Abstract
Late embryogenesis abundant (LEA) proteins are members of a large group of hydrophilic, glycine-rich proteins found in plants, algae, fungi, and bacteria known collectively as hydrophilins that are preferentially expressed in response to dehydration or hyperosmotic stress. Group 2 LEA (dehydrins or responsive to abscisic acid) proteins are postulated to stabilize macromolecules against damage by freezing, dehydration, ionic, or osmotic stress. However, the structural and physicochemical properties of group 2 LEA proteins that account for such functions remain unknown. We have analyzed the structural properties of a recombinant form of a soybean (Glycine max) group 2 LEA (rGmDHN1). Differential scanning calorimetry of purified rGmDHN1 demonstrated that the protein does not display a cooperative unfolding transition upon heating. Ultraviolet absorption and circular dichroism spectroscopy revealed that the protein is in a largely hydrated and unstructured conformation in solution. However, ultraviolet absorption and circular dichroism measurements collected at different temperatures showed that the protein exists in equilibrium between two extended conformational states: unordered and left-handed extended helical or poly (l-proline)-type II structures. It is estimated that 27% of the residues of rGmDHN1 adopt or poly (l-proline)-type II-like helical conformation at 12°C. The content of extended helix gradually decreases to 15% as the temperature is increased to 80°C. Studies of the conformation of the protein in solution in the presence of liposomes, trifluoroethanol, and sodium dodecyl sulfate indicated that rGmDHN1 has a very low intrinsic ability to adopt α-helical structure and to interact with phospholipid bilayers through amphipathic α-helices. The ability of the protein to remain in a highly extended conformation at low temperatures could constitute the basis of the functional role of GmDHN1 in the prevention of freezing, desiccation, ionic, or osmotic stress-related damage to macromolecular structures.
Group 2 LEA proteins or dehydrins or responsive to abscisic acid (RAB) proteins were originally identified as the “D-11” family of LEA proteins in developing cotton (Gossypium hirsutum) embryos (Baker et al., 1988; Dure et al., 1989; Hughes and Galau, 1989). Dehydrins appear to be ubiquitously expressed in gymnosperms (Jarvis et al., 1996; Richard et al., 2000) and angiosperms (Campbell and Close, 1997; Close, 1997). Immunological surveys have also detected dehydrin-related proteins in algae, yeast, and cyanobacteria (Close and Lammers, 1993; Campbell and Close, 1997; Close, 1997; Li et al., 1998; Mtwisha et al., 1998). Group 2 LEA proteins form a subset of evolutionarily conserved Gly-rich, hydrophilic proteins associated with adaptation to hyperosmotic conditions (Garay-Arroyo et al., 2000). Dehydrins are induced typically in maturing seeds or vegetative tissues following salinity, dehydration, cold, or freezing stress or abscisic acid (ABA) treatment (Close, 1996, 1997; Campbell and Close, 1997). Numerous studies have reported a positive correlation between the accumulation of group 2 LEA transcripts or proteins and cold acclimation, chilling or freezing, drought, and salinity tolerance (Houde et al., 1995; Sarhan et al., 1997; Whitsitt et al., 1997; Cellier et al., 1998; Danyluk et al., 1998; Ismail et al., 1997, 1999b; Borovskii et al., 2000, 2002; Tabaei-Aghdaei et al., 2000; Zhu et al., 2000). In addition, dehydrins show association with several quantitative trait loci intervals associated with adaptive traits such as winter hardiness and retention of yield under drought stress (for review, see Campbell and Close, 1997; Choi et al., 1999).
Selected dehydrins are induced preferentially by specific stresses, whereas others are constitutively expressed (Bravo et al., 1999; Nylander et al., 2001). Some dehydrins exhibit distinct tissue- and cell-type-specific expression patterns in unstressed plants. After stress, the expression patterns of some dehydrins become more generalized (Egerton-Warburton et al., 1997; Nylander et al., 2001). For many group 2 LEA proteins, the highest accumulation is observed within vascular tissues (Godoy et al., 1994; Houde et al., 1995; Danyluk et al., 1998; Bravo et al., 1999; Nylander et al., 2001). Others exhibit cellular localization patterns to specific cell types (e.g. pollen sacs, guard cells, and root meristems), suggesting that these proteins may act as regulators of osmotic potential by functioning as water attractants in cells that experience conditions of high osmolarity (Nylander et al., 2001). Dehydrins have been found to localize to the nucleus, cytoplasm, plasma membrane, mitochondria, or vacuole (Sarhan et al., 1997; Danyluk et al., 1998; Borovskii et al., 2000, 2002; Heyen et al., 2002). Dehydrins have been hypothesized to confer dehydration and/or freezing tolerance by stabilizing proteins and membranes through detergent or reverse chaperone activities (Close, 1996, 1997; Ismail et al., 1999a). A dehydrin-like protein from castor bean (Ricinus communis) was recently found to function as an iron transport protein that is thought to facilitate phloem-mediated long-distance transport of micronutrients (Krüger et al., 2002). A vacuole-associated, calcium-binding, dehydrin-related protein from celery (Apium graveolens) binds 100-fold more calcium when phosphorylated and may function as a calcium buffer or as a calcium-dependent chaperone (Heyen et al., 2002). Metal binding by dehydrins is consistent with the ability to purify such proteins by metal ion affinity chromatography (Svensson et al., 2000).
Group 2 LEA proteins are generally highly hydrophilic, lack Trp and most often Cys residues, and contain a high proportion of charged and polar amino acids and a low proportion of nonpolar, hydrophobic residues. This polar/charged amino acid compositional bias explains the high temperature solubility of this and other dehydrins. Dehydrins are distinguished from other LEA proteins by a highly conserved Lys-rich 15-amino acid sequence motif (consensus = EKKGIMDKIKEKLPG) referred to as the K segment (Close, 1996, 1997; Campbell and Close, 1997; Cuming, 1999). K segments are usually located in the C terminus and maybe repeated one to 11 times. In addition, the majority of group 2 LEA proteins contain another conserved sequence (consensus = V/TDE/QYGNP) or Y segment located in the N terminus. Many dehydrins also contain Ser tract repeats (S segment) that can undergo phosphorylation and are thought to participate in nuclear localization (Goday et al., 1994; Godoy et al., 1994). K segments are predicted to form class A amphipathic α-helices (Dure, 1993; Close, 1996). The presence of amphipathic helices suggests that group 2 LEA proteins might act as an interface between with hydrophobic surfaces of membrane phospholipids and the cytosol in plant cells. Furthermore, dehydrins might also interact with exposed hydrophobic surfaces of partially denatured proteins to prevent protein-protein aggregation under conditions of low protoplasmic water activity arising from dehydration or freezing stress conditions (Close, 1996, 1997; Campbell and Close, 1997). The majority of dehydrin polypeptides are composed of domains rich in Gly and polar amino acids (Φ-segments) that are interspersed typically between K segments. These highly polar, hydrophilic Φ-segments have been proposed to interact with hydrophobic surfaces of cytoplasmic or nuclear macromolecules to prevent their coagulation (Campbell and Close, 1997).
Despite the many predicted roles of group 2 LEA proteins, few in vitro functional analyses have been reported. Purified maize (Zea mays) G50 dehydrin reportedly has potent cryoprotective activity, especially in the presence of compatible solutes (Close, 1996). In addition, group 2 LEA proteins have shown positive enzyme (LDH) cryoprotective effects in vitro (Kazuoka and Oeda, 1994; Houde et al., 1995). An Arabidopsis LEA-like protein (COR15a) was also shown to reduce freeze damage to the inner membranes of the chloroplast envelope (Steponkus et al., 1998). Furthermore, the overexpression of a tomato (Lycopersicon esculentum) group 2 LEA protein (le4) in yeast can partially ameliorate the detrimental effects of ionic and freezing stress (Zhang et al., 2000).
