Abstract
Expression of cytosolic and plastid acetyl-coenzyme A carboxylase (ACCase) gene families at the mRNA level was analyzed in developing wheat (Triticum aestivum) plants. The major plastid ACCase mRNA level is high in the middle part of the plant and low in roots and leaf blades. An alternative plastid ACCase transcript initiated at a different promoter and using an alternative 5′ splice site for the first intron accumulates to its highest level in roots. Cytosolic ACCase mRNA also consists of two species, one of which is present at approximately a constant level, whereas the other accumulates to a high level in the lower sheath section. It is likely that different promoters are also responsible for the two forms of cytosolic ACCase mRNA. The abundances of cytosolic and plastid ACCase mRNAs in the sheath section of the plant are similar. ACCase protein level is significantly lower in the leaf blades, in parallel with changes in the total ACCase mRNA level. Homoeologous ACCase genes show the same expression patterns and similar mRNA levels, suggesting that none of the genes was silenced or acquired new tissue specificity after polyploidization.
Acetyl-CoA carboxylase (ACCase) directs the flow of carbon from photosynthesis to primary and secondary metabolites in plants. One ACCase isozyme supplies malonyl-CoA for de novo fatty acid biosynthesis in plastids. A second ACCase isozyme supplies malonyl-CoA for fatty acid elongation and flavonoid biosynthesis in the cytosol. Their activity is expected to be fine tuned, responding to the demand for specific metabolites determined by plant development and by the environment. Our study focused on correlations between major developmental processes in the young wheat (Triticum aestivum) plant and expression of the ACCase gene families. The wheat leaf provides a succession of cells of different age and characteristics, from dividing cells of the leaf meristem located at the leaf base, to cells undergoing rapid enlargement accompanied by chloroplast maturation, to fully photosynthetically active cells in the leaf blade. These cells can be contrasted with photosynthetically inactive cells in the roots.
ACCase and its product, malonyl-CoA, play a key role in regulating metabolite flux through the fatty acid biosynthetic and degradative pathways, and are important regulators of carbon allocation and energy homeostasis. In mammals, ACCase is regulated by multiple mechanisms including tissue-specific promoters, reversible phosphorylation, and feedback by a number of metabolites (Kim, 1997; Munday and Hemingway, 2001). In Escherichia coli, transcription of ACCase genes is correlated with the rate of cellular growth, overexpression of all four subunits of ACCase increasing the rate of fatty acid biosynthesis (Davis et al., 2000). The expression of the ACCase gene in yeast (Saccharomyces cerevisiae) is coordinated with phospholipid metabolism (Haslacher et al., 1993). For plants, it has been suggested that plastid ACCase activity controls flux through the de novo fatty acid biosynthetic pathway (Ohlrogge and Jaworski, 1997; Rawsthorne, 2002). Inhibitors of plastid ACCase are potent herbicides (Gornicki and Haselkorn, 1993; Zagnitko et al., 2001).
Previous studies on ACCase in plants addressed such questions as tissue-specific expression (Hawke and Leech, 1987; Bao et al., 1997; Ke et al., 1997; Caffrey et al., 1998; Thelen et al., 2001; O'Hara et al., 2002), the role of phosphorylation (Savage and Ohlrogge, 1999), light and redox status of the enzyme (Sasaki et al., 1997; Kozaki et al., 2000), mRNA editing (Sasaki et al., 2001), and protein biotinylation (Alban et al., 2000). Possible sources of acetyl-CoA for plastid and cytosolic ACCase and related problems of carbon partitioning in plastid metabolism have been investigated (Bao et al., 2000; Eastmond and Rawsthorne, 2000; Ke et al., 2000; Ohlrogge et al., 2000; Rawsthorne, 2002). Most of these studies focused on the expression of the de novo fatty acid biosynthetic pathway in seeds accumulating oil.
Expression of the cytosolic ACCase involved in very long-chain fatty acid and flavonoid biosynthetic pathways is tissue specific. In leek (Allium porrum) leaves, it is more abundant in the epidermis than in the mesophyll layer (Caffrey et al., 1998). Expression of cytosolic ACCase increased significantly in alfalfa (Medicago sativa) cell cultures and in common bean (Phaseolus vulgaris) leaves and cell cultures treated with fungal elicitors (Shorrosh et al., 1994; Garcia-Ponce and Rocha-Sosa, 2000). It was suggested that this increase is triggered by the demand for malonyl-CoA needed to synthesize flavonoid phytoalexins as part of the defense response.
In wheat, as in other grasses, both plastid and cytosolic ACCase are nuclear-encoded multidomain enzymes of eukaryotic origin. The structure of wheat ACCases and their genes as well as the evolution of the gene family in grasses have been described recently (Gornicki et al., 1994, 1997; Podkowinski et al., 1996; Faris et al., 2001; Huang et al., 2002a, 2002b). The plastid ACCase gene (Acc-1) has a single copy and the cytosolic ACCase gene (Acc-2) has more than one copy in each of the homoeologous chromosomes of hexaploid wheat. Gene loss and silencing of some gene copies observed in natural (Faris et al., 2001) and newly synthesized (Kashkush et al., 2002) wheat polyploids may have a significant effect on specific expression of those genes.
All current evidence points to ACCase as catalyzing a key regulatory step in plant metabolism, with expression regulated at both transcriptional and posttranscriptional levels. In this paper, we analyze mRNA steady-state levels for plastid and cytosolic ACCase, as well as ACCase protein levels, in different sectors of young wheat plants.
RESULTS AND DISCUSSION
All Three Homoeologous Acc-1 Genes Are Transcriptionally Active in Leaves of Hexaploid Wheat, Each Utilizing Two Alternative Promoters and 5′ Splice Sites of the Leader Intron
Three Acc-1 genes, encoding the plastid ACCase, are found in hexaploid wheat, one on each of the group 2 homoeologous chromosomes (Gornicki et al., 1997). Transcriptional activity of individual Acc-1 genes was determined using a reverse transcription (RT)-PCR-cloning approach targeting mRNA leaders (Fig. 1). Further information about wheat Acc genes and their transcripts as well as sequences and target sites of primers used in PCR-based experiments are provided in the Supplemental Data Tables I and II (www.plantphysiol.org). First, to gain information about the 5′ ends of the transcripts, multiple 5′-RACE clones prepared as described previously (Gornicki et al., 1997; Podkowinski et al., 1996) were analyzed. In addition, a primer-walking RT-PCR cloning approach, using pairs of gene-specific primers, was employed. An approximate position of the transcription start site of the genes was deduced from the sequences of those cDNA clones that extended furthest upstream. These sequences are contained in the GenBank files identified in Supplemental Data Table I. Introns within the leaders were identified by comparing cDNA and genomic sequences.
