Abstract
We extend our analysis of the transcriptional reorganization that occurs when the native tobacco, Nicotiana attenuata, is attacked by Manduca sexta larvae by cloning 115 transcripts by mRNA differential display reverse transcription-polymerase chain reaction and subtractive hybridization using magnetic beads (SHMB) from the M. sexta-responsive transcriptome. These transcripts were spotted as cDNA with eight others, previously confirmed to be differentially regulated by northern analysis on glass slide microarrays, and hybridized with Cy3- and Cy5-labeled probes derived from plants after 2, 6, 12, and 24 h of continuous attack. Microarray analysis proved to be a powerful means of verifying differential expression; 73 of the cloned genes (63%) were differentially regulated (in equal proportions from differential display reverse transcription-polymerase chain reaction and SHMB procedures), and of these, 24 (32%) had similarity to known genes or putative proteins (more from SHMB). The analysis provided insights into the signaling and transcriptional basis of direct and indirect defenses used against herbivores, suggesting simultaneous activation of salicylic acid-, ethylene-, cytokinin-, WRKY-, MYB-, and oxylipin-signaling pathways and implicating terpenoid-, pathogen-, and cell wall-related transcripts in defense responses. These defense responses require resources that could be made available by decreases in four photosynthetic-related transcripts, increases in transcripts associated with protein and nucleotide turnover, and increases in transcripts associated with carbohydrate metabolism. This putative up-regulation of defense-associated and down-regulation of growth-associated transcripts occur against a backdrop of altered transcripts for RNA-binding proteins, putative ATP/ADP translocators, chaperonins, histones, and water channel proteins, responses consistent with a major metabolic reconfiguration that underscores the complexity of response to herbivore attack.
Plants are known to exhibit large phenotypic changes when confronted with various abiotic and biotic insults, and these changes are thought to increase plant fitness if the insults continue over time. The mechanisms responsible for these examples of adaptive phenotypic plasticity are largely unknown, but analyses of responses with Arabidopsis clearly indicate that a large proportion of the transcriptome is involved (Maleck et al., 2000; Schenk et al., 2000; Sasaki et al., 2001). Transcriptional responses to environmental stresses are exceptionally complicated because they require a deep understanding of both the environmental parameters that determine a plant's fitness and metabolism sensu lato. Plant responses to herbivore attack, for example, involve the activation of direct and indirect defenses and tolerance responses, which can be specific to the attacking herbivore, as has been demonstrated in the Nicotiana attenuata-Manduca sexta plant herbivore system (Baldwin, 2001; Baldwin et al., 2001).
When attacked by the nicotine-tolerant specialist M. sexta, N. attenuata “recognizes” the attack, as evidenced by alterations in a number of its wound- and jasmonate (JA)-elicited responses. The induced JA levels that are normally proportional to the amount of mechanical wounding erupt into a JA burst that increases concentrations 2 to 10 times that of wound-induced levels and is propagated throughout the damaged leaf ahead of the rapidly foraging herbivore (Schittko et al., 2000; Ziegler et al., 2001). Wounding and JA elicitation caused by wounding do not provoke ethylene emissions, but M. sexta attack produces a rapid ethylene burst, which is sustained during larval feeding (Kahl et al., 2000). The ethylene burst suppresses the wound- and JA-induced accumulation of nicotine biosynthetic genes, NaPMT1 and 2, and the associated nicotine accumulation (Winz and Baldwin, 2001). The ethylene burst does not, however, suppress the release of volatile organic compounds, which function as powerful indirect defenses in nature by attracting a generalist predator to the feeding herbivore (Kessler and Baldwin, 2001) and are also elicited by larval feeding but not by mechanical wounding (Halitschke et al., 2000; Kahl et al., 2000). In summary, at a phenotypic level of analysis, M. sexta attack of N. attenuata results in a down-regulation of a major direct defense, nicotine, which is demonstrably effective against mammalian herbivores, and an up-regulation of an indirect defense, the release of predator-attracting volatiles, which in turn is demonstrably effective against insect herbivores. Because nicotine can be sequestered by M. sexta for its own defense against parasitoids, the M. sexta-induced changes likely represent an adaptive tailoring of N. attenuata's wound response.
To understand the transcriptional basis of these M. sexta-induced changes in defense strategies, we used differential display reverse transcription (DDRT)-PCR with one arbitrary primer to gain an unbiased view of approximately 5% of the M. sexta-induced transcriptome. This analysis identified 53 individual sequences, of which 49 were detectable on RNA gel blots, and differential expression was verified for 27 (Hermsmeier et al., 2001). Here, we provide a second installment in the analysis of the M. sexta-altered transcriptome by continuing the DDRT-PCR analysis with six additional random primers, each with 10 anchor primers. Because DDRT-PCR provides sequence from the 3′-untranslated region of genes, which tends to be highly gene- and species-specific and, therefore, diminishes the probability of finding homology with genes of similar function in the data bases, a subtractive hybridization using magnetic beads (SHMB; Sharma et al., 1993) was used to complement the analysis with sequences more likely to originate from the open reading frame (ORF) of M. sexta-induced N. attenuata genes. All genes cloned by DDRT-PCR and SHMB (53 from Hermsmeier et al., 2001; 115 from this study; and 10 “control” genes) were spotted as cDNAs on glass slide microarrays (see Table I). We examined the transcriptional changes of the cloned transcripts by hybridizing the microarrays with fluorescently labeled transcripts from plants massively attacked by M. sexta larvae for 2, 6, 12, and 24 h and provide full-length sequences of two genes that catalyze the early and final steps in the biosynthesis of terpenoid-derived defense metabolites: 3-hydroxy-3-methylglutaryl CoA reductase (HMGR) and 5-epi-aristolochene synthase (EAS). The analysis highlights the extent to which metabolism is reconfigured during herbivore attack.
Table I.
Genes cloned by DDRT-PCR (DD/arbitrary primer no.) and subtractive hybridization with magnetic beads (SHMB) that exhibited nonsignificant (between 0.5 and 1.50) expression ratios in the microarray analysis and had similarity to genes in the databases
| Clone | Length | Method | Peak at h | Expression Ratio | Type | Accession No. | Sequence Similarity | E Value |
|---|---|---|---|---|---|---|---|---|
| bp | ||||||||
| DH017 | 1028 | SHMB | 24 | 1.33 | IIb | CA591807 | Matrix attachment regions-binding protein (MARBPF; AB059832) | 0.00 |
| DH108 | 453 | SHMB | 12 | 0.80 | III | CA591793 | PSII (NtPII10; X70088) | 0.00 |
| RE322 | 590 | DD/R5 | 24 | 1.25 | Ia | CA591771 | Transcription factor NtWRKY2 (AB020590) | 1 × 10–120 |
| DH123 | 371 | SHMB | 24 | 1.21 | IIa | CA591796 | Cytosolic glyceraldehyde-3-phosphophate dehydrogenase (GapC; M14419) | 2 × 10–94 |
| RE283 | 184 | DD/R5 | 12 | 0.63 | III | CA591769 | PSII oxygen-evolving complex, 23-kD polypeptide (X58910) | 1 × 10–73 |
| RD161 | 234 | DD/R4 | 12 | 1.47 | III | CA591756 | Actin gene Sfa 15B (X03076) | 1 × 10–47 |
| RB142 | 107 | DD/R2 | 24 | 1.13 | IIb | CA591717 | 25S ribosomal RNA gene (X13557) | 1 × 10–43 |
| RF064 | 161 | DD/R6 | 24 | 1.20 | IIb | CA591783 | 26S ribosomal RNA gene (AF479172) | 2 × 10–43 |
| RD131 | 190 | DD/R4 | 12 | 1.30 | III | CA591754 | Signal recognition particle 7S RNA (Z29099) | 3 × 10–35 |
| DH270 | 307 | SHMB | 6, 24 | 1.20, 1.23 | IV | CA591818 | Type 2 metallothionein-like protein (U35225) | 6 × 10–31 |
| RN021 | 389 | DD/R14 | 24 | 1.43 | Ia | CA591773 | Beta-alanine synthase (Y19104) | 3 × 10–30 |
| RN032 | 337 | DD/R14 | 12 | 0.54 | III | CA591774 | Polypeptide of PSII (X85038) | 1 × 10–17 |
| RC231 | 276 | DD/R3 | 12 | 1.23 | III | CA591748 | myb1 gene (AF248962) | 8 × 10–17 |
Genes are listed in order of decreasing E value from BLAST queries. Expression patterns are defined as: gradual (Type Ia) or abrupt (Type Ib) increases; initial decreases followed by either steady (Type IIa) or abrupt increases (Type IIb); an initial increase followed by a decrease (Type III); an increase, a decrease, and finally an increase (Type IV; both peaks are given if equal) as plants were continuously attacked by Manduca sexta larvae over 24 h. Down-regulated genes have the opposite patterns.
RESULTS
We cloned a total of 115 transcripts by DDRT-PCR and SHMB from N. attenuata plants under continuous attack from 20 first instar M. sexta larvae for 24 h. The cDNAs of all cloned transcripts, in addition to eight previously characterized M. sexta-induced genes, were spotted on Lys-coated glass slide microarrays and hybridized with Cy3- and Cy5-labeled mRNA probes isolated from N. attenuata plants subjected to attack from the same number of larvae but harvested at 2, 6, 12, and 24 h after the start of the attack to fully characterize the response.
Utility and Limitations of the Microarrays
Due to variation in the spot placement and shape on these Lys slides, background corrections for each spot were performed manually, and not all spots could be used. We arbitrarily defined cDNAs with mean (of a maximum of eight replicate spots, range 5–8) expression ratios of ≤0.5 or ≥1.50 as being differentially expressed (down- and up-regulated, respectively). These thresholds are higher than those used in the companion paper (Halitschke et al., 2003; ≤0.75 or ≥1.25) because sample size constraints did not allow for the use of the statistical criteria in determining differential expression that were used in the companion paper, but lower than those of other studies (2.0 and 2.5; Reymond et al., 2000) that used only a spot per cDNA and, hence, had no means of determining within-array variance.
The cDNAs from eight N. attenuata genes known to respond to herbivore attack, which had been previously analyzed by RNA blots, were used to monitor the entire experimental process and determine whether the microarrays provided the same results as the northern-blot analysis. Previous work (Halitschke et al., 2001; Hermsmeier et al., 2001; Schittko et al., 2001; Winz and Baldwin, 2001; Ziegler et al., 2001; Glawe et al., 2003) had established that transcripts of Thr deaminase, proteinase inhibitor (PI), allene oxide synthase (AOS), alpha-dioxygenase (α-DIOX), hydroperoxide lyase (HPL), and putrescine N-methyl transferase (PMT) were strongly up-regulated, whereas transcripts of ribulose-1,5-biphosphate carboxylase (RuBPCase) were down-regulated after M. sexta larvae attack, compared with unattacked plants. Microarray analysis (Figs. 1 and 2) confirmed these results, thereby establishing the utility of the procedure for this system. It should be noted that although some of these control genes tended to be strongly regulated in both the array analysis and previous northern analyses, others, such as AOS, which are strongly regulated on northern blots within 30 min of elicitation (Ziegler et al., 2001), were found to be significantly regulated only at the 24-h harvest with the microarray (Fig. 1), suggesting that the arbitrary thresholds may exclude differentially regulated genes. Hence, to avoid disposing of potentially valuable information, we provide a list of genes with significant similarity to known genes in the database that were not differentially regulated by the established criteria (Table I).
Figure 1.
