, the higher apparent rate of synonymous substitution was observed for one pair of genes in maize and one pair of genes in Arabidopsis, despite the very different GC-contents of genes in these organisms.
There are two potential models consistent with all of the available data from paralogous R2R3 Myb genes. It is possible that there are differences in the rate of mutation in different regions of transcription units, a plausible hypothesis considering the involvement of some proteins in both transcription and in DNA repair (Lehmann, 1998). However, we believe a simpler model might be the impact of gene conversions in the conserved regions corresponding to the MYB domains of these regions. This model would explain the more limited divergence of these regions at both synonymous and non-synonymous sites without invoking different rates of mutation. This model would also explain the loss of introns in the conserved region of some R2R3 Myb genes, including OsMYB-IF and ZmMyb-B2 (Rabinowicz and Grotewold, 2000), because this loss could involve gene conversion with the products of reverse transcription after splicing.
Although a general characteristic of the R2R3 Myb gene family is the striking divergence of C-terminal regions, this feature is by no means unique to R2R3 Myb genes. For example, MADS box genes, another large family of genes encoding transcriptional regulators in the green plants (Theissen et al., 1996; Purugganan, 1997), also show extreme sequence divergence in the C termini. Unlike the R2R3 MYB proteins where the C-terminal region can be very large, the divergent C-terminal region of MADS-box proteins usually corresponds to less than one-third of the entire protein. Extending these types of studies to either to a genome-wide scale for Myb genes or other gene families should provide more power to test among distinct evolutionary models. However, detailed analyses of specific genes, such as those conducted here, may provide information that can be difficult to find when conducting large-scale comparative genomic studies.
CONCLUSIONS
The expansion of the R2R3 Myb genes early in the history of plants makes it one of the largest families of regulatory proteins known. Evolutionary studies of this gene family have been hindered by the ancient nature of gene duplications. Here, we took advantage of several recently duplicated R2R3 Myb genes to establish some basic patterns of R2R3 Myb gene evolution. We established a pathway that explains the origin of plant specific R2R3 Myb genes from widely distributed three-MYB repeat genes. We established that the divergent C-terminal regions of R2R3 MYB proteins are subject to purifying selection and found that these regions sometimes show higher numbers of synonymous substitutions than the MYB domain. In fact, sliding window analyses indicated the substantial heterogeneity in the accumulation synonymous and non-synonymous substitutions across the coding regions of all pairs of R2R3 MYB proteins investigated. Together, our studies provide the most in-depth analysis of the sequence divergence of recently duplicated R2R3 Myb genes and suggest novel models for the function and evolution of these genes.
MATERIALS AND METHODS
Screening of Bacterial Artificial Chromosome (BAC) Libraries
The expressed sequence tag (EST) clone corresponding to ZmMyb-IF35 was obtained by searching a proprietary EST database at Pioneer Hi-Bred International (Johnston, IA). After completely sequencing the EST, we established that the 1,577-bp full-length clone encoded a putative protein of 344 amino acids with an amino-terminal R2R3 MYB domain. A probe corresponding to the carboxyl-terminal region of ZmMyb-IF35 cDNA and excluding the amino-terminal MYB domain region was used to screen a maize (Zea mays) B73 inbred BAC library (Incyte Genomics Inc., Palo Alto, CA). Four BAC clones were recovered (91c08, 235g24, 165j16, and 145c17) and were restriction digested to isolate full-length genomic clones. EcoRI was used to digest 165j16 BAC clone and a 6-kb fragment was subcloned into pBluescript II SK− (Stratagene, La Jolla, CA), and the presence of the full-length MYB domain was verified by PCR. Sequencing was carried out using primers flanking the two introns of ZmMyb-IF35. A full-length genomic clone of ZmMyb-IF25 in pBluescript II SK− (Stratagene) was generated by digesting the 145c17 BAC clone with XbaI and inserting a 9-kb fragment into the vector.