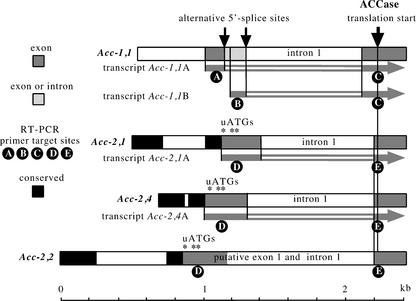
Figure 1.
Transcripts of Acc-1 and Acc-2 genes. Approximate transcription start sites were defined by 5′ ends of the longest cDNAs. The structure of the two transcript types (A and B) found for all three Acc-1 genes is illustrated using the structure of the Acc-1,1 gene. Putative promoter sequences conserved among Acc-2 genes and approximate position of upstream AUG codons (*, uATG) found in the leaders of Acc-2 genes (possible translation starts for small peptides) are shown. Primers targeted to sites A through E are shown in Table II (Supplemental Data). Transcription of gene Acc-2,2 in leaves was not confirmed by RT-PCR but the gene's promoter was shown to be transcriptionally active in transient expression experiments (E. Zuther and J. Jelenska, unpublished data).
We have confirmed that all three homoeologous Acc-1 genes are transcribed in wheat leaves. We have also confirmed the presence of two transcript types (A and B) for each of the three Acc-1 genes, characterized by different 5′ ends and utilization of different 5′ splice sites for the first intron as illustrated in Figure 1 for gene Acc-1,1. Such alternative splicing products were previously detected for one of the genes (Gornicki et al., 1997).
The two transcript types differ as follows: The leader of transcript Acc-1B starts further downstream, corresponding to an apparent start within the first intron of transcript Acc-1A (Fig. 1). Different 5′ splice sites are used for the removal of the leader intron from the two transcript types. These alternative 5′ splice sites are offset in the three different Acc-1 genes by 109 to 130 nucleotides. The two types of transcripts share the 3′ splice site of the first intron. This arrangement suggests that the two types are transcribed from different promoters. Leaders of both mRNA types are at least 160 to 180 nucleotides, are G + C rich (especially within their 5′ half), and have no AUG codons upstream of the ACCase translation start. They share an 87-nucleotide 3′-terminal sequence. The translation start codon context fits the consensus derived for other genes from monocots with A at position −3 and G at position +4 (Joshi et al., 1997).
Multiple Copies of the Acc-2 Gene Are Transcriptionally Active in Leaves of Hexaploid Wheat
For the cytosolic ACCase, we previously found five Acc-2 genes in hexaploid wheat (Faris et al., 2001). These genes were mapped to the group 3 homoeologous chromosomes and to chromosome 5D. Transcription products of these genes were studied using the same approach as described above for the Acc-1 genes. The presence of the previously identified transcripts of genes Acc-2,1 and Acc-2,4 (Podkowinski et al., 1996) in wheat leaves was confirmed in this study (Supplemental Data Table I). Our phylogenetic analysis suggested that Acc-2,1 and Acc-2,4 were orthologs (Faris et al., 2001). An additional transcript was identified (named Acc-2,6A, Supplemental Data Table I), but it could not be assigned to gene Acc-2,3 or Acc-2,6 present on chromosome 3B (Faris et al., 2001), because no overlapping sequences are available (Supplemental Data Table I).
Gene Acc-2,2 appears to be a paralog of the Acc-2 genes described above (Faris et al., 2001). A transcript corresponding to this gene has not yet been identified, but the promoter of this gene was active in wheat protoplasts using promoter fusions with a β-glucuronidase reporter (E. Zuther and J. Jelenska, unpublished data), suggesting that Acc-2,2 is expressed in tissues other than those found in leaves and roots. We also do not know the nature of the Acc-2 gene (or genes) localized on chromosome 5D, except that it (or they) hybridize under stringent conditions with cDNA probes for the other Acc-2 genes described above. They could be pseudogenes. These genes have not yet been cloned; therefore, their transcription status could not be determined by gene-specific RT-PCR.
The leaders of the three Acc-2 transcripts characterized have similar leader sequences and identical positions of the first intron located within the leader (Podkowinski et al., 1996). The leaders are approximately 300 nucleotides long and G + C rich. Each contains three AUG triplets and two or three open reading frames (ORFs) 26 to 99 amino acids long. In each of these Acc-2 gene leaders, two of the small ORFs terminate in the vicinity of the ACCase initiation codon. The translation start codon context for the ACCases fits the consensus derived for other genes from monocots, but the context of the AUG codons that could start the small peptides encoded in the leader shows little similarity to this consensus, suggesting that the short leader ORFs are not translated efficiently (Joshi et al., 1997). Similar structural features are also present at a corresponding location in the Acc-2,2 gene.
There is no sequence similarity between Acc-1 and Acc-2 genes upstream of the conserved biotin carboxylase domain that is located at the N terminus of mature ACCase. These sequence differences reflect acquisition of the plastid-targeting signal by the Acc-1 genes and divergence of function of the Acc-1 and Acc-2 promoters (Huang et al., 2002b). The 5′ end of the Acc-2 genes consists of two conserved blocks separated by a variable element (Fig. 1; Faris et al., 2001), which could be an indication of the first step toward divergence of function of Acc-2 orthologs or even homoeologs.
Northern-Blot Analysis and Quantitative RT-PCR Provide Consistent Expression Patterns of the ACCase Gene Family, Individual Homoeologs and Paralogs, and Alternative Promoters
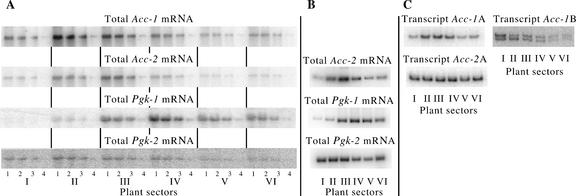
Application of gene-specific cDNA probes and stringent hybridization conditions prevented cross hybridization between mRNAs encoding plastid and cytosolic isozymes, but allowed simultaneous detection of all transcripts of each type. Northern-blot analysis revealed distinct ACCase mRNA expression patterns in six sectors of young wheat plants (Fig. 2A). RT-PCR (Fig. 2B) and real-time RT-PCR measurements reflected the relative level of specific individual mRNAs. The quantitative results of all three methods were consistent in all experiments where such comparisons were made (Fig. 2).