Mean (±sd) expression ratios from microarrays with eight replicate cDNA spots of partial sequences of N. attenuata Thr deaminase (TD; note break in y axis and that only + sd are shown), proteinase inhibitor (PI), PMT, and RuBPCase small subunit genes hybridized with fluorescently labeled probes derived from M. sexta-attacked or control N. attenuata plants (harvested 2, 6, 12, or 24 h after the start of attack). Shaded area represents arbitrarily defined zone of nonsignificant changes in expression.
Figure 2.
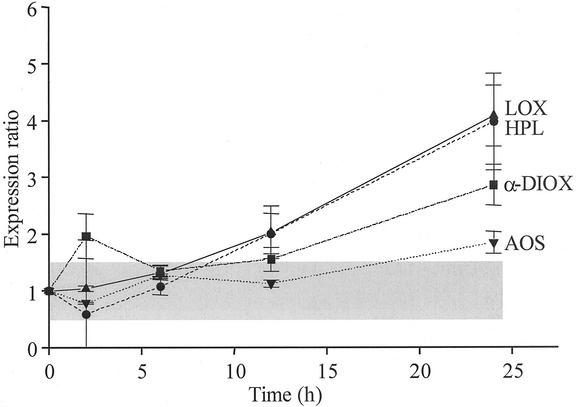
Mean (±sd) expression ratios from microarrays with eight replicate cDNA spots with partial sequences of genes mediating N. attenuata's oxylipin cascade (lipoxygenase [LOX], HPL, AOS, and α-DIOX) hybridized with fluorescently labeled probes derived from M. sexta-attacked or control N. attenuata plants (harvested 2, 6, 12, or 24 h after the start of attack). Shaded area represents arbitrarily defined zone of nonsignificant changes in expression.
Transcriptional changes, measured on the microarrays for each gene during the 24 h of continuous M. sexta larvae attack, were categorized as being: gradual (Type Ia) or abrupt (Type Ib) increases over the 24-h time course; an initial decrease followed by either steady (Type IIa) or abrupt increases (Type IIb); an initial increase followed by a decrease (Type III); an increase, followed by a decrease and an increase (Type IV). Down-regulated genes were similarly classified (Tables I and II). It should be noted that significant expression ratios (or lack thereof) in one of the four harvests with these arrays should be viewed as an indication of differential expression, an indication that should be confirmed with additional northern-blot analysis.
Table II.
Genes cloned by DDRT-PCR (DD/arbitrary primer no.) and subtractive hybridization with magnetic beads (SHMB) that exhibited significant (≤0.5 and ≥1.50) expression ratios in the microarray analysis and had similarity to genes in the databases
| Clone | Length | Method | Peak at h | Expression Ratio | Type | Accession No. | Sequence Similarity | E Value |
|---|---|---|---|---|---|---|---|---|
| bp | ||||||||
| DH120 | 394 | SHMB | 24 | 2.38 | IIa | AF542543 | 3-Hydroxy-3-methylglutaryl-CoA reductase (X63649) | 0 |
| DH164 | 452 | SHMB | 24 | 1.58 | IIa | AF542544 | 5-Epi-aristolochene synthase mRNA (AF272244) | 1 × 10–168 |
| DH114 | 372 | SHMB | 2 | 0.40 | Ia | CA591794 | 26S Ribosomal RNA gene spacer (Y08427) | 1 × 10–156 |
| DH083 | 607 | SHMB | 6, 24 | 1.66, 168 | IV | CA591811 | Tumor-related protein (D26464) | 1 × 10–137 |
| RN254 | 487 | DD/R14 | 24 | 1.90 | Ia | CA591779 | Ubiquitin carrier protein (Ubc-E2) mRNA (L23762) | 1 × 10–120 |
| DH099 | 482 | SHMB | 24 | 3.34 | Ia | CA591812 | Basic pathogenesis-related protein (PR1; X14065) | 1 × 10–115 |
| RN161 | 248 | DD/R14 | 24 | 2.11 | Ia | CA591777 | Transformer-2-like SR ribonucleoprotein (RNP; Y09506) | 1 × 10–113 |
| DH104 | 594 | SHMB | 24 | 1.66 | IIa | CA591822 | Thiazole biosynthetic enzyme precursor (NM 124858) | 1 × 10–86 |
| DH054 | 224 | SHMB | 24 | 2.04 | Ia | CA591790 | Sulfite reductase (AB010717) | 2 × 10–73 |
| RB271 | 484 | DD/R2 | 12 | 2.33 | IV | CA591719 | Xyloglucan endo-transglycosylase (X82685) | 2 × 10–77 |
| RB521 | 484 | DD/R2 | 24 | 3.44 | IV | CA591731 | Xyloglucan endo-transglycosylase (X82685) | 1 × 10–75 |
| RB061 | 230 | DD/R2 | 24 | 2.88 | Ia | CA591715 | 13-Lipoxygenase clone H3 (X96406) | 2 × 10–72 |
| RC191 | 233 | DD/R3 | 24 | 2.70 | Ia | CA591745 | ant Gene for ATP/ADP translocator (X62123) | 8 × 10–72 |
| RB131 | 427 | DD/R2 | 24 | 1.51 | IIb | CA591716 | Allene oxide synthase (AJ457080) | 3 × 10–65 |
| DH162 | 461 | SHMB | 24 | 2.21 | Ia | CA591801 | RNA-binding Gly-rich protein (RGP-1a; D16204) | 1 × 10–61 |
| RF113 | 561 | DD/R6 | 24 | 2.31 | IIa | CA591788 | Cytokinin-induced (cig2) mRNA (AB031321) | 9 × 10–57 |
| RB493 | 231 | DD/R2 | 2 | 1.91 | IIa | CA591728 | (Zymonaonas mobilis) rrnB operon (AF088897) | 1 × 10–55 |
| DH283 | 353 | SHMB | 12 | 1.64 | III | CA591810 | Chaperonin 60 (X70868) | 2 × 10–55 |
| DH124 | 586 | SHMB | 24 | 1.63 | IIa | CA591814 | (Nicotiana tabacum) GTP-binding protein (Ran-A1) mRNA (L16767) | 5 × 10–44 |
| RF071 | 255 | DD/R6 | 12 | 0.50 | IIa | CA591784 | Ribulose-1,5-biphosphate carboxylase small subunit (M32420) | 1 × 10–33 |
| RB012 | 289 | DD/R2 | 24 | 1.51 | IIa | CA591712 | Alpha-amylase (amy21) mRNA (M81682) | 1 × 10–27 |
| DH219 | 341 | SHMB | 24 | 1.76 | Ia | CA591804 | Histone H3 (PcH3–20) gene (M77494) | 1 × 10–18 |
| DH193 | 359 | SHMB | 24 | 4.96 | Ib | CA591816 | Major intrinsic protein (MIP) 2 (Y18312) | 7 × 10–18 |
| DH182 | 627 | SHMB | 24 | 3.65 | Ib | CA591802 | Ser carboxypeptidase cp-b (AJ 251970) | 2 × 10–16 |
| RB481 | 535 | DD/R2 | 24 | 1.79 | Ia | CA591726 | (Arabidopsis) putative protein (MN_127327) | 1 × 10–11 |
| DH138 | 295 | SHMB | 24 | 2.09 | IIa | CA591799 | 60S ribosomal protein (NM_114851) | 2 × 10–11 |
| RB304 | 425 | DD/R2 | 24 | 1.67 | IIa | CA591720 | (Lotus japonicus) genomic DNA, chromosome 6 (AP004526) | 2 × 10–09 |
| RE234 | 326 | DD/R5 | 2 | 1.69 | IIa | CA591767 | Plastidic aldolase-like protein (AB027001) | 3 × 10–08 |
| RE112 | 345 | DD/R5 | 24 | 4.38 | Ia | CA591761 | Ser protease sbt4b (AJ006480) | 6 × 10–06 |
| RC144 | 342 | DD/R3 | 12 | 1.83 | IIb | CA591742 | VFNT Pto locus (AF220603) | 7 × 10–06 |
| DH061 | 399 | SHMB | 24 | 1.70 | Ia | CA591791 | (Arabidopsis) putative protein, mRNA (MN_117913) | 9 × 10–06 |
| DH126 | 302 | SHMB | 12 | 0.43 | IIa | CA591815 | ATP-binding cassette (ABC) transporter protein 1 (MN_125882) | 1 × 10–3 |
| RC095 | 496 | DD/R3 | 24 | 2.41 | Ia | CA591780 | Ubiquitin carrier protein (Ubc) mRNA (L23762) | 0.040 |
Genes are listed in order of decreasing E value from the BLAST queries. Expression patterns are defined as: gradual (Type Ia) or abrupt (Type Ib) increases; initial decreases followed by either steady (Type IIa) or abrupt increases (Type IIb); an initial increase followed by a decrease (Type III); an increase, a decrease, and finally an increase (Type I;: both peaks are given if equal) as plants were continuously attacked by M. sexta larvae over 24 h. Down-regulated genes have the opposite patterns.
Comparison of DDRT-PCR and SHMB
From six rounds of DDRT-PCR with six different arbitrary primers (each with 10 anchor primers), we cloned and sequenced 84 different transcripts that ranged in size from 107 to 535 bp. Of these, 46 transcripts (60%), including 17 with similarity to known genes, had an expression ratio of greater than or equal to 1.50, predominantly at the 24-h harvest. Two transcripts had an expression ratio ≤ 0.5 at 12 and 24 h (Table II). Nine expressed sequence tags (ESTs) with significant similarity to known genes (Table I) and 24 ESTs with no significant similarity to known genes did not show differential expression as defined. Hence, we could not confirm differential expression in 40% of the transcripts identified by DDRT-PCR, and 73% had no similarity to genes in the databases (data not shown).
From SHMB, we sequenced 33 of 60 clones (from one-fourth of the final ligation volume, presumably representing 25% of induced transcripts) that ranged in size from 137 to 937 bp. Twenty-two (73%) of these transcripts, which included 15 with similarity to known genes (Table II), showed expression ratios > 1.50 predominantly at the 24-h harvest. Two transcripts (Table II), including one with similarity to 25S ribosomal RNA gene, had an expression ratio smaller than 0.5 at the 2-h harvest. In addition, five transcripts with significant similarity to known genes (Table I) and four transcripts with no significant similarity did not show any differential expression. Hence, we could not confirm differential expression in 27% of the sequenced transcripts identified by SHMB, and 44% had no similarity to genes in the databases.
Overall, a total of 73 transcripts (63% of the cloned genes) were confirmed to be differentially expressed (down- or up-regulated) with the microarrays (Tables II and III). Of these, 32% of the transcripts exhibited a Type Ia, 8% a Type Ib, 41% a Type IIa, 7% a Type IIb, 8% a Type III, and 4% a Type IV expression pattern over the time course of the experiment. Twenty-four differentially expressed transcripts (32%) had similarity to known genes or putative proteins. The 14 ESTs with significant sequence similarity to known genes that could not be confirmed to be differentially expressed (Table I) tended to be down-regulated at the 2-, 6-, and 12-h harvests, whereas exhibiting modest up-regulation (expression ratios between ≥1.0 and <1.5) at the 24-h harvest. In summary, the proportion of transcripts cloned by DDRT-PCR and SHMB that could be confirmed to be differentially expressed was similar between the two procedures, but as expected (Appel et al., 1999), SHMB produced a greater proportion of clones with sequences with significant similarity to genes in the databases. These genes could be crudely categorized as being involved in oxylipin signaling, transcriptional regulation, terpenoid biosynthesis, antimicrobial defense, and the remodeling of cell walls and metabolism.
