Abstract
We have developed a computer based method to identify candidate single nucleotide polymorphisms (SNPs) and small insertions/deletions from expressed sequence tag data. Using a redundancy-based approach, valid SNPs are distinguished from erroneous sequence by their representation multiple times in an alignment of sequence reads. A second measure of validity was also calculated based on the cosegregation of the SNP pattern between multiple SNP loci in an alignment. The utility of this method was demonstrated by applying it to 102,551 maize (Zea mays) expressed sequence tag sequences. A total of 14,832 candidate polymorphisms were identified with an SNP redundancy score of two or greater. Segregation of these SNPs with haplotype indicates that candidate SNPs with high redundancy and cosegregation confidence scores are likely to represent true SNPs. This was confirmed by validation of 264 candidate SNPs from 27 loci, with a range of redundancy and cosegregation scores, in four inbred maize lines. The SNP transition/transversion ratio and insertion/deletion size frequencies correspond to those observed by direct sequencing methods of SNP discovery and suggest that the majority of predicted SNPs and insertion/deletions identified using this approach represent true genetic variation in maize.
The development of high-throughput methods for the detection of single nucleotide polymorphisms (SNPs) and small indels (insertion/deletions) has led to a revolution in their use as molecular markers. SNPs are increasingly becoming the marker of choice in genetic analysis and are used routinely as markers in agricultural breeding programs (Gupta et al., 2001). They also have many uses in human genetics, such as for the detection of alleles associated with genetic diseases and the identification of individuals (Nikiforov et al., 1994). SNPs are invaluable as a tool for genome mapping, offering the potential for generating very high-density genetic maps, which can be used to develop haplotyping systems for genes or regions of interest (Rafalski, 2002). The low mutation rate of SNPs also makes them excellent markers for studying complex genetic traits and as a tool for the understanding of genome evolution (Syvanen, 2001).
Unlike random amplified polymorphic DNAs and RFLPs, SNPs are direct markers because sequence information provides the exact nature of the allelic variants. They are far more prevalent than microsatellites and, therefore, may provide a high density of markers near a locus of interest. Recent evidence has shown that when comparing human DNA from two individuals, SNPs are found on average every 1 to 2 kb (Clifford et al., 2000; Deutsch et al., 2001). Limited work has been carried out to examine the occurrence of SNPs in plants, although these preliminary studies have indicated that SNPs appear to be even more abundant in plant systems than in the human genome. Germano and Klein (1999) identified five SNPs in 1 kb of nDNA of Picea rubens and Picea mariana, and also discovered SNPs in the chloroplasts of these species. Recently, Coryell et al. (1999) identified two SNPs in approximately 400 bp of sequence in soybean (Glycine max). In maize (Zea mays), they have been found even more frequently, with one SNP approximately every 48 bp and every 130 bp in 3′-untranslated regions and coding regions, respectively (Tenaillon et al., 2001; Rafalski, 2002). Mogg et al. (2002) amplified and sequenced the flanking region of 97 previously characterized microsatellite primer sets in 11 maize inbred lines. The sequencing results indicated that the flanking regions of maize microsatellites show increased levels of polymorphism when compared with other regions of the genome, with SNPs in these regions found on average every 40 bp.
As with the majority of molecular markers, one of the limitations of SNPs is the initial cost associated with their development. A variety of approaches have been adopted for the discovery of novel SNP markers. The conversion of microsatellite markers, identifying the relatively abundant nucleotide polymorphisms surrounding simple sequence repeats has the advantage that many of these markers have already been characterized (Mogg et al., 2002). A further approach identifies SNPs in overlapping genomic sequence, a product of the large genome sequencing programs (Taillon-Miller et al., 1998; Dawson et al., 2001). This later method requires experimental confirmation to preclude errors associated with cloning and sequencing procedures but provides the greatest potential for cost-effective SNP discovery because it uses preexisting sequence data. With the development of high-throughput sequencing technology, large amounts of data have been submitted to the various DNA databases that may be suitable for data mining and SNP discovery. In particular, expressed sequence tag (EST) sequencing programs have provided a wealth of information, identifying novel genes from a broad range of organisms and providing an indication of gene expression level in particular tissues (Adams et al., 1995). EST sequence data may provide the richest source of biologically useful SNPs due to the relatively high redundancy of gene sequence, the diversity of genotypes represented within databases, and the fact that each SNP would be associated with an expressed gene (Picoult-Newberg et al., 1999). The major drawback to this approach is that the relatively high sequence error naturally associated with EST programs may lead to the identification of false positives. We have attempted to overcome these difficulties by developing software for the automated detection of SNPs within EST data. A conservative approach was followed to limit the potential errors associated with cloning and sequencing so that only polymorphisms represented by two or more sequences were considered. For each polymorphism, two associated measurements of confidence in the validity of SNPs were also calculated. The frequency of occurrence of a polymorphism at a particular locus provides a primary measure of confidence in the SNP representing a true polymorphism and is referred to as the SNP redundancy score. The cosegregation of multiple SNPs within an alignment to define a haplotype provides a second measure of confidence in SNP validity and is referred to as the cosegregation score.