Figure 2.
Northern-blot and RT-PCR analysis of specific mRNAs. A, Representative autoradiograms from northern blots used for quantitation. Ten, 5, 2.5, and 0.5 μg of total RNA extracted from each plant sector was analyzed (lanes 1–4). B, Representative autoradiograms from RT-PCR experiments measuring total mRNA for each gene. C, Representative autoradiograms from RT-PCR experiments measuring specific types of mRNA for genes Acc-1 and Acc-2. In each case, only the size-specific product is shown. Autoradiograms presented in this figure represents different experiments; therefore, they do not reveal directly the relative levels of different gene transcripts.
The RT-PCR methods also allowed analysis of specific transcripts of the same gene and transcripts of homoeologs and paralogs. To distinguish the two transcript types found for the Acc-1 genes, the upstream primer was targeted to a site present in one transcript but not in the other (Fig. 1). Three different-sized products predicted for the minor transcript of genes Acc-1,1, Acc-1,2, and Acc-1,3 were resolved into two bands (Fig. 2C). The two Acc-1 transcript types (A and B) have a strikingly different expression pattern (Fig. 3C).
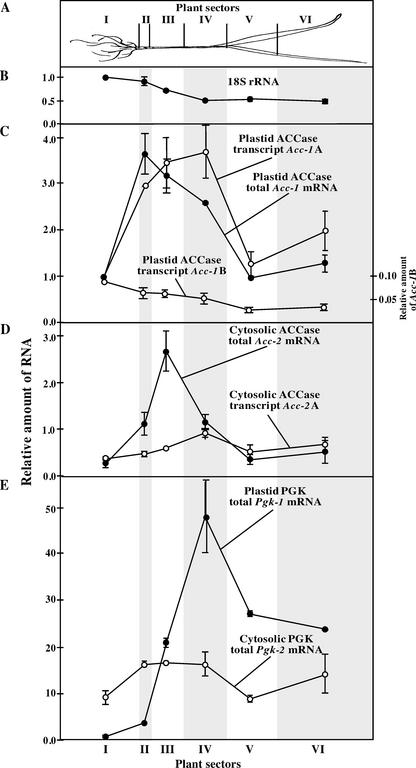
Figure 3.
Relative steady-state levels of specific mRNAs in six sectors of young wheat plants. A, Six sectors of wheat plant; B, Relative amount of 18S rRNA determined by real-time RT-PCR in equal amounts of total RNA from wheat sectors. The level of 18S rRNA in sector I was taken as 1. C, Relative level of total plastid ACCase mRNA (average of northern-blot and real-time RT-PCR measurements), and relative levels of transcripts Acc-1A and Acc-1B (measured by RT-PCR); note different scale for Acc-1B. D, Relative level of the total cytosolic ACCase mRNA (average of northern-blot, RT-PCR, and real-time RT-PCR measurements) and level of transcript Acc-2A (measured by RT-PCR); E, Relative level of the total plastid and cytosolic 3-phosphoglycerate kinase (PGK) mRNA (average of northern blot and RT-PCR measurements). All results were normalized for the amount of 18S rRNA present in each sector (shown in B). The level of total plastid ACCase mRNA in sector I was taken as 1 (C) for this and for all other mRNAs (C and D). This value corresponds to 300 to 600 molecules of mRNA per cell. Note different scale in E.
Targeting RT-PCR to different regions of the Acc-2 gene transcripts, the leader sequence (Fig. 1) and the coding sequence, also revealed significant differences in the expression pattern (Fig. 3D). Apparently, more than one type of Acc-2 transcript is made in wheat leaves. RT-PCR measurements based on the amplification of the mRNA coding region corresponded very well to the pattern detected by northern analysis (averaged in Fig. 3D). This similarity was expected because both the hybridization probe and RT-PCR primers targeted conserved sequences and are expected to detect all Acc-2 transcripts. RT-PCR targeting the known leader sequences gave a different expression pattern, suggesting that this assay detected only a subset of those transcripts. Finally, application of gene-specific primers in real-time RT-PCR allowed measurements of transcripts of individual homoeologs and paralogs (Fig. 4).
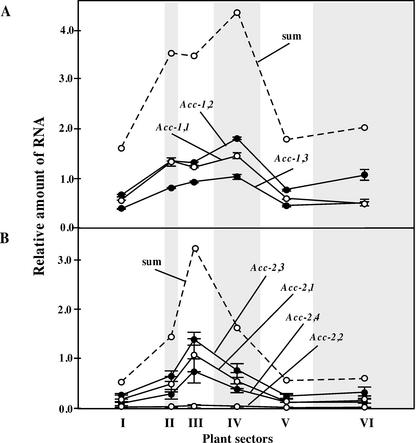
Figure 4.
Relative steady-state level of gene-specific plastid and cytosolic ACCase mRNAs in six sectors of young wheat plants. A, The total transcript level of Acc-1,1; Acc-1,2, and Acc-1,3 genes; B, the total transcript level of Acc-2,1, Acc-2,3, Acc-2,4, and Acc-2,2 genes. Results of multiple gene-specific real-time RT-PCR measurements were averaged. Plant sectors are shown in Figure 3A. These results were normalized for the amount of 18S rRNA present in each sector. The level of total plastid ACCase mRNA in sector I measured as shown in Figure 3C was taken as 1. The scale is the same as in Figure 3, C and D. The dashed line shows a sum of all gene-specific mRNA levels.
The steady-state levels of specific mRNAs measured in total RNA from different plant sectors were normalized relative to the abundance of 18S rRNA (with 18S rRNA in sector I taken as 1). In this way, the effect of the contribution of chloroplast rRNA to total RNA is eliminated. Multiple pair-wise comparisons of northern-blot, quantitative RT-PCR, and real-time RT-PCR measurements were used to estimate the relative abundance of specific transcripts of different genes. For the calculations of the relative mRNA levels shown in Figures 3 and 4, the amount of total plastid ACCase mRNA in sector I was taken as 1. The level of 18S rRNA in sector I was 4.5 ± 1.2 × 104 times higher than the level of Acc-1 transcripts in the same sector. From the published number of ribosomes per cell at the base of a 7-d-old wheat leaf, approximately 20 × 106 (Dean and Leech, 1982), we estimate that the value taken as 1 for the calculation of the relative levels of all transcripts in different plant sectors (Figs. 3 and 4) corresponds to 300 to 600 molecules of ACCase mRNA per cell, including mesophyll, epidermal, and vascular cells.
