Abstract
The model legume Medicago truncatula contains at least six apyrase-like genes, five of which (MtAPY1;1, MtAPY1;2, MtAPY1;3, MtAPY1;4, and MtAPY1;5) are members of a legume-specific family, whereas a single gene (MtAPY2) has closer homologs in Arabidopsis. Phylogenetic analysis has revealed that the proteins encoded by these two plant gene families are more similar to yeast (Saccharomyces cerevisiae) GDA1 and to two proteins encoded by newly described mammalian genes (ENP5 and 6) than they are to mammalian CD39- and CD39-like proteins. Northern analyses and analyses of the frequencies of expressed sequence tags (ESTs) in different cDNA libraries suggest that in roots, leaves, and flowers, the more highly expressed genes are MtAPY1;3/MtAPY2, MtAPY1;3/MtAPY1;5 and MtAPY1;2/MtAPY1;3 respectively. In roots, at least four of the MtAPY1 genes are induced transiently within 3 to 6 h by a stress response that seems to be ethylene independent because it occurs after treatment with an ethylene synthesis inhibitor and also in the skl ethylene-insensitive mutant. This response also occurs in roots of the following symbiotic mutants: dmi1, dmi2, dmi3, nsp, hcl, pdl, lin, and skl. No evidence was obtained for a rapid, transient, and specific induction of the MtAPY genes in roots in response to rhizobia or rhizobial lipochitooligosaccharidic Nod factors. Thus, our data suggest that the apyrase-like genes, which in several legumes have been implicated to play a role in the legume-rhizobia symbiosis (with some members being described as early nodulin genes), are not regulated symbiotically by rhizobia in M. truncatula.
The establishment of the legume-rhizobia symbiosis involves a signal exchange between the specific plant and bacterial couples leading to recognition, nodule organogenesis, and controlled infection. On the bacterial side, plant recognition is determined by a set of genes essential for nodulation and host range that specify the production of lipochitooligosaccharidic signals referred to as Nod factors. At pico-micromolar concentrations, these signals can initiate many of the responses characteristic of the rhizobia themselves such as calcium spiking, root hair deformations, and gene expression (Dénarié et al., 1996; Downie and Walker, 1999). How these LCO signals are perceived by the plant is the subject of much research, and genetic, molecular, and biochemical approaches are being pursued (Cullimore et al., 2001; Geurts and Bisseling, 2002).
One candidate for a role in Nod factor perception is a lectin isolated from roots of Dolichos biflorus called lectin nucleotide phosphohydrolase (Db-LNP). This protein binds Nod factors preferentially from the D. biflorus symbiont and with apparently much higher affinity than chitin fragments (Etzler et al., 1999). Db-LNP is present on the surface of root hairs and relocates to the root hair tips after addition of rhizobia (Kalsi and Etzler, 2000). Antisera raised against the protein inhibit root nodulation by rhizobia. Together these results suggest that Db-LNP plays an important role in nodulation through the binding of Nod factors. A role in nodulation is also supported by work on soybean (Glycine soja) showing that a specific antiserum raised to a similar soybean protein inhibits nodulation of this legume (Day et al., 2000).
Db-LNP is related to a large family of eukaryotic proteins that includes the mammalian CP39- and CD39-like proteins and the yeast (Saccharomyces cerevisiae) protein GDA1 (Roberts et al., 1999). All these proteins contain a highly conserved domain with four regions characteristic of nucleotide phosphohydrolases (Handa and Guidotti, 1996). Where the catalytic activity has been characterized, it can be described as an apyrase (ATP diphosphohydrolases, EC 3.6.1.5.); however, the activity is usually nonspecific for various nucleotide tri- and diphosphates. Apyrase-like proteins have been well studied in animals, where they have been shown to be located either extracellularly, associated with the plasma membranes, the Golgi, or the endoplasmic reticulum and to be involved in such diverse functions as neurotransmission, nucleotide recycling, membrane permeability/transport, and glycosylation (Plesner, 1995; Komoszynski and Wojtczak, 1996). In yeast, GDA1 is essential for correct protein glycosylation and cell wall synthesis (Abeijon et al., 1993).
Before the cloning of Db-LNP, the only two plant sequences belonging to the apyrase family were from potato (Solanum tuberosum; Handa and Guidotti, 1996), and pea (Pisum sativum; Hsieh et al., 1996). The pea protein was originally described as a nuclear ATPase (Matsumoto et al., 1984; Hsieh et al., 1996), but recent studies suggest that it may be associated also with the plasma membrane and involved in mobilizing phosphate from extracellular ATP and in resistance to xenobiotics (Thomas et al., 1999, 2000). It has also been found to be associated with the cytoskeleton (Shibata et al., 1999).
The report that Db-LNP is involved in nodulation in D. biflorus has stimulated recent research on this protein in legumes and now clones and sequences are available from soybean, alfalfa (Medicago sativa), Medicago truncatula, Lotus japonicus, and pea (Roberts et al., 1999; Day et al., 2000; Cohn et al., 2001; Shibata et al., 2001). These sequences have been shown to fall into a legume-specific clade containing Db-LNP and another clade that also contains two Arabidopsis apyrase-like proteins (Roberts et al., 1999). Roberts et al. (1999) have shown that the legume-specific alfalfa and pea proteins bind to hog blood group A + H-Sepharose and, hence, can be referred to as LNPs. In soybean, the legume-specific GS52 protein appears to be located in the plasma membrane, whereas the nonspecific GS50 protein seems to be associated with the endomembranes. GS52 is also induced by rhizobial root inoculation and, therefore, has been termed an early nodulin (Day et al., 2000).
We are interested in the perception of Nod factors, particularly in Medicago spp., and have characterized two Nod factor-binding sites (NFBS1 and NFBS2) in plant extracts (Bono et al., 1995; Gressent et al., 1999). To establish whether the apyrases could represent one of these sites or an additional one, we initiated a study of the apyrase-like genes in the model legume M. truncatula. The small genome, inbreeding genetic system, and ability to transform present a number of advantages for the functional analysis of a gene in this legume (Barker et al., 1990; Cook, 1999). Moreover, expressed sequence tag (EST) databases provide a rich source of clones and expression data (Bell et al., 2001; Journet et al., 2002). A previous study (Cohn et al., 2001) has described four genes in this legume and three of them (Mtapy1, Mtapy3, and Mtapy4) belong to the legume-specific clade and are tightly linked on linkage group 7, whereas Mtapy2 is more similar to the Arabidopsis sequences and is located on linkage group 2. Mtapy1 and Mtapy4, but not Mtapy3, have been shown to be expressed in roots and to be induced within 3 h by inoculation with Sinorhizobium meliloti. Mtapy1 did not seem to be expressed in roots of two symbiotic mutants, dmi1 and pdl, leading to the suggestion that apyrases play a role early in the nodulation response before the involvement of root cortical cell division leading to the nodule structure (Cohn et al., 2001). Our studies provide additional information on the structure and expression of the apyrase gene families in M. truncatula and reveal some fundamental differences to the previously published report.
RESULTS
Characterization of M. truncatula cDNA Clones Related to Apyrases
Using primers to conserved regions of plant apyrase genes, a M. truncatula apyrase fragment of 386 bp was obtained by reverse transcriptase (RT)-PCR to root mRNA. This fragment was used to screen about 300,000 clones from each of two cDNA libraries made from rhizobial-inoculated roots (Szybiak-Strozycka et al., 1995.) or 4-d-old nodules. The nodule library has been used to provide EST sequences for the M. truncatula databases (Journet et al., 2002). Fifty-three clones were identified, some of which were plaque purified and converted into plasmid form. Sequencing of some of these clones and PCR experiments using specific primers on the others resulted in the assignment of the clones to five different genes. A sixth gene was identified from a flower cDNA clone and close analysis of The Institute for Genomic Research (TIGR) M. truncatula databases revealed an additional three clones related to this sequence that had been assigned to a cluster of clones related to one of the other genes. One of the longest cDNA clones of each gene was completely sequenced on both strands and the sequences compared with each other (Table I). Five of the sequences are greater than 70% and 77% identical at the protein and DNA levels, respectively, in the coding regions. The other sequence is less than 63% identical at the protein level but shows up to 73% DNA identity to the other clones in the coding region. Comparison with M. truncatula sequences in the databases revealed that two of the clones corresponded to the Mtapy2 and Mtapy4 genes (Cohn et al., 2001). Two other clones represent genes that had not been described previously (Table II). The remaining two clones were identical in either their 5′ or 3′ moiety to Mtapy1 (Cohn et al., 2001). Inspection of the cluster containing Mtapy1 in the TIGR M. truncatula Gene Index database (Quackenbush et al., 2001) revealed that all the ESTs are homologous to either the 5′ moiety or the 3′ moiety (i.e. they are homologous to one or the other of our cDNA clones), but none of them span a potential fusion site. Thus, our two clones represent two different genes and the Mtapy1 cDNA probably represents a chimera produced during the 3′- and 5′-RACE used to obtain this clone. Together, these data suggest that M. truncatula contains at least six apyrase-like genes (Table II).