To assess this software, we have applied it to maize EST data collated at ZmDB as part of the Maize Gene Discovery Project (Gai et al., 2000). Analysis of these data and experimental verification of 264 of the identified SNPs indicate that this method is not only efficient at detecting SNPs between alleles from different maize genotypes but also identifies expressed paralogous genes present in the maize genome. Allelic SNP data is of use for current mapping and genotyping programs, whereas the ability to differentiate between duplicate paralogous genes is important for the true alignment of genomic sequence data for the forthcoming maize genome sequencing program (Bennetzen et al., 2002).
RESULTS
cDNA Assembly
Expressed maize sequences (102,551) consisting of a total of over 46 million nucleotides were retrieved from ZmDB (Gai et al., 2000). These sequences are derived from a variety of cDNA libraries produced as part of the maize gene discovery program or collated from GenBank. The maize inbred lines OH43, W23, B73, W64a, and BMS represent 32%, 23%, 20%, 6%, and 5% of the sequences, respectively, with the remaining 14% having no defined varietal identification. All sequences were retrieved in FASTA format and assembled into contigs using d2 cluster and cap3 using default values of 80% similarity for d2cluster and 95% for cap3. These sequences aligned into 13,247 contigs and 19,112 singletons. A total of 6,107 of the contigs (46%) contained four or more sequence reads, the minimum required for redundancy-based SNP detection (Table I).
Table I.
Cluster profile of maize EST data
| Cluster Size | Frequency |
|---|---|
| 1 | 15,506 |
| 2 | 4,662 |
| 3 | 2,478 |
| 4 | 1,422 |
| 5 | 966 |
| 6 | 647 |
| 7 | 518 |
| 8 | 351 |
| 9 | 274 |
| 10 | 234 |
| 11 | 198 |
| 12 | 170 |
| 13 | 140 |
| 14 | 118 |
| 15 | 95 |
| 16 | 89 |
| 17 | 70 |
| 18 | 69 |
| 19 | 50 |
| 20 | 61 |
| 21 | 36 |
| 22 | 34 |
| 23 | 35 |
| 24 | 32 |
| 25 | 37 |
| 26 | 23 |
| 27 | 20 |
| 28 | 20 |
| 29 | 23 |
| 30 | 20 |
| 31 | 15 |
| 32 | 20 |
| 33 | 17 |
| 34 | 12 |
| 35 | 10 |
| 36 | 9 |
| 37 | 16 |
| 38 | 15 |
| 39 | 9 |
| 40 | 12 |
| 41 | 13 |
| 42 | 8 |
| 43 | 4 |
| 44 | 12 |
| 45 | 11 |
| 46 | 8 |
| 47 | 2 |
| 48 | 8 |
| 49 | 3 |
| 50 | 5 |
| 51+ | 146 |
A total of 102,551 expressed maize sequences were aligned using d2 cluster and cap3. Minimum similarity thresholds of 80% and 95% were used for d2 cluster and cap3, respectively, and a minimum overlap of 100 bases was specified for cap3.
Identifying Candidate SNPs
Of the 6,107 contigs containing four or more sequence reads, 3,479 (57%) alignments contained candidate SNPs with a redundancy score of two or greater. Although only 22% of contigs of four reads contained candidate SNPs, this proportion increased rapidly with 75% of 10 read contigs and 89% of 15 read contigs containing candidate SNPs (Fig. 1). Alignments representing over 50 sequence reads contained a disproportionate number of SNPs due to random errors accumulating redundancy scores greater than one. Therefore, these larger contigs were ignored in calculating SNP abundance.
Figure 1.
Abundance of maize EST contigs containing candidate SNPs in relation to contig size. The frequency of contigs that contained SNPs was calculated for all alignments of increasing numbers of sequence reads.
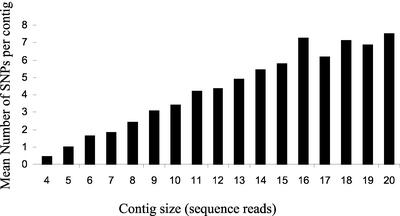
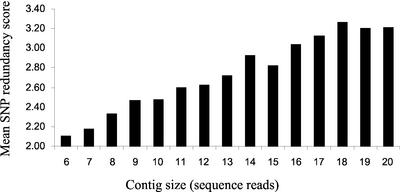
A total of 13,122 candidate SNPs were detected with SNP abundance increasing with increasing contig size (Fig. 2). This relates to approximate SNP frequencies of one per 600 bp of aligned sequence for five read contigs to one per 100 bp of aligned sequence for 20 read contigs. SNP scores ranged from two to 20, with mean SNP score increasing in direct proportion to the number of sequences in an alignment (Fig. 3).
