Abstract
Microtubules in interphase plant cells form a cortical array, which is critical for plant cell morphogenesis. Genetic studies imply that the minus end-directed microtubule motor kinesin-like calmodulin-binding protein (KCBP) plays a role in trichome morphogenesis in Arabidopsis. However, it was not clear whether this motor interacted with interphase microtubules. In cotton (Gossypium hirsutum) fibers, cortical microtubules undergo dramatic reorganization during fiber development. In this study, cDNA clones of the cotton KCBP homolog GhKCBP were isolated from a cotton fiber-specific cDNA library. During cotton fiber development from 10 to 21 DPA, the GhKCBP protein level gradually decreases. By immunofluorescence, GhKCBP was detected as puncta along cortical microtubules in fiber cells of different developmental stages. Thus our results provide evidence that GhKCBP plays a role in interphase cell growth likely by interacting with cortical microtubules. In contrast to fibers, in dividing cells of cotton, GhKCBP localized to the nucleus, the microtubule preprophase band, mitotic spindle, and the phragmoplast. Therefore KCBP likely exerts multiple roles in cell division and cell growth in flowering plants.
Microtubules exist in a cortical array at interphase in somatic plant cells. In fast growing interphase cells, microtubules of the cortical array are highly dynamic compared with the interphase array of animal cells (Hush et al., 1994) and undergo constant reorganization/reorientation during cell growth and in response to external and internal cues (Lloyd, 1994). Reorganization of cortical microtubules is believed to be an energy-dependent event (Wymer et al., 1996). The orientation of cortical microtubules usually parallels and may play a role in the orientation of the newly deposited cellulose microfibrils in the cell wall (Lloyd and Chan, 2002).
Cotton (Gossypium hirsutum) fibers (also called trichomes) are single cells differentiated from the epidermis of the ovule. Unlike tip-growing cells of pollen tubes and root hairs, cotton fibers elongate by a diffuse growing mechanism (Tiwari and Wilkins, 1995). Microtubules in the cotton fiber undergo several phases of reorganization during fiber development (Seagull, 1992). At the initiation stage, or 1 to 3 days post anthesis (DPA), microtubules assume a random organization in the cell cortex. At the elongation stage, or 7 to 19 DPA, cortical microtubules align in a parallel manner, perpendicular to the axis of elongation. At the onset of secondary cell wall synthesis, or approximately 24 DPA, parallel microtubules align in a steeply pitched manner. Early during secondary wall synthesis, the density of cortical microtubules begins to increase. The molecular mechanisms underlying such a dramatic reorganization are not clear. Nevertheless, the orientation of cortical microtubules is mirrored by the orientation of the cellulose microfibrils in both the primary and secondary cell wall (Seagull, 1992). It is known that an organized microtubule array is critical for microfibrils to assume their orientation patterns (Seagull, 1990).
Very little has been learned about molecular mechanisms underlying growth/shrinkage and reorientation of cortical microtubules in plant cells. Recently, a genetic study in Arabidopsis has revealed that the zwichel mutations result in defects in trichome stalk expansion and branching (Oppenheimer et al., 1997; Reddy and Day, 2000). The ZWICHEL gene encodes kinesin-like calmodulin-binding protein (KCBP; Oppenheimer et al., 1997), a kinesin-related motor protein, and KCBP was originally isolated by a screen for calmodulin-binding proteins (Reddy et al., 1996). Immunostaining results with dividing cells from several different plant species using an anti-KCBP antibody have revealed that the protein colocalizes with microtubules in the preprophase band, spindle, and the phragmoplast (Bowser and Reddy, 1997; Smirnova et al., 1998). However the localization of KCBP in trichomes has not been confirmed, and it has not been clear how this protein exerts its role in interphase cells.
Because of the rapid microtubule-dependent growth of cotton fibers, we hypothesized that microtubule-based motors likely participated in rapid reorganization of microtubules and transport of various materials and organelles. In this study, we report the identification of the cotton KCBP homolog GhKCBP from a cotton fiber-specific cDNA library. Furthermore, we present results showing that in this differentiated cell type, GhKCBP localizes to the cortical microtubules.
RESULTS
Isolation of GhKCBP cDNA
To identify cotton kinesin cDNA sequences, available plant kinesin sequences were used to compare with cotton fiber expressed sequencing tag (EST) sequences. One sequence from the Clemson University cotton EST database (http://www.genome.clemson.edu/projects/cotton/est/) was found to match the KCBP sequence significantly. Probes based on this sequence were used to screen a 21-DPA cotton fiber-specific library (Pear et al., 1996). Two clones were isolated that overlapped and together covered the full-length coding region of GhKCBP. On the basis of the cDNA sequence (GenBank accession no. AY216263), GhKCBP contains 1,209 amino acids with a calculated molecular mass of 133 kD and pI of 7.77. Its amino acid sequence is 72% identical to the Arabidopsis KCBP (Reddy et al., 1996).