Oxylipin Signaling and Transcriptional Regulation
Two transcripts identified by DDRT-PCR (RB061 and RB131) with similarity to potato (Solanum tuberosum) LOX (Royo et al., 1996) and tomato (Lycopersicon esculentum) AOS (Sivasankar et al., 2000), respectively, as well as HPL (J. Zeigler and R. Halitschke, unpublished data) were strongly up-regulated by M. sexta attack (Fig. 2; Table II). The 13-lipoxygenase catalyzes the dioxygenation of fatty acids with a 1,4-pentadiene structure to produce, among other products, 13-hydroperoxy linoleic acid, which is a substrate for AOS and HPL, initiating the biosynthesis of JA or volatile C6 compounds.
Interestingly, the 427-bp fragment of AOS cloned by DDRT-PCR (RB131) had greater similarity (86%) to the tomato gene than to the NaAOS previously cloned from N. attenuata (80%). Southern-blot analysis of N. attenuata genomic DNA digested with HindIII and XbaI, for which one recognition site per enzyme exists within the NaAOS ORF, revealed a complex banding pattern consistent with the existence of two genes coding for AOS in the N. attenuata genome (Ziegler et al., 2001). NaAOS originated from a screen of a cDNA library constructed from equal quantities of M. sexta-attacked N. attenuata leaves from seven genotypes, including the genotype used in this study. Although the microarray revealed that both NaAOS (Fig. 2) and RB131 had Type IIa expression patterns, the sequence differences between RB131 and NaAOS are likely too substantial to reflect allelic differences, and it is possible that RB131 represents a fragment from the second aos in the N. attenuata genome.
Although transcripts of LOX, AOS, and HPL exhibited coordinated Type I or II patterns of expression (Fig. 2), α-DIOX, which catalyzes the alpha-oxidation of fatty acids to hydroperoxy fatty acids and may be involved in signal generation (Sanz et al., 1998; Hamberg et al., 1999), exhibited a Type IV pattern of expression (Fig. 2). Prior work with α-DIOX demonstrated that transcripts increased rapidly in leaves attacked by M. sexta larvae and that fatty acid amino acid conjugates (FACs) in the oral secretions were responsible for up-regulating the wound-induced increase (Halitschke et al., 2001; Schittko et al., 2001). The type IV expression patterns are likely to reflect the interplay between wound-induced and oral secretion-mediated increases during massive caterpillar attack.
The clones coding putative transcription factors (RE322 encoding 590 bp of a WRKY transcription factor and RC231 encoding 276 bp of a putative MYB transcription factor) were found to be down-regulated both on DDRT-PCR display gels and at the 2- and 6-h harvests with the microarray. However, in later harvests (12 h for RE322 and 24 h for RC231), both tended to be up-regulated but not with expression ratios above the arbitrarily defined threshold of 1.5 (Table I). WRKY transcription factors occur in large gene families and are known to regulate a plethora of different genes, including pathogen- and wound-induced gene expression, by binding to W-box promoter elements in a variety of plant species (Eulgem et al., 2000). WRKY transcription factors are known to bind to W-box elements in PR1 genes (a homolog of which was cloned by SHMB; Table II) and regulate their expression after salicylic acid (SA) induction and pathogen elicitation (Rushton et al., 1996). Although many WRKY factors are known to be wound induced, few if any studies have found them to be induced by herbivore attack or JAs. Similarly, the expression of MYB transcription factors are known to be induced by tobacco mosaic virus and bacterial pathogen infection and SA treatment, with subsequent induction of PR genes (Yang and Klessig, 1996), and are generally involved in the regulation of phenylpropanoid metabolism, cell shape, and hormone signal transduction (Martin and PazAres, 1997). More recently, a novel MYB was found in rice (Oryza sativa) that is JA inducible (Lee et al., 2001).
Terpenoid Biosynthesis
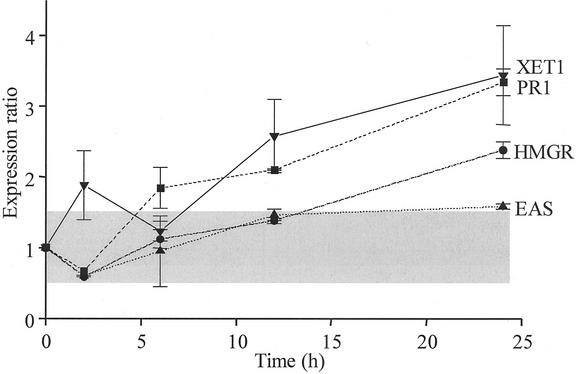
The SHMB analysis provided the DH120 and DH164 clones that code for HMGR (Genschik et al., 1992) and EAS (Mandujano-Chavez et al., 2000; Bohlmann et al., 2002), respectively. The enzymes catalyze the early and final steps in terpenoid biosynthesis, in which acetyl CoA and acetoacetyl CoA are converted to HMG CoA by 3-hydroxy-3-methylglutaryl CoA synthase, reduced to mevalonate by HMGR, and subsequently converted to isoprenoid pyrophosphate, the universal precursor for isoprenoids. EAS is a terpenoid synthase that catalyzes the cyclization of farnesyl diphosphate to the sesquiterpenoid (5-epi-aristolochene) for the subsequent formation of bicyclic sesquiterpenoid phytoalexin capsidiol (Bohlmann et al., 2002). The expression of both genes after M. sexta attack was highly coordinated: Both were initially down-regulated with subsequent increases in expression as herbivore attack proceeded, but the expression ratios for HMGR were greater than that of EAS at the 24-h harvests (Fig. 3).
Figure 3.
Mean (±sd) expression ratios from microarrays with eight replicate cDNA spots with partial sequences of N. attenuata xyloglucan (XG) endo-transglycosylase (XTH1), basic pathogenesis-related protein (PR1), HMGR, and EAS genes hybridized with fluorescently labeled probes derived from M. sexta-attacked or control N. attenuata plants (harvested 2, 6, 12, or 24 h after the start of attack). Shaded area represents arbitrarily defined zone of nonsignificant changes in expression.
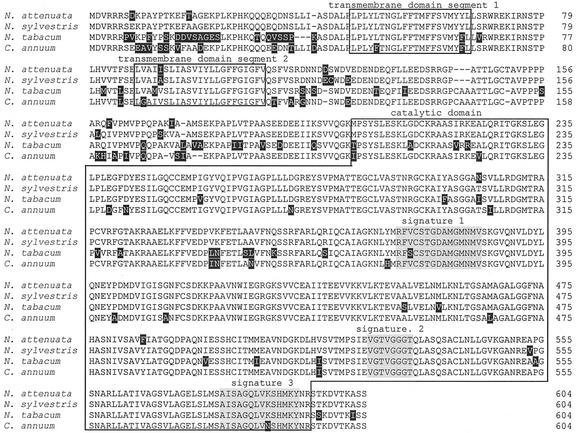
Because HMGR was one of the first transcripts shown to be differentially regulated in potato after attack by M. sexta larvae (Korth and Dixon, 1997) and is known to exist in a gene family and thought to play an essential role in regulating substrate flux into the cytosolic pathway of terpenoid biosynthesis (Korth et al., 2000), we amplified a 2,000-bp HGMR cDNA from N. attenuata by PCR with primer HMGf1 (5′-CGGCAATCTTACCGGTGAAA) derived from the HMGR cDNA sequence of Nicotiana sylvestris and primer HMGr1 (5′-TGAGATAGCTGACATGAGGG) derived from clone DH120. The DNA sequence contains an ORF of 1,812 nucleotides (AF542543) and encodes 604 amino acids with a calculated molecular mass of 65,125 D. The amino acid sequence showed 95% homology to N. sylvestris HMGR, 87% to pepper (Capsicum annuum), and 85% to N. tabacum (Fig. 4). The deduced protein consists of two transmembrane domains in the N-terminal region and a C-terminal catalytic domain of 349 amino acid residues containing the three highly conserved signatures located in the center of the catalytic domain, in a Gly-rich region, and in a region containing a His residue thought to be essential for catalytic activity (Fig. 4).
Figure 4.
Alignment of deduced amino acid sequences of N. attenuata HMGR (AF542543), N. sylvestris (S24760), N. tabacum (AAB87727), and pepper (Q9XEL8). Missing amino acids are indicated by dashes, and different amino acids are indicated by black shading. A transmembrane domain with two segments located in the N-terminal region and a 349-amino acid residue catalytic domain containing three highly conserved signatures, located in the center of the catalytic domain (1), in a Gly-rich region (2), and in a region containing a His residue (3), are indicated by boxes and gray shading.
Prior work on N. attenuata identified three new copies of EAS (NaEAS-12, -34, and -37) by screening a cDNA library of N. attenuata leaves from plants originating from seven geographically distinct populations, including the genotype used in this study, all of which had been attacked by M. sexta larvae (Bohlmann et al., 2002). To determine whether the EAS cloned by SHMB was the same as one of the previously cloned ones, a 1,950-bp cDNA (NaEASutah; AF542544) was generated by PCR with primer EPIf1 (5′-AATACACTCATCTTTAATTAG) derived from the N. tabacum ESA cDNA sequence and primer EPIr1 (5′-CACTAGCTTCAAGAATTTTAG) derived from clone DH164. The sequence contains an ORF of 1,647 nucleotides and encodes 548 amino acids of a calculated molecular mass of 62,895 D (Fig. 5), which showed 92% similarity to N. tabacum gene and 92% to 93% similarity to the previously cloned N. attenuata genes. Structurally, the enzyme is organized into two domains and there are two Mg2+-binding sites located in the C-terminal domain (Starks et al., 1997; Fig. 5). Elicitor-induced EASs from N. tabacum are known to occur in small gene families (Facchini and Chappell, 1992), and recently an EAS from corn (Zea mays; stc1; (Shen et al., 2000) has been shown to be induced by volicitin, the fatty acid-amino acid conjugate found in Spodoptera exigua oral secretions that elicits the release of volatiles in this species.
Figure 5.
Alignment of deduced amino acid sequences of EAS from different genotypes of N. attenuata (EASutah [AF542544], EAS12 [AF484123], EAS34 [AF484124], and EAS37 [AF484124]) and N. tabacum (5EAS). Amino acid differences are indicated by black shading. The gene consists of two domains: domain 1 (enclosed in box with a dotted line), which bisects domain 2 into two parts (both enclosed in boxes with a solid lines). Two conserved Mg2+-binding sites located in C-terminal portion of domain 2 are indicated by gray shading.
Cell Wall Remodeling
A total of 20 sequences from the DDRT-PCR analysis with arbitrary primer 2 could be assembled into two contigs of the same length (484 bp), representing clones RB271 and RB521, which had 88.4% similarity to tomato XG endo-transglycosylase/hydrolyase (tXET-B1; Arrowsmith and Desilva, 1995; renamed XTH by Yokoyama and Nishitani, 2002). Both exhibited a Type IV expression pattern (Fig. 3). XTHs are thought to function in the cleavage and concomitant transfer of XG molecules into plant cell walls by transglycosylation and may mediate the loosening of cell walls during growth (Emons and Mulder, 2000).