Figure 2.
Abundance of candidate SNPs identified within contigs in relation to contig size for maize EST data. The mean SNP abundance was calculated for all alignments of increasing numbers of sequence reads.
Figure 3.
A measurement of SNP redundancy score in relation to contig size for maize EST data. The mean SNP redundancy score was calculated for all candidate SNPs identified in alignments of increasing numbers of sequence reads.
Along with the SNP redundancy score, a further measure of confidence of SNP validity was calculated based on the cosegregation of SNP pattern between multiple SNP loci within an alignment. SNPs between sequences representing divergence between two genes (orthologs or paralogs) would be expected to cosegregate defining a haplotype at multiple loci within an alignment, whereas sequencing errors would occur randomly between haplotypes. Where several candidate SNPs are detected within an alignment, the majority of SNPs cosegregate with a haplotype.
Validation of Candidate SNPs
A total of 264 candidate SNPs from 27 loci were validated using direct sequencing of PCR products. The SNPs were chosen based on a range of redundancy and cosegregation scores and predicted expression of multiple genes. Of the 264 candidate SNPs, 241 (91%) were shown to be true polymorphisms (Table II). Of the 23 candidate SNPs that were shown to be false, 22 (96%) had an SNP score of 2. Overall, 130 of the candidate SNPs had an SNP score of 2, of which 108 (83.1%) were shown to be true polymorphisms. The average weighted cosegregation value of the validated SNPs was 57.5%, with scores ranging from 6% to 100%. In all cases, the validated SNPs with low SNP, cosegregation, and weighted cosegregation scores were in contigs where many different haplotypes were present (Fig. 4). Frequently, one of these haplotypes was represented by a single sequence only and, therefore, could only be confirmed by sequence validation. The average weighted cosegregation score in the false SNPs was 9.5%, ranging from 4% to 24%, and the highest cosegregation score was 4/17.
Table II.
Details of the 27 loci genotyped and SNPs validated
| SNP Report | Predicted Gene No. | Predicted SNPs | Verified SNPs | Comments |
|---|---|---|---|---|
| 5 | 2 | 13 | 13 | Both genes amplified and SNPs between verified |
| 17 | 1 | 21 | 20 | The false SNP had low redundancy and cosegregation scores |
| 58 | 1 | 10 | 10 | |
| 72 | 1 | 4 | 4 | |
| 92 | 1 | 5 | 5 | |
| 136 | 1 | 12 | 12 | |
| 165 | 1 | 8 | 7 | The false SNP had low redundancy and cosegregation |
| 186 | 1 | 3 | 3 | |
| 187 | 1 | 11 | 10 | The false SNP had low redundancy and cosegregation |
| 194 | 1 | 9 | 8 | The false SNP had low redundancy and cosegregation |
| 196 | 1 | 8 | 8 | |
| 214 | 1 | 8 | 6 | The two false SNPs had low redundancy and cosegregation scores |
| 220 | 1 | 8 | 6 | The two false SNPs had low redundancy and cosegregation scores |
| 229 | 1 | 19 | 16 | |
| 231 | 1 | 9 | 9 | |
| 238 | 1 | 12 | 10 | The two false SNPs had low redundancy and cosegregation scores |
| 241 | 1 | 13 | 12 | The false SNP had low redundancy and cosegregation |
| 246 | 1 | 9 | 9 | |
| 260 | 1 | 6 | 5 | The false SNP had low redundancy and cosegregation |
| 301 | 1 | 8 | 7 | The false SNP had low redundancy and cosegregation |
| 325 | 1 | 9 | 9 | |
| 331 | 1 | 6 | 6 | |
| 365 | 2 | 16 | 16 | Both genes amplified and SNPs between verified |
| 429 | 2 | 9 | 7 | Both genes amplified and multiple haplotypes identified |
| 494 | 1 | 17 | 13 | Three SNPs could not be verified because the original lines are not documented. One false SNP had low redundancy and cosegregation scores |
| 531 | 1 | 6 | 5 | The false SNP had low redundancy and cosegregation |
| 578 | 2 | 5 | 5 | Both genes amplified and SNPs between verified |
Candidate SNPs from 27 loci (264), with a range of redundancy and cosegregation scores, were validated in the four inbred maize lines B73, W23, OH43, and W64a. The candidate SNPs that could not be verified had low redundancy and cosegregation scores. Where multiple genes were predicted in a sequence alignment and candidate SNPs were predicted between these genes, these SNPs were verified.