Mapping of ZmMyb-IF35 and ZmMyb-IF25
The following primers were designed from the 5′ sequences of ZmMyb-IF25 and ZmMyb-IF35: IF25, forward, TTTGGTCTGGTGATCAAATCAATG; IF25, reverse, AGGTGCAACTGCAAGAAATGC; IF35, forward, GCAATCCCTTCTCGCCCTTT; and IF35, reverse, CTGCTTGGGAGAGGAGATCGAG. These primer pairs were used to amplify the corresponding regions from genomic DNA of 12 inbred lines: B73, Mo17, GT119, T218, sm1-stock, A619, Mp708, W23c2whp1, NC7A, Tx501, CO159, and Tx303. The PCR reaction conditions were: 1× PCR buffer, 0.4 mm dNTPs, 50 ng each of SSR primers (forward and reverse), 0.3 units of AmpliTaq Gold (Perkin-Elmer Applied Biosystems, Foster City, CA), and 50 ng of genomic DNA, in a total volume of 15 μL. Thermocycling conditions were: 95°C, 1′; 65°C, 1′; and 72°C, 30′ for one cycle and then a 1°C decrement for the annealing temperature, each repeated once, until the annealing temperature is 55°C followed by 95°C 1′, 55°C 1′, and 72°C 30′ for 30 cycles. The amplification products were resolved on 3.5% (w/v) agarose gels. Genotypes for mapping ZmMyb-IF35 were determined using a size polymorphism of the amplification products between B73 and Mo17; likewise, genotypes for mapping ZmMyb-IF25 were determined using a size polymorphism between T218 and GT119. Linkage maps for placing ZmMyb-IF25 and ZmMyb-IF35 were constructed using MAPMAKER/EXP (version 3.0, Whitehead Institute, Cambridge, MA). Starting with a framework map for each population, the “assign” and “build” commands were used to identify the chromosome and place the locus within the framework order.
Sequence Analysis
The analyses of the sequences obtained were conducted using the Oxford Molecular MacVector 6.0 (Accelrys Inc., San Diego) and MacDNAsis (version 2.0, Hitachi Ltd., San Bruno, CA). Sequences used to examine the evolution of MYB domains were aligned using ClustalW (Thompson et al., 1994) and trimmed to exclude sequences outside of the MYB domain. Phylogenetic analyses of these sequences were conducted by weighted neighbor joining (Bruno et al., 2000) of distance estimates obtained using the WAG model of sequence evolution (Whelan and Goldman, 2001). Previous analyses have suggested significant variance in rates across sites for MYB domains (Rabinowicz et al., 1999), and this variance was accommodated using a Γ distribution with a shape parameter (α) estimated from the data by maximum likelihood (α = 0.74 for the 46-taxon alignment and α = 0.74 for the 127-taxon alignment). Addition of this parameter resulted in a highly significant improvement to model fit using the likelihood ratio test (Goldman and Whelan, 2000).
We examined confidence in clades using the bootstrap (Felsenstein, 1985), applying “seqboot” from the PHYLIP package (http://evolution.genetics.washington.edu). Distance estimates were calculated using TREE-PUZZLE 5.0 (http://www.tree-puzzle.de) and trees were identified using weighbor 1.2 (http://www.t10.lanl.gov/billb/weighbor/index.html). Bayesian analyses were also conducted to examine confidence in clades, using a version of MrBayes 2.01 (Huelsenbeck and Ronquist, 2001; http://morphbank.ebc.uu.se/mrbayes/) that had been modified to allow use of the WAG+ Γ model of amino acid evolution (specific modifications available upon request from E.L. Braun). The Bayesian analyses used four Markov chains with default heating and were run for 106 generations. The Markov chains appeared to converge rapidly, and the first 2 × 105 generations were discarded as “burn-in.”
Reconciled trees, which show gene duplications given a specific species tree (Goodman et al., 1979), were displayed using GeneTree 1.3.0 (Page, 1998; available from http://taxonomy.zoology.gla.ac.uk/rod/genetree/genetree.html).
All of the R2R3 Myb genes from grasses examined by this study show the extreme codon bias characteristic of other nuclear encoded genes in grasses (Murray et al., 1989), with a third codon position GC content of >80%. In contrast, the Arabidopsis R2R3 Myb genes investigated show a much more limited codon bias, as is generally the case for eudicot genes (Murray et al., 1989). For this reason, KS and KA were calculated using the YN00 model of coding sequence evolution (Yang and Nielsen, 2000) because this model accommodates compositional bias and variable transition to transversion ratios. The YN00 program distributed with the PAML package (http://abacus.gene.ucl.ac.uk/software/paml.html) was used for these calculations. When estimates of KS and KA were calculated for short segments of the R2R3 Myb coding regions, the transition-transversion parameter and the base composition at each codon position were fixed at the values estimated from the complete sequences. This was accomplished by modifying the YN00 program to allow input of these values from a data file.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to J. Marcela Hernandez and Katherine Houchins for excellent technical assistance and Rob Fields for assistance in the assembly of data sets for computer analyses. Anusha P. Dias acknowledges Rasil Warnakulasooriya for helpful discussions. We thank Pioneer Hi-Bred International and Dr. Wes Bruce for the ZmMyb-IF35 cDNA. We also appreciate the comments and suggestions of the two anonymous reviewers.