Like homologous proteins from other plants, GhKCBP has a kinesin motor domain located toward the C terminus of the polypeptide followed by a Ca2+/calmodulin-binding site. GhKCBP contains a neck sequence (RKRYFNTIEDMKGK) that is conserved among C terminus motor kinesins, and determines a minus end-directed motile activity (Endow and Waligora, 1998; Higuchi and Endow, 2002). The Arabidopsis KCBP has already been demonstrated to bear a minus end-directed motility (Song et al., 1997). The N-terminal part of the GhKCBP polypeptide is 81% identical to an ATP-independent microtubule-binding site defined in Arabidopsis KCBP (Narasimhulu and Reddy, 1998).
GhKCBP Polypeptide from Cotton Fibers
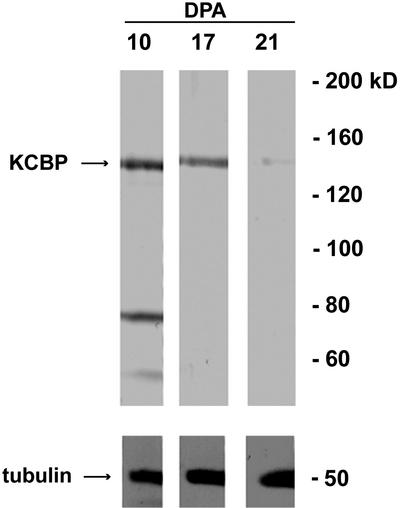
Antibodies were raised in two rats against a GST-fusion protein containing amino acids 561 to 867 of GhKCBP, a part of the protein that is unique to KCBP. Affinity-purified antibodies were used in an immunoblotting experiment. Antibodies from both rats recognized a band of approximately 140 kD (Fig. 1). Therefore we concluded that GhKCBP is an expressed protein in the cotton fiber. When the same protein blot was probed with an antibody against α-tubulin, a single band of approximately 50 kD was revealed (Fig. 1). Interestingly, there was consistent decrease of GhKCBP protein level from 10 to 21 DPA (Fig. 1). It has been shown that the α-tubulin level shows some increase reflecting higher intensity of cortical microtubules at later stages of fiber development (Seagull, 1992; Whittaker and Triplett, 1999).
Figure 1.
GhKCBP immunoblotting in cotton fibers. Equal amounts of fiber total proteins were separated by SDS-PAGE and transferred to nitrocellulose. The upper part of the blot was probed with anti-GhKCBP antibodies, and the lower part was probed with an anti-α-tubulin antibody. Note that the GhKCBP level decreased from 10 to 21 DPA, whereas the tubulin level at least remained similar or showed some increase.
GhKCBP Localization in Cotton Fibers
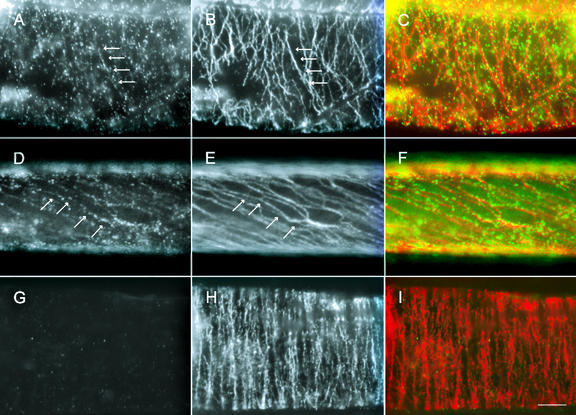
Because the purified antibodies specifically recognized GhKCBP in immunoblotting experiments, they were then used for immunolocalization experiments in intact cotton fibers. At 10 DPA, GhKCBP was localized in a uniform punctate pattern in the cell cortex (Fig. 2A). In the same fiber cell, microtubules appeared in a transverse parallel array (Fig. 2B). When the two signals were superimposed, the punctate GhKCBP signal was clearly along the microtubules (Fig. 2C). At 24 DPA when secondary wall cellulose synthesis approached its maximum, parallel microtubules were steeply pitched at the cell cortex (Fig. 2E). Punctate GhKCBP signal still appeared along these microtubules (Fig. 2, D and F). We have also noticed that there was a GhKCBP signal not associated with cortical microtubules (Fig. 2, A and D). The signal could be due to the detection of soluble forms of GhKCBP. To verify whether the localization was specific for the GhKCBP antigen, purified antibodies were pre-absorbed with the antigen used for antibody production and applied for localization. Although in the same cell whose cortical microtubules were clearly detected, GhKCBP was not detected (Fig. 2, G–I). Therefore, we conclude that the punctate signal reflected GhKCBP localization in these fixed fibers.