Biochemical or physiochemical evidence for the proposed protective mechanisms of group 2 LEA proteins remains limited. Circular dichroism (CD) analyses of a purified G50 dehydrin from maize kernels showed approximately 75% random and 15% α-helical conformation consistent with structural prediction programs (Ceccardi et al., 1994). NMR spectral analysis studies of purified, recombinant dehydrin-related LEA (DSP16) from Craterostigma plantagineum suggests that DSP16 forms a highly mobile unordered conformation in equilibrium with preferentially extended substructures having different conformational states (Lisse et al., 1996). This largely unfolded structure lacks a well-defined three-dimensional structure with folded hydrophobic regions. Far-UV CD spectra indicated the presence of some α-helical content that was enhanced by the presence of the structure-making cosolvent, trifluoroethanol (TFE). A 35-kD dehydrin isolated from cowpea (Vigna unguiculata) similarly adopted greater α-helical structure in the presence of SDS (Ismail et al., 1999a). In addition, both the maize G50 and cowpea 35-kD LEA proteins exhibit significant hydrophobic characteristics as shown by their interaction with hydrophobic-interaction chromatography columns used in their purification and putative interaction with copurifying proteins (Ceccardi et al., 1994; Ismail et al., 1999). Such in vitro hydrophobic interactions are postulated to be mediated by amphipathic α-helical structures formed by K segments, which may form the basis of group 2 LEA proteins ability stabilize protein or membrane structures in freezing or desiccating tissues (Close, 1997; Ismail et al., 1999a). More detailed structural examination of group 2 LEA proteins is necessary to fully explain their functional roles in vivo. To this end, we have analyzed the structural properties of a recombinant group 2 LEA protein from soybean (Glycine max; rGmDHN1) using differential scanning calorimetry (DSC) and UV absorption and CD spectroscopy as a function of temperature and in the presence of detergent, TFE, and phospholipid bilayers.
RESULTS
Overexpression and Purification of rGmDhn1
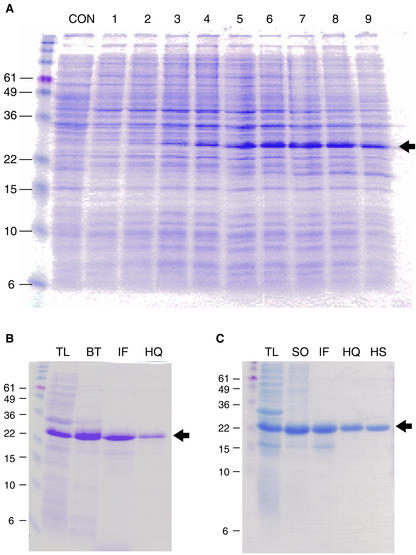
To study the structural and physicochemical characteristics of a group 2 LEA protein from soybean, large (milligram) quantities of the recombinant protein (rGmDHN1) were produced and purified from Escherichia coli. The soybean Dhn1 cDNA encodes a protein with a predicted amino acid sequence identical to the maturation-associated protein 9 (MAT9; GenBank accession no. M94012) isolated from soybean seeds (Chyan and Kriz, 1992). Affinity tag sequences were removed from the pET30a expression vector to express the native protein. From total cell lysates of E. coli BL21 (DE3) plysS cells, we observed the induction of the putative rGmDHN1 gene product with an apparent molecular mass of approximately 27 to 28 kD (Fig. 1A). The apparent molecular mass of the expressed protein was 4 to 5 kD larger than the predicted molecular mass of 23.7 kD, consistent with the previous observations of aberrant electrophoretic mobility of dehydrins (Close et al., 1989; Ceccardi et al., 1994). Maximum accumulation of rGmDHN1 occurred 240 min (OD600 = 1.5) after IPTG induction. Growth rates of noninduced E. coli cells and cells induced for rGmDHN1 overexpression showed similar growth rates, suggesting that GmDHN1 accumulation was not detrimental to cell growth (data not shown).
Figure 1.
Expression and purification of a soybean group 2 LEA protein from E. coli. A, rGmDHN1 accumulation at different times after isopropylthio-β-galactoside (IPTG) induction. Protein overexpression was induced with 0.1 mm IPTG when the optical density (OD600) reached 0.8. Cells were collected at various time points, and soluble proteins were extracted. Con, No IPTG; 1, induction point, 120 min; 2, 135 min; 3, 150 min; 4, 180 min; 5, 210 min; 6, 240 min; 7, 270 min; 8, 300 min; and 9, 360 min. Approximately 10 μg of protein was loaded in each lane. B, Composite purification steps of rGmDHN1 treated with boiling. Approximate amount of protein loaded in each lane is indicated in parentheses. TL, Total lysate (10 μg); BT, boiling treated (5 μg); IF, IEF (2 μg); HQ, high Q (1.5 μg). C, Composite purification steps of rGmDHN1 without boiling. TL, Total lysate (10 μg); SO, 40% (w/v) (NH4)2SO4 salting-out (10 μg); IF, IEF (2 μg); HQ, high Q (2 μg); HS, high S (1.5 μg). Arrows designates location of rGmDHN1 on gel. The relative mass of prestained molecular mass standards is designated in kilodaltons.
Initial purification of rGmDHN1 took advantage of the unique property of rGmDHN1 solubility following exposure to high temperatures. Subsequent purification steps included preparative isoelectric focusing (IEF) in the pH 5.5 to 7.0 range consistent with the calculated pI of rGmDHN1 (6.08), followed by anion-exchange chromatography, which was necessary to achieve a high degree of purification (Fig. 1B). To determine whether rGmDHN1 structure was altered by heat denaturation, rGmDHN1 protein was also purified without boiling. Precipitation at 40% (w/v) (NH4)2SO4 was substituted for heat and was used as the initial purification step. Preparative IEF and anion-exchange column chromatography steps followed; however, it was necessary to include an additional cation-exchange column chromatography step to achieve homogeneity (Fig. 1C). Recombinant proteins purified using heat or (NH4)2SO4 showed identical electrophoretic mobility after analysis by native or denaturing SDS-PAGE (data not shown). The identity of the proteins purified by either strategy was confirmed by matrix-assisted laser-desorption ionization time of flight mass spectrometry (MS; data not shown). The observed molecular mass for rGmDHN1 was 23.578 kD, which is nearly identical to the predicted molecular mass 23.583 kD (without N-terminal f-Met).
Microenvironment of rGmDHN1 Tyr Residues: Temperature Effects
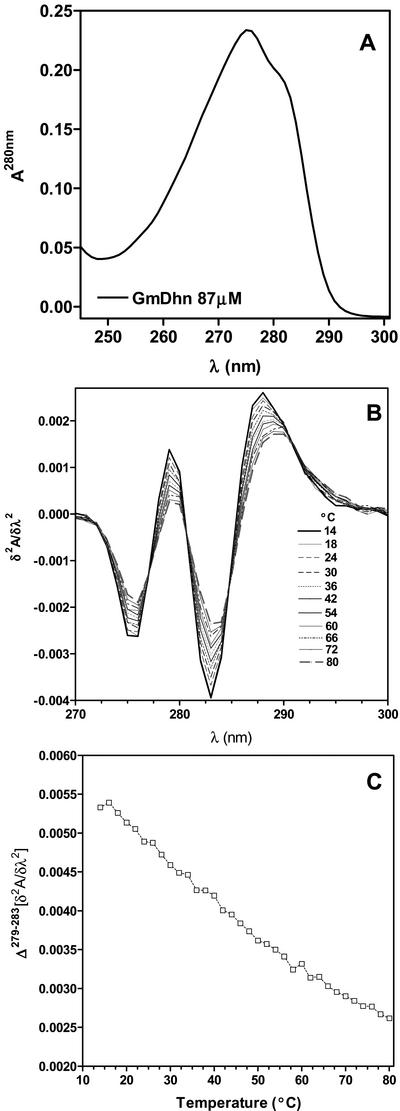
rGmDHN1 has 18 Tyr and no Trp residues. Therefore, the near-UV absorption spectrum of rGmDHN1 is dominated by the absorption of the Tyr residues. In the near UV, the absorption spectrum of rGmDHN1 shows a maximum at 275 nm (Fig. 2A) that is characteristic of the absorption by Tyr and confirms the absence of Trp. The absorption spectrum of a tyrosyl residue is sensitive to the polarity of its environment, and therefore the intensity and shape of the absorption band can be used to determine the degree of hydration of Tyr. The spectral features affected by the hydration are conveniently monitored by the second derivative spectra rather than the zero order spectra (Ragone et al., 1984; Demchenko, 1986; Soulages and Bendavid, 1998; Soulages et al., 2002). The second derivative spectra of rGmDHN1 as a function of temperature are shown in the Figure 2B. The spectral changes that occur when the temperature is increased from 14°C to 80°C indicate that the protein becomes more hydrated as the temperature is increased. This increase suggests that heating promotes a structural transition involving an unfolding process. In addition to the changes in the shape and intensity of the second derivative spectra, another important feature observed in this study is the presence of several crossover points (identical second derivative points). The fact that these crossover points remain unchanged over the entire range of temperatures studied indicates that there is equilibrium between two conformational states. Using the values of the derivative at 279 and 283 nm, it is observed that between 14°C and 80°C, there is a continuous change in the shape and intensity of the Tyr absorption (Fig. 2C). However, because the spectral changes do not reach a plateau at either the low- or the high-temperature ends of the curve, it is not possible for us to estimate the limiting values corresponding to the each of the two involved states. Thus, although the change in the second derivative is proportional to the population of the states, we are not able to estimate the relative change in the population of the conformational states. Despite this limitation, the absence of plateaus indicates that within the temperature range studied, none of the two states become fully populated. This fact, as well as the slope of the plot in Figure 2C, indicates that the cooperativity of the conformational transition observed by UV absorption is low. It must be noted that given the large number of Tyr residues the UV absorption study is sampling most of the polypeptide chain.