Table I.
Percentage sequence identity between the proteins and corresponding cDNA regions of the M. truncatula apyrase-like genes
| MtAPY1;1 | MtAPY1;2 | MtAPY1;3 | MtAPY1;4 | MtAPY1;5 | MtAPY2 | |
|---|---|---|---|---|---|---|
| MtAPY1;1 | — | 85 | 84 | 75 | 70 | 59 |
| — | 91a | 89 | 83 | 78 | 71 | |
| MtAPY1;2 | 85 | — | 91 | 73 | 71 | 63 |
| 91 | — | 94 | 79 | 79 | 73 | |
| MtAPY1;3 | 84 | 91 | — | 73 | 72 | 61 |
| 89 | 94 | — | 81 | 79 | 72 | |
| MtAPY1;4 | 75 | 73 | 73 | — | 70 | 56 |
| 83 | 79 | 81 | — | 77 | 69 | |
| MtAPY1;5 | 70 | 71 | 72 | 70 | — | 60 |
| 78 | 79 | 79 | 77 | — | 71 | |
| MtAPY2 | 59 | 63 | 61 | 56 | 60 | — |
| 71 | 73 | 72 | 69 | 71 | — |
The percentages in italics refer to the cDNA.
Table II.
Nomenclature for genes of the M. truncatula apyrase-like gene families
| New Gene Name | GenBank cDNA Accession Nos. | Corresponding Genea |
|---|---|---|
| MtAPY1;1 | AY180377 | 3′ Moiety of Mtapy1 cDNA |
| MtAPY1;2 | AY180378 | New |
| MtAPY1;3 | AY180379 | 5′ Moiety of Mtapy1 cDNA Mtapy3 (only 3′ sequence available) |
| MtAPY1;4 | AY180380 | Mtapy4 |
| MtAPY1;5 | AY180381 | New |
| MtAPY2 | AY180382 | Mtapy2 |
From Cohn et al. (2001)
Phylogenetic Analysis of Apyrase-Like Sequences
BLAST analyses were used to identify the closest relatives of the M. truncatula apyrase-like proteins from other legumes, Arabidopsis, potato, human (Homo sapiens), bovine (Bos taurus), mouse (Mus musculus), chicken (Gallus gallus), and yeast. All of the proteins identified, including all six M. truncatula ones, are predicted to contain the GDA1/CD39 (nucleoside phosphatase) domain as described in the Pfam database (Bateman et al., 2002). This prediction was validated by alignment of the sequences where the four “apyrase-conserved regions” (Handa and Guidotti, 1996) are clearly highly conserved (data not shown).
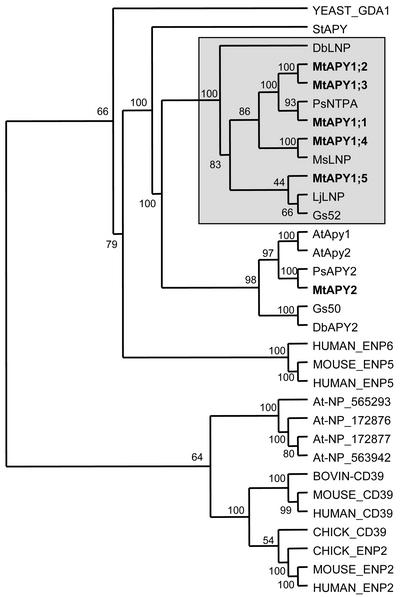
The complete alignment and a truncated alignment (missing the divergent and different length N- and C-terminal regions) were used in various phylogenetic analyses using the PHYLIP package of programs (Felsenstein, 1993). These analyses involved both neighbor-joining and parsimony methods. Only minor variations in tree topology were seen with the different program and the description below is based on conclusions from all the analyses. A representative analysis is shown in Figure 1.
Figure 1.
Phylogenetic tree of various apyrase-related proteins of M. truncatula (MtAPY), Arabidopsis (At), potato (St), soybean (Gs), D. biflorus (Db), pea (Ps), L. japonicus (Lj), chicken, human, bovine, mouse, and yeast. The accession numbers of each sequence are cited in “Materials and Methods.” Alignment of the amino acid sequences was performed using ClustalX and the phylogenetic analysis used the PROTDIST (with 100 bootstrap analyses) and KITSCH neighbor-joining program of the PHYLIP package. The number of times each node is supported by the bootstrap analysis is indicated. The tree is rooted arbitrarily. The MtAPY sequences are shown in bold and the legume-specific clade is boxed.
The M. truncatula proteins described in this article fall into two clades, one of which seems to be legume-specific (as previously pointed out by Roberts et al., 1999) and includes the legume LNPs. This clade contains five of the six M. truncatula sequences. The other clade contains only one M. truncatula sequence and is not legume specific because it also contains the two Arabidopsis sequences previously described as Atapy1 and Atapy2. The relative similarity of the proteins in the legume-specific clade is not clear as shown by the low bootstrap values of some of the nodes, and in most analyses the Db-LNP protein is the most divergent. In the nonlegume-specific family, the pea and M. truncatula proteins are more similar to the Arabidopsis proteins than they are to the soybean and D. biflorus sequences, thus questioning the evolutionary relationship between these proteins. In most analyses the potato protein is the most divergent of the plant apyrase-like sequences.
The phylogenetic analyses also revealed that the closest relatives of the plant apyrase-like proteins are two recently described human and mouse sequences, annotated as ENP5 and ENP6 (ectonucleoside triphosphate diphosphohydrolases), and the yeast protein GDA1. The animal CD39- and CD39-like genes (now renamed as ENP1 and ENP2) are clearly more distantly related and have closer plant relatives in the form of four proteins predicted from the Arabidopsis genome sequencing project. Interestingly, BLAST analysis to M. truncatula EST and other databases did not reveal any close M. truncatula relatives to these proteins.
A Modified Nomenclature for the M. truncatula Apyrase-Like Genes
A phylogenetic analysis using the cDNA sequences related to the plant proteins described above revealed a similar phylogenetic tree topology to the tree obtained with the protein sequences (data not shown). Therefore, the classification of the M. truncatula apyrase-like sequences into two clades is supported by analyses at both the protein and DNA levels and also by the high bootstrap values obtained.
Due to the clear separation of the M. truncatula genes and their encoded proteins into two phylogenetic classes, it is proposed to name these two type of genes APY1 (the legume-specific class) and APY2, with the members of the APY1 class being designated as members of a multigene family (Table II). This nomenclature follows the guidelines of the Commission on Plant Gene Nomenclature (Price and Reardon, 2001) and the guidelines for genetic nomenclature for M. truncatula (VandenBosch and Frugoli, 2001). Moreover, it involves a minimum number of changes to the names given by Cohn et al. (2001), but eliminates the problem caused by their potentially chimeric cDNA clone. The prefix Mt for M. truncatula is used to distinguish between similarly named but not necessarily orthologous genes of Arabidopsis and other plant species.
Differential Organ Expression of the M. truncatula Apyrase-Like Genes by Northern and in Silico Analysis
An “in silico” analysis of the frequencies of specific ESTs in different libraries in the M. truncatula databases can be used to elucidate gene expression patterns (Journet et al., 2002). Such an analysis was performed on the 134,477 M. truncatula ESTs available in the September 2001 version of the Functional Genomics in M. truncatula web site (Journet et al., 2002) from the sequencing of clones from 31 different cDNA libraries. BLAST interrogation of the GenBank Medicago ESTs revealed 116 sequences related to the apyrase-like cDNAs. Twenty-five of them were considered redundant because they had homologs in the same bank starting at the same nucleotide and, hence, may have arisen during amplification of the libraries used for sequencing. The frequency of expression of each gene in 12 different conditions (see Web site of Journet et al., 2002) was calculated from the number of specific and total ESTs in banks grouped into these conditions (Table III). Three of the genes, MtAPY1;1, MtAPY1;2, and MtAPY1;4, appear to be poorly expressed with only one to three ESTs each. Interestingly all three MtAPY1;2 ESTs arose from flower libraries, whereas two of the three MtAPY1.1 ESTs came from roots infected with arbuscular mycorrhizae. The MtAPY2 gene, with 14 ESTs, seemed to be expressed in many organs but with a relatively higher number of ESTs from the stem library. The MtAPY1;3 gene shows the highest level of expression with 38 representatives. This gene and MtAPY1;5 show a remarkable induction in leaves after insect herbivory stress (18 and 26 ESTs from this library), which is not seen in leaves infected by a fungus. MtAPY1;3 was the only other gene with representatives (three) in the flower library, whereas MtAPY1;5 was relatively overrepresented in the stem libraries with four ESTs. Only two ESTs were found in libraries made predominantly from nodule tissue (one each of MtAPY1;3 and MtAPY2), suggesting that the genes are poorly expressed in this organ. No evidence was obtained for induction by phosphate starvation (data not shown).