It is important to note that RT-PCR cloning experiments fail to identify transcripts that are significantly less abundant in a particular tissue (e.g. leaves; this study), transcripts of genes whose sequence differs at primer target sites, or transcripts that do not contain the upstream primer target sites at all. In the latter cases, PCR-related artifacts such as template switching (cDNA-cDNA or cDNA-genomic DNA) during PCR may pose a problem. Identification of all transcripts of all gene copies is essential to overcome these problems. 5′-RACE cloning experiments yielded important sequence information, although full-length transcripts of the mRNA 5′ ends are difficult to achieve for Acc genes, whose leader sequences are G/C rich.
Developmental Status of Different Parts of a Young Wheat Plant
A two-leaf wheat plant (Fig. 3A) includes roots, the sheath and the blade of the first and the second leaf, and the younger leaves developing inside the sheath. In terms of plant development, age, and status of leaf cells, the six sectors used in this study can be described as follows: Sector I is a pool of all roots; sector II includes leaf meristem sections of all leaves, dividing and young cells at the base of the leaves; sectors III and IV consist of the lower and the upper part of the sheath of the first leaf (including the ligule and auricle section in sector IV), respectively, as well as the corresponding fragments of the younger leaves growing inside the sheath; and sectors V and VI include, respectively, the lower and the upper part of the blades of the first two leaves.
The nature of the organs changes gradually from sink to source, from photosynthetically inactive roots and leaf meristems to less photosynthetically active leaf sheath and developing middle part of a young leaf, to fully photosynthetically active leaf blades. The number of mature chloroplasts in a cell, and the corresponding amount of chloroplast rRNA, increases during early development of the first wheat leaf (Dean and Leech, 1982), reflecting the photosynthetic capacity of the leaf sections. It was shown previously that Rubisco also accumulates in chloroplasts of developing leaves, reaching the highest level near the leaf tip (Dean and Leech, 1982). In our experiments, the relative level of cytosolic 18S rRNAs in total RNA measured by real-time RT-PCR (Fig. 3B) decreased from 1 in roots (sector I) to 0.5 in the leaf blade (sectors V and VI). This decrease in the relative contribution of cytosolic rRNAs to total RNA is due to the increased contribution of chloroplast rRNAs to the total RNA from the upper sectors of the wheat plant.
Specific ACCase mRNAs Reach Maximum Levels at Different Stages of Wheat Development
The northern-blot, RT-PCR, and real-time RT-PCR experiments produced several consistent results (Figs. 3 and 4). First, total plastid ACCase mRNA showed the highest levels at the base of the leaves and in the sheath section of the plant (sectors II–IV), and 3-fold lower levels in roots (sector I) and in the leaf blade (sectors V and VI). Second, the level of transcript Acc-1B is highest in roots (sector I) where it accounts for approximately 10% of the total plastid ACCase mRNA, and it is 3-fold lower in the leaf blade (sectors V and VI). Third, total cytosolic ACCase mRNA showed the highest level above the base of the leaves (sector III) and a 5- to 10-fold lower level in roots (sector I) and in the leaf blade (sectors V and VI). Fourth, cytosolic ACCase mRNA consists of two components, one of which is present at approximately a constant level in all parts of the plant and another that accumulates to a high level in the lower sheath section of the plant. Fifth, the maximum levels of total cytosolic and plastid ACCase mRNAs in the sheath section of the plant are very similar.
The expression pattern of plastid ACCase mRNAs, by revealing their different tissue specificity, supports our conclusion that the two transcripts (Acc-1A and Acc-1B) are transcribed from different promoters. We also postulate that different promoters and alternative splicing of the leader intron are responsible for the specific expression of the two types of cytosolic ACCase mRNA (Acc-2A and Acc-2B). The structure of the second transcript type (Acc-2B) is not known because the corresponding cDNA has not yet been cloned and sequenced. Deletion analysis of the promoter of the Acc-2,1 gene in transient expression experiments in wheat protoplasts revealed a high transcriptional activity even after a large 5′ end fragment of the gene, including the part coding for the known mRNA leader, was deleted (E. Zuther and J. Jelenska, unpublished data). This result suggested the existence of a second promoter located further downstream and within the region encoding the first intron of transcript Acc-2A. Clearly, two promoters with different specificity work in concert, one to support expression of cytosolic ACCase in all organs and the other to boost the expression of this enzymatic activity in tissues that have an increased demand for cytosolic malonyl-CoA.
Homoeolog-specific expression of Acc genes was assessed by real-time RT-PCR (Fig. 4). Individual Acc-1 genes showed the expected level (approximately one-third of the total) and the same expression pattern as the total plastid ACCase mRNA (Fig. 3C). Three of the Acc-2 genes, presumably homoeologs, also showed the expected expression level and pattern as anticipated based on the total cytosolic mRNA measurements (Fig. 3D). However, the Acc-2,2 gene, a paralog of the other three Acc-2 genes, was not expressed in young wheat plants at any significant level (Fig. 4). This observation explains why the corresponding cDNA could not be cloned from leaf RNA.
We were interested in determining whether expression of cytosolic or plastid ACCase was induced by the wheat fungal pathogen Erysiphe graminis f. sp. tritici. The level of ACCase specific mRNAs measured by northern blot did not increase in leaves of wheat plants infected with the fungus (data not shown). Treatment of wheat plants with salicylic acid or 2,6-dichloroisonicotinic acid did not cause any increase either.