Footnotes
This work was supported in part by the National Science Foundation (grant nos. MCB–9974474 and MCB–9896111 to E.G.), by the U.S. Department of Agriculture (postdoctoral fellowship no. USDA 1999–01582 to E.L.B.), and by the U.S. Department of Agriculture-Agricultural Research Service (research funds to M.D.M.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.012047.
LITERATURE CITED
- Alvarez-Valin F, Jabbari K, Bernardi G. Synonymous and nonsynonymous substitutions in mammalian genes: intragenic correlations. J Mol Evol. 1998;46:37–44. doi: 10.1007/pl00006281. [DOI] [PubMed] [Google Scholar]
- Barrier M, Robichaux RH, Purugganan MD. Accelerated regulatory gene evolution in an adaptive radiation. Proc Natl Acad Sci USA. 2001;98:10208–10213. doi: 10.1073/pnas.181257698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun EL, Grotewold E. Newly discovered plant c-myb-like genes rewrite the evolution of the plant mybgene family. Plant Physiol. 1999a;121:21–24. doi: 10.1104/pp.121.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun EL, Grotewold E. Diversification of the R2R3 Mybgene family and the segmental allotetraploid origin of the maize genome. Maize Genet Coop Newsl. 1999b;73:26–27. [Google Scholar]
- Braun EL, Matulnik T, Dias AP, Grotewold E. Transcription factors and metabolic engineering: novel applications for ancient tools. Rec Adv Phytochem. 2001;35:79–109. [Google Scholar]
- Bruno WJ, Socci ND, Halpern AL. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol Biol Evol. 2000;17:189–197. doi: 10.1093/oxfordjournals.molbev.a026231. [DOI] [PubMed] [Google Scholar]
- Chopra S, Athma P, Peterson T. Alleles of the maize Pgene with distinct tissue specificities encode Myb-homologous proteins with C-terminal replacements. Plant Cell. 1996;8:1149–1158. doi: 10.1105/tpc.8.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone KC, Cocciolone SM, Burr FA, Burr B. Maize anthocyanin regulatory gene pl is a duplicate of c1that functions in the plant. Plant Cell. 1993;5:1795–1805. doi: 10.1105/tpc.5.12.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolphin K, Belshaw R, Orme CDL, Quicke DLJ. Noise and incongruence: interpreting results of the incongruence length difference test. Mol Phylogenet Evol. 2000;17:401–406. doi: 10.1006/mpev.2000.0845. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Bulmer M. Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res. 1993;21:4599–4603. doi: 10.1093/nar/21.19.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence-limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Cone KC, Fromm ME. Identification of functional domains in the maize transcriptional activator C1: comparison of wild-type and dominant inhibitor proteins. Genes Dev. 1991;5:298–309. doi: 10.1101/gad.5.2.298. [DOI] [PubMed] [Google Scholar]
- Goldman N, Whelan S. Statistical tests of gamma-distributed rate heterogeneity in models of sequence evolution in phylogenetics. Mol Biol Evol. 2000;17:975–978. doi: 10.1093/oxfordjournals.molbev.a026378. [DOI] [PubMed] [Google Scholar]
- Goodman M, Czelusniak J, Moore GW, Romero-Herrera AE, Matsuda G. Fitting the gene lineage into its species lineage: a parsimony strategy illustrated by cladograms constructed from globin sequences. Syst Zool. 1979;28:132–168. [Google Scholar]
- Grotewold E, Athma P, Peterson T. Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of Myb-like transcription factors. Proc Natl Acad Sci USA. 1991;88:4587–4591. doi: 10.1073/pnas.88.11.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond B, Bowen B, Peterson T. The Myb-homologous Pgene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL. Identification of the residues in the Myb domain of C1 that provide the specificity of the interaction with the bHLH cofactor R. Proc Natl Acad Sci USA. 2000;97:13579–13584. doi: 10.1073/pnas.250379897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helentjartis T, Weber D, Wright S. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics. 1988;118:353–363. doi: 10.1093/genetics/118.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kranz H, Scholtz K, Weisshaar B. c-MYB oncogene-like genes encoding three MYB repeats occur in all major plant lineages. Plant J. 2000;21:231–235. doi: 10.1046/j.1365-313x.2000.00666.x. [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Lin H-L, Leyva A, Meissner R, Petroni K, Urzainiqui A, Bevan M, Martin C et al. Towards the functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Lee M, Sharopova N, Beavis WD, Grant D, Katt M, Blair, Hallauer A. Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol Biol. 2002;48:453–461. doi: 10.1023/a:1014893521186. [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development. 2001;128:1539–1546. doi: 10.1242/dev.128.9.1539. [DOI] [PubMed] [Google Scholar]