Figure 2.
UV absorption spectroscopy of rGmDHN1. A, UV absorption spectrum of rGmDHN1 in 50 mm phosphate buffer, pH 7. B, Effect of temperature on the second derivative spectrum of rGmDHN1. The spectra shown, reading downward at 283 nm, represent decreasing temperatures from 14°C to 80°C. The temperatures are indicated in the figure. C, The temperature-induced changes are represented using the difference between the derivative values at 279 and 283 nm: Δ(279–283) [δ2ε/δλ2].
Thermal Stability of rGmDHN1
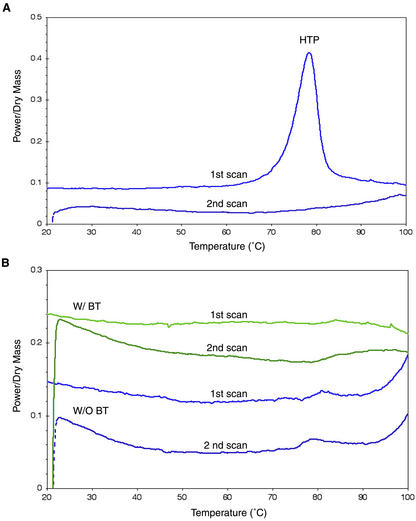
DSC was used to investigate further the presence of potential cooperative structural transitions of rGmDHN1. The DSC scans in Figure 3A show a typical denaturation pattern of a heat-labile protein. BSA undergoes an irreversible thermal denaturation around 78°C as indicated by a high-temperature peak in the first scan and its disappearance in the second (Leprince and Vertucci, 1995). In contrast, DSC scans of rGmDHN1, purified with or without boiling treatment, showed no detectable high-temperature peak up to 100°C (Fig. 3B). These results are consistent with the UV absorption studies, which suggested rGmDHN1 to be highly hydrated and to lack highly cooperative folding transitions.
Figure 3.
Heating thermograms of bovine serum albumin (BSA) and rGmDHN1 using DSC. DSC heat scans of BSA (A) and rGmDHN1 (B) with (green) and without (blue) boiling treatment (BT) were performed at 10°C min−1 from 0°C to 100°C. Plots of each scan were offset slightly for clarity. HTP, High-temperature peak.
Secondary Structure of rGmDHN1: Effect of Temperature
Earlier investigations of a dehydrin-like protein (Dsp16) from the resurrection plant C. plantagineum showed that this protein underwent temperature- and denaturant-dependent structural transitions as determined by CD spectroscopy (Lisse et al., 1996). To investigate possible temperature-dependent changes in the structural properties of rGmDHN1, the secondary structure of rGmDHN1 in solution was investigated using CD spectroscopy. The far-UV CD spectra of rGmDHN1 at several temperatures are shown in the Figure 4A. At any one of the temperatures studied, the CD spectrum of rGmDHN1 shows a strong negative band at 197 nm, which is typically found in highly unfolded proteins or unordered structures (Woody, 1992). Therefore, the common spectral deconvolution analyses, which are based on data sets of proteins rich in α-helical or β-sheet, cannot be applied to estimate the structural composition of a predominantly unordered protein (Bienkiewicz et al., 2000), such as rGmDHN1.
Figure 4.
Effect of temperature on the secondary structure of rGmDHN1. A, Near-UV CD spectra of rGmDHN1 obtained in buffer sodium phosphate at pH 7 at the temperatures indicated in the figure. The spectra shown, reading upward at 197 nm, represent increasing temperatures: 12, 20, 30, 40, 50, 60, 70, and 80°C. B, The temperature-induced changes in the CD spectrum of rGmDHN1 are represented using the difference between the CD intensities at 220 and 197 nm. The CD intensity at 208 nm is also represented to illustrate the presence of the isodichroic point.
Despite the apparent unordered conformation of the rGmDHN1, increasing the temperature produced a gradual increase in the dichroic negative band centered between 210 and 230 nm and a concomitant decrease in the intensity of the negative band centered at 197 nm (Fig. 4, A and B). The observed decrease in intensity of the dichroic band at 197 nm and the increase in intensity of the band near 220 nm prompted us to consider the possibility of a temperature-induced folding process rather than unfolding. However, the fact that, as shown by UV absorption, the hydration of the protein increases as the temperature is raised indicated that the increase in temperature was actually driving an unfolding process. Besides the changes in the two negative bands discussed above, it is also evident that at 208 nm and −1.85 m−1cm−1, there is an isodichroic point (Fig. 4, A and B). This point is maintained in the temperature range studied, 12°C to 80°C, suggesting the presence of an equilibrium between two conformational states. This observation is consistent with the isosbestic points observed by UV absorption and provides further support for the existence of a two-state equilibrium.
Evidence of an Extended Helix Conformation
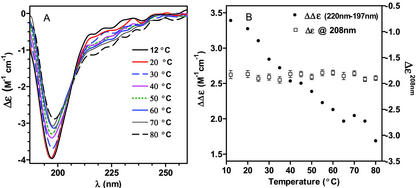
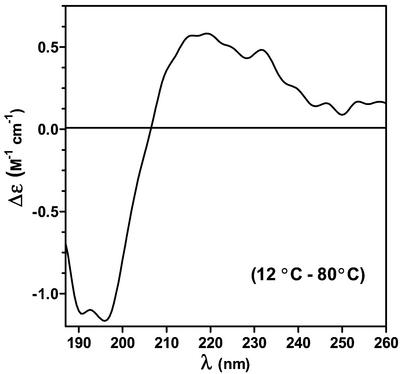
The CD difference spectrum (Δ12°C–80°C) of rGmDHN1 was determined as shown in Figure 5. This difference spectrum is characterized by an intense negative CD-band centered at 200 nm and a positive CD band above 200 nm. The positive band indicates that the contribution of α-helical structures to the CD spectrum is absent or very low. Otherwise, we should expect a decrease in α-helical content as the temperature increases and a negative CD band above 200 nm in the difference spectrum. Interestingly, these two features observed in the difference spectrum are present in the CD spectra of peptides rich in poly (l-Pro)-type II (PII) structures (Woody, 1992; Park et al., 1997; Fox et al., 1999; Bienkiewicz et al., 2000; Soulages et al., 2002). This similarity suggested that the spectral changes observed in rGmDHN1 could be due to a temperature-induced extended helix/unordered transition. The difference spectrum shows a maximum at 215 nm, which is coincident with the maximum observed in Pro-poor peptides adopting PII-like conformations. Additional support for this conformation arises from the coordinates of the isodichroic point observed in Figure 4, A and B (at 208 nm and −1.85 m−1cm−1). These coordinates are coincident with the coordinates observed in model polypeptides and proteins that undergo PII/unordered transitions (Tiffany and Krimm, 1968; Tiffany, 1975; Woody, 1992; Siligardi and Drake, 1995; Park et al., 1997; Fox et al., 1999; Bienkiewicz et al., 2000; Soulages et al., 2002).
Figure 5.
Evidence of PII structure. The CD component that disappears upon heating is illustrated in the graph by the CD difference spectrum obtained by subtracting the CD spectrum obtained at 80°C from the CD spectrum obtained at 12°C.
Park et al. (1997) have investigated a series of host/guest peptides and suggested limiting values of ellipticity for the PII and the unordered structures of +9,580 and −5,560° cm2 dmol−1 at 220 nm. Using these suggested values we can estimate the fractions of unfolded and PII conformations in rGmDHN1 at different temperatures. To perform this calculation, we also assumed that the α-helical content of the protein was nearly zero at high and low temperatures. This assumption appears to be well supported by the maintenance of the isodichroic point along the entire temperature range studied. If this transition would involve three states, unordered, α-helical, and PII, one should not expect the maintenance of the isodichroic point. Using these limiting values, and the ellipticity of rGmDHN1 at pH 7.0 and 12°C, we inferred that this protein contains 27% of its residues in an extended-helix conformation. This fraction is reduced to 15% at 80°C.