Table III.
In silico expression analysis of the M. truncatula apyrase-like genes by EST analysis
| Origin of Banksa | No. of Banks | No. of ESTs | Specific Gene ESTs as Normalized No.b(or Actual No.)
|
|||||
|---|---|---|---|---|---|---|---|---|
| MtAPY1;1 | MtAPY1;2 | MtAPY1;3 | MtAPY1;4 | MtAPY1;5 | MtAPY2 | |||
| Roots (various) | 9 | 23,885 | 0 | 0 | 0.04 (1) | 0 | 0 | 0.08 (2) |
| Roots (mycorrhizal) | 2 | 12,246 | 0.02 (2) | 0 | 0 | 0 | 0 | 0.04 (3) |
| Roots (rhizobial) | 3 | 9,116 | 0 | 0 | 0.44 (4) | 0.11 (1) | 0 | 0.11 (1) |
| Nodules | 5 | 17,172 | 0 | 0 | 0.06 (1) | 0 | 0 | 0.06 (1) |
| Stems | 1 | 10,314 | 0 | 0 | 0.10 (1) | 0 | 0.39 (4) | 0.48 (5) |
| Leaves (various) | 3 | 19,008 | 0 | 0 | 0.32 (6) | 0 | 0.05 (1) | 0 |
| Leaves (fungal) | 1 | 6,003 | 0 | 0 | 0.17 (1) | 0 | 0 | 0 |
| Leaves (insect) | 1 | 9,921 | 0 | 0 | 1.81 (18) | 0 | 2.61 (26) | 0.10 (1) |
| Plantlets | 2 | 9,444 | 0.11 (1) | 0 | 0.33 (3) | 0 | 0 | 0 |
| Flowers | 1 | 3,404 | 0 | 0.88 (3) | 0.88 (3) | 0 | 0 | 0 |
| Seeds/pods | 3 | 5,038 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cell cultures | 1 | 8,926 | 0 | 0 | 0 | 0 | 0.11 (1) | 0.11 (1) |
| Total | 32 | 134,477 | 0.022 (3) | 0.022 (3) | 0.281 (38) | 0.007 (1) | 0.236 (32) | 0.103 (14) |
The names and origins of the banks are described in “Materials and Methods.”
Expressed as no. of specific ESTs per 1,000 ESTs.
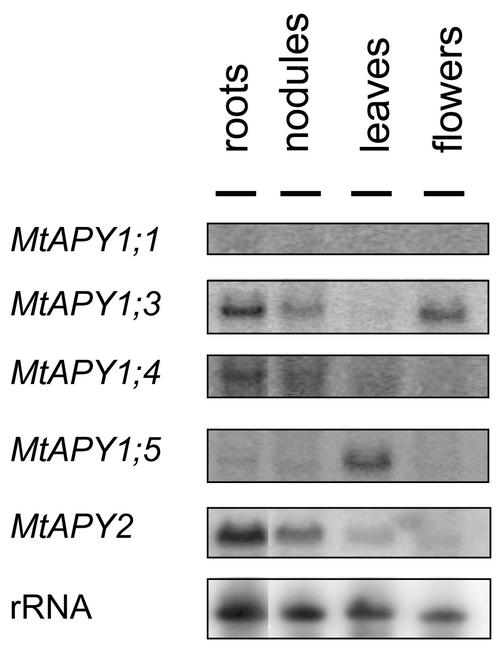
Northern analysis of root, nodule, leaf, and flower RNA was used to validate part of the “in silico” analysis (Fig. 2). For this experiment, specific probes corresponding to the 3′-untranslated regions of the cDNAs were used, which in Southern experiments with the different apyrase clones were shown to be gene specific (data not shown). Analysis of the abundance of mRNA of each individual MtAPY gene confirmed that MtAPY1;3 and MtAPY2 are generally the more highly expressed genes. The mRNA of MtAPY1;1 and MtAPY1;4 could hardly be detected by northern analysis, and although evidence was obtained that MtAPY1;2 is expressed in flowers (data not shown), the probe for this gene also cross hybridized with MtAPY1;3. In the four organs examined, the northern analysis confirmed that MtAPY1;3 is relatively highly expressed in flowers compared with the other genes and that MtAPY1;5 has a very specific and relatively high expression in leaves (Fig. 2). In roots and nodules, MtAPY1;3 and MtAPY2 were the more highly expressed genes as predicted even from the low number of apyrase-like ESTs from these organs.
Figure 2.
Northern analysis of the expression of the M. truncatula apyrase-like genes in various organs. Roots, nodules, leaves, and flowers were harvested from aeroponically grown plants and used for RNA extraction. Ten micrograms of total RNA was run on agarose gels, blotted onto nylon membranes, and hybridized with radiolabeled specific 3′ probes from the cDNAs. The blots were exposed for different times depending on the level of hybridization.
The Apyrase Genes Are Induced in Roots But Not Specifically by Rhizobia or Rhizobial Nod Factors
Cohn et al. (2001) have shown that certain M. truncatula apyrase genes are transiently induced within 6 h postinoculation with S. meliloti and have stated that data using rhizobial mutants suggest that only rhizobia that are able to produce Nod signals are able to induce the transcription of these genes. To directly investigate whether addition of Nod factors induces the apyrase-like genes of M. truncatula, we grew plants in aeroponic containers, starved them of a fixed nitrogen source for 4 d, and then added 10−9 m Nod factors to the root systems. To assess the potential diurnal variation in expression of the apyrase-like genes, some plants were harvested at various times during the day before the Nod factor addition. The expression of the apyrase-like genes were analyzed by northern analysis using a probe that cross hybridizes well with all the M. truncatula apyrase genes of the legume-specific (APY1) family (Fig. 3). Surprisingly, a strong and transient increase in APY1 mRNA was observed 6 to 8 h after the start of the harvesting without addition of Nod factors. The following day when Nod factors were added, an increase was also observed but was weaker than before. In contrast, two early nodulin genes, MtENOD11 (Journet et al., 2001) and MtRip1 (Cook et al., 1995), showed induced expression only after addition of Nod factors and not during the preceding day. It is noteworthy that in accordance with previous studies, MtRip1 has a much higher basal level of expression than MtENOD11, but for both genes the effect of Nod factor addition was clear. In other experiments when separate aeroponic chambers were used, both control and Nod factor-treated plants showed a similar transient induction of APY1 genes, peaking at about 6 h after the start of harvesting (data not shown), whereas MtENOD11 was induced only after addition of Nod factors. Thus APY1 genes show a transient induction response that does not require Nod factors.
Figure 3.
Effect of Nod factors on the expression of the M. truncatula APY1 genes in roots of aeroponically grown plants. Samples were taken at various hours during Day 4 after nitrogen starvation and then at Day 5 after addition of 10−9 m NodSm factors (+NF). T = 0 refers to the first harvest of the day (at 10 am). Northern analysis was performed on blots, prepared as described in Figure 2, and hybridized with a probe for the MtAPY1 genes. The expression of the nodulins ENOD11 and Rip1 was used as controls for Nod factor induction.
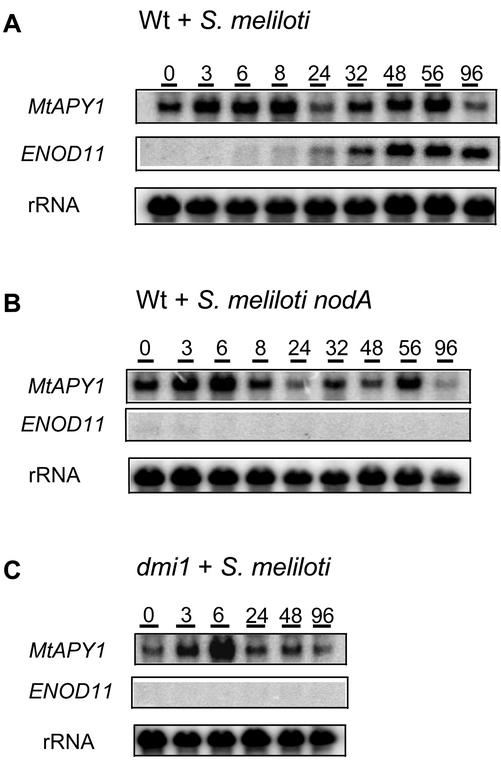
Thus, we decided to test whether, in our system, the apyrase-like genes are induced by S. meliloti as proposed by Cohn et al. (2001) and to see whether such an induction was dependent on the initiation of the symbiosis. The expression of MtENOD11 was used as a control for symbiotic induction. Using wild-type plants with a wild-type S. meliloti strain, MtENOD11 induction was observed at 6 h postinoculation, with its mRNA increasing in abundance up to the end of the experiment (96 h postinoculation). When one of the two partners was incapable of forming the symbiosis (the S. meliloti nodA mutant or the M. truncatula dmi1 mutant), no induction of this gene was observed. However, the APY1 mRNA was transiently induced by 6 h postinoculation in all three plant-bacterial couples and, thus, occurred irrespective of the initiation of the symbiosis (Fig. 4). Therefore, in our aeroponic chambers the level of apyrase mRNA does increase within 6 h of the start of harvesting, but this induction is independent of the addition of either rhizobia or Nod factors.