Antimicrobial-Associated Genes
SHMB provided three clones that are implicated in pathogen resistance. One of these, DH099, has similarity to the basic-type pathogenesis-related protein PR1 of N. tabacum (Payne et al., 1989), and the microarray analysis demonstrated that it was strongly up-regulated by M. sexta attack (Fig. 3). Basic PR1s are intracellular proteins associated with viral, fungal, and bacterial infections and are strongly elicited by ethylene (Kitajima and Sato, 1999). Therefore, the observed PR1 response is expected, given that M. sexta attack is known to elicit an ethylene burst in N. attenuata (Kahl et al., 2000). SHMB also provided DH083, which had similarity to genetic tumor-related cDNA in an interspecific hybrid (F1) between Nicotiana glauca and Nicotiana langsdorffii (Fujita et al., 1994) and DH270, which had similarity to a Nicotiana plumbaginifolia type 2 metallothtionein-like protein-encoding gene (Table I). Transcripts of both exhibited a Type IV expression pattern; however, DH270's increases were never higher than a 1.23 expression ratio (Table I), whereas DH083 attained 1.66 and 1.68 at the 6- and 24-h harvests (Table II). Some type 2 metallothioneins are thought to function as potent metal chelators (Giritch et al., 1998), but others play roles in different cell death pathways, including senescence and the hypersensitive response (HR) after pathogen attack (Butt et al., 1998). Pathogen recognition is implicated in the up-regulation of clone RC144, which has similarity to a putative pto gene (D.T. Lavelle, G.E.D. Oldroyd, D. Dalhbeck, B.J. Staskawicz, and R.W. Michelmore, unpublished data). The pto gene complex codes for protein kinases that mediate resistance against Pseudomonas syringae pv tomato infections and is correlated with HR (Loh and Martin, 1995). Clone DH126 (Table I) has similarity with a putative Arabidopsis ABC transporter (C.D. Town, B.J Haas, R. Maiti, L.I. Hannick, A.P. Chan, C.M. Ronning, R.K. Smith Jr, C.Y. Yu, J.R. Wortman, O. White et al., unpublished data). The superfamily of ABC transporter genes code for ATP-driven membrane associated efflux pumps that export a range of cytotoxic compounds. In N. tabacum, transcripts for a ABC transporter are reported to be JA elicited (Sasabe et al., 2002) and in tomato, an ABC transporter called pti3 is known to interact with pto in two-hybrid screens.
Remodeling of Metabolism
The remaining clones with similarity to known genes reflect the extent to which metabolism (sensu lato) is reorganized, presumably to make resources available for regrowth-, repair-, and defense-related processes after massive herbivore attack.
Five photosynthesis-related genes were identified by DDRT-PCR and SHMB, but only one RF071; Table I) was found to be significantly down-regulated by M. sexta attack (Fig. 1). RF071 codes for a transcriptionally active pseudogene of the small subunit gene of RuBPCase complex, which catalyzes the photosynthetic fixation of CO2 through the Calvin cycle (Oneal et al., 1987). The down-regulation of this gene complements that of the functional RuBPCase small subunit of Rubisco gene found in the Hermsmeier et al. (2001) study. Three photosynthetic-related transcripts (DH108, PSII; NtPII10, Zhou et al., 1993; RE283, 23-kD polypeptide of PSII oxygen-evolving complex, Hua et al., 1991; and RN032, 6.1-kD polypeptide of PSII, Lorkovic et al., 1995) all tended to be down-regulated with the lowest expression ratio at the 12-h harvest (Table I).
The down-regulation of genes related to photosynthesis may allow attacked plants to reinvest resources into other processes; a similar reinvestment function may be played by RN021 and DH182, both of which could allow for the recovery of amino acids invested in pyrimidine and protein synthesis, respectively. RN021, which has similarity to transcripts for beta-Ala synthase (C. Chevalier, J. Joubes, J. Petit, and P. Raymond, unpublished data), an enzyme that catalyzes the third and final step of pyrimidine catabolism to produce beta-Ala, was slightly up-regulated. DH182 was strongly up-regulated (Table II) and exhibits homology to barley (Hordeum vulgare) and Arabidopsis Ser carboxypeptidase (Dal Degan et al., 1994.). A member of this gene family, Ser CPII (BRS1), plays a regulatory role in the brassinosteroid (BR) signaling (Li et al., 2001), which is mediated by BR1 receptor in BR-insensitive (BRI1) mutants. In tomato, type I Ser CPs are among the “late wound-inducible” genes. Elicited by JAs and systemin, they may be involved in general protein turnover rather than signaling (Moura et al., 2001). Clone RE112, with similarity to a Ser protease of the subtilase gene family (Meichtry et al., 1999), could be playing a role in signaling because related members of this family are thought to process the wound hormone systemin. Two clones, RN254 and RC095, with sequence similarity to the tomato ubiquitin carrier protein (ubc) or alternatively, ubiquitin-conjugating enzyme (E2; D.M. Bird and M.A. Wilson, unpublished data), which is responsible for recognizing and tagging appropriate targets for the main non-lysosomal route for intracellular protein degradation in response to stress (Jesenberger and Jentsch, 2002), were found to be gradually up-regulated by herbivore attack. The ubiquitin protein-conjugating system plays a pivotal role in the ubiquitin-dependent proteosome pathway in the regulation of apoptosis or programmed cell death, the central process of the HR that is an important means of limiting the spread of pathogens in plants. However, because an HR is not observed after M. sexta attack, E2 is more likely involved in selective protein turnover.
Although alterations in these transcripts may directly or indirectly help a plant to meet its amino acid or more generally, nitrogen requirements for the necessary metabolic reconfiguration, Glc demands might be met by the hydrolysis of starch and oligosaccharides, as is suggested by the up-regulation of RB012, a transcript homologous to alpha-amylase (K. Gausing and T. D. Kreiberg, unpublished data). Alpha-amylase is normally GA inducible, but in this case it is gradually up-regulated during a massive herbivore attack. The expression of DH123, a 371-bp fragment with similarity to the cytosolic glyceraldehyde-3-phosphophate dehydrogenase (Shih et al., 1986) tended to be down-regulated (at the 2-, 6-, and 12-h harvests). Glyceraldehyde-3-phosphophate dehydrogenase participates in carbohydrate metabolism and is known to be elicited by SA and pathogen infection in potato (Laxalt et al., 1996) and by anaerobic conditions in corn (Manjunath and Sachs, 1997). RE234, which corresponds to plastidic aldolase (aldP; Yamada et al., 2000), was up-regulated shortly after the start of herbivore attack. aldP is involved in photosynthetic carbon reduction and catalyzes the synthesis of Fru-1,6 bis-phosphate from d-glyceraldehyde-3-phosphate and dihydroxyacetone phosphate. AldP transcripts are up-regulated after salt stress in various Nicotiana spp. (Yamada et al., 2000), and small changes in AldP activity by antisense-mediated gene silencing have dramatic effects on photosynthesis (Haake et al., 1998). The changes in energy metabolism elicited by herbivore attack will likely require increases in the exchange of ADP and ATP between the cytosol and mitochondria. RC191 corresponds to potato adenine nucleotide translocase (ANT) gene for ADP/ATP translocator (Emmermann et al., 1991) and is strongly up-regulated in herbivore-attacked plants (Table II). ANT is a translocator protein essential for the formation of the mitochondria permeability transition pore and is the most abundant protein of the inner mitochondrial membrane.
Transcripts for another mitochondria protein, DH283, a chaperonin of the 60-kD heat shock protein (Hsp) family (Prasad et al., 1990; Tsugeki et al., 1992), are thought to be important in the folding and assembly of multimeric proteins in the mitochondria. This is the second Hsp cloned from N. attenuata that is elicited by M. sexta attack. The first was AW191822, which has similarity to luminal-binding proteins and was also up-regulated by herbivore attack (Hermsmeier et al., 2001). Other researchers have examined the production of Hsp in N. attenuata and found evidence for systemic elicitation by methyl JA of an Hsp70 and smaller Hsps (16–23 kD; Hamilton and Coleman, 2001). Clone RF113 has sequence similarity to a family of cytokinin-induced transcripts (cig2) that are specifically up-regulated by cytokinins and function as GDP/GTP exchange factors (eIF2b) and regulate translation initiation.
Two chloroplast-localized transcripts were cloned by SHMB and found to be significantly up-regulated on the microarray. The DH104 fragment corresponds to the Arabidopsis biosynthetic enzyme of the thiamine precursor thiazole (Ribeiro et al., 1996), and the DH054 fragment corresponds to N. tabacum sulfite reductase (Yonekura-Sakakibara et al., 1998). Thiamine functions as cofactor for two enzyme complexes of pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase in the citric acid cycle (Belanger et al., 1995). Sulfite reductase catalyzes the six electron reductions of sulfite to sulfide and nitrite to ammonia using electrons donated from ferredoxin.
One of the more strongly up-regulated transcripts by M. sexta attack was a DH193, which has similarity to an MIP2 from potato (G. Leggewie, L. Willmitzer, and J.W. Riesmeier, unpublished data). MIPs are a superfamily of membrane channel proteins, some of which are known to function as aquaporins or water or neutral solute facilitators and tend to be induced by water or salt stress in various Nicotiana spp. (Yamada et al., 1997; Smart et al., 2001).
Given that M. sexta attack results in large-scale remodeling of metabolism, perhaps it is not surprising to find several genes for DNA- and RNA-binding proteins, RNPs, and ribosomal RNA, which together suggest a remodeling of the transcriptional machinery. DH017, which tended to be up-regulated, has similarity to an N. tabacum matrix attachment regions-binding protein (M. Maeshima and S. Fujiwara, unpublished data), which is thought to play multifunctional roles in chromatin organization and may control the accessibility of promoters to factors required for transcription (Hatton and Gray, 1999). Similarly, DH219, which has similarity to the H3 class of histones (S.C. Wu, P. Gregersen, and K. Hahlbrock, unpublished data), proteins known to organize chromatin and nucleosome structure and influence the fundamental nuclear processes of transcription, replication, and DNA repair, was strongly up-regulated by M. sexta attack. RN161 was significantly up-regulated and has similarity to the transformer-2-like protein (Petitot et al., 1997) that is a Ser-/Arg-rich RNP family thought to play a important role in the regulation of constitutive and alternative splicing of nuclear pre-mRNA. A similar function has been attributed to DH162, a transcript with similarity to the N. sylvestris RNA-binding Gly-rich protein (RGP-1a; Hirose et al., 1993). Both of these putative RNA-binding transcripts exhibited Type Ia expression patterns, which increased steadily as herbivore attack progressed. Two ribosomal RNA genes (DH114 and RB493) were found to show different expression patterns in the time course. The internal transcribed spacer of 26S ribosomal RNA gene (DH114) was significantly down-regulated at the 2-h harvest, whereas RB493, which had similarity to Z. mobilis rrnB operon and 23S ribosomal RNA genes, was significantly up-regulated at the same harvest. RD131, with similarity to the 7S RNA RNP complex of signal recognition particles (Riedel et al., 1995) that mediates the targeting of proteins to the endoplasmic reticulum, was not significantly regulated at any harvest but tended to increase with herbivore attack from an initial down-regulated state. Interpretations of differential regulation of ribosomal RNA species with microarrays that use cDNAs reverse transcribed from mRNA species as probes should be viewed with caution because it is unknown whether the ribosomal RNA is quantitatively amplified.
DISCUSSION
Experimental Approach
We cloned 115 transcripts from the insect-responsive transcriptome of N. attenuata by SHMB and DDRT-PCR using six of the possible 26 arbitrary primers (Liang et al., 1993) thereby extending the initial DDRT-PCR analysis of this plant-insect interaction (Hermsmeier et al., 2001) from approximately 4% to 26% of the herbivore-induced transcriptome. The proportion of the transcriptome covered by the SHMB analysis is more difficult to estimate, but given that no gene was cloned by both procedures, a large number of genes were probably involved. Both SHMB and the DDRT-PCR, unlike other fingerprinting techniques, provide an unbiased view of the transcriptional changes elicited during a plant-insect interaction. These two procedures, however, differ in their ability to simultaneously detect transcripts that are induced and repressed by the interaction. Only DDRT-PCR allows for this possibility in a single experiment. This advantage of DDRT-PCR is balanced by the higher proportion (76% versus 45%) of clones without significant similarities to known genes in BLAST queries, a distinct disadvantage of a procedure that utilizes poly(A+)-rich anchor primers and, as a consequence, delivers sequence from the 3′-untranslated region of induced genes (Appel et al., 1999). This disadvantage will presumably decrease as the number of sequences available in the databases increases. An additional difficulty of DDRT-PCR is the high rate of apparent false-positives and the labor-intensive verification procedures for detecting differential expressions (Appel et al., 1999). Moreover, the most commonly used verification procedure, the northern blot, may not be sufficiently sensitive to detect differential expression in rarely expressed transcripts: a putative advantage of the DDRT-PCR procedure.