Figure 2.
Immunolocalization of GhKCBP in cotton fibers. Fibers of 10 DPA (A–C and G–I) and 21 DPA (D–F) were stained with anti-GhKCBP (A, D, and G) and anti-α-tubulin (B, E, and H). Pseudocolored images (C, F, and I) showed anti-GhKCBP in green and anti-α-tubulin in red. Note that GhKCBP signal could be easily traced along transverse cortical microtubules at 10 DPA, and along steeply pitched cortical microtubules at 21 DPA (arrows). In control experiments, fibers were stained with depleted anti-GhKCBP (G) and anti-α-tubulin (H). See “Materials and Methods” for methods of depletion. Scale bar = 10 μm.
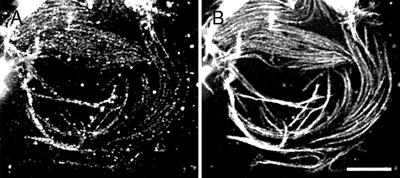
To ask whether GhKCBP localization was dependent on the cell wall, cotton fiber cytoplasts were processed for an immunostaining experiment. GhKCBP was clearly still present along randomly oriented microtubules in the cytoplast (Fig. 3, A and B). Thus GhKCBP localization along microtubules was independent of an intact cell wall.
Figure 3.
Immunolocalization of GhKCBP in cotton cytoplasts. Cytoplasts were stained with anti-GhKCBP (A) or anti-α-tubulin (B). Scale bar = 5 μm.
GhKCBP Localization in Dividing Cells
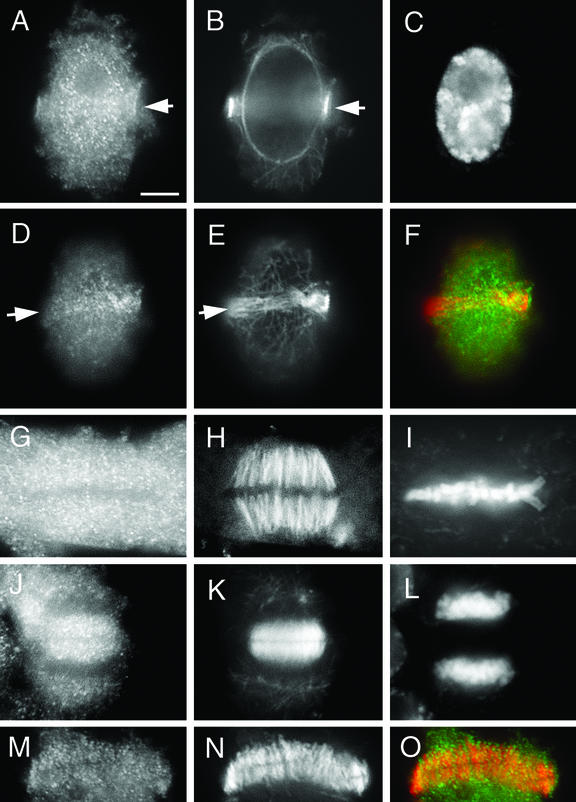
Because previous studies only revealed KCBP localization in dividing cells (Bowser and Reddy, 1997; Smirnova et al., 1998), we examined whether GhKCBP was also expressed in dividing cells in cotton. Root meristematic cells were chosen for localization experiments. At prophase when microtubules appeared in a cortical preprophase band (Fig. 4, B and E), GhKCBP associated with the preprophase band and the nucleus (Fig. 4, A, D, and F). Again, GhKCBP appeared in a punctate pattern along preprophase band microtubules (Fig. 4D). The nuclear localization of GhKCBP persisted until the nuclear envelope broke down (data not shown). When the mitotic spindle was assembled, GhKCBP appeared relatively abundant in the cytoplasm, and some puncta appeared to be along kinetochore fibers (Fig. 4, G–I). As the phragmoplast microtubule array was assembled, abundant GhKCBP signal was detected in the phragmoplast (Fig. 4, J–L). In an isolated phragmoplast, punctate GhKCBP signal was clearly present among the microtubules (Fig. 4, M and O). Thus, consistent with findings in Arabidopsis suspension cells and Haemanthus spp. endosperm cells, GhKCBP is also present in mitotic microtubule arrays in cotton meristematic cells.