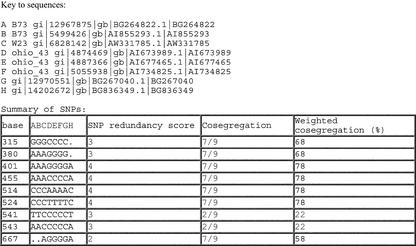
Figure 4.
AutoSNP summary report 246. This report depicts nine candidate SNPs, identifying their base position in the sequence alignment along with measures of confidence of SNP validity. The key relates the aligned sequences to original GenBank sequence identification and also identifies the maize line (where available) derived from the GenBank annotation. The SNP redundancy score measures the minimum number of sequences that represent a polymorphism. The cosegregation score is a measure of the number of SNPs in the alignment that share the same pattern of polymorphism between aligned sequences. The weighted cosegregation score corrects for missing data in the EST alignments that may otherwise bias the cosegregation score. In this example, all SNPs were verified as true polymorphisms.
Analysis of Base Changes
Candidate SNPs were categorized according to nucleotide substitution as either transitions (C/T or G/A) or transversions (C/T, A/G, C/A, or T/G; Table III). There was a relative increase in the proportion of transitions over transversions. We also observe a relative increase in frequency of C/A and its reverse complement, T/G transversions compared with C/G and A/T transversions (Table III).
Table III.
Nucleotide substitution frequencies for candidate SNPs identified in maize EST data
| Transitions | 6,640 |
| C/T | 3,277 |
| G/A | 3,363 |
| Transversions | 6,482 |
| C/G | 1,212 |
| A/T | 1,022 |
| C/A | 2,025 |
| T/G | 2,223 |
| Total | 13,122 |
Candidate polymorphisms (14,832) were identified with an SNP redundancy score of 2 or greater. Of these, 13,122 were nucleotide substitutions. The absolute frequencies of the different types of nucleotide substitutions were calculated.
Analysis of Insertions/Deletions
As well as nucleotide transitions and transversions, 1,710 insertion/deletion (indels) were identified as having a redundancy score of two or greater. These occur as single nucleotides or strings of up to 26 bases (Table IV). The frequency of indels does not decrease exponentially with their increase in length but displays a relative increase in frequency of six-, eight-, 10-, 12-, and 15-base indels. Analysis of indel sequences indicated a bias toward A and T nucleotides for both single-base and longer indels. Significantly, there was also an underrepresentation of CG dinucleotide indels compared with other dinucleotide indels (Table V).
Table IV.
Prevalence of candidate indels identified in maize EST data
| Indel Size | Frequency | Total |
|---|---|---|
| bp | % | |
| 1 | 1,014 | 59.30 |
| 2 | 230 | 13.45 |
| 3 | 168 | 9.82 |
| 4 | 84 | 4.91 |
| 5 | 48 | 2.81 |
| 6 | 72 | 4.21 |
| 7 | 25 | 1.46 |
| 8 | 34 | 1.99 |
| 9 | 6 | 0.35 |
| 10 | 11 | 0.64 |
| 11 | 3 | 0.18 |
| 12 | 5 | 0.29 |
| 13 | 1 | 0.06 |
| 14 | 1 | 0.06 |
| 15 | 4 | 0.23 |
| 16 | 0 | 0.00 |
| 17 | 1 | 0.06 |
| 18 | 2 | 0.12 |
| 26 | 1 | 0.06 |
Candidate polymorphisms (14,832) were identified with an SNP redundancy score of 2 or greater. Of these, 1710 were small indels. The absolute frequencies of different size indels was calculated, as well as a percentage of the total no. of indels (1,710).
Table V.
The frequency of single and dinucleotide indel sequences predicted from the alignment of 102,551 maize EST sequences
| Indel | Frequency |
|---|---|
| A | 318 |
| C | 207 |
| G | 186 |
| T | 303 |
| AA | 25 |
| AC | 14 |
| AG | 18 |
| AT | 23 |
| CA | 12 |
| CC | 7 |
| CG | 3 |
| CT | 16 |
| GA | 15 |
| GC | 11 |
| GG | 6 |
| GT | 9 |
| TA | 15 |
| TC | 15 |
| TG | 17 |
| TT | 24 |
Of the 1,710 candidate indels identified with an SNP redundancy score of 2 or greater, 1,014 were 1-bp indels and 230 were 2-bp indels. The absolute frequencies of the different 1- and 2-bp indel compositions were calculated, demonstrating the relatively low frequency of C or G indels.