Comparison of ACCase and PGK mRNA Levels
Expression of genes encoding PGKs was determined in a similar way to provide a reference for the ACCase mRNA measurements. In plants, PGK is found both in the cytosol and in plastids, consistent with the subcellular localization of glycolysis and the Calvin cycle in which PGK participates. These two pathways and their regulation in plants were reviewed recently (Plaxton, 1996; Dennis et al., 1997; Schnarrenberger and Martin, 1997; Givan, 1999). The origin of plant PGKs (Martin and Schnarrenberger, 1997) and the evolution of PGK genes in grasses (Huang et al., 2002b) also has been analyzed. The chloroplast and cytosolic PGK isogenes (Pgk-1 and Pgk-2, respectively) are both nuclear genes of bacterial (endosymbiont) origin. We have shown previously that the plastid Pgk-1 gene is present in single copy in each homoeologous chromosome set of hexaploid wheat: A (GenBank accession no. AF343475), B (GenBank accession no. AF343480), and D (GenBank accession no. AF343478; Huang et al., 2002a, 2002b). Only two copies of the cytosolic Pgk-2 gene have been identified so far in hexaploid wheat (GenBank accession nos. AF343450 and AF343449; Huang et al., 2002b). They are likely to be orthologs present on different homoeologous chromosomes, but the total copy number of the Pgk-2 gene in wheat is not yet known (Huang et al., 2002b).
Northern-blot and RT-PCR experiments targeting conserved coding regions of the Pgk genes were designed to simultaneously detect transcripts of all three orthologs of Pgk-1 and separately the two known copies of Pgk-2. The expression pattern of cytosolic and chloroplast PGK mRNA is distinct (Fig. 3E) and significantly different from the ACCase mRNA expression patterns. The level of cytosolic PGK mRNA remains constant in different plant sections. Plastid PGK mRNA peaks in the upper sheath section of the plant (sector IV) and then decreases 2-fold in the leaf blade; the level of this mRNA was 100-fold lower in roots (sector I). These relative levels agree with the anticipated high demand for plastid PGK during chloroplast maturation and a demand for cytosolic PGK for glycolysis operating throughout plant development at a constant level. The cytosolic and plastid PGK mRNAs accumulate to 5- and 15-fold higher levels, respectively, than the ACCase mRNAs.
Peak Levels of mRNA Support ACCase Protein Accumulation in Young Wheat Plants
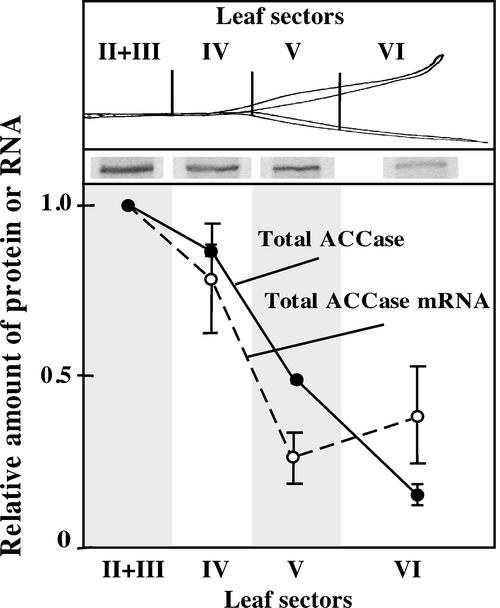
The mRNA measurements revealed high transcript levels for both Acc genes in the lower part of the leaf (Fig. 3, sectors II–IV), suggesting an increased demand for both cytosolic and plastid ACCase activity in these sectors. This observation was followed up by measuring the amount of plastid plus cytosolic ACCase, both 250-kD biotinylated peptides, in different leaf sectors. The total ACCase protein was 5 times more abundant at the bottom of the leaf (Fig. 5, sectors II and III) than in its upper part. These differences are consistent with the decreasing levels of total ACCase mRNA in the upper leaf sections (Fig. 5). These results could also be correlated with the distribution of ACCase activity in sections of 7-d-old wheat leaves (Hawke and Leech, 1987): high in the lower part of the leaf and low at the leaf tip, and with a higher level of incorporation of acetate into fatty acids in leaf slices and in isolated chloroplasts from the midsection of a young wheat plant (Bolton and Harwood, 1978). Chloroplast ACCase is the major isozyme present in protein extracts from entire wheat leaves (P. Gornicki, unpublished data). This observation is in agreement with the results shown in Figure 3, where plastid ACCase mRNA is significantly more abundant than cytosolic ACCase mRNA in all leaf sectors except sector III.
Figure 5.
Relative ACCase protein levels in different sectors of young wheat plants. The amount of ACCase was measured in equal amounts of total protein extracted from different wheat leaf sectors. The amount of ACCase in sector II + III was taken as 1. The inset shows the ACCase band (a 240-kD biotinylated protein) revealed with radiolabeled streptavidin from one of the two blots used for the quantitation shown. Relative amount of total ACCase mRNA (cytosolic and plastid) was estimated from results shown in Figure 3. Leaf sections used in this experiment were prepared independently but they corresponded to similar sections used for RNA preparation for northern-blot and RT-PCR analysis (Fig. 3).
In plant source organs, carbon, energy, and reducing power needed for fatty acid biosynthesis can be derived from light energy and photosynthesis in chloroplasts. In heterotrophic cells of the sink and storage organs, however, these resources are derived from imported carbohydrates. ACCase directs acetate derived from the cytosolic and/or plastid carbohydrate pools into fatty acid biosynthesis. The carbohydrate pools, in turn, are supplied by the Calvin cycle in chloroplasts and drained by cytosolic and plastid glycolysis to feed fatty acid biosynthesis and other biosynthetic pathways. The developmental changes in accumulation of specific RNAs described in this paper reflect the demand for metabolites required by these processes. These differences can be expected to be even more profound in specific cell types. Utilization of alternative promoters and splicing of the first intron play an important role in regulation of expression of the ACCase genes. It is important to point out that many enzymatic steps and processes occur between synthesis of malonyl-CoA and accumulation of the final products of the pathways: membrane and storage lipids, cuticular wax, and flavonoids.
The high steady-state level of the plastid ACCase mRNA correlates well with the expected high demand for de novo fatty acid biosynthesis in dividing cells and during chloroplast biogenesis, as well as demand for long chain fatty acids for very long-chain fatty acid biosynthesis. The high steady-state level of the cytosolic ACCase mRNA level reflects a high demand for fatty acid elongation, needed for the synthesis of cuticular wax, which is deposited on the surface of developing leaves to protect them against water loss and environmental stresses (Post-Beittenmiller, 1996). Deposition of wax on the leek leaf begins above the base meristem and cell elongation zone, coinciding with an increased level of fatty acid elongase activity (Rhee et al., 1998). These results are consistent with the ACCase expression patterns in wheat reported in this paper. Leaf development in leek, a monocot plant, and in grasses is similar. Malonyl-CoA from the pool produced by the cytosolic ACCase is also used for biosynthesis of flavonoids including pigments, signaling, and defense compounds (Shirley, 1996). It is possible that the high level of the cytosolic ACCase expression in the midsection of the wheat leaf is needed to support flavonoid accumulation, which would add another layer of protection against environmental stresses (e.g. exposure to UV light) to the emerging plant. The high demand for the chloroplast PGK activity in photosynthetically active organs is consistent with the high levels of this specific mRNA at the base of the leaf blade, where biosynthesis of this enzyme is expected to be high. Maturing chloroplasts require the full complement of enzymes involved in CO2 assimilation, including the chloroplast PGK, which participates in the Calvin cycle. On the contrary, the demand for the cytosolic PGK activity, reflecting a demand for cytosolic glycolytic activity, is uniform in different plant sectors.