Figure 4.
Immunolocalization of GhKCBP in cotton cells undergoing mitotic cell division. Cotton root cells were stained by immunoflurescence techniques for GhKCBP (A, D, G, J, and M) or for α-tubulin (B, E, H, K, and N) or were stained with DAPI to detect DNA (C, I, and L). Pseudocolored images (F and O) show anti-GhKCBP in green and anti-α-tubulin in red. GhKCBP could be detected in the microtubule preprophase band (arrow, A, B, and D–F). Abundant GhKCBP signal was present in metaphase cells (G–I). GhKCBP clearly was present among phragmoplast microtubules (J, K, M, to O). Scale bar = 10 μm.
DISCUSSION
The kinesin-related protein KCBP is conserved at least in algae and flowering plants (Abdel-Ghany et al., 2000; Abdel-Ghany and Reddy, 2000). A related protein was also identified in sea urchin (Rogers et al., 1999). Our results indicated that GhKCBP clearly associated with cortical microtubules in cotton fibers, which supported the notion that KCBP plays a role in interphase cells, as suggested by genetic studies of trichome morphogenesis (Oppenheimer et al., 1997).
Our work and previous studies (Seagull, 1992) with fixed cells at different stages of fiber development clearly indicate that reorientation of cortical microtubules take place, although we have not directly observed microtubule reorganization in living cotton fibers. It has been recognized that cortical microtubule reorganization requires energy (Wymer et al., 1996). Such an energy requirement could be due to the roles of motor proteins. On the basis of our localization data, we suggest that GhKCBP plays a role in reorganization of cortical microtubules during cotton fiber development. Because KCBP has a nu-cleotide-independent microtubule-binding site outside the motor domain (Narasimhulu and Reddy, 1998), it is reasonable to presume that microtubules could be a cargo of this motor. KCBP could contribute to organizing cortical microtubules as they move against each other by such a minus end-directed motor would converge their minus ends. Newly converged microtubules could assume new orientation depending on how microtubules were stabilized at a certain direction.
Alternatively, GhKCBP may contribute to microtubule stability directly. One possibility is that KCBP acts as a microtubule stabilizer. KCBP could participate in stabilizing cortical microtubules in a certain orientation so that a cell could undergo proper elongation and morphogenesis. This hypothesis is supported by the fact that the phenotype of reduced branches of the zwichel mutation can be suppressed by the microtubule stabilization agent taxol (Mathur and Chua, 2000).
Conversely, KCBP may act as a microtubule destabilizer. A C terminus motor in yeast, Kar3p, has been shown to be able to destabilize microtubules at their minus ends (Endow et al., 1994). Loss of the Kar3p protein in yeast increases the number of cytoplasmic microtubules (Huyett et al., 1998). If GhKCBP has a similar function in microtubule stabilization, we could expect that a lower level of GhKCBP would be detected at later stages of fiber development. In our immunoblotting experiments, there was a significant reduction of GhKCBP protein level at 21 DPA when secondary wall synthesis was about to initiate, whereas α-tubulin content increased at 21 DPA compared with earlier stages (Whittaker and Triplett, 1999). At this stage, cortical microtubule intensity has begun to increase (Seagull, 1992).
It is noteworthy that at later stages of trichome development, the basal region of the trichome has fewer cortical microtubules than the apical region (Folkers et al., 2002). If KCBP plays a role in microtubule destabilization, it is conceivable that increased level of KCBP toward the basal region would cause such a phenomenon. As a matter of fact, in the zwichel mutant, differential distribution of microtubule network in the tip region versus basal region is not as obvious as in the wild type (compare Fig. 2J and Fig. 2C in Mathur and Chua, 2000).
In Arabidopsis, KCBP interacts with the protein kinase KIPK and the ANGUSTIFOLIA protein, a protein that is required for trichome branching (Day et al., 2000; Folkers et al., 2002). Although the function of KIPK is unknown, ANGUSTIFOLIA plays a role in cortical microtubule reorganization in the trichome because angustifolia mutants have even distribution of cortical microtubules in the trichome, whereas in the wild-type plants, a denser microtubule network can be observed toward the trichome tip (Folkers et al., 2002). If loss of ANGUSTIFOLIA leads to a loss of KCBP activity, cortical microtubules at the basal region would survive.