- Leech MJ, Kammerer W, Cove DJ, Martin C, Wang TL. Expression of myb-related genes in the moss, Physcomitrella patens. Plant J. 1993;3:51–61. doi: 10.1046/j.1365-313x.1993.t01-3-00999.x. [DOI] [PubMed] [Google Scholar]
- Lehmann AR. Dual functions of DNA repair genes: molecular, cellular, and clinical implications. Bioessays. 1998;20:146–155. doi: 10.1002/(SICI)1521-1878(199802)20:2<146::AID-BIES7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Lipsick JS. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- Loguercio LL, Zhang J-Q, Wilkins TA. Differential regulation of six novel MYB-domain genes defines two distinct expression patterns in allotetraploid cotton (Gossypium hirsutumL.) Mol Gen Genet. 1999;261:660–671. doi: 10.1007/s004380050009. [DOI] [PubMed] [Google Scholar]
- Murray EE, Lotzer J, Eberle M. Codon usage in plant genes. Nucleic Acids Res. 1989;17:477–498. doi: 10.1093/nar/17.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2gene encodes an R2R3 Myb domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Berlin: Springer-Verlag; 1970. [Google Scholar]
- Page RDM. GeneTree: comparing gene and species phylogenies using reconciled trees. Bioinformatics. 1998;14:819–820. doi: 10.1093/bioinformatics/14.9.819. [DOI] [PubMed] [Google Scholar]
- Page RDM, Holmes EC. Molecular Evolution: A Phylogenetic Approach. Oxford: Blackwell Science Ltd.; 1998. [Google Scholar]
- Paz-Ares J, Ghosal D, Weinland U, Peterson PA, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to mybproto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6:3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD. The MADS-box floral homeotic gene lineages predate the origin of seed plants: phylogenetic and molecular clock estimates. J Mol Evol. 1997;45:392–396. doi: 10.1007/pl00006244. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. Molecular analysis of the anthocyanin2gene of petunia and its role in the evolution of flower color. Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Braun EL, Wolfe AD, Bowen B, Grotewold E. Maize R2R3 Mybgenes: sequence analysis reveals amplification in higher plants. Genetics. 1999;153:427–444. doi: 10.1093/genetics/153.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz PD, Grotewold E. A novel reverse-genetic approach (SIMF) identifies Mutator insertions in new Mybgenes. Planta. 2000;211:887–893. doi: 10.1007/s004250000402. [DOI] [PubMed] [Google Scholar]
- Reichmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Curr Opin Plant Biol. 2000;3:423–434. doi: 10.1016/s1369-5266(00)00107-2. [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR. Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol. 1998;46:74–83. doi: 10.1007/pl00006285. [DOI] [PubMed] [Google Scholar]
- Sainz MB, Goff SA, Chandler VL. Extensive mutagenesis of a transcriptional activation domain identifies single hydrophobic and acidic amino acids important for activation in vivo. Mol Cell Biol. 1997;17:115–122. doi: 10.1128/mcb.17.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharopova N, McMullen MD, Schultz L, Schroeder S, Sanchez-Villeda H, Gardiner J, Bergstrom D, Houchins K, Melia-Hancock S, Musket T et al. Development and mapping of SSR markers for maize. Plant Mol Biol. 2002;48:463–481. doi: 10.1023/a:1014868625533. [DOI] [PubMed] [Google Scholar]
- Smith NG, Hurst LD. The effect of tandem substitutions on the correlation between synonymous and nonsynonymous rates in rodents. Genetics. 1999;153:1395–1402. doi: 10.1093/genetics/153.3.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Theissen G, Kim JT, Saedler H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol. 1996;43:484–516. doi: 10.1007/BF02337521. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;11:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Zhang P, Chopra S, Peterson T. A segmental gene duplication generated differentially expressed myb-homologous genes in maize. Plant Cell. 2000;12:2311–2322. doi: 10.1105/tpc.12.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.