MATERIALS AND METHODS
RNA from Wheat (Triticum aestivum) Sectors
Wheat var. Hard Red Winter Tam107 plants, at a two-leaf stage and approximately the same size, were divided into six sectors as shown in Figure 3A. RNA from each sector was prepared as previously described (Podkowinski et al., 1996). RNA concentration was determined spectrophotometrically and the RNA preparations were evaluated on ethidium bromide-stained gels run in parallel and under the same conditions as for northern blots. Quantitation used ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The amount of RNA on stained gels varied between 89% and 117% of an average for the six sectors. The contribution of full-length plastid and cytosolic rRNAs to total RNA was estimated from areas under peaks assigned to rRNAs based on their size. Two independent RNA quantitations were averaged. The contribution of the chloroplast rRNAs to total RNA increased from unmeasurable (no rRNA peaks observed above the background) in roots (sector I) to approximately 10% in the leaf blade (sectors V and VI). This estimate was based on the observed increase in the contribution of all of the chloroplast rRNA-size RNAs from 10% to 20%. The corresponding contribution of the large cytosolic rRNAs to total RNA decreased from 50% in roots (sector I) to 40% in the leaf blade (sectors V and VI).
cDNA Probes and Northern-Blot Analysis
The Acc-1-specific probe was a 3.4-kb BamHI cDNA fragment (Nikolskaya et al., 1999), and the Acc-2-specific probe was a 3.6-kb SacI-BamHI cDNA (Joachimiak et al., 1997). The Pgk-1-specific probe was a 0.95-kb fragment of cDNA (GenBank accession no. X15233) cloned by RT-PCR (using primers ASp23 and ASp21, Supplemental Data Table II), and the Pgk-2-specific probe was a 0.46-kb fragment of cDNA (GenBank accession no. X15232) cloned by RT-PCR (using primers JP14 and JP15, Supplemental Data Table II). Hybridization probes were labeled with 32P using a Random Primed DNA Labeling Kit (Boehringer Mannheim/Roche, Basel). Four different amounts of RNA from each plant sector (0.5, 2.5, 5, and 10 μg) were separated by electrophoresis on formaldehyde-agarose gels, transferred to GeneScreen Plus membranes (DuPont, Wilmington, DE), and hybridized according to the manufacturer's protocol. All northern blots were processed under the same conditions. The hybridization signal was quantitated using a STORM 860 PhosphorImager (Molecular Dynamics) and ImageQuant software (Molecular Dynamics). Each series of bands was quantitated twice using two different methods of background subtraction (“local average” and “manual”). Relative band intensities were calculated independently for each background subtraction method, taking the maximum value for each series of RNAs from six plant sectors as one and averaging the results of all series of measurements. Lanes containing 2.5 and 5 μg of total RNA gave consistently proportional hybridization signals and were used for quantitation. The signal in the 10-μg lane was in many cases beyond the linear range, and the signal in the 0.5-μg lane was in some cases too weak for reliable quantitation.
RT-PCR and Real-Time RT-PCR
The C. therm. Polymerase One-Step RT-PCR System (Boehringer Mannheim/Roche) was used following the manufacturer's protocol. The RT was 30 min at 60°C using gene-specific downstream primers. First strand cDNA prepared from 10 ng of total RNA, 0.3 or 0.6 μm primers, and 1 to 3 μCi of [α-32P]dCTP were used in a 20-μL PCR reaction. Amplification was for 20 to 26 cycles: denaturation at 94°C for 30 to 45 s, annealing at 60°C for 30 to 40 s, and elongation at 72°C for 1 to 2 min. Three to 5 μL of each PCR product was separated on a 10% (w/v) Tris-borate/EDTA-polyacrylamide gel (Bio-Rad Laboratories, Hercules, CA). After drying the gels, radioactive PCR products were detected and quantitated by phosphor imaging as described above. Primer combinations and amplification conditions that yielded only the specific product were selected for this analysis. The relative strength of the signal for transcripts of each gene in different RNA preparations remained similar for different combinations of specific primers and was independent of the number of amplification cycles within the 22- to 26-cycles range. In separate experiments, RT-PCR products obtained with multiple combinations of primers targeting leader sequences were cloned into the pGEM-T Easy vector (Promega, Madison, WI) and sequenced. New cDNA sequences were deposited in GenBank with the accession numbers AF438767 through AF438775.
Specific primers were designed for each gene family and for the alternative splicing products of the Acc-1 gene (Supplemental Data Table II) based on genomic and cDNA sequences identified by their GenBank accession numbers in Supplemental Data Table I. The primer target sites flanked introns to eliminate any PCR-amplified fragments of genomic DNA, a possible minor component in the RNA preparations, which were expected to be significantly longer. The gene-specific downstream primers were targeted to conserved stretches of coding sequences. The upstream primers were targeted to the leader sequences identified by 5′-RACE and primer-walking RT-PCR experiments. In the primer-walking experiments, a series of primers targeting genomic sequences around an anticipated transcription start site were used (not shown). In quantitative RT-PCR experiments targeting coding regions, both the downstream and the upstream primers were targeted to conserved sequences. The Pgk-1 and Pgk-2 primers targeting conserved exon sites (Huang et al., 2002b) were designed based on available gene sequences (GenBank accession nos. AF343475, AF343478, AF343480, AF343449, and AF343450).