To minimize the labor associated with verification, the clones were arrayed as cDNAs, and differential expression (arbitrarily defined as having an expression ratio of ≤0.5 or ≥1.50) was verified for 73 clones, with approximately equivalent proportions being derived from the SHMB and DDRT-PCR procedures. Hence, by these criteria, the rate of false-positives did not differ between the two display procedures. Eight N. attenuata genes, whose transcriptional responses after M. sexta attack had been characterized previously by northern-blot analysis, were included on the microarray, and in all cases, their expression ratios were consistent with the patterns observed in the northern analyses. The microarrays not only allowed for the verification of differential expression in plants that were under continuous attack for 24 h, the time when the SHMB and DDRT-PCR analyses were performed, but by analyzing expression patterns after 2, 6, and 12 h of continuous attack, they documented the ontogeny of the differential expression patterns. In some cases, these expression patterns suggest functional associations between previously unassociated genes. This approach yielded a number of insights into the transcriptional changes that occur during the interaction. However, it should be noted that these responses require confirmation by northern-blot analysis.
Oxylipin Signaling
JA elicitation of N. attenuata is known to confer dramatic induced resistance in both field (Baldwin, 1998) as well as laboratory (van Dam et al., 2000, 2001a) trials with M. sexta larvae. Moreover, M. sexta attack is known to result in a JA burst and increases in NaAOS transcripts (Ziegler et al., 2001). The coordinated increases in LOX, AOS, and HPL transcripts observed in this study (Fig. 2) are consistent with those reported from other species (Reymond et al., 2000; Sasaki et al., 2001) and are correlated with M. sexta attack-induced changes in JA and C6 volatiles but are too slow to account for their induced changes in metabolites (Kessler and Baldwin, 2001; Ziegler et al., 2001). Although the importance of these genes in plant-herbivore interactions is being convincingly demonstrated with plants deficient in their expression (for review, see Blee, 2002), the function of the transcriptional changes remains enigmatic.
The microarray analysis provided evidence for the simultaneous activation of SA- (DH099), ethylene- (PI), cytokinin- (RF113), and JA (Fig. 2)-signaling pathways during massive herbivore attack. The co-activation of numerous signal cascades in response to various biotic and abiotic stresses has been found in numerous studies using Arabidopsis microarrays (Maleck et al., 2000; Schenk et al., 2000; Moran and Thompson, 2001; Sasaki et al., 2001; Chen et al., 2002) and suggests that what had been described previously as linear signal cascades associated with particular elicitors are in fact a network of interacting cascades. Moreover, the inhibition of the JA cascade by the SA cascade may not be occurring during caterpillar attack as suggested by the strong up-regulation of transcripts for PR-1 (Fig. 3), a hallmark signature of SA signaling (Payne et al., 1989), in conjunction with those for PMT and PIs, signatures of JA signaling (Van Dam et al., 2001b; Winz and Baldwin, 2001).
An additional intriguing correspondence was observed between expression patterns of the oxylipin biosynthetic genes and those of clones RN254 and RC095, which had similarity to the tomato ubiquitin-conjugating enzyme (E2). The JA-insensitive coi1 mutant of Arabidopsis, which is defective in most JA-mediated defense signaling, has recently been shown to result from a single amino acid change in the F-box motif of the COI protein and abolishes the formation of a ubiquitin-ligase complex (Xu et al., 2002). In the ubiquitin-dependent proteolytic pathway, ubiquitin is linked to particular substrates to activate targeting via the sequential actions of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). Although the targeting of particular proteins for degradation is thought to be mediated by the E3 complex, the other two are essential for the function of the complex, and it is unclear which elements may be limiting during periods of large metabolic reconfigurations. Another F-box protein within the E3 complex has been reported recently to mediate a novel form of SA-mediated pathogen resistance in the Arabidopsis son1 mutant (Kim and Delaney, 2002), suggesting that both JA- and SA-mediated signaling involves the specific degradation of particular proteins.
Defense Responses
The “ask the plant” experimental approach used in this study provided transcripts that had not been associated previously with herbivore attack and new insights into previously characterized M. sexta- and MeJA-induced transcripts. The microarray analysis suggested dramatic increases in PI transcripts and more modest increases in PMT transcripts (Fig. 1), consistent with previous work (Baldwin, 2001; Van Dam et al., 2001b; Glawe et al., 2003). Interestingly, the down-regulation of the wound-induced increase in PMT transcripts and nicotine accumulation, which results from an ethylene burst that is produced when single caterpillars or their oral secretions are applied to mechanical wounds (Winz and Baldwin, 2001), evolves into a sustained increase when plants are massively attacked by larvae (Fig. 1). The down-regulation of the nicotine response is thought to be an optimization of defense responses because the plant switches from using a metabolically demanding direct defense, which could be coopted by a nicotine-tolerant herbivore for its own defensive purposes, to a metabolically inexpensive but effective indirect defense (Baldwin, 2001). Because this analysis did not include plants that had suffered the same amount of damage but were not exposed to caterpillar-derived signals, it is difficult to determine the degree to which the nicotine response was down-regulated. However, it is clear that the down-regulation was not complete, suggesting that during massive herbivore attack plants can readjust their defense responses if, for example, the loss of their entire canopy is imminent.
In addition to providing new kinetic information on previously characterized transcripts, the analysis suggested that previously uncharacterized transcripts underpin responses that had been phenotypically characterized. The dramatic up-regulation of HMGR and the more modest up-regulation of EAS may reflect the metabolic commitment to terpenoid-based indirect defenses, which are demonstrably effective in nature for N. attenuata (Kessler and Baldwin, 2001). In addition, the analysis provided a number of transcripts (PR1, metalothionein, PTO, and ABC transporters) that have been associated with defense against microbes. The up-regulation of these transcripts suggests that more attention should be given to the direct effect of these defensive proteins on insect herbivores or their potential indirect effects by inhibiting the microbial endosymbionts frequently found in insect herbivores.
The most frequently sequenced clones from the DDRT-PCR were the two thigmomorphogentically responsive clones with similarity to XTHs, whose expression increased (3.44-fold) during massive herbivore attack with a Type IV expression pattern (Fig. 3). Reducing XTH enzyme activity is thought to strengthen cell walls (Herbers et al., 2001) because this enzyme cuts XG polymers and inserts glucan subunits into existing cell wall polymers. The resulting average length of the XGs in the cell walls decreases. Decreasing the number of long linear cell wall polymers is thought to diminish the strength of cell wall fibers, so it is unlikely that the herbivore-induced increase represents a strengthening of cell walls as a defensive response against herbivore attack (Herbers et al., 2001; but see Braam et al., 1996). Cell walls represent a source of carbohydrate-based elicitors, and if XTH transfers an XG to water or other XGs not linked to the matrix of the cell wall, these transcripts might conceivably influence defensive signaling in a manner analogous to the cev1 mutant of Arabidopsis. Cev1 is defective in cellulose synthase regulation and exhibits constitutive expression of ethylene and JA response genes (Ellis et al., 2002). The extracellular matrix of plant cell walls clearly represents a rich source of paracrine elicitors (Brownlee, 2002); XTH up-regulation may play a role in generating these elicitors. Alternatively, XTH may be involved in loosening cell walls (Braam et al., 1996) in response to herbivore attack. By allowing plants to alter their shape and potentially speed the recovery of the photosynthetic canopy lost to herbivores, XTH may increase a plant's tolerance to herbivore attack.
Tolerance and Other “Civilian” Responses to Herbivory
Plant responses that decrease the amount of tissue lost to herbivores (defensive traits) are only one means of minimizing the fitness consequences of herbivore attack. Responses that minimize the fitness consequences of losing tissue to herbivores (tolerance responses) represent an equally effective but largely unstudied means of coping with herbivore attack. Mobilizing limiting resources from tissues that are about to be consumed into parts that are less likely to be eaten by herbivores (petioles, stems, and below-ground tissues) may represent such a mechanism. Such mobilization may share components with the metabolic mobilization of resources required for the production of defense traits. The resources required for the resistance traits could be made available by decreases in the five photosynthetic-related transcripts, increases in transcripts associated with protein, and nucleotide turnover and increases in transcripts associated with carbohydrate metabolism. Whether or not these transcriptional changes play a role in balancing the plant's resource budgets remains to be determined.
CONCLUSION
M. sexta attack results in a large-scale transcriptional changes in N. attenuata genes that are collectively consistent with a reconfiguration of metabolism that reduces photosynthetic activity, slows growth, and increases a diversity of defense traits. Numerous signal cascades appear to be involved in coordinating the responses. These coordinated changes point to the existence of central herbivore-activated regulators of metabolism, which in turn are activated by minute amounts of FACs in M. sexta's oral secretions (Schittko et al., 2000, 2001; Halitschke et al., 2001). In a companion paper, we use microarrays to examine the proportion of the M. sexta-induced transcriptome that is elicited by FACs.
MATERIALS AND METHODS
Plant Growth
An inbred line of Nicotiana attenuata Torr. ex Wats. originally collected from southwestern Utah in 1988 was used for all experiments and was the same genotype used by Hermsmeier et al. (2001). Seeds were germinated in potting soil after soaking with a 1:50 (w/v) dilution of liquid smoke (House of Herbs, Passaic, NY). One-week-old seedlings were transferred to 28-L communal hydroponic boxes with a nutrient solution consisting of 0.292 g L−1 of Peter's Hydrosol (W.R. Grace, Fogelsville, PA) and 0.193 g L−1 of Ca(NO3)2. After an adaptation period of 5 d, seedlings were transferred to individual 1-L hydroponic chambers containing a no-nitrogen hydroponic solution (Baldwin et al., 1994). Nitrogen was added after transfer by adding 2 mL of a 1 m KNO3 to each chamber and 1 mL a day before placing larvae on plants. Plants were placed in a growth chamber with a photoperiodic cycle programmed for a 16-h light period at 32°C and an 8-h dark period at 28°C with 65% constant relative humidity. Forty of the most similar looking plants in the rosette stage of growth were chosen for the display experiments, and in a separate experiment, 80 plants were chosen for the microarray experiment.
Insect Rearing and Plant Treatments
The eggs of Manduca sexta (Lepidoptera, Sphingidae) from Carolina Biological Supply (Burlington, NC) were hatched at 28°C. For the display experiment, 20 first instar larvae were placed on each of 20 plants at 12 pm (6 h into the light cycle), with one to three larvae per leaf, depending on the leaf size. After 24 h of feeding, the larvae and frass were removed, and 20 attacked and 20 control plants were harvested, separated into shoots and roots, immediately placed in liquid nitrogen, and stored at −80°C until used for DDRT-PCR and SHMB. Plants used in the microarray experiment were grown and treated identically as those used in the display experiment, except that 10 attacked and 10 control plants were harvested 2, 6, 12, and 24 h, respectively, after larvae were placed on plants.