Because of its presence in dividing cells, the role of KCBP should not be limited to interphase cells. However, in the zwichel mutants, there is no defect in cell division (Oppenheimer et al., 1997). Such a phenomenon could be due to functional redundancy among C terminus motor kinesins in plants. For example, at least three other C terminus motor kinesin-related proteins are known to be expressed in plants and to associate with mitotic microtubule arrays (Liu et al., 1996; Mitsui et al., 1996, 1994, 1993). Therefore, in the zwichel mutants, the function of KCBP could be compensated by other C terminus motor kinesins. It is noteworthy that a mutation at the KATA locus also did not cause a mitotic phenotype; instead, it has a meiotic phenotype, only on the male side (Chen et al., 2002). Thus mitosis may require more than one C terminus motor kinesin. In a highly differentiated cell like the trichome or the cotton fiber, it is possible that no other C terminus motor kinesin can compensate for the loss of KCBP. A phenotype is thus shown.
It is clear that KCBP activities are precisely regulated in plant cells. Constitutive activation of its motor activity in Tradescantia spp. stamen hair cells by microinjection of anti-KCBP calmodulin-binding peptide causes early mitotic onset and a delay in cytokinesis (Vos et al., 2000). However, there was no effect on anaphase progression, indicating that KCBP is probably naturally activated in anaphase. Therefore, it is likely that even when KCBP is abundantly expressed, it may not be constitutively active.
It should be noted that KCBP is encoded by a single gene in Arabidopsis. The cotton species G. hirsutum probably has multiple homologs of KCBP. This is not only because of the size of its genome, but also because of the complexity of its genetic background. The GhKCBP reported here might be one of several expressed homologous proteins.
In summary, our data support the notion that GhKCBP plays a role in interphase microtubule reorganization by directly interacting with the cortical microtubules. Regulation of cortical microtubule dynamics must be important for cell morphogenesis in the cotton fiber and other interphase cells in higher plants.
MATERIALS AND METHODS
Plant Materials
Cotton (Gossypium hirsutum cv Coker 130) plants were grown under greenhouse conditions. Flowers were marked at anthesis, and at indicated times, the bolls were collected for further experiments. For root tip squashes, cotton seeds were germinated in vermiculite, and root tips were collected from cotton seedlings.
Cloning of the GhKCBP cDNA
An EST clone showing sequence similarity to AtKCBP was found in the on-line database (Clemson cotton database, http://www.genome.clemson.edu/projects/cotton/est/). PCR primers KCBP-F (5′-GAACATCTT-ACAAAAGATG-3′) and KCBP-R (5′-TTCTCCCAACAATTGATTAGTGT-3′), designed according to the EST sequence, were used for amplification of the region encoding amino acids 332 to 398 from a 21-DPA cotton (cv Acala SJ-2) fiber-specific cDNA library (Pear et al., 1996). This fragment was purified and used as the probe to screen the cDNA library by using a digoxigenin-labeling method according to manufacturer's procedure (Roche Diagnostics, Indianapolis). Two overlapping clones were identified in the screen and sequenced. Together, the clones covered 3,976 bp of the GhKCBP cDNA. The sequence also conforms with the Kozak consensus start sequence. The two cDNA clones were spliced together in the pBluescript SK+ (Stratagene, La Jolla, CA) vector and used for subsequent studies.
Antibody Production and Purification
A glutathione S-transferase (GST)-KCBP fusion was constructed by ligating a XbaI-NcoI fragment (encoding amino acids 561–867) into the pGEX-KG vector (Guan and Dixon, 1991). The GST-fusion proteins were expressed in BL21(DE3) pLys S cells (Novagen, Madison, WI). The GST-KCBP fusion protein was not soluble; therefore it was purified from inclusion bodies with B-PER extraction reagent as described by the manufacturer (Pierce, Rockford, IL). Polyclonal antibodies were raised against the GST-KCBP fusion protein in rats at a commercial facility (Antibodies Inc., Davis, CA).
To purify antibodies specifically against GhKCBP, a BamHI/HinDIII fragment was excised from the GST-KCBP construct. The fragment was ligated into the pQE-30 vector (Qiagen, Valencia, CA). The 6X-His-KCBP fusion protein was expressed in M15pREP4 cells and purified by using the Talon resin (BD Biosciences Clontech, Palo Alto, CA) at denatured condition. Renatured fusion protein was coupled to an agarose matrix using the AminoLink Plus Coupling Gel (Pierce) as instructed. Specific antibodies were eluted with 100 mm Gly, pH 2.7, and neutralized immediately with 1.0 m Tris, pH 8.0. These purified antibodies were used for subsequent analyses.