Real-time RT-PCR experiments using the ABI Prism 7700 Sequence Detection System (PE-Applied Biosystems, Foster City, CA) were carried out according to the manufacturer's protocols. First-strand cDNA was prepared using random hexamers and MultiScribe Reverse Transcriptase (PE-Applied Biosystems). The same cDNA preparation was used as a template in real-time PCR experiments designed to detect transcripts of different genes. The PCR amplification was initiated by incubation at 95°C for 10 min followed by 40 cycles: 15 s at 95°C and 1 min at 60°C. cDNA prepared from 20 ng of total RNA from different plant sectors was used as template, except for 18S rRNA amplification where the cDNA was used at a 1:100 (v/v) dilution. A standard curve was determined for each experiment using Acc-1-specific primers (AccIF and AccIR) and three or four 2-fold serial dilutions of cDNA prepared from sector I RNA. Each measurement was repeated in at least three independent PCR experiments with the same or different pairs of primers, each with duplicates or three 2-fold serial dilutions of cDNA, except for gene-specific amplification of the Acc-1 transcripts, for which an average of two measurements is shown. The results were analyzed using Sequence Detector 1.7a software (PE-Applied Biosystems).
Real-time PCR probes (Taq-Man Probes, PE-Applied Biosystems) and PCR primers were designed according to the Sequence Detection System manufacturer's recommendations (Supplemental Data Table II). The probes were designed to reveal all known copies of each gene, but to eliminate any cross hybridization between Acc-1 and Acc-2 genes by selecting targets with four or five mismatches. The universal upstream primers (AccIF and Acc2F) targeted two adjacent exons to avoid amplification of any genomic DNA contaminating the RNA preparations. To add specificity, the Acc-1 universal primers were designed to work only with all known Acc-1 genes but not with the Acc-2 genes, and vice versa. Gene-specific primers were designed with one or two mismatches near the 3′ ends of the primers. All the primers were spaced such that the amplified products were similar in size and within the recommended range (50–160 nucleotides), except for one product, with primers AccIF and 1,3B/R, which was 259 nucleotides long.
Western Analysis of Biotinylated Proteins
The steady-state level of biotinylated proteins was measured by quantitation of western blots probed with [35S]-streptavidin as described previously (Gornicki and Haselkorn, 1993). Total protein was measured by the Bradford method.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jörn Gorlach for providing northern blots for wheat plants treated with salicylic acid, 2,6-dichloroisonicotinic acid, and Erysiphe graminis f. sp. tritici, and Sean Callahan for suggestions on the manuscript.
Footnotes
This work was supported by the Consortium for Plant Biotechnology Research (grant), by the Monsanto Co. (grant), and by the German Academic Exchange Service (fellowship to E.Z.).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.013169.
LITERATURE CITED
- Alban C, Job D, Douce R. Biotin metabolism in plants. Annu Rev Plant Physiol Mol Biol. 2000;51:17–47. doi: 10.1146/annurev.arplant.51.1.17. [DOI] [PubMed] [Google Scholar]
- Bao X, Shorrosh BS, Ohlrogge JB. Isolation and characterization of an Arabidopsis biotin carboxylase gene and its promoter. Plant Mol Biol. 1997;35:539–550. doi: 10.1023/a:1005881006620. [DOI] [PubMed] [Google Scholar]
- Bao XM, Focke M, Pollard M, Ohlrogge J. Understanding in vivo carbon precursor supply for fatty acid synthesis in leaf tissue. Plant J. 2000;22:39–50. doi: 10.1046/j.1365-313x.2000.00712.x. [DOI] [PubMed] [Google Scholar]
- Bolton P, Harwood JL. Fatty acid synthesis by slices from developing leaves. Planta. 1978;138:223–228. doi: 10.1007/BF00386815. [DOI] [PubMed] [Google Scholar]
- Caffrey JJ, Choi JK, Wurtele ES, Nikolau BJ. Tissue distribution of acetyl-CoA carboxylase in leaves of leek (Allium porrum L.) J Plant Physiol. 1998;153:265–269. [Google Scholar]
- Davis MS, Solbiati J, Cronan JE. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem. 2000;275:28593–28598. doi: 10.1074/jbc.M004756200. [DOI] [PubMed] [Google Scholar]
- Dean C, Leech RA. Genome expression during normal leaf development. Plant Physiol. 1982;69:904–910. doi: 10.1104/pp.69.4.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Huang Y, Negm FB. Glycolysis, the pentose pathway and anaerobic respiration. In: Dennis DT, Turkin DH, Lefebvre DD, Layzell DB, editors. Plant Metabolism. Harlow, Essex, UK: Addison-Wesley Longman; 1997. pp. 105–123. [Google Scholar]
- Eastmond PJ, Rawsthorne S. Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryo. Plant Physiol. 2000;122:767–774. doi: 10.1104/pp.122.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris J, Sirikhachornkit A, Haselkorn R, Gill B, Gornicki P. Chromosome mapping and phylogenetic analysis of the cytosolic acetyl-CoA carboxylase loci in wheat. Mol Biol Evol. 2001;18:1720–1733. doi: 10.1093/oxfordjournals.molbev.a003960. [DOI] [PubMed] [Google Scholar]
- Garcia-Ponce B, Rocha-Sosa M. The octadecanoic pathway is required for pathogen-induced multi-functional acetyl-CoA carboxylase accumulation in common bean (Phaseolus vulgaris L.) Plant Sci. 2000;157:181–190. doi: 10.1016/s0168-9452(00)00285-5. [DOI] [PubMed] [Google Scholar]
- Givan CV. Evolving concepts in plant glycolysis: two centuries of progress. Biol Rev. 1999;74:277–309. [Google Scholar]
- Gornicki P, Faris J, King I, Podkowinski J, Gill B, Haselkorn R. Plastid-localized acetyl-CoA carboxylase of bread wheat is encoded by a single gene on each of the three ancestral chromosome sets. Proc Natl Acad Sci USA. 1997;94:14179–14185. doi: 10.1073/pnas.94.25.14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornicki P, Haselkorn R. Wheat acetyl-CoA carboxylase. Plant Mol Biol. 1993;22:547–552. doi: 10.1007/BF00015984. [DOI] [PubMed] [Google Scholar]