DDRT-PCR
Procedures follow closely those described in Hermsmeier et al. (2001) with minor modifications. Total RNAs of shoots and roots were extracted separately from 5-g aliquots of 20 attacked and control plants. Genomic DNA was removed by adding 20 units of RNase free DNase I (Life Technologies, Eggenstein, Germany) for each 100-μL reaction volume containing 100 μg of total RNA. DNA-free RNAs were adjusted to a 1:1 (root:shoot) concentration for each reverse transcription (RT). First strand cDNAs were synthesized with 400 ng of purified total RNAs and 25 μm anchor primers A1(T12AA), A2(T12AC), A3(T12AG) A4(T12CA), A5(T12 CC), A6(T12CG), A7(T12GA), A8(T12GC), A9(T12GG), and A10(T12GT) (MWG Biotech, Munich), 200 units of SuperScript-II reverse transcriptase (Life Technologies), and 200 μm dNTPs, respectively. The reactions of each anchor primer that did not receive reverse transcriptase served as quality controls for potential RNA contamination by residual genomic DNA, which was later amplified in the DDRT-PCR procedure. DDRT-PCR was performed with each RT reaction with Platinum Taq polymerase (Life Technologies), dNTPs including α-33P labeled dCTP (NEN Life Science, Zaventem, Belgium), arbitrary primers R2(TGGATTGGTC), R3(CTTTCTACCC), R4(TTTTGGCTCC), R5(GGAACCAATC), R6(AAACTCCGTC), and R14(GATCAAGTCC) in combination with anchor primers A1 to A10, respectively. Thermocycling parameters were: denaturation at 94°C for 2 min for activation, followed by 40 cycles of 30 s of denaturation at 94°C, 120 s of annealing at 40°C, and 30s of extension at 72°C. The PCR amplification products were separated on a 6% (w/v) polyacrylamide denaturing gel. Gels were dried on Whatman 3MM paper (Whatman, Clifton, NJ) and exposed to Kodak Biomax MR film (Amersham Pharmacia Biotech, Freiburg, Germany). The developed films were used as templates to excise the differential (both amplified and suppressed) bands from the display gel. The cDNAs were eluted from the gel by incubating gel slices in 150 μL of water for 10 min at 25°C, 15 min at 100°C, then transferring to 4°C, followed by centrifugation at 13,000 rpm for 2 min, and the cDNAs were recovered from the supernatant. To clone the eluted cDNA fragments, the TOPO TA cloning kit (Invitrogen, CH Groningen, The Netherlands) was employed directly with the PCR products that were re-amplified by using corresponding primers and the PCR temperature program given above. Plasmid DNA was isolated with NucleoSpin plasmid kit (Macherey-Nagel, Düren, Germany) for sequencing. Plasmid inserts were sequenced on an ABI Prism 377 XL DNA sequencer with the Big Dye terminator kit (PE-Applied Biosystems, Weiterstadt, Germany) and analyzed with the Lasergene software package (DNASTAR, Madison WI).
SHMB followed closely the protocol of Sharma et al. (1993) using Dynabeads as described in “Method 1” of the manufacturer's instructions (Dynal Biotech, Hamburg, Germany). The mRNAs from attacked (tester) and control (driver) plants were isolated with paramagnetic oligo(dT)25 beads (Dynabeads) from 100 μg of total RNAs treated with DNase I (Life Technologies). Driver mRNA on the beads was directly converted to the complementary first strand cDNA using SuperScript-II reverse transcriptase and rTth reverse polymerase (Life Technologies) according to the manufacturer's instructions. Tester mRNA was then eluted from the beads and hybridized to the driver cDNA, which was immobilized on the Dynabeads by the RT reaction. After three stringent hybridizations (each for 24 h) in 4.5× SSPE and 0.1% (w/v) SDS buffers at 68°C and removal of the subtracted mRNA, 20 μL of fresh oligo(dT)25 beads was used for collecting the mRNA left after subtraction. The eluted mRNA was reverse transcribed to the first strand cDNA using SuperScript-II reverse transcriptase (Life Technologies) and oligo(dT)21 (MWG Biotech). Subsequently, DNA polymerase I and RNase H were used for second strand synthesis. Double-strand cDNA was blunt ended with T4 DNA polymerase (Amersham Pharmacia Biotech) and subsequently treated with T4 polynucleotide kinase for cloning to pUC18 vector prepared by restriction with SmaI enzyme.
Fabrication of cDNA Microarray. Fabrication of cDNA Microarray
The cDNAs cloned in the pCR2.1-TOPO (Hermsmeier et al., 2001) and pUC18 vectors were PCR amplified with the following primers derived from vector sequences close to the insert: TOP5-20, 5′-CAGTGTGCTGGAATTCGCCC-3′; TOP6-21, 5′-GGATATCTGCAGAATTCGCCC-3′; SMA1-19, 5′-GAATTCGAGCTCGGTACCC-3′; SMA4-23, 5′-CAGGTCGACTCTAGAGGATCCCC-3′; SMA3-22, 5′-TACGAATTCGAGCTCGGTACCC-3′; and SMA2-20, 5′-GTCGACTCTAGAGGATCCCC-3′. For pCR2.1-TOPO, TOP5-20, and TOP6-21 were used. For pUC18, primer pairs SMA3-22 and SMA2-20 and SMA1-19 and SMA4-23 were used.
For the preparation of the well-characterized control genes, plasmid pNATGUS3 (Krügel et al., 2002) digested with BstEII and NcoI was used as a vector to clone the following N. attenuata gene PCR fragments digested with the same enzymes: pi, primers, PIA1-34 (5′-GCGGCGGGTCACCGTACTTTAGTGATGATGGAAC-3′) and PIA2-32 (5′-GCGGCGCCATGGCTTACAACCCTTCGTGCCTG-3′); template, chromosomal DNA of N. attenuata; pmt1, primers, PMT6-36 (5′-GCGGCGGGTCACCGGTACCAACACAAATGGCTCTAC-3′) and PMT7-31 (5′-GCGGCGCCATGGAGCCCTTAAAGACTTGACG-3′); template, pmt1 cDNA cloned on plasmid pBI121-ASPMT (Voelckel et al., 2001); aos, primers, AOS1-35 (5′-GCGGCGGGTCACCGTGTTCTTTCTTATCTTGATCC-3′) and AOS2-31 (5′-GCGGCGCCATGGAAGTAGGAAAACCAAGAAC-3′); template, chromosomal DNA of N. attenuata; xet, primers, XET1-32 (5′-GCGGCGGGTCACCATTCACAGCTTCTTACAGG-3′) and XET2-33 5′-GCGGCGCCATGGCCTTGAACGCTTGCATTCAGG-3′); template, RB271 (this publication); and wrky, primers, TFN1-34 (5′-GCGGCGGGTCACCGGAACCAATCATGGAATTATC-3′) and TFN2-31 5′-GCGGCGCCATGGTGGGACAATTTGGGAAAG-3′); template, RE322 (this publication), yielding plasmids pNATPI1, pNATPMT1, pNATAOS1, pNATXET1, and (with wrky fragment) pNATTFN1, respectively. Afterward, the N. attenuata control gene PCR products for spotting onto the chip were synthesized as follows: pi, hpl, pmt1, aos, xet, and wrky with primers ASV5-21 (5′-GGAGAAACTCGACCGGTCACC-3′) and ASV6-22 (5′-CTACAAATCTATCTCTCCATGG-3′); templates pNATPI1, pNATHPL1 (Krügel et al., 2002), pNATPMT1, pNATAOS1, pNATXET1, and pNATTFN1, respectively; 3′ region of lox with primers LOX4-22 (5′-CTTTGGCGTTTTGATTTGGAAG-3′), ASV6-22, template pNATLOX1 (Krügel et al., 2002), and 5′ region of lox with primers ASV5-21, LOX3-21 (5′-CCAGTGCGACAACGTCTTGGG-3′), and template pNATLOX1. For each cDNA, two PCR fragments, with 5′-Aminolink C6 modification (Sigma-ARK, Darmstadt, Germany) on either strand, were synthesized. Even-numbered fragments (Table I) carry the Aminolink modification at primers TOP5-20, SMA4-23, or ASV6-22, whereas odd numbered fragments carry the modification at primers TOP6-21, SMA3-22, or ASV5-21.
PCR products were purified by a PCR purification kit (QIAquick, Qiagen, Hilden, Germany) following the manufacturer's instructions. Agarose gel electrophoresis was performed to confirm the purity, and the concentration of the amplified products was determined spectrophotometrically. Commercially available Lys-coated slides (PL-25C Poly-l-Lys slides, CEL Associates, Inc., Houston) were used. Before spotting, all the cDNA samples were concentrated through a micron-MultiScreen-PCR (Millipore, Bedford, MA) to approximately 0.5 to 1.0 μg μL−1 with spotting solution from Telechem (CEL Associates).
All the cDNA samples were arrayed four times (so that each gene was represented by eight spots) on the slides by a robot equipped with four printing tips (OmniGrid Microarrayer, Genemachine, San Carlos, CA). A list of genes on the microarray is in Table I. The spotted DNA on slides was hydrated in 1× SSC buffer for 1 to 5 min and snap dried at 140°C for 3 s, followed by cross-linking with a Stratalinker-2400 apparatus (Stratagene, La Jolla, CA). To prevent intrinsic fluorescence, the surface of the slides was treated with a blocking solution containing 5.5 g of succinic anhydride dissolved in 335 mL of 1-methyl-2-pyrrlidinone, mixed in 15 mL of 1 m NaBorate prepared with boric acid, and adjusted with NaOH to pH 8.0. Finally, spotted DNA on slides was denatured in boiling water for 2 min, rinsed with ethanol, and dried by centrifugation. After prehybridization processing, sample slides were hybridized with Cy3- or Cy5-labeled random primers (9 mer) to examine qualitative characteristics of the microarrays.
Microarray hybridization and quantification: Poly(A+) RNAs were isolated from 100 μg of total RNA (adjusted to a 1:1 [root:shoot] concentration) with Dynabeads Oligo(dT)25 (Dynal Biotech) and used for RT. To synthesize the first strand, 2 μg of poly(A+) RNAs was mixed with 4 μg of random hexamer (pdN6, Sigma), 4 μg oligo(dT)22 (Sigma) in 15 μL, and incubated at 65°C for 10 min. Subsequently, 0.6 μL of 50× dUTP/dNTPs [10 μL of each 100 mm dATP, dGTP, and dCTP; 6 μL of 100 mm dTTP; and 4 μL of 100 mm dUTP [5-(3-Aminoallyl)-2′-deoxyuridine 5′-triphosphate sodium salt, Sigma], 6 μL of 5× buffer, 3 μL of dithiothreitol (0.1 m), 1.9 μL of SuperScript (RNase II Hfree) reverse transcriptase, and 3.5 μL of water were added to a volume of 30 μL and incubated at 42°C for 2 h. cDNA/mRNA hybrids were hydrolyzed with 10 μL of NaOH (1 n) and 10 μL of EDTA (0.5 m) and incubated at 65°C for 15 min after neutralization with 25 μL (1 m) Tris (pH 7.4).
The cDNA mixtures were cleaned with a Microcon 30 concentrator (YM-30, Millipore) and dried in a speed vac. The pellets of both induced and control sample were resuspended in 9 μL of NaHCO3 buffer (0.5 m, pH 9.0) and added to the dried aliquot of monofunctional NHS-ester Cy3 dye and to Cy5 dye (Amersham Pharmacia Biotech), respectively, for labeling at room temperature in darkness. After 1.5 h, the Cy3 and Cy5 reactions were quenched with 4.5 μL of hydroxylamine (4 m) and mixed. After purification with Qiaquick PCR purification kit (Qiagen), the eluted products were dried in a speed vac. The labeling efficiency of the cDNA probe was checked by a spectrophotometer at a wavelength of 200 to 700 nm.