Effect of α-Helix Inducing Cosolvents and Lipids on rGmDHN1 Secondary Structure
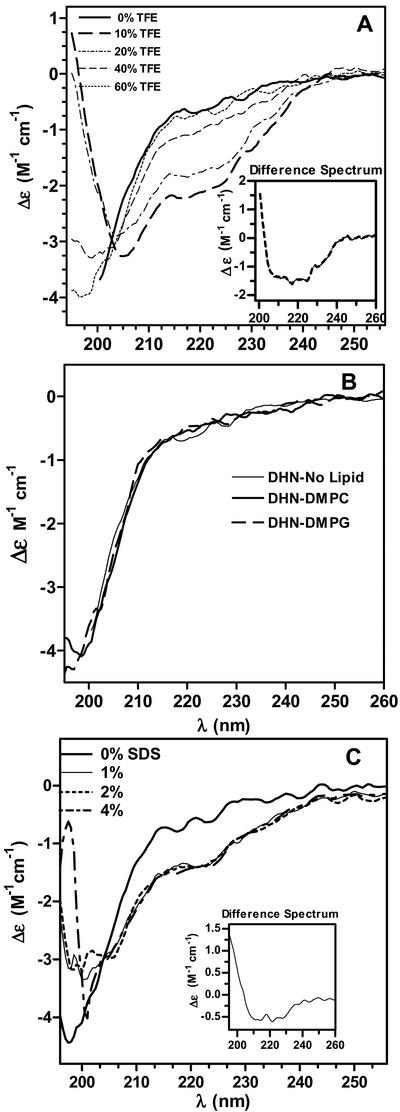
The LEA-like COR15am protein from Arabidopsis has been postulated to interact in vivo and alter the intrinsic curvature of the monolayers that compose the inner membrane of the chloroplast envelope (Steponkus et al., 1998). This alteration is thought to decrease its propensity to undergo lamellar-to-hexagonal II phase transitions during freeze-induced dehydration at low temperatures (−5°C to −8°C), thereby reducing membrane leakage within the chloroplast envelope. Because the COR15am protein is predicted to be composed largely of amphipathic α-helices, this interaction is thought to be mediated by amphipathic α-helices that can have a strong effect on the intrinsic curvature of such monolayers. To investigate the potential for rGmDHN1 to mediate protection against freezing damage using a similar mechanism, we studied the tendency of rGmDHN1 to adopt α-helical conformations through the addition of the helix-promoting cosolvent TFE. The CD spectra of rGmDHN1 at several concentrations of TFE are shown in the Figure 6A. TFE induced the formation of α-helix in rGmDHN1. This is evident at 40% and 60% (v/v) TFE. For clarity, we have included the difference spectrum of rGmDHN1 (spectrum in the presence of 60% [v/v] TFE minus spectrum in 50 mm phosphate buffer at pH 7.0) in Figure 6A. From the difference spectrum or from the ellipticity at 220 nm, it was estimated that at 60% (v/v) TFE rGmDHN1 contains 14.8% of α-helical structure. This was estimated by the method of Chen et al. (1972). Given the limited fraction of α-helical structure observed at a relatively high concentration of TFE, it can be concluded that GmDHN1 has a very low intrinsic tendency to adopt α-helical conformations in solution.
Figure 6.
Effect of α-helix-inducing cosolvents and lipids on the secondary structure of rGmDHN1. A, Effect of increasing concentrations of TFE on the near-UV CD spectrum of rGmDHN1. The concentrations of TFE corresponding to individual spectra are indicated in the figure. Inset, Difference spectrum calculated as the difference between the spectrum of rGmDHN1 in 60% (v/v) TFE minus the spectrum of the protein in 0% (v/v) TFE. B, CD spectra of rGmDHN1 (3 μm) incubated with multilamellar liposomes of DMPC or DMPG at a 400 to 1 lipid to protein molar ratio. The spectra of rGmDHN1 shown were obtained after a 2-h incubation period at the temperatures of the corresponding thermotropic transitions of the lipids (DMPC, 24°C; DMPG, 23.4°C). C, CD spectra of rGmDHN1 (3 μm) in the presence of SDS micelles. The concentrations of SDS in grams per 100 mL are indicated in the figure. The data shown in all the panels were obtained in 50 mm sodium-phosphate buffer pH 7.4. CD spectra were acquired at 25°C.
Previous studies have suggested that closely related dehydrins in vivo might contain K-segment α-helical structures in a lipid-bound state (Close, 1996; Ismail et al., 1999a). To investigate the potential interaction of rGmDHN1 with lipid membranes, we acquired the CD spectra of the protein in the presence of liposomes of dimyristoyl phosphatidylcholine (DMPC) and in the presence of dimyristoyl phosphatidylglycerol (DMPG). None of these lipids promoted a change in the CD spectrum of rGmDHN1 (Fig. 6B), suggesting that no lipid-protein interaction takes place in solution or, otherwise, that the interaction does not occur through amphipathic α-helices. It must be noted that we studied the interaction with zwitterionic and anionic liposomes at the temperature of the lipid thermotropic transition of the lipids (23.9°C for DMPC and 23.4°C for DMPG). Under these conditions, the interaction of amphipathic α-helical proteins and peptides with the lipid membrane is highly favored (Swaney and Chang, 1972; Soulages et al., 2001). To directly compare our studies with those carried out with a group 2 LEA protein from cowpea (Ismail et al., 1999a), we also determined the effect of SDS in the conformation of rGmDHN1. The results of this study are shown in Figure 6C and indicate that SDS promotes the formation of α-helical structure. Analysis of the spectrum in the presence of SDS by the method of Chen et al. (1972) indicates that rGmDHN1 acquires a maximum of 7.4% of α-helical content. Increasing the concentration of SDS from 1% to 4% did not modify further the structure of the protein. Because SDS does not adopt a bilayer structure, its use as a means to investigate the potential interaction between the protein and cell membranes may not be as relevant as the use of liposomes. However, as with TFE, SDS provides a means to test the intrinsic ability of the protein to adopt α-helical structure. The results obtained with SDS and TFE confirm that the primary structure of rGmDHN1 imposes strong restrictions to the formation of α-helices in solution. This is supported by the fact that this protein contains nearly 25% of residues characterized by their inability to adopt α-helical conformations (50 Gly residues and four Pro residues).
DISCUSSION
Previous in vivo functional studies of a group 2 LEA protein from tomato (le4) have shown that this protein can ameliorate the detrimental effects of ionic (KCl) or freezing stress, but not NaCl or osmotic (sorbitol) stress in yeast cells (Zhang et al., 2000). Despite these observations, the mechanisms by which this protein acts to afford such protection remain largely unknown. Possible clues to the functional roles of group 2 LEA proteins can also be gathered from detailed in vitro biochemical, physiochemical, and structural analyses. To gain a better understanding of the potential mechanistic basis of group 2 LEA protein function, we have conducted detailed structural analysis of a recombinant Y2K type soybean LEA protein. To conduct such studies, we purified large amounts of this protein from E. coli by taking advantage of its heat stability properties. A simple heat denaturation of crude E. coli lysates resulted in a significant enrichment in protein purification. Preparative IEF and ion-exchange chromatography steps followed this initial heating step to attain homogeneous preparations of the recombinant protein (Fig. 1).
Potential Physiological Roles of PII Structures
The studies of the structure of rGmDHN1 showed that the protein has a predominantly unordered conformation in solution. However, the examination of the UV absorption and CD spectral properties of the purified protein over a range of temperatures clearly showed the existence of a temperature-dependent structural transition. Analysis of the second-derivative UV absorption spectrum for rGmDHN1 suggested that its Tyr residues become more hydrated at higher temperature, and the presence of isosbestic points along the entire temperature range studied, 14°C to 80°C, indicated that the temperature-dependent changes in hydration of the Tyr residues were due to a structural transition involving two states (Fig. 2). The structural change detected by UV absorption did not reach a plateau at either the high- or low-temperature ends of the curve, indicating that the structural transition is characterized by a very low cooperativity. Moreover, the study of the thermal behavior of rGmDHN1 by DSC showed no sharp endothermic peaks, from 10°C to 100°C, indicating the absence of a highly cooperative unfolding transition (Fig. 3). This result was independent of whether the protein was purified by heat denaturation or (NH4)2SO4 precipitation.