Figure 4.
Effect of rhizobial inoculation on the expression of the M. truncatula APY1 genes in roots of aeroponically grown plants. Wild-type (Wt) or mutant dmi1 plants were starved of nitrogen for 4 d and then inoculated with either wild-type or nodA S. meliloti at T = 0. Samples were harvested at various hours after inoculation. Blots were prepared as described in Figure 2 and hybridized with a probe for the MtAPY1 genes or for ENOD11.
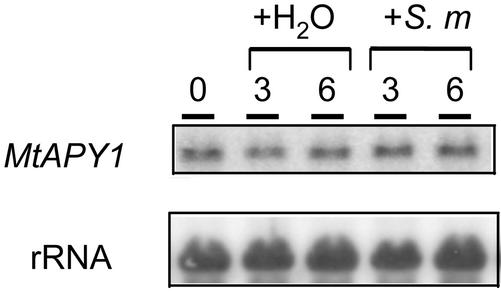
In the final experiment of this series, plants were grown in growth pouches and inoculated with rhizobia or water mock inoculated, taking care to disturb the plants as little as possible during the experimentation. The roots from several pouches were harvested either 3 or 6 h later (Fig. 5). In this experiment, no change in the level of APY1 mRNA was seen by 6 h after treatment, indicating that the apyrase genes are not appreciably induced by rhizobia during this time frame.
Figure 5.
Effect of rhizobial inoculation on the expression of the M. truncatula APY1 genes in roots of plants grown in growth pouches. Plants were grown in growth pouches without a nitrogen source for 8 d before inoculation with S. meliloti or mock inoculation with water and roots were harvested 3 or 6 h later. Blots were prepared as described in the legend to Figure 2 and hybridized with a probe for the MtAPY1 genes.
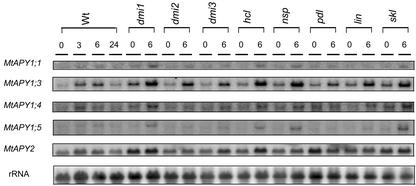
To examine whether all the MtAPY genes are subject to the induction response we measured, in an induction experiment with wild-type plants, the abundance of mRNA of each individual MtAPY1 gene by northern analysis using the gene-specific 3′-untranslated region probes (Fig. 6). The results indicated that all four of the tested MtAPY1 genes show an induction response; however, the mRNA of the MtAPY1;3 gene was clearly severalfold more abundant in roots than the mRNA of the other MtAPY1 genes. Cohn et al. (2001) showed induction of MtAPY1;1 (Mtapy1) and MtAPY1;4 (Mtapy4) genes but failed to detect MtAPY1;3 (Mtapy3) mRNA in roots. However, we found that the 3′ primer used for MtAPY1;3 detection in their RT-PCR approach is beyond the polyadenylation point of the mRNAs that we have detected for this gene in root cDNA clones and, thus, would not detect all the mRNA derived from this gene.
Figure 6.
Expression of the apyrase-like genes in roots of aeroponically grown symbiotic mutants of M. truncatula. Wild-type (Wt) or various mutant plants were starved of nitrogen for 4 d and then treated with 10−9 m NodSm factors and samples were harvested at T = 0, 3, 6, and 24 h. For the mutants, only the T = 0 and 6 h samples are shown. Blots were prepared as described in Figure 2 and hybridized with specific 3′ probes from the various APY1 or the APY2 cDNAs.
Cohn et al. (2001) have also reported that two nodulation deficient mutants of M. truncatula, dmi1 and pdl, do not express apyrases to any detectable level, whereas another mutant, lin, showed wild-type basal levels and rhizobial induction of apyrase mRNA. We used these three mutants and several other symbiotic mutants (Table IV) to look at both basal levels of apyrase mRNA in roots and the levels of apyrase mRNA at 6 h during an induction experiment. It was clear from this experiment that MtAPY1;3 is the most highly expressed gene in the roots of all the mutants and that all the mutants showed the induction response (Fig. 6). Although the level of abundance of the mRNA corresponding to the MtAPY1;1, MtAPY1;4, and MtAPY1;5 genes were toward the limit of detection, these genes also appeared to be expressed in all the mutants and to show the induction response. The level of mRNA of MtAPY1;2 could not be detected specifically. The MtAPY2 gene was clearly the second most highly expressed apyrase gene in roots of all the mutants and did not show a consistent induction response, in accordance with the results of Cohn et al. (2001). Thus, our experiments did not show any clear differences in the expression of the apyrase genes in any of the symbiotic mutants; all the mutants, even those that are deficient in almost all symbiotic responses, showed the induction phenomenon that appears to occur for MtAPY1;1, MtAPY1;3, MtAPY1;4, MtAPY1;5, and not MtAPY2.
Table IV.
Plant lines and rhizobial strains used in this study
| Designation | Line/Strain | Relevant Characteristicsa | Reference/Source |
|---|---|---|---|
| M. truncatula | |||
| Wild type | Jemalong A17 | Nod+Fix+ with S. meliloti | Penmetsa and Cook (1997) |
| dmi1 | Y6 | Has+, Inf−, Ccd−, and Nod− | Penmetsa and Cook (1997); Catoira et al. (2000) |
| dmi2 | TR25 | Has+, Inf−, Ccd−, and Nod− | Catoira et al. (2000) |
| dmi3 | TRV25 | Has+, Inf−, Ccd−, and Nod− | Catoira et al. (2000) |
| hcl | B56 | Hab+, Inf−, Ccd+, and Nod− | Catoira et al. (2001) |
| nsp | B85 | Hab+, Inf−, Ccd−, and Nod− | Catoira et al. (2000) |
| pdl (Poodle) | BC1F4 | Hab+, Inf+, Ccd−, and Nod− | R.V. Penmetsa and D.R. Cook (unpublished data) |
| lin (Lumpy infections) | M4 | Hab+, Inf+/−, Ccd+, and Nod− | R.V. Penmetsa and D.R. Cook (unpublished data) |
| skl | BC1F4 | Nod+++, ethylene insensitive | Penmetsa and Cook (1997) |
| S. meliloti | |||
| Wild type | GMI6526 | 2011(pXLGD4), Nod+Fix+ on M. truncatula | Ardourel et al. (1994) |
| NodA | GMI6702 | 2011nodA::Tn5#2208(pXLGD4). Hab−, Inf−, Ccd−, and Nod−on M. truncatula | Debellé et al. (1986) |
Responses to rhizobia: root hair swelling (Has) or branching (Hab), infection threads (Inf), cortical cell divisions (Ccd), and nodulation (Nod).
The Induction of Apyrases in Roots Is Stress-Related But Is Not Mediated by Ethylene
The previous experiments have shown a rapid induction of APY1 mRNA in the aeroponic chambers that occurs independently of rhizobial addition. Several experiments were carried out to investigate this response further.
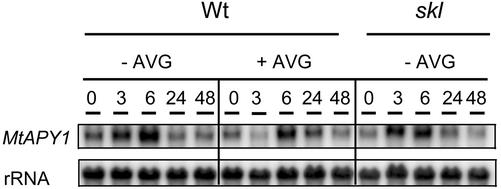
In the first experiment, we investigated whether this response was mediated by ethylene. Two mechanisms were used to inhibit ethylene responses: Either the ethylene synthesis inhibitor, l-alpha-(2-aminoethoxyvinyl)-Gly (AVG), was included in the medium of wild-type plants, or the mutant skl was used that is ethylene insensitive (Penmetsa and Cook, 1997). The results in Figure 7 show that the level of APY1 mRNA increased by 6 h whether or not ethylene signaling had been inhibited; thus, the induction response is independent of ethylene.
Figure 7.
Role of ethylene in the induction of the M. truncatula APY1 genes in roots of aeroponically grown plants. Wild-type (Wt) plants grown either in the presence or absence of an ethylene inhibitor (AVG) or plants of the mutant defective in ethylene perception (skl) were starved of nitrogen for 4 d and harvesting was started at 10 h (T = 0). Northern analysis was performed on blots prepared as described in Figure 2 and hybridized with a probe for the MtAPY1 genes.