The probe solution was prepared by resuspending the dried pellets in a hybridization buffer consisting of 2 μL of poly(A+)(22) (10 μg μL−1, MWG Biotech), 5 μL of 20× SSC, 2 μL of yeast-tRNA (1.25 μg μL−1, Life Technologies), 0.6 μL of 10% (w/v) SDS, and 20.4 μL of distilled water for a final volume of 30 μL. The probe solution (after heating at 95°C for 2 min) was hybridized to the microarray, which was denatured in boiling water for 2 min, dried before use, and covered with a silanized coverslip. Hybridization was carried out for 12 h in a hybridization chamber (a 50-mL falcon tube supplied with 2 mL of 20× SSC on Whatman paper) and placed in a hybridization oven at 55°C to 58°C. After hybridization, the slides were immediately washed, initially with a solution of 1× SSC and 0.1% (w/v) SDS for 15 min, then with a solution of 1× SSC for 5 min, before being dried by centrifugation (3 min at 1,000 rpm).
A ScannArray-3000 (GSI Lumonics, Watertown, MA) was used to scan the hybridized cDNA with sequential scanning for Cy5 cDNA and then for Cy3cDNA at a maximum resolution of 10 μm pixel−1 with a 16-bit depth. The hybridization images were evaluated using the program AIDA Image Analyzer (Raytest Isotopenmeβgräte GmbH, Straubenhardt, Germany). Each image was overlaid with a grid to assess the signal strength from each spot. The background correction was manually calculated around each spot with a depth of 2 pixels. To calculate a microarray-specific normalization factor, the measured Cy5 and Cy3 fluorescence intensities were ranked independently and after discarding the 12.5% maximum and minimum values, the remaining 75% of the values were summed. The array-specific normalization factor was obtained by dividing the calculated sum of Cy3 values by those of the Cy5 values. The ratios of normalized fluorescence values for Cy3 and Cy5 of each individual spot for which an adequate background correction could be determined (typically five–six of the eight replicate spots for each gene) were used to calculate the mean and (sds) for each cDNA. We arbitrarily defined cDNAs with mean expression ratios of ≤0.5 and ≥1.50 as being differentially expressed (down- and up-regulated, respectively).
ACKNOWLEDGMENTS
We thank Susan Kutschbach for the DNA preparation; Thomas Hahn and Dominika Schnabelrauch for sequencing; Evelyn Claussen for assistance with the figures; Anja Paschold, Kristine Brathen, and Katja Schenke for technical assistance; and Rayko Halitschke, Claudia Voelckel, and Emily Wheeler for helpful comments on the manuscript.
Footnotes
This work was supported by the Max Planck Gesellschaft.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.018176.
LITERATURE CITED
- Appel M, Bellstedt DU, Fresshoff PM. Differential display of eukaryotic mRNA: meeting the demands of the new millennium? J Plant Physiol. 1999;154:561–570. [Google Scholar]
- Arrowsmith DA, Desilva J. Characterization of 2 tomato fruit-expressed cDNAs encoding xyloglucan endo-transglycosylase. Plant Mol Biol. 1995;28:391–403. doi: 10.1007/BF00020389. [DOI] [PubMed] [Google Scholar]
- Baldwin IT. Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA. 1998;95:8113–8118. doi: 10.1073/pnas.95.14.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT. An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiol. 2001;127:1449–1458. [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT, Halitschke R, Kessler A, Schittko U. Merging molecular and ecological approaches in plant-insect interactions. Curr Opin Plant Biol. 2001;4:351–358. doi: 10.1016/s1369-5266(00)00184-9. [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Karb MJ, Ohnmeiss TE. Allocation of 15N from nitrate to nicotine: production and turnover of a damage-induced mobile defense. Ecology. 1994;75:1703–1713. [Google Scholar]
- Belanger FC, Leustek T, Chu BY, Kriz AL. Evidence for the thiamine biosynthetic pathway in higher-plant plastids and its developmental regulation. Plant Mol Biol. 1995;29:809–821. doi: 10.1007/BF00041170. [DOI] [PubMed] [Google Scholar]
- Blee E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 2002;7:315–321. doi: 10.1016/s1360-1385(02)02290-2. [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Stauber EJ, Krock B, Oldham NJ, Gershenzon J, Baldwin IT. Gene expression of 5-epi-aristolochene synthase and formation of capsidiol in roots of Nicotiana attenuata and N. sylvestris. Phytochemistry. 2002;60:109–116. doi: 10.1016/s0031-9422(02)00080-8. [DOI] [PubMed] [Google Scholar]
- Braam J, Sistrunk ML, Polisensky DH, Xu W, Purugganan MM, Antosiewicz DM, Campbell P, Johnson KA. Life in a changing world: TCH gene regulation of expression and responses to environmental signals. Physiol Plant. 1996;98:909–916. [PubMed] [Google Scholar]
- Brownlee C. Role of the extracellular matrix in cell-cell signalling: paracrine paradigms. Curr Opin Plant Biol. 2002;5:396–401. doi: 10.1016/s1369-5266(02)00286-8. [DOI] [PubMed] [Google Scholar]
- Butt A, Mousley C, Morris K, Beynon J, Can C, Holub E, Greenberg JT, Buchanan-Wollaston V. Differential expression of a senescence-enhanced metallothionein gene in Arabidopsis in response to isolates of Peronospora parasitica and Pseudomonas syringae. Plant J. 1998;16:209–221. doi: 10.1046/j.1365-313x.1998.00286.x. [DOI] [PubMed] [Google Scholar]
- Chen WQ, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou GZ, Whitham SA et al. Expression profile matrix of Arabidopsistranscription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Degan F, Rocher A, Cameron-Mills V, Von WD. The expression of serine carboxypeptidases during maturation and germination of the barley grain. Proc Natl Acad Sci USA. 1994;91:8209–8213. doi: 10.1073/pnas.91.17.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C, Karafyllidis I, Wasternack C, Turner JG. The Arabidopsismutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell. 2002;14:1557–1566. doi: 10.1105/tpc.002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmermann M, Braun HP, Schmitz UK. The Adp Atp translocator from potato has a long amino-terminal extension. Curr Genet. 1991;20:405–410. doi: 10.1007/BF00317069. [DOI] [PubMed] [Google Scholar]
- Emons AMC, Mulder BM. How the deposition of cellulose microfibrils builds cell wall architecture. Trends Plant Sci. 2000;5:35–40. doi: 10.1016/s1360-1385(99)01507-1. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Chappell J. Gene family for an elicitor-induced sesquiterpene cyclase in tobacco. Proc Natl Acad Sci USA. 1992;89:11088–11092. doi: 10.1073/pnas.89.22.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Kouchi H, Ichikawa T, Syono K. Cloning of cDNAs for genes that are specifically or preferentially expressed during the development of tobacco genetic tumors. Plant J. 1994;5:645–654. doi: 10.1111/j.1365-313x.1994.00645.x. [DOI] [PubMed] [Google Scholar]
- Genschik P, Criqui MC, Parmentier Y, Marbach J, Durr A, Fleck J, Jamet E. Isolation and characterization of a cDNA-encoding a 3-hydroxy-3-methylglutaryl coenzyme A reductase from Nicotiana sylvestris. Plant Mol Biol. 1992;20:337–341. doi: 10.1007/BF00014504. [DOI] [PubMed] [Google Scholar]
- Giritch A, Ganal M, Stephan UW, Baumlein H. Structure, expression and chromosomal localisation of the metallothionein-like gene family of tomato. Plant Mol Biol. 1998;37:701–714. doi: 10.1023/a:1006001701919. [DOI] [PubMed] [Google Scholar]
- Glawe GA, Zavala JA, Kessler A, van Dam NM, Baldwin IT. Ecological costs and benefits correlated with trypsin protease inhibitor production in Nicotiana attenuata. Ecology. 2003;84:79–90. [Google Scholar]
- Haake V, Zrenner R, Sonnewald U, Stitt M. A moderate decrease of plastid aldolase activity inhibits photosynthesis, alters the levels of sugars and starch, and inhibits growth of potato plants. Plant J. 1998;14:147–157. doi: 10.1046/j.1365-313x.1998.00089.x. [DOI] [PubMed] [Google Scholar]
- Halitschke R, Gase K, Hui D, Schmidt D, Baldwin IT (2003) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: VI Microarray analysis reveals that most herbivore-specific transcriptional changes are mediated by fatty acid-amino acid conjugates. Plant Physiol (in press) [DOI] [PMC free article] [PubMed]
- Halitschke R, Kessler A, Kahl J, Lorenz A, Baldwin IT. Ecophysiological comparison of direct and indirect defenses in Nicotiana attenuata. Oecologia. 2000;124:408–417. doi: 10.1007/s004420000389. [DOI] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol. 2001;125:711–717. doi: 10.1104/pp.125.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Castresana C. Alpha-oxidation of fatty acids in higher plants: identification of a pathogen-inducible oxygenase (PIOX) as an alpha-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J Biol Chem. 1999;274:24503–24513. doi: 10.1074/jbc.274.35.24503. [DOI] [PubMed] [Google Scholar]
- Hamilton EW, Coleman JS. Heat-shock proteins are induced in unstressed leaves of Nicotiana attenuata(Solanaceae) when distant leaves are stressed. Am J Bot. 2001;88:950–955. [PubMed] [Google Scholar]
- Hatton D, Gray JC. Two MAR DNA-binding proteins of the pea nuclear matrix identify a new class of DNA-binding proteins. Plant J. 1999;18:417–429. doi: 10.1046/j.1365-313x.1999.00468.x. [DOI] [PubMed] [Google Scholar]
- Herbers K, Lorences EP, Barrachina C, Sonnewald U. Functional characterisation of Nicotiana tabacumxyloglucan endotransglycosylase (NtXET-1): generation of transgenic tobacco plants and changes in cell wall xyloglucan. Planta. 2001;212:279–287. doi: 10.1007/s004250000393. [DOI] [PubMed] [Google Scholar]
- Hermsmeier D, Schittko U, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: I. Large-scale changes in the accumulation of growth- and defense-related plant mRNAs. Plant Physiol. 2001;125:683–700. doi: 10.1104/pp.125.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Sugita M, Sugiura M. CDNA structure, expression and nucleic acid-binding properties of 3 RNA-binding proteins in tobacco: occurrence of tissue-specific alternative splicing. Nucleic Acids Res. 1993;21:3981–3987. doi: 10.1093/nar/21.17.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Dube SK, Barnett NM, Kung SD. Nucleotide sequence of gene Oee2-a and its complementary DNA encoding 23 Kda polypeptide of the oxygen-evolving complex of photosystem II in tobacco. Plant Mol Biol. 1991;17:551–554. doi: 10.1007/BF00040655. [DOI] [PubMed] [Google Scholar]
- Jesenberger V, Jentsch S. Deadly encounter: ubiquitin meets apoptosis. Nat Rev Mol Cell Biol. 2002;3:112–121. doi: 10.1038/nrm731. [DOI] [PubMed] [Google Scholar]
- Kahl J, Siemens DH, Aerts RJ, Gabler R, Kuhnemann F, Preston CA, Baldwin IT. Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore. Planta. 2000;210:336–342. doi: 10.1007/PL00008142. [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. Defensive function of herbivore-induced plant volatile emissions in nature. Science. 2001;291:2141–2144. doi: 10.1126/science.291.5511.2141. [DOI] [PubMed] [Google Scholar]
- Kim HS, Delaney TP. ArabidopsisSON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell. 2002;14:1469–1482. doi: 10.1105/tpc.001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima S, Sato F. Plant pathogenesis-related proteins: molecular mechanisms of gene expression and protein function. J Biochem. 1999;125:1–8. doi: 10.1093/oxfordjournals.jbchem.a022244. [DOI] [PubMed] [Google Scholar]