The study of the temperature dependence of the CD spectrum of GmDHN1 confirmed the conclusions reached in the UV absorption study. The changes observed in the CD spectrum of rGmDHN1 indicated that the presence of a structural transition characterized by a low cooperatively (Fig. 4). The CD study also showed that the temperature-dependent spectral changes were consistent with a structural transition involving two conformational states. Because only two states were apparently involved in the temperature-induced transition, the difference spectrum between a low and a high temperature provided the spectrum of the component that disappeared upon heating. Using this approach it was found that in aqueous solution, rGmDHN1 was present as an equilibrium mixture between PII and truly unordered conformations. The identification of the PII structure was based on the comparison of the difference spectra of rGmDHN1 with the CD data reported for other proteins and polypeptides for which a PII structure has been inferred (Woody, 1992; Park et al., 1997; Bienkiewicz et al., 2000).
Intrinsically unstructured proteins, such as hydrophilins and group 1 and 2 LEA proteins, participate in many cellular functions (Wright and Dyson, 1999). Among such proteins, the left-handed PII helical conformation is a common and important structural component. This structure plays a role in several biochemical processes including signal transduction, transcription, cell motility, and the immune response (Creamer, 1998; Stapley and Creamer, 1999). PII helices are major features of collagens (Pauling and Corey, 1951) and plant cell wall proteins (Ferris et al., 2001). PII structures are also found in the antifreeze glycoproteins of polar fish (Lane et al., 1998, 2000). Antifreeze glycoproteins are known to prevent ice crystal formation by promoting cooperative hydrogen bonding with water over its length (Yeh and Feeney, 1996). It has been estimated that 2% of all residues in known protein structures are found in PII helices at least four residues long (Adzhubei and Sternberg, 1993; Stapley and Creamer, 1999). The fraction of residues in PII conformation would be even higher (approximately 10%) if the conformation of individual residues were evaluated (Sreerama and Woody, 1994).
We have recently shown that a group 1 LEA protein from soybean (GmD-19) contains a significant proportion of PII structure at room temperature (Soulages et al., 2002). Previous hydrodynamic, CD and NMR spectroscopy studies showed that group 2 LEA proteins possess little apparent defined secondary structure (Ceccardi et al., 1994; Lisse et al., 1996; Ismail et al., 1999). Far-UV CD analysis of a purified 42-kD maize dehydrin estimated a 75% unordered and 15% α-helical content (Ceccardi et al., 1994). Similar results were obtained for a 35-kD dehydrin from cowpea (Ismail et al., 1999a) and a recombinant Dsp16 derived from the resurrection plant C. plantagineum (Lisse et al., 1996). With the exception of our recent report (Soulages et al., 2001), PII structures have not been identified as important structural elements of LEA proteins. However, the overall similarity among the physical properties of many hydrophilins suggests that PII structures may be a common structural feature of not only group 1 and 2 LEA proteins, but also other hydrophilic, Gly-rich proteins belonging to the hydrophilin superfamily of proteins.
Large regions of unordered conformation are considered to promote the efficient interaction of group 1 LEA proteins with water and to aid in their ability to prevent cellular water loss (McCubbin et al., 1985; Eom et al., 1996). Because PII helices also have a large solvent-exposed area, the extent of the polypeptide-water interaction would not be expected to change drastically as the protein undergoes a transition from a truly unordered conformation to a poly(Pro) II helix. Even though the biochemical roles of PII structures have not been firmly established in plants, the fact that decreasing the temperature induces an increase in the structural organization of the protein without compromising the interaction of the protein with the solvent suggests a role for LEA proteins in preserving the cellular integrity under stress conditions that could affect the content of cellular water or its physical state. Although the content of PII structure increases significantly as the temperature decreases (Fig. 5), there is no increase in the apparent content of α-helical or β-structures, which would decrease the number of potential interactions between the solvent and the protein backbone. Compared with an unordered structure, the α-helical or β-sheet structures represent a dramatic decrease in the number of H-bond interactions between the solvent and the polypeptide backbone. However, the extended helical conformation of the PII structure of the protein backbone also remains available for interaction with the solvent.
rGmDHN1 α-Helical Conformation and Lipid Membrane Interaction
The majority of group 2 LEA proteins are predicted to contain amphipathic α-helix along K-segments (Dure et al., 1989; Dure, 1993; Close, 1997). Amphipathic α-helical structures of LEA proteins are thought to interact with and to protect membranes or partially denatured proteins. As indirect evidence for such hydrophobic interaction, a dehydrin-like protein, COR15am, has been shown to alter the intrinsic curvature of the inner membrane of the chloroplast envelope (Steponkus et al., 1998). However, direct evidence for the formation of such hydrophobic interactions involving group 2 LEA has not been obtained. GmDHN1 from soybean has one K-segment (13 residues long) near the C-terminal of the polypeptide chain. From the CD spectrum in aqueous solution, we inferred that rGmDHN1 does not contain α-helical regions. Assuming that the K-segment of rGmDHN1 would adopt an α-helical conformation, we should have estimated an α-helical content close to 6.2%. On the other hand, confirming that rGmDHN1 has a limited ability to adopt α-helical conformations, it was observed that in the presence of TFE and SDS, the protein contains 15% and 7% of α-helical structure, respectively (Fig. 6, A and C). If adopted under physiological conditions, this small fraction of α-helix is unlikely to be adequate to promote membrane interactions. The incubation of rGmDHN1 with an excess of liposomes made with two different phospholipids, one zwitterionic and the other negatively charged, did not induce the formation of α-helix that is expected to occur in unordered peptides that interact with membranes through amphipathic α-helices (Fig. 6B). Therefore, we have found little evidence to suggest that rGmDHN1 can interact with cell membranes through lipid-protein interactions in solution.
Despite these observations, it remains possible that under conditions of increasing ionic content or dehydration, the formation of amphipathic α-helices with GmDHN1 may still play a physiological role in vivo. Solution ionic strength has been shown to reversibly influence the structure of pea (Pisum sativum) group 1 LEA protein (Russouw et al., 1995, 1997). For the group 2 LEA-like Dsp16 protein from C. plantagineum, increasing the NaCl concentration from 0.1 to 2 m resulted in an increase in α-helical content from 6% to 15% (Lisse et al., 1996). Increasing ionic and Suc content within the cytoplasm would be expected to accompany the dehydration process during embryo desiccation. Furthermore, recent studies of a group 3 (D-7) LEA-like protein from cattail (Typha latifolia) pollen show that this protein assumes an entirely unordered conformation in solution, but upon drying, the protein assumes a largely α-helical structure as measured by Fourier transform infrared spectroscopy (Wolkers et al., 2001). The extent of α-helical content (and extended β-sheet structures) was dependent upon the rate of drying and the presence of Suc. These results suggest that LEA protein structure can be strongly influenced by their immediate environment. Although group 1 and 2 LEA proteins have a much lower predicted α-helical content than group 3 LEA proteins, increases in α-helical content arising from changes in ion or sugar concentrations or hydration status could contribute to functional interactions with other biomolecules. α-Helical structures were found to compose approximately 40% of the overall protein secondary structures in dry cattail pollen (Wolkers and Hoekstra, 1995). Similar α-helical contributions to overall protein secondary structure have also been observed in maize embryos (Wolkers et al., 1998a) and carrot (Daucus carota) somatic embryos (Wolkers et al., 1998b) consistent with earlier predictions of a functional contribution of α-helical conformations by several LEA proteins (Dure et al., 1989; Dure, 1993). However, such studies cannot reveal the contribution made to the overall structural conformation content by a specific group of LEA proteins. The extended PII helical and unordered conformations of GmDHN1 may alternatively retain a high degree of exposed surface area and make a significant contribution to water-peptide interactions as postulated for group 1 LEA proteins (Soulages et al., 2002). Efficient interactions with water predict the ability to slow water loss during dehydration. During embryo or pollen desiccation and in the dry state, group 2 LEA proteins might also play a role in the formation of tightly hydrogen-bonded networks, in concert with carbohydrates, to provide stability to macromolecular structures through the inhibition of cellular membrane fusion and the denaturation of cytoplasmic proteins, and to limit the diffusion of free radicals (Oliver et al., 2001; Wolkers et al., 2001).