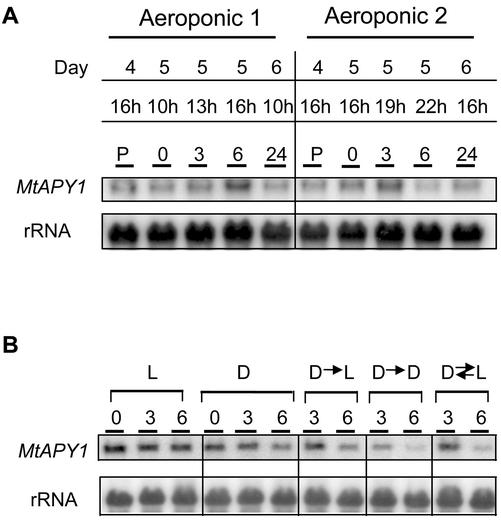
Another possible reason for the increased expression of the APY1 genes during our experiments could be an endogenous rhythm. In all our experiments, plants were grown in a 16-h-light/8-h-dark regime and the zero time point was taken at 10 am Greenwich Mean Time (10 h), 6 h into the light period. In the following experiment, two aeroponic chambers were used (Fig. 8A). The first samples were harvested at 4 pm (corresponding to a normal 6-h induced time), and then the next day more samples were harvested at 0, 3, 6, and 24 h, starting at either 10 am or at 4 pm. (In this experiment, care was taken not to disturb unduly the root systems to be harvested at later time points.) The results show that the level of APY1 mRNA is not at the high level at 4 pm unless preceded by other harvests. Moreover, although an increase in APY1 mRNA was observed at the 3-h time point when harvesting was started at 4 pm, this was not maintained at 6 h. One reason for this result could be that by this time the plants were already in the dark period; thus, the induction response could be influenced by light.
Figure 8.
Effect of time of harvesting and the light regime on the expression of the M. truncatula APY1 genes in roots. A, Aeroponically grown plants were starved of nitrogen for 4 d and the roots harvested at different times during the daylight periods of 16-/8-h diurnal cycles (light period from 4–20 h). The first harvest on Day 5 is T = 0. B, Plants were grown in diurnal cycles (16/8 h) in growth pouches with the root systems either in the light (L) or the dark (D). At the start of harvesting, the exposure to light of some of the plants with root systems in the dark was altered: transfer of root systems to light (D-L), transfer of shoots to dark (D-D), and transfer of roots systems to light and shoots to dark (D ⇌ L). Roots were harvested at 3 and 6 h starting at 10 h (T = 0). Northern analysis was performed on blots prepared as described in Figure 2 and hybridized with a probe for the MtAPY1 genes.
To investigate this possibility further, plants were grown in growth pouches in a 16-h-light/8-h-dark regime with their root systems either exposed to the light or not (Fig. 8B). Root systems were then harvested at 10 am (T = 0) and 3 and 6 h later and subjected to northern analysis. It is clear that after prolonged growth with the root systems in the light the level of root APY1 mRNA is higher than if the roots are kept in the dark. Moreover, the level of APY1 mRNA did not increase during the day of harvesting; in fact, the dark-grown root systems seemed to decrease at the 6-h time point. If the roots of plants with dark-grown root systems were exposed to the light at 10 am, no increase in APY1 mRNA was observed either 3 or 6 h later, thus suggesting that the induction response is not due to short-term exposure of the root systems to light. If similarly grown plants were covered in black plastic to also place the shoots in the dark, the level of APY1 mRNA decreased even further at the 6-h time point. Finally, if the roots were switched to the light and the shoots to the dark, the level of the mRNA again decreased by 6 h. These experiments show that exposure of the shoots or the roots to light does have some influence on the level of root APY1 mRNA, but that it does not lead to short-term increases and, therefore, probably is not the cause of the induction of the APY1 mRNA in the aeroponic chambers.
DISCUSSION
The report that a D. biflorus apyrase-like protein (Db-LNP) binds Nod factors and that antibodies raised against it inhibit root nodulation (Etzler et al., 1999) suggests that this protein plays an important role in the establishment of the symbiosis between this legume and rhizobia. If these proteins play an essential rather than facultative role in nodulation, it would be expected that Db-LNP orthologs play similar roles in other legume species. Day et al. (2000) have shown that apyrase-specific antisera inhibit nodulation in soybean and that a soybean apyrase-like gene is induced early during nodulation. However, D. biflorus and G. soja both belong to the Phaseoleae and produce determinate nodules in which cell division begins in the outer cortex. Many other species of importance to temperate agriculture (for example, those belonging to the genera Medicago, Pisum, Vicia, and Trifolium) are taxonomically more distant and produce indeterminate nodules in which cell division begins in the inner cortex and pericycle (Gualtieri and Bisseling, 2000). Thus, it is important to establish whether apyrase-like proteins play an important symbiotic role in these species. Among these species is the model legume M. truncatula, in which genetic and genomic tools have been developed (Cook, 1999).
As a first step to a functional analysis of the role of apyrase-like proteins in indeterminate nodulation, we have characterized the gene families encoding these proteins in M. truncatula. Through isolation of cDNA clones from root and nodule libraries, we have identified five genes related to plant apyrase-like proteins and an additional gene has been identified from the M. truncatula EST databases. Phylogenetic analysis has revealed that the proteins encoded by five of these genes fall into the legume-specific class of sequences identified by Roberts et al. (1999), whereas the sixth gene is more similar to two Arabidopsis proteins. We have confirmed and refined the position of one of the legume-specific genes on linkage group 7 (A. Kereszt, A. Niebel, J. Cullimore, and T. Huguet, unpublished data), which has been shown previously to be part of a cluster of four tightly linked genes, whereas the other gene is located on linkage group 2 (Cohn et al., 2001). Based on the phylogenetic analysis and in accordance with the guidelines for naming plant genes (Price and Reardon, 2001), we propose to denote the five genes as members of the MtAPY1 gene family (MtAPY1;1, 1;2, 1;3, 1;4, and 1;5), whereas the other gene is referred to as MtAPY2. APY is preferred as the gene mnemonic to LNP because APY is the precedent to LNP in the literature and the four regions of the apyrase domain are clearly very highly conserved (data not shown), whereas the position of the putative lectin domain has yet to be defined.
Do these six genes represent the total number of apyrase-like genes in M. truncatula? This question may only be definitively answered by the complete sequencing of the whole genome. Cohn et al. (2001) identified four of these genes by a combination of library screening, RACE experiments, and analysis of the M. truncatula EST banks. The possibility raised here that the gene they named Mtapy1 may be based on a chimeric clone of MtAPY1:1 and MtAPY1;3 illustrates the dangers of using RACE to elongate clones related to multigene families. By renaming the genes as members of the MtAPY1 multigene family, we have kept as close as possible to their preceding nomenclature while eliminating the confusion caused by their potentially chimeric clone. Close analysis of the current EST banks (TIGR version 5.0) has revealed that there may be an additional APY1 and an APY2 gene. Moreover, a 6-bp deletion near the encoded C terminus in several clones that are otherwise identical may suggest that there is an additional gene closely related to MtAPY1;1. However, at present we cannot exclude that these variants have arisen either by posttranscriptional modification or are alleles due to polymorphisms in the seed stocks. In pea, which is closely related to Medicago spp., two different types of cDNA and genomic clones differing only in six point mutations near the N terminus have been identified, but again, whether they are alleles or different genes is not clear (Shibata et al., 2001). These clones are interestingly putative orthologs to MtAPY1;1, suggesting that this gene may be particularly susceptible to variation.
In addition to the MtAPY1;1 and pea gene orthologs, it is also clear that the alfalfa gene named MsLNP (Roberts et al., 1999) is the ortholog of MtAPY1;4 (Fig. 1). In relation to the aim of this work, this analysis failed to discover a clear single ortholog of Db-LNP in M. truncatula, which would be the likeliest candidate to play a similar symbiotic role. At present, studies of the symbiotic role of Db-LNP-related genes in M. truncatula would need to include all five members of the MtAPY1 family.
The phylogenetic analyses (Fig. 1) also revealed that the closest mammalian homologs of the plant apyrases are not CD39- and CD39-like genes (now named ENP1 and ENP2) as previously described (Roberts et al., 1999) but are a recently reported group of apyrase-like proteins from rat, human, and mouse named ENP5 and ENP6 (Chadwick and Frischauf, 1998). Comparative studies of these related proteins may lead to new ideas of their function in different organisms.
Another way in which a potential symbiotic function of a gene is inferred is by its expression pattern. In this respect, it is interesting to note that the soybean GS52 gene has been described as an early nodulin gene (Day et al., 2000), and that two of the M. truncatula genes, Mtapy1 (MtAPY1;1 or 1;3) and Mtapy4 (MtAPY1;4) have been reported to be induced by rhizobia (Cohn et al., 2001). In this latter paper, transient increases in the abundance of mRNA of the two genes were observed at 3 and 6 h postinoculation and data not shown suggested that this increase was dependent on the ability of the rhizobia to produce Nod signals. Such an expression pattern could be consistent with the encoded protein acting in Nod signal perception because there are examples in the literature of receptor genes being induced by their ligands (Tata, 2000; Beuschlein et al., 2001).