Protein Extraction and Immunoblotting
For isolation of total protein from fibers, bolls were harvested at 10, 17, and 21 DPA. Ovules were excised from the bolls with a scalpel and immediately frozen in liquid nitrogen. The frozen fibers were chipped away from the rest of the ovules. Fibers (about 2 g per sample) were ground in liquid nitrogen with a mortar and a pestle, and the powder was transferred to a tube with 20 mL of 20% (w/v) trichloroacetic acid solution. The sample was centrifuged at 31 kg at 4°C for 10 min. Pellets were washed for three times with 80% (v/v) acetone in 25 mm Tris (pH 7.5) and centrifuged. Pellets were suspended in 1 mL of 2× sample buffer (2% [v/v] β-mercaptoethanol, 2% [w/v] SDS, and 40 mm Tris, pH 6.8). Samples were boiled for 3 min and cooled on ice. Insoluble material was pelleted, and the supernatant was separated by 7.5% (w/v) SDS-PAGE. The proteins were then transferred to nitrocellulose for immunoblotting analysis.
Nitrocellulose blots were blocked with 5% (w/v) milk in phosphate-buffered saline (PBS) for 30 min at room temperature. Excess milk was washed off with washing solution (0.05% [v/v] Tween 20 and PBS). Blots were incubated in primary antibody from 3 h to overnight in a humid chamber at room temperature. Blots were then given five washes, for 5 min each, with washing solution. Alkaline-phosphatase conjugated anti-rat IgG secondary antibodies (Sigma-Aldrich, St. Louis) were then applied for 1 h at room temperature. Blots were washed five more times with washing solution, then twice with distilled water. Finally, they were incubated in nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrate solution (Bio-Rad, Hercules, CA). Blots were scanned with a SNAPSCAN 1212 u scanner (Agfa, Ridgefield Park, NJ), and images were composed in Photoshop 7.0 (Adobe Systems, San Jose, CA).
Immunolocalization
Antibodies Used
Primary antibodies were affinity-purified rat anti-GhKCBP and rabbit anti-tubulin (Sharp et al., 1999). Secondary antibodies were fluorescein isothiocyanate-conjugated goat anti-rat (Sigma-Aldrich), and Texas Red X-conjugated goat anti-rabbit (Molecular Probes, Eugene, OR).
Immunolocalization in Cotton Fibers
Cotton bolls were collected from the greenhouse. Ovules with the fibers attached were dissected out with a scalpel and incubated in fixative of 4% (w/v) paraformaldehyde and 0.1% (w/v) glutaraldehyde in 50 mm PIPES, 1 mm MgSO4, and 5 mm EGTA, pH 6.9 (PME), containing 0.1% (v/v) Tween-80, and 0.3 m mannitol for 1 h at room temperature. After three washes in PME, the fibers were incubated with 1.5% (w/v) Cellulase “Onozuka” RS (Yakult Pharmaceutical, Tokyo) and 0.1% (w/v) Macerozyme R-10 (Yakult Pharmaceutical) in PME for 15 to 30 min for 10-DPA fibers and for 45 min to 1 h for 24-DPA fibers at RT. Fibers were washed again three times in PME then incubated for 1 h in 1% (v/v) Triton X-100 and PME at room temperature. After three more washes in PME, fibers were moved to methanol at −20°C for 10 min. Then they were removed and rehydrated in PBS. Fibers were cut off the ovules onto poly-l-Lys (Mr > 300,000; Sigma-Aldrich)-coated slides and incubated overnight at 4°C in primary antibody. Slides were washed with PBS followed by secondary antibody incubation for 1 h. Slides were mounted with 50% (v/v) glycerol and sealed with nail polish before observation.
Immunolocalization in Cotton Cytoplasts
Cytoplasts isolated from cotton bolls of different ages were collected from the greenhouse according to Andersland et al. (1998). In brief, the ovules with fibers attached were dissected out and incubated for 3 h with 1.5% (w/v) Cellulase-RS and 0.1% (w/v) Macerozyme R-10 in 0.6 m mannitol, pH 5.7, containing 10 μm Paclitaxel (Sigma-Aldrich) on a rocker. The released cytoplasts were filtered through a cotton sieve and collected by centrifugation. After being washed with 0.6 m mannitol containing 10 μm Paclitaxel, the cytoplasts were placed onto slides freshly coated with poly-l-Lys. After 1 min, excess liquid was removed and replaced with fixative of 4% (w/v) paraformaldehyde and 0.1% (w/v) glutaraldehyde in PME for 45 min in a humid chamber. Slides were then washed three times with PME and incubated for 10 min in 0.5% (v/v) Triton X-100 in PME. After three washes with PME, slides were incubated in 100% (v/v) methanol at −20°C for 10 min. They were then rehydrated immediately with PBS before application of primary antibody. Slides were incubated in primary antibody for 3 h to overnight. After washing with PBS, secondary antibodies were applied. Slides were then washed and mounted before observation.