Differentiation between Paralogs and Orthologs
The identification of multiple cosegregating SNPs within an alignment of EST sequences allows the accurate prediction of sequence haplotypes. Comparison of predicted haplotypes with known maize lines from which the sequences were derived allows the identification of predicted orthologous genes. By examining multiple SNPs from 100 random alignments representing over 1,450 sequence reads, a total of more than 250 haplotypes were observed. In 66 of these alignments, each of the haplotypes cosegregated with the maize lines from which the ESTs were derived, whereas for 34 alignments, the segregation of haplotypes indicated the expression of multiple genes within a single maize line.
DISCUSSION
SNPs are becoming the marker of choice for molecular genetic analysis. As with other molecular markers, their discovery and characterization is both expensive and laborious. Of the methods applied for the discovery of SNPs, the mining of sequence data sets should provide the cheapest source of abundant SNPs (Gu et al., 1998; Taillon-Miller et al., 1998; Buetow et al., 1999; Picoult-Newberg et al., 1999). Although every effort is made to produce and submit sequence of only the highest quality, the high-throughput nature of the sequencing programs inevitably leads to the submission of inaccuracies. The electronic filtering of these data to identify potentially biologically relevant polymorphisms is thereby hampered by the false calling of these bases. Previous methods used to identify SNPs in aligned sequence data has relied on the comparison of sequence trace files to filter out polymorphisms, where the base calling within one or more of the traces is of dubious quality and, therefore, likely to be due to sequence error rather than representative of a true polymorphism (Kwok et al., 1994; Garg et al., 1999; Marth et al., 1999). This method, although suitable for comparing genomic sequence, is limited by the requirement of sequence trace file data and does not distinguish errors incorporated during the reverse transcription of mRNA. For highly redundant data sets compiled from a variety of sources with a limited availability of sequence trace files, this means of filtering sequence errors from true polymorphisms is not feasible. However, the redundant nature of these EST data sets does permit the selection of polymorphisms that occur multiple times within a set of aligned sequences. The frequency of occurrence of a polymorphism at a particular locus provides a measure of confidence in the SNP representing a true polymorphism and is referred to as the SNP redundancy score. By examining SNPs that have a redundancy score of two or greater, i.e. two or more of the aligned sequences represent the polymorphism, the vast majority of sequencing errors are removed. Although some true genetic variation is also ignored due to its presence only once within an alignment, the high degree of redundancy within the data permits the rapid identification of large numbers of SNPs from data collated from a variety of sources.
We have applied this SNP detection method to maize sequence data compiled at ZmDB that consist both of sequences derived from the maize gene discovery program and those submitted to the DNA sequence repositories by other researchers. The 102,551 EST sequences were processed using stringent parameters to limit the alignment of multiple genes from gene families and identify polymorphisms between homologs from different maize lines. In removing the 54% of the total contigs, which contained less than four aligned sequence reads from the analysis, we greatly restrict the number of potentially polymorphic loci that may be detected. However, this sacrifice is necessary if we are to use redundancy to measure confidence in the validity of SNPs from the remaining loci. Analysis of larger data sets would increase the proportion of contigs containing more than four sequence reads and, therefore, would identify SNPs in a greater number of genes. The observation that the proportion of contigs that contain SNPs increases with contig size (Fig. 1) suggests that the number of SNP loci identified would increase with larger data sets. The mean number of SNPs identified per locus also increases with increasing number of sequences aligned (Fig. 2). This suggests that larger data sets with increased contig sizes would provide a greater number of SNPs per locus, whereas the increase in mean SNP score with contig size (Fig. 3) indicates that larger data sets would also provide a greater confidence in the validity of the predicted polymorphisms. Together, these results indicate that, although we have identified a large number of candidate SNPs in maize, these only represent a small proportion of the total genetic variation between maize expressed sequences.
Although using a redundancy-based approach to distinguish between sequence errors and true SNPs is highly efficient, the nonrandom nature of sequence error may lead to certain sequence errors within complex DNA structures being repeated between runs. Therefore, errors at these loci would have a relatively high SNP redundancy score and appear as confident SNPs. To identify these sequence errors and distinguish them from true SNPs, a further measure of SNP confidence was also calculated. Sequencing errors at complex loci are random between runs, whereas SNPs that represent divergence between homologous genes would cosegregate with haplotype. A cosegregation score based on the frequency of an SNP pattern occurring at multiple loci in an alignment allows ready identification of non-cosegregating SNPs. Weighting this score to account for the number of SNP loci and missing sequence data within an alignment further permits comparison of cosegregation scores across alignments. The SNP score and cosegregation score together provide a means for estimating confidence in the validity of SNPs within aligned sequences.