In this paper, we have confirmed that the abundance of mRNA of the MtAPY1 genes increases transiently in roots after inoculation with rhizobia in aeroponic chambers; an increase was seen at 3 and 6 h and then decreased by 24 h (Fig. 4). These results are similar to those obtained by Cohn et al. (2001), who also used aeroponic chambers for some of their experiments. However, we found that no such response to rhizobia was seen if the plants were grown in growth pouches (Fig. 5). Further analysis of this surprising result showed that the response in aeroponic chambers was not dependent on Nod factors (Fig. 3), the ability of the rhizobia to produce Nod factors (Fig. 4), or the addition of the rhizobia itself: A similar response occurred without any additions. Thus, we have not found any evidence to support the notion that the expression of the apyrase-like genes in M. truncatula are induced transiently by rhizobia or by Nod factors in the early stages of the legume-rhizobia symbiosis. Therefore, our studies do not support a description of MtAPY1;1 and MtAPY1;4 as ENOD (early nodulin) genes.
We also examined the levels of apyrase-like mRNA and the induction response in roots of a number of symbiotic mutants (Fig. 6). Some of these are mutated in components of a Nod factor signal transduction pathway (dmi1, dmi2, and dmi3) and others have later defects in nodulation (hcl, nsp1, pdl, lin, and skl). The results showed that there was no great difference in the levels of MtAPY1 mRNA in roots of any of the mutants compared with wild type and that the induction response occurred in all the mutants. Although the mRNA of some of the individual MtAPY1 genes was near the limit of detection, it seems that at least four of the MtAPY1 genes show an induction response and that this occurred in all the mutants. Thus, most of the MtAPY1 genes are affected by the induction response and studies with plant mutants suggests that this induction occurs independently of the establishment of the symbiosis. Our results are in contrast to those of Cohn et al. (2001), who failed to detect MtAPY1 mRNA in the dmi1 and pdl mutants and suggested that MtAPY1 expression is correlated with the formation of nodule primordia. We have no explanation for the discrepancy between the two studies.
If the induction response is not due to rhizobia or rhizobial Nod factors, what could it be due to? The following possibilities were examined: (a) an endogenous rhythm/diurnal variation, (b) exposure of the roots to light, or (c) stress related to the harvesting of multiple samples from the aeroponic chambers. Because the higher level of mRNA is not observed at different times of the day unless it is preceded by at least one harvest, the response does not appear to be due either to an endogenous rhythm or diurnal variation. Moreover, in plants grown in pouches short term (3–6 h), exposure of the roots to light did not lead to an increase in the APY1 mRNA levels, suggesting that light is not the key factor triggering the induction although it may modulate the degree or maintenance of the response (Fig. 8). Together, these results suggest that the induction response seems to be related to the harvesting of multiple samples from the aeroponic chambers.
In most experiments, the plants were densely planted in the aeroponic chambers, and it is possible that harvesting of some samples caused physical damage to the remaining roots; hence, the APY1 gene induction could be a direct wounding response. Consistent with this possibility is the fact that APY1;3 and APY1;5 ESTs are highly overrepresented in the leaf herbivory library compared with leaf libraries prepared from either pooled developmental stages, phosphate-starved plants, or fungus-infected plants (Table III), suggesting that the APY1 genes may be induced by tissue damage. However, in Figure 8A care was taken not to physically damage the remaining plants in the aeroponic systems and yet induction still occurred, albeit at a lower level. Alternative explanations are that the induction is due to mechanostimulation (see Johnson et al., 1998) due to movement of the root systems in the aeroponic containers during harvesting, or induction by volatiles (see Baldwin et al., 2002) released from the harvested plants. Whatever the stimulus, it is clear that the induction response is independent of ethylene because induction occurred both in wild-type plants treated with an inhibitor of ethylene synthesis and in skl mutant plants that are insensitive to ethylene (Fig. 7). Ethylene independence is also a characteristic of certain responses to mechanostimulation (Johnson et al., 1998) and elicitation of some defense genes (Yamamoto et al., 1999).
Expression via both EST and northern analysis have shown that, in the conditions studied, MtAPY1;3, MtAPY1;5, and MtAPY2 are clearly more highly expressed than MtAPY1;1, MtAPY1;2, and MtAPY1;4, whose mRNAs are rare (less than 0.003% of total ESTs). MtAPY2 mRNAs have been detected in most organs studied; hence, this gene shows a rather constitutive expression, whereas the MtAPY1 genes show a more differential organ expression (MtAPY1;3 and MtAPY1;2 are higher in flowers and MtAPY1;5 is higher in leaves). MtAPY1;3, with MtAPY2, is the most highly expressed MtAPY gene in roots, in contrast to the results of Cohn et al. (2001) who failed to detect MtAPY1;3 mRNA in this organ. The observation that all five MtAPY1 genes can be transiently induced in roots suggests that this ethylene-independent stress-like response is imposed on other controls regulating the qualitative and quantitative expression of these genes. What is the purpose of this response? Our data have shown clearly that it is not specific to the rhizobial symbiotic interaction and, thus, may have a role in other processes. Thomas et al. (2000) have shown that Arabidopsis plants overexpressing apyrases are more resistant to xenobiotics; hence, one possibility is that a stress induction may be in anticipation of toxin exposure.
In conclusion, M. truncatula contains at least five (MtAPY1) genes, which from phylogenetic analysis are members of a legume-specific clade including the D. biflorus Db-LNP. In addition, there is a MtAPY2 gene that is more similar to Arabidopsis apyrase-like genes. Expression studies have shown that the MtAPY1 genes are induced in roots by stress and that this response is not mediated by ethylene. No evidence has been found for a transient induction of these genes by rhizobia or Nod factors. Whether these genes could, however, play a role in the establishment of the symbiosis, independently of a symbiotic-specific regulation of expression, is under investigation.
MATERIALS AND METHODS
Growth Conditions and Plant Material
All plant lines and bacterial strains are listed in Table IV. Medicago truncatula Gaernt. cv Jemalong lines A17 or J5 (these lines are probably identical; T. Huguet, personal communication) were grown in aeroponic chambers (unless otherwise indicated) in a 16-h photoperiod on a medium containing 5 mm NH4NO3 for 2 weeks essentially as described by Journet et al. (2001). The media were then replaced by similar media lacking a nitrogen source, and the plants were then starved of nitrogen for 4 d before either inoculation with Sinorhizobium meliloti (Table IV), treatment with 10−9 m NodSm factors, or other treatments. Plant mutant lines were obtained from Prof. D. Cook (University of California, Davis), Dr. G. Duc (INRA, Dijon, France), or Dr. J. Dénarié (INRA, Toulouse, France) as indicated in Table IV and grown in the same way. Treatment with the ethylene inhibitor AVG involved the addition of 0.88 μm AVG 17 h before the first harvest. Note that the plants for different time points of an experiment were grown in the same aeroponic chamber; thus, all the root systems were exposed to light and potential stress during the few minutes of harvesting. In most experiments, about 150 plants were grown in an area of 0.16 m2 and roots corresponding to at least five plants were harvested for each sample and frozen immediately in liquid nitrogen and stored at −80°C. In the experiment in Figure 8A, the plants were grouped in five lots of 25 plants and one lot was harvested per time point. In some experiments (Figs. 5 and 8B), plants were grown in growth pouches for 8 d using media lacking a nitrogen source essentially as described by Vernoud et al. (1999). After the various treatments, roots of plants from several whole pouches were harvested for each time point.
Isolation and Characterization of cDNA Clones
RT-PCR was used to obtain a fragment corresponding to nucleotides 173 through 559 of MtAPY1;3, and this fragment was radiolabeled with 32P and used to screen about 300,000 plaque-forming units of each of two M. truncatula cDNA libraries prepared, in the Lambda ZAP vector (Stratagene, La Jolla, CA), from either 4-d old nodules or from roots inoculated with S. meliloti and harvested 6, 24, and 48 h later. The phage were eluted from agar plugs containing the hybridizing phage, and these were used either for purification of the hybridizing phage or for PCR analysis. Purified positive phage were converted by in vivo excision to plasmid form into the vector pBluescript SK− (Stratagene) and characterized either by PCR using specific gene primers or sequencing. At least one cDNA clone encoding full-length proteins of each of the identified six apyrase-like genes was sequenced on both strands.