In conclusion, we suggest that GmDHN1 can interact efficiently with water due to its hydrophilicity and its ability to adopt extended helical conformation and unordered structures at very low temperatures. Future in vitro biochemical and physicochemical analyses, including examination of the protective and hydration properties of purified, rGmDHN1, and overexpression studies in vivo are under way and should provide important clues about the functional roles that group 2 LEA proteins play in combating macromolecular destabilization due to temperature- and dehydration-related stresses.
MATERIALS AND METHODS
Cloning and Expression of Soybean (Glycine max) Group 2 LEA
The soybean group 2 (GmDhn1) LEA cDNA coding region (684 bp; GenBank accession no. U10111 and AAA18834.1; N. Maitra and J.C. Cushman, unpublished data) was amplified using ULTma DNA polymerase (Promega, Madison, WI) and gene-specific primers containing NcoI (5′-CATGCCATGGCAAGTTATCAAAAGC-3′) or EcoRI (5′-CGGAATTCCTACTTGTCACTGTGTC-3′) restriction sites. The amplified fragment was digested with NcoI/EcoRI and ligated into the NcoI/EcoRI sites of the pET30a Escherichia coli expression vector (Novagen, Madison, WI). His- and S-tag sequences (157 bp) were then removed from the cloning vector by inverse PCR using Pfu polymerase (Invitrogen, Carlsbad, CA) and outward facing primers (5′-GGCGCGCCCTCCTTCTTAAAGTTAAACAA-3′; 5′-GGCGCGCCATGGCAAGTTATCAAAAGCA-3′) containing AscII restriction sites. The amplification product was digested with AscII and religated with T4 ligase. The integrity of the cloned insert was confirmed by automated DNA sequencing. The resulting pET30a::Dhn1 construct was introduced into E. coli BL21 (DE3) plysS cells and grown in 2× yeast-tryptone medium under kanamycin selection (50 μg mL−1) at 37°C with vigorous agitation (300 rpm). Recombinant GmDHN1 protein expression was induced by adding IPTG to a final concentration of 0.1 mm when cells reached an OD600 of 0.8. Cells were harvested when the culture reached an OD600 of 1.5. Expression of the recombinant protein (rGmDHN1) was confirmed by 12% (w/v) SDS-PAGE and Coomassie Brilliant Blue staining. Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Purification of rGmDHN1 from E. coli
Bacterial cells were harvested by centrifugation at 5,000g and resuspended in B-PER Bacterial Extraction buffer (Pierce, Rockford, IL) in the presence of Complete Mini protease inhibitor cocktail (Roche Diagnostics, Indianapolis). The soluble cell lysate extract was sonicated to reduce viscosity, heated-denatured in boiling water for 10 min., and clarified by centrifugation at 26,500g for 20 min. The clarified supernatant was concentrated using a Centricon Plus-80, Mr cut off 5,000 polycarbonate centrifugal filter (Amicon, Beverly, MA) and dialyzed overnight against 10 mm Tris-HCl, pH 7.5, using dialysis membrane tubing (SnakeSkin Mr cut off 3,000, Pierce). The dialyzed extracts were subjected to preparative IEF in the presence of ampholytes (2% [v/v], pH range 5–7; Bio-Rad Laboratories, Hercules, CA) using a Rotofor Cell (Bio-Rad Laboratories) at 15-W constant power for 4 h. Fractions were surveyed by 12% (w/v) SDS-PAGE, and fractions containing rGmDHN1 protein were pooled and stored at −20°C.
Further purification of the pooled IEF fractions was conducted using anionic exchange column chromatography on an High-Q column (Bio-Rad Laboratories) in 10 mm Tris-HCl, pH 7.5, eluted by a 0 to 250 mm NaCl gradient at 1 mL min−1 flow rate. Collected fractions (2 mL) were analyzed by SDS-PAGE, pooled, and stored as the final purified proteins. rGmDHN1 was alternatively purified without heat denaturation by 40% (w/v) ammonium sulfate precipitation. Pellets were resuspended with 10 mm Tris-HCl, pH 7.5, and desalted using a Centricon Plus-80, Mr cut off 5,000 polycarbonate centrifugal filter (Amicon) and dialyzed extensively (SnakeSkin Mr cut off 7000) before preparative IEF and anion-exchange column chromatography. Cation exchange column chromatography using a High-S column (Bio-Rad Laboratories) equilibrated with 10 mm sodium-acetate, pH 4.8, was used as a final purification step. Recombinant GmDHN1 was eluted using a 0 to 500 mm NaCl gradient at 1 mL min−1 flow rate, and fractions containing the purified protein were pooled and desalted by dialysis as described above.
MS
Molecular mass determination of rGmDHN1 was performed by matrix-assisted laser-desorption ionization time of flight MS (Proflex, Bruker, Karlsruhe, Germany) using sinapinic acid as a matrix. Samples were prepared using a ZipTipC18 reverse phase column (Millipore, Bedford, MA) following manufacturer's instructions. After desalting, samples were dissolved in 50% (v/v) acetonitrile and 0.1% (v/v) trifluoroacetic acid and then dried by gentle heating before MS analysis. Cytochrome C (molecular mass, 12,384 D) was used as a calibration standard.
DSC and Thermal Analysis
Three milligrams of lyophilized protein powder was loaded on a DSC volatile samples pan and hydrated under 80% (v/v) relative humidity controlled by saturated KNO3 at room temperature. Thermal events were measured from 0°C to 100°C at a rate of 10°C min−1 using a differential scanning calorimeter (DSC-4 and DSC-7, PerkinElmer Instruments, Norwalk, CT) calibrated for temperature using methylene chloride (−95°C) and indium (156°C) standards and for energy with indium (28.54 J g−1) as previously described (Leprince and Vertucci, 1995). Helium gas was used for purging at a rate of 20 mL min−1. To standardize the dry mass of each sample, heat flow in every DSC scan was divided by the sample dry weight.
UV Absorption Spectroscopy
UV absorption spectra were recorded with an HP 8453 diode array spectrophotometer (Hewlett Packard, St. Paul). The concentration of rGmDHN1 was calculated from the absorbance of the samples at 280 nm in the presence of 6 m guanidinium-HCl (εTyr = 1,285 cm−1 m−1; Pace et al., 1995). Second derivatives were calculated by the Savitzky-Golay differentiation technique using a filtering length of 9. The temperature dependence of the difference between the second derivatives at 283 and 279 nm was determined in 50 mm phosphate buffer solutions. The sample temperature was modified and controlled by a Peltier temperature-controlled cell holder.
CD Analysis
CD spectra were acquired with a CD-spectropolarimeter (model J715, Jasco, Easton, MD) using a 0.1-cm-path-length cell over the 184- to 260-nm range. The temperature was controlled by a circulating water bath (model RTE 111, Neslab, Newington, NH) and determined directly into the cell using a thermocouple. CD spectra were acquired every 1 nm with 2 s averaging time per point and a 1-nm band pass. Quadruplicate average spectra were corrected for the blank and smoothed.
ACKNOWLEDGMENTS
We thank Sue Ann Hudiburg and Janet Rogers (Oklahoma State University) for the synthesis of oligonucleotides and automated DNA sequencing services, respectively. We also thank Dr. David Quilici (University of Nevada) for providing MS data.
Footnotes
This work was supported in part by the U.S. Department of Agriculture National Research Initiative-Competitive Grants Program (grant no. 98–35100–10216 to J.C.C.), by the National Institutes of Health (grant no. GM 55622 to J.L.S.), and by the Nevada Agricultural Experiment Station (publication no. 0302382).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.015891.