- Korth KL, Dixon RA. Evidence for chewing insect-specific molecular events distinct from a general wound response in leaves. Plant Physiol. 1997;115:1299–1305. doi: 10.1104/pp.115.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth KL, Jaggard DAW, Dixon RA. Developmental and light-regulated post-translational control of 3-hydroxy-3-methylglutaryl-CoA reductase levels in potato. Plant J. 2000;23:507–516. doi: 10.1046/j.1365-313x.2000.00821.x. [DOI] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology. 2002;12:177–183. [Google Scholar]
- Laxalt AM, Cassia RO, Sanllorenti PM, Madrid EA, Andreu AB, Daleo GR, Conde RD, Lamattina L. Accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase RNA under biological stress conditions and elicitor treatments in potato. Plant Mol Biol. 1996;30:961–972. doi: 10.1007/BF00020807. [DOI] [PubMed] [Google Scholar]
- Lee MW, Qi M, Yang YO. A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol Plant-Microbe Interact. 2001;14:527–535. doi: 10.1094/MPMI.2001.14.4.527. [DOI] [PubMed] [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC. BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2001;98:5916–5921. doi: 10.1073/pnas.091065998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Averboukh L, Pardee AB. Distribution and cloning of eukaryotic messenger-RNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 1993;21:3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y-T, Martin GB. The disease-resistance gene PTO and the fenthion-sensitivity gene FEN encode closely related functional protein kinases. Proc Natl Acad Sci USA. 1995;92:4181–4184. doi: 10.1073/pnas.92.10.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic ZJ, Schroder WP, Pakrasi HB, Irrgang KD, Herrmann RG, Oelmuller R. Molecular characterization of psbw, a nuclear-encoded component of the photosystem II reaction center complex in spinach. Proc Natl Acad Sci USA. 1995;92:8930–8934. doi: 10.1073/pnas.92.19.8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thalianaduring systemic acquired resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Mandujano-Chavez A, Schoenbeck MA, Ralston LF, Lozoya-Gloria E, Chappell J. Differential induction of sesquiterpene metabolism in tobacco cell suspension cultures by methyl jasmonate and fungal elicitor. Arch Biochem Biophys. 2000;381:285–294. doi: 10.1006/abbi.2000.1961. [DOI] [PubMed] [Google Scholar]
- Manjunath S, Sachs MM. Molecular characterization and promoter analysis of the maize cytosolic glyceraldehyde 3-phosphate dehydrogenase gene family and its expression during anoxia. Plant Mol Biol. 1997;33:97–112. doi: 10.1023/a:1005729112038. [DOI] [PubMed] [Google Scholar]
- Martin C, PazAres J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- Meichtry J, Amrhein N, Schaller A. Characterization of the subtilase gene family in tomato (Lycopersicon esculentumMill.) Plant Mol Biol. 1999;39:749–760. doi: 10.1023/a:1006193414434. [DOI] [PubMed] [Google Scholar]
- Moran PJ, Thompson GA. Molecular responses to aphid feeding in Arabidopsisin relation to plant defense pathways. Plant Physiol. 2001;125:1074–1085. doi: 10.1104/pp.125.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura DS, Bergey DR, Ryan CA. Characterization and localization of a wound-inducible type I serine-carboxypeptidase from leaves of tomato plants (Lycopersicon esculentumMill.) Planta. 2001;212:222–230. doi: 10.1007/s004250000380. [DOI] [PubMed] [Google Scholar]
- Oneal JK, Pokalsky AR, Kiehne KL, Shewmaker CK. Isolation of tobacco SSU genes: characterization of a transcriptionally active pseudogene. Nucleic Acids Res. 1987;15:8661–8677. doi: 10.1093/nar/15.21.8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G, Middlesteadt W, Desai N, Williams S, Dincher S, Carnes M, Ryals J. Isolation and sequence of a genomic clone encoding the basic form of pathogenesis-related protein-1 from Nicotiana tabacum. Plant Mol Biol. 1989;12:595–596. doi: 10.1007/BF00036973. [DOI] [PubMed] [Google Scholar]
- Petitot AS, Blein JP, Pugin A, Suty L. Cloning of two plant cDNAs encoding a beta-type proteasome subunit and a transformer-2-like SR-related protein: early induction of the corresponding genes in tobacco cells treated with cryptogein. Plant Mol Biol. 1997;35:261–269. doi: 10.1023/a:1005833216479. [DOI] [PubMed] [Google Scholar]
- Prasad TK, Hack E, Hallberg RL. Function of the maize mitochondrial chaperonin Hsp60-specific association between Hsp60 and newly synthesized F1-atpase alpha-subunits. Mol Cell Biol. 1990;10:3979–3986. doi: 10.1128/mcb.10.8.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro A, Praekelt U, Akkermans ADL, Meacock PA, vanKammen A, Bisseling T, Pawlowski K. Identification of agthi1, whose product is involved in biosynthesis of the thiamine precursor thiazole, in actinorhizal nodules of Alnus glutinosa. Plant J. 1996;10:361–368. doi: 10.1046/j.1365-313x.1996.10020361.x. [DOI] [PubMed] [Google Scholar]
- Riedel L, Putz A, Hauser MT, Luckinger R, Wassenegger M, Sanger HL. Characterization of the signal recognition particle (srp) RNA population of tomato (Lycopersicon esculentum) Plant Mol Biol. 1995;27:669–680. doi: 10.1007/BF00020221. [DOI] [PubMed] [Google Scholar]
- Royo J, Vancanneyt G, Perez AG, Sanz C, Stormann K, Rosahl S, SanchezSerrano JJ. Characterization of three potato lipoxygenases with distinct enzymatic activities and different organ-specific and wound-regulated expression patterns. J Biol Chem. 1996;271:21012–21019. doi: 10.1074/jbc.271.35.21012. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996;15:5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Sanz A, Moreno JI, Castresana C. PIOX, a new pathogen-induced oxygenase with homology to animal cyclooxygenase. Plant Cell. 1998;10:1523–1537. doi: 10.1105/tpc.10.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe M, Toyoda K, Shiraishi T, Inagaki Y, Ichinose Y. cDNA cloning and characterization of tobacco ABC transporter: NtPDR1 is a novel elicitor-responsive gene. FEBS Lett. 2002;518:164–168. doi: 10.1016/s0014-5793(02)02697-2. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, Awai K, Amagai M, Kuwata C, Tsugane T, Masuda T et al. Monitoring of methyl jasmonate-responsive genes in Arabidopsisby cDNA macroarray: self-activation of jasmonic acid biosynthesis and cross talk with other phytohormone signaling pathways. DNA Res. 2001;8:153–161. doi: 10.1093/dnares/8.4.153. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsisrevealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Hermsmeier D, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: II. Accumulation of plant mRNAs in response to insect-derived cues. Plant Physiol. 2001;125:701–710. doi: 10.1104/pp.125.2.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Preston CA, Baldwin IT. Eating the evidence? Manduca sexta larvae cannot disrupt specific jasmonate induction in Nicotiana attenuataby rapid consumption. Planta. 2000;210:343–346. doi: 10.1007/PL00008143. [DOI] [PubMed] [Google Scholar]
- Sharma P, Lonneborg A, Stougaard P. PCR-based construction of subtractive cDNA library using magnetic beads. BioTechniques. 1993;15:610–611. [PubMed] [Google Scholar]
- Shen BZ, Zheng ZW, Dooner HK. A maize sesquiterpene cyclase gene induced by insect herbivory and volicitin: characterization of wild-type and mutant alleles. Proc Natl Acad Sci USA. 2000;97:14807–14812. doi: 10.1073/pnas.240284097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih MC, Lazar G, Goodman HM. Evidence in favor of the symbiotic origin of chloroplasts: primary structure and evolution of tobacco glyceraldehyde-3-phosphate dehydrogenases. Cell. 1986;47:73–80. doi: 10.1016/0092-8674(86)90367-3. [DOI] [PubMed] [Google Scholar]
- Sivasankar S, Sheldrick B, Rothstein SJ. Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol. 2000;122:1335–1342. doi: 10.1104/pp.122.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart LB, Moskal WA, Cameron KD, Bennett AB. MIP genes are down-regulated under drought stress in Nicotiana glauca. Plant Cell Physiol. 2001;42:686–693. doi: 10.1093/pcp/pce085. [DOI] [PubMed] [Google Scholar]
- Tsugeki R, Mori H, Nishimura M. Purification, cDNA cloning and northern-blot analysis of mitochondrial chaperonin 60 from pumpkin cotyledons. Eur J Biochem. 1992;209:453–458. doi: 10.1111/j.1432-1033.1992.tb17309.x. [DOI] [PubMed] [Google Scholar]
- van Dam NM, Hadwich K, Baldwin IT. Induced responses in Nicotiana attenuata affect behavior and growth of the specialist herbivore Manduca sexta. Oecologia. 2000;122:371–379. doi: 10.1007/s004420050043. [DOI] [PubMed] [Google Scholar]
- Van Dam NM, Hermenau U, Baldwin IT. Instar-specific sensitivity of specialist Manduca sexta larvae to induced defences in their host plant Nicotiana attenuata. Ecol Entomol. 2001a;26:578–586. [Google Scholar]
- Van Dam NM, Horn M, Mares M, Baldwin IT. Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J Chem Ecol. 2001b;27:547–568. doi: 10.1023/a:1010341022761. [DOI] [PubMed] [Google Scholar]
- Voelckel C, Krugel T, Gase K, Heidrich N, van Dam NM, Winz R, Baldwin IT. Anti-sense expression of putrescine N-methyltransferase confirms defensive role of nicotine in Nicotiana sylvestris against Manduca sexta. Chemoecology. 2001;11:121–126. [Google Scholar]
- Winz RA, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: IV. Insect-induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol. 2001;125:2189–2202. doi: 10.1104/pp.125.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Komori T, Hashimoto A, Kuwata S, Imaseki H, Kubo T. Differential expression of plastidic aldolase genes in Nicotianaplants under salt stress. Plant Sci. 2000;154:61–69. doi: 10.1016/s0168-9452(00)00188-6. [DOI] [PubMed] [Google Scholar]
- Yamada S, Komori T, Myers PN, Kuwata S, Kubo T, Imaseki H. Expression of plasma membrane water channel genes under water stress in Nicotiana excelsior. Plant Cell Physiol. 1997;38:1226–1231. doi: 10.1093/oxfordjournals.pcp.a029109. [DOI] [PubMed] [Google Scholar]
- Yang YO, Klessig DF. Isolation and characterization of a tobacco mosaic virus-inducible myb oncogene homolog from tobacco. Proc Natl Acad Sci USA. 1996;93:14972–14977. doi: 10.1073/pnas.93.25.14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Ashikari T, Tanaka Y, Kusumi T, Hase T. Molecular characterization of tobacco sulfite reductase: enzyme purification, gene cloning, and gene expression analysis. J Biochem. 1998;124:615–621. doi: 10.1093/oxfordjournals.jbchem.a022156. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. A comparative expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 2002;42:1025–1033. doi: 10.1093/pcp/pce154. [DOI] [PubMed] [Google Scholar]
- Zhou XR, Costantino P, De Paolis A. Isolation and sequence analysis of cDNA clones encoding the precursor of the 10-kDa polypeptide of photosystem II from tobacco. J Genet Breed. 1993;47:273–276. [Google Scholar]
- Ziegler J, Keinanen M, Baldwin IT. Herbivore-induced allene oxide synthase transcripts and jasmonic acid in Nicotiana attenuata. Phytochemistry. 2001;58:729–738. doi: 10.1016/s0031-9422(01)00284-9. [DOI] [PubMed] [Google Scholar]