Immunolocalization in Cotton Root Tip Cells
Root tip cells were processed for immunolocalization as previously described (Palevitz, 1988).
For immunolocalization controls, 6X-His-KCBP fusion protein was separated on a 12% (w/v) SDS-PAGE and transferred to nitrocellulose. The blots were stained with Ponceau S to visualize the protein. The fusion protein band was cut out and blocked in 5% (w/v) dry milk PBS. Purified anti-KCBP antibodies were incubated with the protein blots for 3 h on a shaker at room temperature. Depleted antibodies were collected and used as controls.
All samples were observed on a microscope equipped with epifluorescence optics (Eclipse E600, Nikon, Melville, NY). Images were acquired with a CCD camera (Hamamatsu Photonics K.K., Tokyo) using the ImageProPlus 4.0 software package (Media Cybernetics, Silver Spring, MD). Modifications of images were performed in Photoshop 7.0 (Adobe Systems).
ACKNOWLEDGMENTS
We are grateful to Drs. David Sharp and Jonathan Scholey for their generous gift of purified rabbit anti-tubulin antibodies.
Footnotes
This work was supported by the Division of Energy Biosciences, U.S. Department of Energy (grant nos. DE–FG–03–01ER15189 to B.L. and DE–FG–03–963ER20238 to D.P.D.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020339.
LITERATURE CITED
- Abdel-Ghany SE, Kugrens P, Reddy ASN. CpKLP1: a calmodulin-binding kinesin-like protein from Cyanophora paradoxa (glaucophyta) J Phycol. 2000;36:686–692. doi: 10.1046/j.1529-8817.2000.00024.x. [DOI] [PubMed] [Google Scholar]
- Abdel-Ghany SE, Reddy ASN. A novel calcium/calmodulin-regulated kinesin-like protein is highly conserved between monocots and dicots. DNA Cell Biol. 2000;19:567–578. doi: 10.1089/104454900439791. [DOI] [PubMed] [Google Scholar]
- Andersland JM, Dixon DC, Seagull RW, Triplett BA. Isolation and characterization of cytoskeletons from cotton fiber cytoplasts. In Vitro Cell Dev Biol Plant. 1998;34:173–180. [Google Scholar]
- Bowser J, Reddy ASN. Localization of a kinesin-like calmodulin-binding protein in dividing cells of Arabidopsis and tobacco. Plant J. 1997;12:1429–1437. doi: 10.1046/j.1365-313x.1997.12061429.x. [DOI] [PubMed] [Google Scholar]
- Chen C, Marcus A, Li W, Hu Y, Calzada J-P V, Grossniklaus U, Cyr RJ, Ma H. The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development. 2002;129:2401–2409. doi: 10.1242/dev.129.10.2401. [DOI] [PubMed] [Google Scholar]
- Day IS, Miller C, Golovkin M, Reddy ASN. Interaction of a kinesin-like calmodulin-binding protein with a protein kinase. J Biol Chem. 2000;275:13737–13745. doi: 10.1074/jbc.275.18.13737. [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtuble motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA, Waligora KW. Determinants of kinesin motor polarity. Science. 1998;281:1200–1202. doi: 10.1126/science.281.5380.1200. [DOI] [PubMed] [Google Scholar]
- Folkers U, Kirik V, Schoebinger U, Falk S, Krishnakumar S, Pollock MA, Oppenheimer DG, Day I, Reddy AR, Juergens G et al. The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J. 2002;21:1280–1288. doi: 10.1093/emboj/21.6.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Higuchi H, Endow SA. Directionality and processivity of molecular motors. Curr Opin Cell Biol. 2002;14:50–57. doi: 10.1016/s0955-0674(01)00293-9. [DOI] [PubMed] [Google Scholar]
- Hush JM, Wadsworth P, Callaham DA, Hepler PK. Quantification of microtuble dynamics in living plant cells using fluorescence redistribution after photobleaching. J Cell Sci. 1994;107:775–784. doi: 10.1242/jcs.107.4.775. [DOI] [PubMed] [Google Scholar]
- Huyett A, Kahana J, Silver P, Zeng X, Saunders W. The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J Cell Sci. 1998;111:295–301. doi: 10.1242/jcs.111.3.295. [DOI] [PubMed] [Google Scholar]