LITERATURE CITED
- Adzhubei AA, Sternberg MJ. Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol. 1993;229:472–493. doi: 10.1006/jmbi.1993.1047. [DOI] [PubMed] [Google Scholar]
- Baker J, Steele C, Dure L., III Sequence and characterization of 6 Lea proteins and their genes from cotton. Plant Mol Biol. 1988;11:277–291. doi: 10.1007/BF00027385. [DOI] [PubMed] [Google Scholar]
- Bienkiewicz EA, Woody Moon AY, Woody RW. Conformation of the RNA polymerase II C-terminal domain: circular dichroism of long and short fragments. J Mol Biol. 2000;297:119–133. doi: 10.1006/jmbi.2000.3545. [DOI] [PubMed] [Google Scholar]
- Borovskii GB, Stupnikova IV, Antipina AA, Downs CA, Voinikov VK. Accumulation of dehydrin-like proteins in the mitochondria of cold-treated plants. J Plant Physiol. 2000;156:797–800. [Google Scholar]
- Borovskii GB, Stupnikova IV, Antipina AA, Vladimirova SV, Voinikov VK. Accumulation of dehydrin-like proteins in the mitochondria of cereals in response to cold, freezing, drought and ABA treatment. BioMed Central Plant Biol. 2002;2:5. doi: 10.1186/1471-2229-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo LA, Close TJ, Corcuera LJ, Guy CL. Characterization of an 80-kDa dehydrin-like protein in barley responsive to cold acclimation. Physiol Plant. 1999;106:177–183. [Google Scholar]
- Campbell SA, Close TJ. Dehydrins: genes, proteins, and associations with phenotypic traits. New Phytol. 1997;137:61–74. [Google Scholar]
- Ceccardi TL, Meyer NC, Close TJ. Purification of a maize dehydrin. Protein Express Purif. 1994;5:266–269. doi: 10.1006/prep.1994.1040. [DOI] [PubMed] [Google Scholar]
- Cellier F, Conéjéro G, Breitler J-C, Casse F. Molecular and physiological responses to water deficit in drought-tolerant and drought-sensitive lines of sunflower. Plant Physiol. 1998;116:319–328. doi: 10.1104/pp.116.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-H, Yang JT, Martinez HM. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry. 1972;11:4120–4131. doi: 10.1021/bi00772a015. [DOI] [PubMed] [Google Scholar]
- Choi D-W, Zhu B, Close TJ. The barely (Hordeum vulgareL.) dehydrin multigene family: sequences, allele types, chromosome arrangements, and expression characteristics of 1 Dhn genes of cv Dicktoo. Theor Appl Genet. 1999;98:1234–1247. [Google Scholar]
- Chyan YJ, Kriz AL. Analysis of maturation-specific proteins genes: structure, desiccation induction, and abscisic acid responsiveness. PhD thesis. Urbana-Champaign: University of Illinois; 1992. [Google Scholar]
- Close TJ. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant. 1996;97:795–803. [Google Scholar]
- Close TJ. Dehydrins: a commonality in the response of plants to dehydration and low temperature. Physiol Plant. 1997;100:291–296. [Google Scholar]
- Close TJ, Kortt AA, Chandler PM. A cDNA-based comparison of dehydration-induced proteins (dehydrins) in barley and corn. Plant Mol Biol. 1989;13:95–108. doi: 10.1007/BF00027338. [DOI] [PubMed] [Google Scholar]
- Close TJ, Lammers PJ. An osmotic stress protein of cyanobacteria is immunologically related to plant dehydrins. Plant Physiol. 1993;101:773–779. doi: 10.1104/pp.101.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer TP. Left-handed polyproline II helix formation is (very) locally driven. Proteins Struct Funct Genet. 1998;33:218–226. [PubMed] [Google Scholar]
- Cuming AC. LEA proteins. In: Shewry PR, Casey R, editors. Seed Proteins. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 753–780. [Google Scholar]
- Danyluk J, Perron A, Houde M, Limin A, Fowler B, Benhamou N, Sarhan F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell. 1998;10:623–638. doi: 10.1105/tpc.10.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko AP. Fluorescence analysis of protein dynamics. Essays Biochem. 1986;22:120–157. [PubMed] [Google Scholar]
- Dure L, III, Crouch M, Harada J, Ho THD, Mundy J, Quatrano R, Thomas T, Sung ZR. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol. 1989;12:475–486. doi: 10.1007/BF00036962. [DOI] [PubMed] [Google Scholar]
- Dure L., III . Structural motifs in Lea proteins. In: Close TJ, Bray EA, editors. Plant Responses to Cellular Dehydration during Environmental Stress. Current Topics in Plant Physiology. Vol. 10. Rockville, MD: American Society of Plant Physiologists; 1993. pp. 91–103. [Google Scholar]
- Egerton-Warburton LM, Balsamo RA, Close TJ. Temporal accumulation and ultrastructural localization of dehydrins in Zea mays. Physiol Plant. 1997;101:545–555. [Google Scholar]
- Eom J, Baker WR, Kintanar A, Wurtele ES. The embryo-specific EMB-1 protein of Daucus carotais flexible and unstructured in solution. Plant Sci. 1996;115:17–24. [Google Scholar]
- Ferris PJ, Woessner JP, Waffenschmidt S, Kilz S, Drees J, Goodenough UW. Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry. 2001;40:2978–2987. doi: 10.1021/bi0023605. [DOI] [PubMed] [Google Scholar]
- Fox DG, Cary PD, Kneale GG. Conformational studies of the C-terminal domain of bacteriophage Pf1 gene 5 protein. Biochim Biophys Acta. 1999;1435:138–146. doi: 10.1016/s0167-4838(99)00209-5. [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem. 2000;275:5668–5674. doi: 10.1074/jbc.275.8.5668. [DOI] [PubMed] [Google Scholar]
- Goday A, Jensen AB, Culianez-Macia FA, Alba MM, Figueras M, Serratosa J, Torrent M, Pages M. The maize abscisic acid-responsive protein Rab17 is located in the nucleus and interacts with nuclear localization signals. Plant Cell. 1994;6:351–360. doi: 10.1105/tpc.6.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy JA, Lunar R, Torres-Shumann S, Morena J, Rodrigo RM, Pintor-Toro JA. Expression, tissue distribution and subcellular localization of dehydrin TAS14 in salt-stressed tomato plants. Plant Mol Biol. 1994;26:1921–1934. doi: 10.1007/BF00019503. [DOI] [PubMed] [Google Scholar]
- Heyen BJ, Alsheikh MK, Smith EA, Torvik CF, Seals DF, Randall SK. The calcium-binding activity of a vacuole-associated, dehydrin-like protein is regulated by phosphorylation. Plant Physiol. 2002;130:675–687. doi: 10.1104/pp.002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde M, Daniel C, Lachapelle M, Allard F, Laliberte S, Sarhan F. Immunolocalization of freezing tolerance associated proteins in the cytoplasm and nucleoplasm of wheat crown tissues. Plant J. 1995;8:583–593. doi: 10.1046/j.1365-313x.1995.8040583.x. [DOI] [PubMed] [Google Scholar]
- Hughes MA, Galau GA. Temporally modular gene expression during cotyledon development. Genes Dev. 1989;3:358–369. doi: 10.1101/gad.3.3.358. [DOI] [PubMed] [Google Scholar]
- Ismail AM, Hall AE, Close TJ. Chilling tolerance during emergence of cowpea associate with a dehydrin and slow electrolyte leakage. Crop Sci. 1997;37:1270–1277. [Google Scholar]
- Ismail AM, Hall AE, Close TJ. Purification and partial characterization of a dehydrin involved in chilling tolerance during seedling emergence of cowpea. Plant Physiol. 1999a;120:237–244. doi: 10.1104/pp.120.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AM, Hall AE, Close TJ. Allelic variation of a dehydrin gene cosegregates with chilling tolerance during seedling emergence. Proc Natl Acad Sci USA. 1999b;96:13566–13570. doi: 10.1073/pnas.96.23.13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis SB, Taylor MA, MacLeod MR, Davies HV. Cloning and characterization of the cDNA clones of three genes that are differentially expressed during dormancy-breakage in the seeds of Douglas fir (Pseudosuga menziesii) J Plant Physiol. 1996;147:559–566. [Google Scholar]
- Kazuoka T, Oeda K. Purification and characterization of Cor85-oligomeric complex from cold-acclimated spinach. Plant Cell Physiol. 1994;35:601–611. [Google Scholar]
- Krüger C, Berkowitz O, Stephan UW, Hell R. A metal-binding member of the late embryogenesis abundant protein family transports iron in the phloem of Ricinus communisL. J Biol Chem. 2002;277:25062–25069. doi: 10.1074/jbc.M201896200. [DOI] [PubMed] [Google Scholar]