- Gornicki P, Podkowinski J, Scappino LA, DiMaio J, Ward E, Haselkorn R. Wheat acetyl-CoA carboxylase: cDNA and protein structure. Proc Natl Acad Sci USA. 1994;91:6860–6864. doi: 10.1073/pnas.91.15.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslacher M, Ivessa A, Paltauf F, Kohlwein S. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- Hawke JC, Leech RA. Acetyl-CoA-carboxylase activity in normally developing wheat leaves. Planta. 1987;171:489–495. doi: 10.1007/BF00392296. [DOI] [PubMed] [Google Scholar]
- Huang S, Sirikhachornkit A, Su XJ, Faris J, Gill B, Haselkorn R, Gornicki P. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA. 2002a;99:8133–8138. doi: 10.1073/pnas.072223799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SX, Sirikhachornkit A, Faris JD, Su XJ, Gill BS, Haselkorn R, Gornicki P. Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol Biol. 2002b;48:805–820. doi: 10.1023/a:1014868320552. [DOI] [PubMed] [Google Scholar]
- Joachimiak M, Tevzadze G, Podkowinski J, Haselkorn R, Gornicki P. Wheat cytosolic acetyl-CoA carboxylase complements an ACC1 null mutation in yeast. Proc Natl Acad Sci USA. 1997;94:9990–9995. doi: 10.1073/pnas.94.18.9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL. Context sequences of translation initiation codon in plants. Plant Mol Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Behal RH, Back SL, Nikolau BJ, Wurtele ES, Oliver DJ. The role of pyruvate dehydrogenase and acetyl-coenzyme A synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiol. 2000;123:497–508. doi: 10.1104/pp.123.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Choi J-K, Smith M, Horner HT, Nikolau BJ, Wurtele ES. Structure of the CAC1 gene and in situ characterization of its expression. Plant Physiol. 1997;113:357–365. doi: 10.1104/pp.113.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-H. Regulation of mammalian acetyl-coenzyme A carboxylase. Annu Rev Nutr. 1997;17:77–99. doi: 10.1146/annurev.nutr.17.1.77. [DOI] [PubMed] [Google Scholar]
- Kozaki A, Kamado K, Nagano Y, Iguchi H, Sasaki Y. Recombinant carboxyltransferase responsive to redox of pea plastidic acetyl-CoA carboxylase. J Biol Chem. 2000;275:10702–10708. doi: 10.1074/jbc.275.14.10702. [DOI] [PubMed] [Google Scholar]
- Martin W, Schnarrenberger C. The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: a case study of functional redundancy in ancient pathways through endosymbiosis. Curr Genet. 1997;32:1–18. doi: 10.1007/s002940050241. [DOI] [PubMed] [Google Scholar]
- Munday MR, Hemingway CJ. The regulation of acetyl-CoA carboxylase: a potential target for the action of hypolipidemic agents. Adv Enzyme Regul. 2001;39:205–234. doi: 10.1016/s0065-2571(98)00016-8. [DOI] [PubMed] [Google Scholar]
- Nikolskaya T, Zagnitko O, Tevzadze G, Haselkorn R, Gornicki P. Herbicide sensitivity determinant of wheat plastid acetyl-CoA carboxylase is located in a 400-amino acid fragment of the carboxyltransferase domain. Proc Natl Acad Sci USA. 1999;96:14647–14651. doi: 10.1073/pnas.96.25.14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara P, Slabas AR, Fawcett T. Fatty acid and lipid biosynthetic genes are expressed at constant molar ratios but different absolute levels during embryogenesis. Plant Physiol. 2002;129:310–320. doi: 10.1104/pp.010956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Pollard M, Bao X, Focke M, Girke T, Ruuska S, Benning C. Fatty acids synthesis: from CO2 to functional genomics. Biochem Soc T. 2000;28:567–574. [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG. Regulation of fatty acid synthesis. Annu Rev Plant Physiol Mol Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- Plaxton WC. The organization and regulation of plant glycolysis. Annu Rev Plant Physiol. 1996;47:185–214. doi: 10.1146/annurev.arplant.47.1.185. [DOI] [PubMed] [Google Scholar]
- Podkowinski J, Sroga GE, Haselkorn R, Gornicki P. Structure of a gene encoding a cytosolic acetyl-CoA carboxylase of hexaploid wheat. Proc Natl Acad Sci USA. 1996;93:1870–1874. doi: 10.1073/pnas.93.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller D. Biochemistry and molecular biology of wax production in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:405–430. doi: 10.1146/annurev.arplant.47.1.405. [DOI] [PubMed] [Google Scholar]
- Rawsthorne S. Carbon flux and fatty acid synthesis in plants. Prog Lipid Res. 2002;41:182–196. doi: 10.1016/s0163-7827(01)00023-6. [DOI] [PubMed] [Google Scholar]
- Rhee Y, Hlousek-Radojcic A, Ponsamuel J, Liu D, Post-Beittenmiller D. Epicuticular wax accumulation and fatty acid elongation activities are induced during leaf development of leeks. Plant Physiol. 1998;116:901–911. doi: 10.1104/pp.116.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Hatano M. Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1997;94:11096–11101. doi: 10.1073/pnas.94.20.11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Ohmori A, Iguchi H, Nagano Y. Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J Biol Chem. 2001;276:3937–3940. doi: 10.1074/jbc.M008166200. [DOI] [PubMed] [Google Scholar]
- Savage LJ, Ohlrogge JB. Phosphorylation of pea chloroplast acetyl-CoA carboxylase. Plant J. 1999;18:521–527. doi: 10.1046/j.1365-313x.1999.00478.x. [DOI] [PubMed] [Google Scholar]
- Schnarrenberger C, Martin W. The Calvin cycle, a historical perspective. Photosynthetica. 1997;33:331–345. [Google Scholar]
- Shirley BW. Flavonoid biosynthesis: “new” functions for an “old” pathway. Trends Plant Sci. 1996;1:377–382. [Google Scholar]
- Shorrosh BS, Dixon RA, Ohlrogge JB. Molecular cloning, characterization, and elicitation of acetyl-CoA carboxylase from alfalfa. Proc Natl Acad Sci USA. 1994;91:4323–4327. doi: 10.1073/pnas.91.10.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Mekhedov S, Ohlrogge JB. Brassicaceae express multiple isoforms of biotin carboxyl carrier protein in a tissue-specific manner. Plant Physiol. 2001;125:2016–2028. doi: 10.1104/pp.125.4.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagnitko O, Jelenska J, Tevzadze G, Haselkorn R, Gornicki P. An isoleucine/leucine residue in the carboxyltransferase domain of acetyl-CoA carboxylase is critical for interaction with aryloxyphenoxypropionate and cyclohexanedione inhibitors. Proc Natl Acad Sci USA. 2001;98:6617–6622. doi: 10.1073/pnas.121172798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.