SNPs may differentiate between duplicate genes within a genome (paralogs) or orthologous genes between maize lines. Where validated SNPs are present in sequences derived from the same line, they must be due to gene duplication and the expression of the resulting paralogous genes. SNPs that define haplotypes that differentiate between maize lines may represent orthologous genes, although it is possible that segregation of these SNPs may be coincidental or reflect the differential expression of paralogous genes found in cDNA libraries produced from different lines. Analysis of SNPs from 100 random contigs suggests that 34 contain multiple genes from the same line. This is in line with other reports of duplicate gene copy number in maize and reflects its ancient allotetraploid origin (Gaut and Doebley, 1997; Gaut, 2001). Estimation of the copy number of closely related genes is essential for the avoidance of errors in whole genome sequence assembly (Eichler, 1998). Therefore, the availability of large numbers of SNPs that differentiate between maize paralogs should assist in the assembly of the sequence data from the proposed maize genome sequencing program (Bennetzen et al., 2002).
The high frequency of transitions detected has been observed in previous SNP discovery programs (Garg et al., 1999; Picoult-Newberg et al., 1999; Deutsch et al., 2001) and reflects the high frequency of the C to T mutation after methylation (Coulondre et al., 1978). The relative abundance of the C/A and its reverse complement, T/G transversions compared with C/G and A/T transversions, was unexpected and remains to be explained.
Along with SNPs, 1,710 predicted indel polymorphisms in maize EST data were identified. The majority of indels (>80%) were three bases or less in length with a disproportionate increase in the frequency of six-, eight-, 10-, 12-, and 15-base indels (Table IV). This distribution is similar to that observed by Bhattramakki et al. (2002), with the exception that we failed to identify large indels, presumably due to larger indels limiting contig assembly. Indels that increase in size by 3 bp suggests selection for the conservation of reading frames within coding sequence. There was no differentiation between coding or non-coding regions during contig assembly or in the screening for indels. Although indels occur most frequently in non-coding regions, our data suggest that coding regions may also be a rich source of indel polymorphisms. Indels may be produced by errors in DNA synthesis, repair, or recombination or be due to the insertion and excision of transposable elements that often leave a characteristic DNA footprint of several nucleotide bases. The relative abundance of eight base indels was also observed in maize by Bhattramakki et al. (2002) and may be due to sequence duplication during insertion and excision of Ac/Ds transposable elements (Sutton et al., 1984). The high frequency of 10 base indels does not correspond to any characterized maize transposon footprint, and the source of these indels remains unexplained. Comparison of indel dinucleotide sequence frequencies reveals a relative bias against CG dinucleotides (Table V). This may reflect methylation and conversion of these sequences (Coulondre et al., 1978).
A selection of SNPs, representing a range of SNP type, redundancy scores, cosegregation scores, and predicted expression of multiple genes, were validated using direct sequencing of PCR products. These SNPs (91%) were verified to be true polymorphisms, demonstrating the ability of the program to predict true polymorphisms. The redundancy and cosegregation scores were also demonstrated to be accurate indicators of valid polymorphisms because all false SNPs had low scores. However, a small proportion of the validated SNPs also had low cosegregation scores; this was due to the presence of many haplotypes in the contig. This suggests that the greater the number of sequences aligned, the more haplotypes are accurately predicted.
CONCLUSION
In total, we have identified over 14,832 candidate sequence polymorphisms in maize EST sequence data, along with two measures of confidence for each predicted polymorphism. Segregation of these SNPs with haplotype along with validation demonstrates that candidate SNPs with high redundancy and cosegregation confidence scores are likely to represent true SNPs. The transition to transversion ratio and indel size frequencies correspond to those observed by direct sequencing methods of SNP discovery and suggest that the majority of predicted SNPs and indels identified using this approach represent true genetic variation in maize.
MATERIALS AND METHODS
Auto SNP Version 1.0
Candidate SNPs were detected using the PERL script Auto_snip version 1.0 available from the authors. Auto_snip clusters and contigs FASTA format sequences by acting as a wrapper for the clustering package d2cluster (Burke et al., 1999) and the contig building package cap3 (Huang and Madan, 1999). Using d2cluster to break the sequences into subgroups for cap3 assembly allows the analysis of more than 100,000 sequence reads on a desktop personal computer (1.3 GHz PIII, 576 Mb RAM) running Red Hat Linux. Auto_snip parses the d2cluster output table and generates a set of cluster output files in multiple FASTA format. These files are then passed as input to cap3 for contig building, after which the AceDB output file is parsed to produce a series of gapped multiple FASTA format files. Contigs containing at least four reads were selected for SNP detection by SNP score. Spacing characters (-) added during sequence alignment were considered as a fifth element in addition to A, C, G, and T. This permits the identification of insertion/deletion polymorphisms between sequences that may be used to differentiate between genes using an SNP-based assay. Where a nucleotide polymorphism was shared between two or more sequences, a candidate SNP was recorded and an SNP score was allocated that was equal to the minimum number of reads that share a common polymorphism. Where several SNPs are present in an alignment, a redundant cosegregation score was calculated for each SNP. This was measured as the frequency of that SNP pattern occurring among each of the SNPs identified in the alignment. This figure was then normalized to the number of sequences and number of SNPs detected in the alignment to produce a standard cosegregation score. An HTML format output file is generated to allow the user to browse through the SNP results. Relevant statistics are written to a summary HTML page and process log file during the clustering, contig building, and SNP detection phases of the analysis. Minimum similarity thresholds of 80% and 95% were used for d2cluster and cap3, respectively, and a minimum overlap of 100 bases was specified for cap3.