Sequence and Phylogenetic Analyses
Sequence data were analyzed using the GCG software (Genetics Computer Group, Madison, WI), and derived protein sequences were analyzed using the SwissProt Expasy server. Sequences showing homology were identified using the National Center for Biotechnology Information BLAST server (Altschul et al., 1997) and recovered from either the GenBank or SwissProt databases. The following sequences (with their accession nos.) were used: PsNTPA (P52914); MsLNP (AAF00611); LjLNP (AAF00609); Gs52 (AF207688); Atapy1 (AF093604); Atapy2 (AAF66599); PsAPY2 (AB071370); Gs50 (AF207687); DbLNP (AAD31285); DbAPY2 (AAF00610); StAPY (P80595); YEAST_GDA1 (P32621); HUMAN_ENP6 (O75354); HUMAN_ENP5 (O75356); MOUSE_ENP5 (Q9W429); At-NP_565293, At-NP_172876, At-NP_172877, At_NP_563942, and BOVIN_CD39 (O18956); MOUSE_CD39 (P55772); HUMAN_CD39 (P49961); CHICK_CD39 (O93295); CHICK_ENP2 (P79784); HUMAN_ENP2 (O55026); and MOUSE_ENP2 (O55026). ClustalX (Thompson et al., 1997) was used to align the sequences, and minor adjustments were made either to improve the alignments or to delete extruding ends using GeneDoc version 1997 (Nicholas and Nicholas, 1997). Phylogenetic analyses were performed either on a 32-sequence amino acid alignment of 632 residues or on a derived truncated alignment of 521 residues using the PHYLIP package of programs (Felsenstein, 1993) on the Pasteur Institute Server (http://bioweb.pasteur.fr/seqanal/phylogeny/). PROTDIST with 100 bootstrap analyses was used to calculate the distance between the sequences before analysis with the neighbor-joining programs NEIGHBOR, KITSCH, and FITCH, and calculation of the consensus tree using CONSENSE. The parsimony method used the PROTPARS program, again with 100 bootstrap analyses. The sequences of the 17 plant cDNAs were similarly aligned and used in phylogenetic analyses either using the above neighbor-joining programs or the DNAPARS parsimony program. All trees were viewed and edited using TREEVIEW (Page, 1996).
Comparisons with M. truncatula ESTs and Electronic Northern Analyses
The full-length cDNAs reported here were used in BLAST analyses with the GenBank Medicago ESTs and the M. truncatula Gene Index Release 4.0 database at TIGR (http://www.tigr.org/tdb/tgi/mtgi/) or the M. truncatula EST database (version September 2001) at the Institut National de la Recherche Agronomique (http://medicago.toulouse.inra.fr/Mt/EST/DOC/MtB.html), and 116 ESTs were identified. Duplicate ESTs of the same gene from the same cDNA library that started at the same nucleotide (potentially arising from the same cloning event) were eliminated from further analyses. Alignments were perused manually and each apyrase-like EST could be classified into one of six clusters centered on the cDNAs. It is noteworthy that the clustering of certain ESTs did not correspond with the database clusters, particularly for the TIGR database. To derive data on the differential expression of the six genes (electronic northern analyses), the 32 cDNA banks were classified into 12 classes of similar origin and the frequency of expression of each gene in each class was calculated by the number of ESTs divided by the total ESTs and expressed per 1,000 ESTs. The 12 classes were slightly modified from that described at http://medicago.toulouse.inra.fr/Mt/EST/DOC/MtB.html and contained the following banks (all are prefixed by Mt): roots, various (NF-RT, RHE, RP, MHRP, KV0, BA, DSIR, MGHG, and HOGA); roots, rhizobial (KV1, KV2, and KV3); roots, mycorrhizal (MHAM and BC); nodules (BB, R108, NF-NR, GVN, and GSVN); stem (NF-ST); leaves (DSLC, NF-LF, and NF-PL); leaves, fungal (DSIL); leaves, insect challenged (NF-IN); plantlets (NF-DT and NF-IR); flowers (NF-FL); seeds/pods (GESD, NF-GS, and GPOD); and cell cultures (NF-EC).
RNA Isolation and Northern Hybridization
The plant material corresponding to a sample was crushed in a mortar and pestle with liquid nitrogen, and aliquots corresponding to about 180 mg fresh weight were used for total RNA isolation using the EXTRACT-ALL kit (Eurobio, Les Ulis, France). The total RNA was dissolved in 30 μL of sterile water, and the quality and quantity of the RNA was assessed by migration on agarose gels with ethidium bromide staining and by spectrophotometry. RNA gels were run in phosphate buffer (Lummerzhein et al., 1994) using 10 μg of total RNA per track, and the RNA was then blotted on to Nytran+ membranes (Schleicher & Schuell, Dassel, Germany). Probes were prepared by labeling with 32P using the Ready-To-Go kit (Amersham Biosciences, Buckinghamshire, UK) using the DNA fragments that were isolated either using restriction enzymes or by PCR. The following fragments were used to produce MtAPY probes: MtAPY1 general probe (whole cDNA of MtAPY1;3), MtAPY2 probe (whole cDNA), MtAPY1;1-specific probe (nucleotides 1,354–1,469), MtAPY1;3-specific probe (nucleotides 1,345–1,444), MtAPY1;4-specific probe (nucleotides 1,323–1,466), and MtAPY1;5-specific probe (nucleotides 1,326–1,588). Probes were also prepared using cDNA fragments of the MtENOD11 (Journet et al., 2001), MtRip1 (Cook et al., 1995), and rRNA genes.
Prehybridization was carried out for at least 2 h using the phosphate-based solution (Lummerzheim et al., 1994). Hybridization was carried out overnight using the same solution and about 2 × 106 cpm mL−1 of labeled probe. The blots were then washed twice in SSC (0.15 m NaCl and 0.015 m sodium citrate) containing 0.1% (w/v) SDS at 65°C and the patterns of hybridization to the blots were analyzed after exposure to a Phosphor screen using a PhosphorImager. Different blots and probes were exposed for different times to emphasize the relative rather than quantitative differences.
Distribution of Materials
Upon request, all novel materials described in this publication (the six MtAPY cDNA clones) will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor. The cDNAs have the following GenBank accession numbers: APY1;1, AY180377; APY1;2, AY180378; APY1;3, AY180379; APY1;4, AY180380; APY1;5, AY180381; and APY2, AY180382.
ACKNOWLEDGMENTS
We thank Jerome Cazot (our laboratory) for his valuable help in the early parts of this work and David Barker (our laboratory) for his helpful comments on the manuscript. We gratefully acknowledge the following persons for sending us biological material: Dr. H. Kuester (Institute of Genetics, University of Bielefeld, Germany) for the flower MtAPY1;2 EST clone and Dr. J. Dénarié (INRA, Toulouse, France), Prof. D. Cook (University of California, Davis), and Dr. G. Duc (INRA, Dijon, France) for M. truncatula mutants.
Footnotes
This work was supported by the European Union (Marie Curie Fellowship no. HPMF–CT–1999–00073 to M.-T.N.G.), by the European Union Research Training Network (grant no. FMRX–CT98–0243), and by the Région Midi-Pyrénées (France).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.010926.