- Lane AN, Hays LM, Feeney RE, Crowe LM, Crowe JH. Conformational and dynamic properties of a 14 residue antifreeze glycopeptide from Antarctic cod. Protein Sci. 1998;7:1555–1563. doi: 10.1002/pro.5560070709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AN, Hays LM, Tsvetkova N, Feeney RE, Crowe LM, Crowe JH. Comparison of the solution conformation and dynamics of antifreeze glycoproteins from Antarctic fish. Biophys J. 2000;78:3195–3207. doi: 10.1016/S0006-3495(00)76856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince O, Vertucci CW. A calorimetric study of the glass transition behaviors in axes of bean seeds with relevance to storage stability. Plant Physiol. 1995;109:1471–1481. doi: 10.1104/pp.109.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Brawley SH, Close TJ. Proteins immunologically related to dehydrins in fucoid algae. J Phycol. 1998;34:642–650. [Google Scholar]
- Lisse T, Bartels D, Kalbitzer HR, Jaenicke R. The recombinant dehydrin-like desiccation stress protein from the resurrection plant Craterostigma plantagineumdisplays no defined three-dimensional structure in its native state. Biol Chem. 1996;377:555–561. doi: 10.1515/bchm3.1996.377.9.555. [DOI] [PubMed] [Google Scholar]
- McCubbin WD, Kay CM, Lane BG. Hydrodynamic and optical properties of the wheat germ Em protein. Can J Biochem Cell Biol. 1985;63:803–811. [Google Scholar]
- Mtwisha L, Brandt W, McCread L, Lindsey GG. HSP12 is a LEA-like protein in Saccharomyces cerevisiae. Plant Mol Biol. 1998;37:513–521. doi: 10.1023/a:1005904219201. [DOI] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol. 2001;45:263–279. doi: 10.1023/a:1006469128280. [DOI] [PubMed] [Google Scholar]
- Oliver AE, Leprince O, Wolkers WF, Hincha DK, Heyer AG, Crowe JH. Non-disaccharide-based mechanisms of protection during drying. Cryobiology. 2001;43:151–167. doi: 10.1006/cryo.2001.2359. [DOI] [PubMed] [Google Scholar]
- Pace NC, Vadjos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Shalongo W, Stellwagen E. The role of PII conformations in the calculation of peptide fractional helix content. Protein Sci. 1997;6:1694–1700. doi: 10.1002/pro.5560060809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L, Corey RB. Structure of fibrous proteins of the collagen-gelatin group. Proc Natl Acad Sci USA. 1951;37:272–281. doi: 10.1073/pnas.37.5.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragone R, Colonna G, Balestrieri C, Servillo L, Irace G. Determination of tyrosine exposure in proteins by second-derivative spectroscopy. Biochemistry. 1984;23:1871–1875. doi: 10.1021/bi00303a044. [DOI] [PubMed] [Google Scholar]
- Richard S, Morency MJ, Drevet C, Jouanin L, Séguin A. Isolation and characterization of a dehydrin gene from white spruce induced upon wounding, drought, and cold stresses. Plant Mol Biol. 2000;43:1–10. doi: 10.1023/a:1006453811911. [DOI] [PubMed] [Google Scholar]
- Russouw PS, Farrant J, Brandt W, Lindsey GG. The most prevalent protein in a heat-treated extract of pea (Pisum sativum) embryos is an LEA group I protein: Its conformation is not affected by exposure to high temperature. Seed Sci Res. 1997;7:117–123. [Google Scholar]
- Russouw PS, Farrant J, Brandt W, Maeder D, Lindsey GG. Isolation and characterization of a heat-soluble protein from pea (Pisum sativum) embryos. Seed Sci Res. 1995;5:137–144. [Google Scholar]
- Sarhan F, Oullet F, Vazquez-Tello A. The wheat wcs120 gene family: a useful model to understand the molecular genetics of freezing tolerance in cereals. Physiol Plant. 1997;101:439–445. [Google Scholar]
- Siligardi G, Drake AF. The importance of extended conformations and, in particular, the PII conformation for the molecular recognition of peptides. Biopolymers. 1995;37:281–292. doi: 10.1002/bip.360370406. [DOI] [PubMed] [Google Scholar]
- Soulages JL, Arrese EL, Chetty PS, Rodriguez V. Essential role of the conformational flexibility of helices 1 and 5 on the lipid binding activity of apolipophorin-III. J Biol Chem. 2001;276:34162–34166. doi: 10.1074/jbc.M105836200. [DOI] [PubMed] [Google Scholar]
- Soulages JL, Bendavid O. The lipid binding activity of the exchangeable apolipoprotein apolipophorin-III correlated with the formation of a partially folded conformation. Biochemistry. 1998;37:10203–10210. doi: 10.1021/bi980622l. [DOI] [PubMed] [Google Scholar]
- Soulages JL, Kim K, Walters C, Cushman JC. Temperature-induced extended helix/random coil transitions in a group 1 late embryogenesis-abundant protein from soybean. Plant Physiol. 2002;128:822–832. doi: 10.1104/pp.010521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N, Vanyaminov SY, Woody RW. Estimation of the number of α-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N, Woody RW. Poly(pro) II helices in globular proteins: identification and circular dichroic analysis. Biochemistry. 1994;33:10022–10025. doi: 10.1021/bi00199a028. [DOI] [PubMed] [Google Scholar]
- Stapley BJ, Creamer TP. A survey of left-handed polyproline II helices. Protein Sci. 1999;8:587–595. doi: 10.1110/ps.8.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF. Mode of action of the COR15a gene of the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1998;95:14570–14575. doi: 10.1073/pnas.95.24.14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson J, Palva ET, Welin B. Purification of recombinant Arabidopsis dehydrins by metal ion affinity chromatography. Protein Expr Purif. 2000;20:169–178. doi: 10.1006/prep.2000.1297. [DOI] [PubMed] [Google Scholar]
- Swaney JB, Chang BC. Thermal dependence of apolipoprotein-A-I phospholipid recombination. Biochemistry. 1972;19:5637–5644. doi: 10.1021/bi00565a028. [DOI] [PubMed] [Google Scholar]
- Tabaei-Aghdaei SR, Harrison P, Pearce RS. Expression of dehydration-stress-related genes in the crowns of wheatgrass species [Lophopyrum elongatum (Host) A. Love and Agropyron desertorum(Fisch. ex Link.) Schult.] having contrasting acclimation to salt, cold and drought. Plant Cell Environ. 2000;23:561–571. [Google Scholar]
- Tiffany ML. A circular dichroism study of charged polypeptides interaction with salts. Physiol Chem Phys. 1975;7:191–207. [PubMed] [Google Scholar]
- Tiffany ML, Krimm S. New chain conformations of poly(glutamic acid) and polylysine. Biopolymers. 1968;6:1379–1382. doi: 10.1002/bip.1968.360060911. [DOI] [PubMed] [Google Scholar]
- Whitsitt MS, Collins RG, Mullet JE. Modulation of dehydration tolerance in soybean seedlings. Plant Physiol. 1997;114:917–925. doi: 10.1104/pp.114.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody RW. Circular dichroism and conformation of unordered polypeptides. Adv Biophys Chem. 1992;2:37–79. [Google Scholar]
- Wolkers WF, Bochicchio A, Selvaggi G, Hoekstra FA. Fourier transform infrared microspectroscopy detects changes in protein secondary structure associated with desiccation tolerance in developing maize embryos. Plant Physiol. 1998a;116:1169–1177. doi: 10.1104/pp.116.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkers WF, Hoekstra FA. Aging of dry desiccation-tolerant pollen does not affect protein secondary structure. Plant Physiol. 1995;109:907–915. doi: 10.1104/pp.109.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkers WF, McCready S, Brandt WF, Lindsey GG, Hoekstra FA. Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim Biophys Acta. 2001;1544:196–206. doi: 10.1016/s0167-4838(00)00220-x. [DOI] [PubMed] [Google Scholar]
- Wolkers WF, Tetteroo FAA, Alberda M, Hoekstra FA. Protective role of umbelliforerose and sucrose in desiccation tolerant carrot (Daucus carota) somatic embryos. Plant Physiol. 1998b;120:153–164. [Google Scholar]
- Wright PR, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Yeh T, Feeney RE. Antifreeze proteins: structures and mechanisms of function. Chem Rev. 1996;96:601–618. doi: 10.1021/cr950260c. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ohta A, Takagi M, Imai R. Expression of plant group 2 and group 3 lea genes in Saccharomyces cerevisiaerevealed functional divergence among LEA protein. J Biochem. 2000;127:611–616. doi: 10.1093/oxfordjournals.jbchem.a022648. [DOI] [PubMed] [Google Scholar]
- Zhu B, Choi DW, Fenton R, Close TJ. Expression of the barley dehydrin multigene family and the development of freezing tolerance. Mol Gen Genet. 2000;264:145–153. doi: 10.1007/s004380000299. [DOI] [PubMed] [Google Scholar]