- Liu B, Cyr RJ, Palevitz BA. A kinesin-like protein, KatAp, in the cells of Arabidopsis and other plants. Plant Cell. 1996;8:119–132. doi: 10.1105/tpc.8.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C. Why should stationary plant cells have such dynamic microtubules? Mol Biol Cell. 1994;5:1277–1280. doi: 10.1091/mbc.5.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C, Chan J. Helical microtubule arrays and spiral growth. Plant Cell. 2002;14:2319–2324. doi: 10.1105/tpc.141030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Chua N-H. Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell. 2000;12:465–477. doi: 10.1105/tpc.12.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui H, Hasezawa S, Nagata T, Takahashi H. Cell cycle-dependent accumulation of a kinesin-like protein, KatB/C, in synchronized tobacco BY-2 cells. Plant Mol Biol. 1996;30:177–181. doi: 10.1007/BF00017812. [DOI] [PubMed] [Google Scholar]
- Mitsui H, Nakatani K, Yamaguchishinozaki K, Shinozaki K, Nishikawa K, Takahashi H. Sequencing and characterization of the kinesin-related genes Katb and Katc of Arabidopsis thaliana. Plant Mol Biol. 1994;25:865–876. doi: 10.1007/BF00028881. [DOI] [PubMed] [Google Scholar]
- Mitsui H, Yamaguchi-Shinozaki K, Shinozaki K, Nishikawa K, Takahashi H. Identification of a gene family (kat) encoding kinesin-like proteins in Arabidopsis thaliana and the characterization of secondary structure of KatA. Mol Gen Genet. 1993;238:362–368. doi: 10.1007/BF00291995. [DOI] [PubMed] [Google Scholar]
- Narasimhulu SB, Reddy ASN. Characterization of microtubule binding domains in the Arabidopsis kinesin-like calmodulin binding protein. Plant Cell. 1998;10:957–965. doi: 10.1105/tpc.10.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Ericson B, Feldmann K, Marks MD. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc Natl Acad Sci USA. 1997;94:6261–6266. doi: 10.1073/pnas.94.12.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palevitz BA. Cytochalasin-induced reorganization of actin in Allium root cells. Cell Motil Cytoskelet. 1988;9:283–298. [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN, Day IS. The role of the cytoskeleton and a molecular motor in trichome morphogenesis. Trends Plant Sci. 2000;5:503–505. doi: 10.1016/s1360-1385(00)01792-1. [DOI] [PubMed] [Google Scholar]
- Reddy ASN, Safadi F, Narasimhulu SB, Golovkin M, Hu X. A novel plant calmodulin-binding protein with a kinesin heavy chain motor domain. J Biol Chem. 1996;271:7052–7060. doi: 10.1074/jbc.271.12.7052. [DOI] [PubMed] [Google Scholar]
- Rogers GC, Hart CL, Wedaman KP, Scholey JM. Identification of kinesin-C, a calmodulin-binding carboxy-terminal kinesin in animal (Strongylocentrotus purpuratus) cells. J Mol Biol. 1999;294:1–8. doi: 10.1006/jmbi.1999.3249. [DOI] [PubMed] [Google Scholar]
- Seagull RW. The effects of microtubule and microfilament disrupting agents on cytoskeletal arrays and wall deposition in developing cotton fibers. Protoplasma. 1990;159:44–59. [Google Scholar]
- Seagull RW. A quantitative electron microscopic study of changes in microtubule arrays and wall microfibril orientation during in-vitro cotton fiber development. J Cell Sci. 1992;101:561–577. [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova EA, Reddy ASN, Bowser J, Bajer AS. Minus end-directed kinesin-like motor protein, Kcbp, localizes to anaphase spindle poles in Haemanthus endosperm. Cell Motil Cytoskelet. 1998;41:271–280. doi: 10.1002/(SICI)1097-0169(1998)41:3<271::AID-CM8>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Song H, Golovkin M, Reddy ASN, Endow SA. In vitro motility of AtKCBP, a calmodulin-binding kinesin protein of Arabidopsis. Proc Natl Acad Sci USA. 1997;94:322–327. doi: 10.1073/pnas.94.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SC, Wilkins TA. Cotton (Gossypium hirsutum) seed trichomes expand via diffuse growing mechanism. Can J Bot. 1995;73:746–757. [Google Scholar]
- Vos JW, Safadi F, Reddy ASN, Hepler PK. The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell. 2000;12:979–990. doi: 10.1105/tpc.12.6.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker DJ, Triplett BA. Gene-specific changes in α-tubulin transcript accumulation in developing cotton fibers. Plant Physiol. 1999;121:181–188. doi: 10.1104/pp.121.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymer CL, Fisher DD, Moore RC, Cyr RJ. Elucidating the mechanism of cortical microtubule reorientation in plant cells. Cell Motil Cytoskelet. 1996;35:162–173. doi: 10.1002/(SICI)1097-0169(1996)35:2<162::AID-CM8>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]