Experimental Validation
Twenty-seven SNP reports were selected for validation of 264 SNPs, based on a range of redundancy scores, cosegregation scores, and predicted multiple copy genes. Genomic DNA was isolated from the four inbred maize (Zea mays) lines OH43, B73, W23, and W64a, using the procedure of Edwards et al. (1991). Amplification of the 27 loci was performed using primers designed to the conserved sequence surrounding the SNPs, using the primer design program PRIMER version 0.5 (Whitehead Institute, Cambridge, MA). Amplifications were carried out in a 25-μL reaction volume containing 25 ng of DNA, 2.5 μL of 10× PCR reaction buffer (Qiagen, Valencia, CA), 15 pmol forward and reverse primers, 200 μm of each dNTP, and 2 units of HotStar Taq polymerase (Qiagen). After an initial hot start at 95°C for 15 min, the following cycling parameters were employed: denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min. After 35 rounds of amplification, a final extension step was performed at 72°C for 10 min. All PCR reactions were performed in a PE9700 DNA thermal cycler (PE Biosystems, Foster City, CA). After amplification, PCR products were purified by electrophoresis and subsequent elution from 1.2% (w/v) agarose gels using a QiaEX II gel extraction kit (Qiagen).
Gel-purified PCR products were sequenced according to the protocol outlined in the DYEnamic ET Dye Terminator kit (Amersham Biosciences, Little Chalfont, Buckinghamshire, UK), using both forward and reverse PCR primers, and analyzed using a MegaBACE 1000 DNA analysis system (Molecular Dynamics, Sunnyvale, CA). To obtain an accurate consensus sequence, individual PCR products were sequenced at least twice using both the forward and reverse primers. Allele sequences from each locus and inbred line were aligned and compared using Sequencher (GeneCode, Ann Arbor, MI), and each of the 264 SNPs was assessed.
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council and the Victorian Bioinformatics Consortum (UK; grant-aided support to IACR-Long Ashton, Investigating Gene Function initiative grant no. IGF12403 to D.E. and G.B., and grant no. D14009 to H.O.). Detailed results from this study are available on-line at www.cerealsdb.uk.net. D.E. is supported by the Victorian Bioinformatics Consortum.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019422.
LITERATURE CITED
- Adams MD, Kerlavage AR, Fleischmann RD, Fuldner RA, Bult CJ, Lee NH, Kirkness EF, Weinstock KG, Gocayne JD, White O et al. Initial assessment of human gene diversity and expression patterns based upon 83-million nucleotides of cDNA sequence. Nature. 1995;377:3. [PubMed] [Google Scholar]
- Bennetzen JL, Chandler VL, Schnable P. National Science Foundation-Sponsored Workshop Report: Maize Genome Sequencing Project. Plant Physiol. 2002;127:1572–1578. doi: 10.1104/pp.127.4.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattramakki D, Dolan M, Hanafey M, Wineland R, Vaske D, Register JC, III, Tingey SV, Rafalski A. Insertion-deletion polymorphisms in 3′ regions of maize genes occur frequently and can be used as highly informative genetic markers. Plant Mol Biol. 2002;48:539–547. doi: 10.1023/a:1014841612043. [DOI] [PubMed] [Google Scholar]
- Buetow KH, Edmonson MN, Cassidy AB. Reliable identification of large numbers of candidate SNPs from public EST data. Nat Genet. 1999;21:323–325. doi: 10.1038/6851. [DOI] [PubMed] [Google Scholar]
- Burke J, Davison D, Hide W. d2_cluster: a validated method for clustering EST and full-length cDNA sequences. Genome Res. 1999;9:1135–1142. doi: 10.1101/gr.9.11.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R, Edmonson M, Hu Y, Nguyen C, Scherpbier T, Buetow KH. Expression-based genetic/physical maps of single nucleotide polymorphisms identified by the cancer genome anatomy project. Genome Res. 2000;10:1259–1265. doi: 10.1101/gr.10.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coryell VH, Jessen H, Schupp JM, Webb D, Keim P. Allele-specific hybridisation markers for soybean. Theor Appl Genet. 1999;101:1291–1298. [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hot spots in Escherichia coli. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Dawson E, Chen Y, Hunt S, Smink LJ, Hunt A, Rice K, Livingston S, Bumpstead S, Bruskiewich R, Sham P et al. A SNP resource for human chromosome 22: extracting dense clusters of SNPs from the genomic sequence. Genome Res. 2001;11:170–178. doi: 10.1101/gr.156901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch S, Iseli C, Bucher P, Antonarakis SE, Scott HS. A cSNP map and database for human chromosome 21. Genome Res. 2001;11:300–307. doi: 10.1101/gr.164901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE. Masquerading repeats: paralogous pitfalls of the human genome. Genome Res. 1998;8:758–762. doi: 10.1101/gr.8.8.758. [DOI] [PubMed] [Google Scholar]
- Gai X, Lal S, Xing L, Brendel V, Walbot V. Gene discovery using the maize genome database ZmDB. Nucleic Acids Res. 2000;28:94–96. doi: 10.1093/nar/28.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg K, Green P, Nickerson DA. Identification of candidate coding region single nucleotide polymorphisms in 165 human genes using assembled expressed sequence tags. Genome Res. 1999;9:1087–1092. doi: 10.1101/gr.9.11.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS. Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res. 2001;11:55–66. doi: 10.1101/gr.160601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germano J, Klein AS. Species specific nuclear and chloroplast single nucleotide polymorphisms to distinguish Picea glauca, P. mariana and P. rubens. Theor Appl Genet. 1999;99:37–49. [Google Scholar]
- Gu Z, Hillier L, Kwok P-Y. Single nucleotide polymorphism hunting in cyberspace. Hum Mutat. 1998;12:221–225. doi: 10.1002/(SICI)1098-1004(1998)12:4<221::AID-HUMU1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Roy JK, Prasad M. Single nucleotide polymorphisms: a new paradigm for molecular marker technology and DNA polymorphism detection with emphasis on their use in plants. Curr Sci. 2001;80:524–535. [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok PY, Carlson C, Yager TD, Ankener W, Nickerson DA. Comparative analysis of human DNA variations by fluorescence-based sequencing of PCR products. Genomics. 1994;23:138–144. doi: 10.1006/geno.1994.1469. [DOI] [PubMed] [Google Scholar]
- Marth GT, Korf I, Yandell MD, Yeh RT, Gu ZJ, Zakeri H, Stitziel NO, Hillier L, Kwok PY, Gish WR. A general approach to single-nucleotide polymorphism discovery. Nat Genet. 1999;23:452–456. doi: 10.1038/70570. [DOI] [PubMed] [Google Scholar]
- Mogg R, Batley J, Hanley S, Edwards D, O'Sullivan H, Edwards KJ. Characterising the flanking regions of Zea mays microsatellites reveals a large number of useful sequence polymorphisms. Theor Appl Genet. 2002;105:532–543. doi: 10.1007/s00122-002-0897-1. [DOI] [PubMed] [Google Scholar]
- Nikiforov TT, Rendle RB, Goelat P, Rogers Y-H, Kotewicz ML, Anderson S, Trainor GL, Knapp MR. Genetic bit analysis: a solid phase method for typing single nucleotide polymorphisms. Nucleic Acids Res. 1994;22:4167–4175. doi: 10.1093/nar/22.20.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picoult-Newberg L, Ideker TE, Pohl MG, Taylor SL, Donaldson MA, Nickerson DA, Boyce-Jacino M. Mining SNPs from EST databases. Genome Res. 1999;9:167–174. [PMC free article] [PubMed] [Google Scholar]
- Rafalski A. Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol. 2002;5:94–100. doi: 10.1016/s1369-5266(02)00240-6. [DOI] [PubMed] [Google Scholar]
- Sutton WD, Gerlach WL, Schwartz D, Peacock WJ. Molecular analysis of Ds controlling element mutations at the Adh1 locus of maize. Science. 1984;223:1265–1268. doi: 10.1126/science.223.4642.1265. [DOI] [PubMed] [Google Scholar]
- Syvanen AC. Genotyping single nucleotide polymorphisms. Nat Rev Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- Taillon-Miller P, Gu ZJ, Li Q, Hillier L, Kwok PY. Overlapping genomic sequences: a treasure trove of single-nucleotide polymorphisms. Genome Res. 1998;8:748–754. doi: 10.1101/gr.8.7.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp mays L.) Proc Natl Acad Sci USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]