LITERATURE CITED
- Abeijon D, Yanagisawa K, Mandon EC, Hausler A, Moreman K, Hirshchberg DB, Robbins PW. Guanosine diphosphatase is required for protein and sphingolipid glycosylation in the Golgi lumen of Saccharomyces cerevisiae. J Biol Chem. 1993;122:307–323. doi: 10.1083/jcb.122.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC, Dénarié J, Truchet G. Rhizobium melilotilipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT, Kessler A, Halitschke R. Volatile signaling in plant-plant-herbivore interactions: What is real? Curr Opin Plant Biol. 2002;5:351–354. doi: 10.1016/s1369-5266(02)00263-7. [DOI] [PubMed] [Google Scholar]
- Barker D, Bianchi S, Blondon F, Dattee Y, Duc G, Essad S, Flament T, Gallusci T, Genier G, Guy P. Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Rep. 1990;8:40–49. [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Dixon RA, Farmer AD, Flores R, Inman J, Gonzales RA, Harrison MJ, Paiva NL, Scott AD, Weller JW et al. The MedicagoGenome Initiative: a model legume database. Nucleic Acids Res. 2001;29:114–117. doi: 10.1093/nar/29.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuschlein F, Fassnacht M, Klink A, Allolio B, Reincke M. ACTH-receptor expression, regulation and role in adrenocortical tumor formation. Eur J Endocrinol. 2001;144:199–206. doi: 10.1530/eje.0.1440199. [DOI] [PubMed] [Google Scholar]
- Bono JJ, Riond J, Nicolaou KC, Bockovich NJ, Estevez VA, Cullimore JV, Ranjeva R. Characterization of a binding site for chemically synthesized lipo-oligosaccharidic NodRm factors in particulate fractions prepared from roots. Plant J. 1995;7:253–260. doi: 10.1046/j.1365-313x.1995.7020253.x. [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Journet EP, Maillet F, Penmetsa V, Rosenberg C, Gough C, Cook D, Dénarié J. Identification of four genes of Medicago truncatulacontrolling steps in Nod factor transduction. Plant Cell. 2000;12:1647–1665. doi: 10.1105/tpc.12.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, Timmers AJ, Maillet F, Galera C, Penmetsa RV, Cook D, Gough C, Dénarié J. The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development. 2001;128:1507–1518. doi: 10.1242/dev.128.9.1507. [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Frischauf AM. The CD39-like gene family: identification of three new human members (CD39L2, CD39L3, and CD39L4), their murine homologues, and a member of the gene family from Drosophila melanogaster. Genomics. 1998;50:357–367. doi: 10.1006/geno.1998.5317. [DOI] [PubMed] [Google Scholar]
- Cohn J, Uhm T, Ramu S, Nam YW, Kim DJ, Penmetsa RV, Wood T, Denny RL, Young ND, Cook DR et al. Differential regulation of apyrase genes from Medicago truncatula. Plant Physiol. 2001;125:2104–2119. doi: 10.1104/pp.125.4.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D. Medicago truncatula: a model in the making! Curr Opin Plant Biol. 1999;2:301–304. doi: 10.1016/s1369-5266(99)80053-3. [DOI] [PubMed] [Google Scholar]
- Cook D, Dreyer D, Bonnet D, Howell M, Nony E, VandenBosch K. Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell. 1995;7:43–55. doi: 10.1105/tpc.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullimore JV, Ranjeva R, Bono JJ. Perception of lipo-chitooligosaccharidic Nod factors in legume. Trends Plant Sci. 2001;6:24–30. doi: 10.1016/s1360-1385(00)01810-0. [DOI] [PubMed] [Google Scholar]
- Day RB, McAlvin CB, Loh JT, Denny RL, Wood TC, Young ND, Stacey G. Differential expression of two soybean apyrases: one of which is an early nodulin. Mol Plant-Microbe Interact. 2000;13:1053–1070. doi: 10.1094/MPMI.2000.13.10.1053. [DOI] [PubMed] [Google Scholar]
- Debellé F, Rosenberg C, Vasse J, Maillet F, Martinez E, Dénarié J, Truchet G. Assignment of symbiotic developmental phenotypes to common and specific (nod) genetic loci of Rhizobium meliloti. J Bacteriol. 1986;168:1075–1086. doi: 10.1128/jb.168.3.1075-1086.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dénarié J, Debellé F, Promé JC. Rhizobiumlipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- Downie JA, Walker SA. Plant responses to nodulation factors. Curr Opin Plant Biol. 1999;2:483–489. doi: 10.1016/s1369-5266(99)00018-7. [DOI] [PubMed] [Google Scholar]
- Etzler ME, Kalsi G, Eqing NN, Roberts NJ, Day RB, Murphy JB. A Nod factor binding lectin with apyrase activity from legume roots. Proc Natl Acad Sci USA. 1999;96:5856–5861. doi: 10.1073/pnas.96.10.5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) Version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- Geurts R, Bisseling T (2002) Rhizobium Nod factor perception and signalling. Plant Cell S239–S249 [DOI] [PMC free article] [PubMed]
- Gressent F, Drouillard S, Mantegazza N, Samain E, Geremia RA, Canut H, Niebel A, Driguez H, Ranjeva R, Cullimore JV et al. Ligand specificity of a high-affinity binding site for lipo-chitooligosaccharidic Nod factors in Medicagocell suspension cultures. Proc Natl Acad Sci USA. 1999;96:4704–4709. doi: 10.1073/pnas.96.8.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualtieri G, Bisseling T. The evolution of nodulation. Plant Mol Biol. 2000;42:181–194. [PubMed] [Google Scholar]
- Handa M, Guidotti G. Purification and cloning of a soluble ATP-diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum) Biochem Biophys Res Commun. 1996;218:916–923. doi: 10.1006/bbrc.1996.0162. [DOI] [PubMed] [Google Scholar]
- Hsieh HS, Tong DG, Thomas C, Roux SJ. Light-modulated abundance of an mRNA encoding a calmodulin-regulated, chromatin-associated NTPases in pea. Plant Mol Biol. 1996;30:135–147. doi: 10.1007/BF00017808. [DOI] [PubMed] [Google Scholar]
- Johnson KA, Sistrunk ML, Polisensky DH, Braam J. Arabidopsis thalianaresponses to mechanical stimulation do not require ETR1 or EIN2. Plant Physiol. 1998;116:643–649. doi: 10.1104/pp.116.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, El-Gachtouli N, Vernoud V, de Billy F, Pichon M, Dedieu A, Morandi D, Barker DG, Gianinazzi-Pearson V. Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscular-containing cells. Mol Plant-Microbe Interact. 2001;14:737–748. doi: 10.1094/MPMI.2001.14.6.737. [DOI] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer MJ, Niebel A, Schiex T, Jaillon O, Chatagnier O et al. Exploring root symbiotic programs in the model legume Medicago truncatulausing EST analysis. Nucleic Acids Res. 2002;30:5579–5592. doi: 10.1093/nar/gkf685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsi G, Etzler K. Localization of a Nod-factor-binding protein in legume roots and factors influencing its distribution and expression. Plant Physiol. 2000;124:1039–1048. doi: 10.1104/pp.124.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoszynski M, Wojtczak A. Apyrases (ATP diphosphohydrolases, EC 3.6.1.5): function and relationship to ATPases. Biochem Biophys Acta. 1996;1310:233–241. doi: 10.1016/0167-4889(95)00135-2. [DOI] [PubMed] [Google Scholar]
- Lummerzheim M, de Oliviera D, Castresana C, Miguens FC, Louzada E, Roby D, van Montagu M, Timmerman B. Identification of compatible and incompatible interactions between Arabidopsis thaliana and Xanthomonas campestris pv. campestrisand characterisation of the hypersensitive response. Mol Plant-Microbe Interact. 1994;6:532–544. [Google Scholar]
- Matsumoto H, Yamaya T, Tanigawa M. Activation of ATPase activity in the chromatin fraction of pea nuclei by calcium and calmodulin. Plant Cell Physiol. 1984;25:191–195. [Google Scholar]
- Nicholas KB, Nicholas HB Jr (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. www.psc.edu/biomed/genedoc/
- Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Cook D. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science. 1997;275:527–530. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
- Plesner L. Ecto-ATPases: identities and functions. Int Rev Cytol. 1995;158:141–214. doi: 10.1016/s0074-7696(08)62487-0. [DOI] [PubMed] [Google Scholar]
- Price CA, Reardon EM. Mendel, a database of nomenclature for sequenced plant genes. Nucleic Acids Res. 2001;29:118–119. doi: 10.1093/nar/29.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. The TIGR gene indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 2001;29:159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts NJ, Brigham J, Wu B, Murphy JB, Volpin H, Philips DA, Etzler ME. A Nod factor-binding lectin is a member of a distinct class of apyrases that may be unique to the legumes. Mol Gen Genet. 1999;262:261–267. doi: 10.1007/s004380051082. [DOI] [PubMed] [Google Scholar]
- Shibata K, Abe S, Davies E. Structure of the coding region and mRNA variants of the apyrase gene from pea (Pisum sativum) Acta Physiol Plant. 2001;23:3–13. doi: 10.1007/s11738-001-0016-y. [DOI] [PubMed] [Google Scholar]
- Shibata K, Morita Y, Abe S, Stankovic B, Davies E. Apyrase from pea stems: purification, characterization and identification of a NTPase from the cytoskeleton fraction of pea stem tissue. Plant Physiol Biochem. 1999;37:881–888. [Google Scholar]
- Szybiak-Strozycka U, Lescure N, Cullimore JV, Gamas P. A cDNA encoding a PR-1-like protein in the model legume Medicago truncatula. Plant Physiol. 1995;107:273–274. doi: 10.1104/pp.107.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata JR. Autoinduction of nuclear hormone receptors during metamorphosis and its significance. Insect Biochem Mol Biol. 2000;30:645–651. doi: 10.1016/s0965-1748(00)00035-7. [DOI] [PubMed] [Google Scholar]
- Thomas C, Rajagopal A, Windsor B, Dudler R, Lloyd A, Roux SJ. A role for ectophosphatase in xenobiotic resistance. Plant Cell. 2000;12:519–533. doi: 10.1105/tpc.12.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Sun Y, Naus K, Lloyd A, Roux SJ. Apyrase functions in plant phosphate nutrition and mobilizes phosphate from extracellular ATP. Plant Physiol. 1999;119:543–551. doi: 10.1104/pp.119.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins D. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch KA, Frugoli J. Guidelines for genetic nomenclature and community governance for the model legume Medicago truncatula. Mol Plant-Microbe Interact. 2001;14:1364–1367. doi: 10.1094/MPMI.2001.14.12.1364. [DOI] [PubMed] [Google Scholar]
- Vernoud V, Journet EP, Barker DG. MtENOD20, a Nod factor-inducible molecular marker for root cortical activation. Mol Plant-Microbe Interact. 1999;12:604–614. [Google Scholar]
- Yamamoto S, Suzuki K, Shinshi H. Elicitor-responsive, ethylene-independent activation of GCC box-mediated transcription that is regulated by both protein phosphorylation and dephosphorylation in cultured tobacco cells. Plant J. 1999;20:571–579. doi: 10.1046/j.1365-313x.1999.00634.x. [DOI] [PubMed] [Google Scholar]