Abstract
The defensins knowledgebase is a manually curated database and information source focused on the defensin family of antimicrobial peptides. The current version of the database holds a comprehensive collection of over 350 defensin records each containing sequence, structure and activity information. A web-based interface provides access to the information and allows for text-based searching on the data fields. In addition, the website presents information on patents, grants, research laboratories and scientists, clinical studies and commercial entities pertaining to defensins. With the rapidly increasing interest in defensins, we hope that the knowledgebase will prove to be a valuable resource in the field of antimicrobial peptide research. The defensins knowledgebase is available at http://defensins.bii.a-star.edu.sg/.
INTRODUCTION AND MOTIVATION
Antimicrobial peptides (AMPs) are important components of the immune system of a wide range of organisms, presenting the first line of defence against a variety of pathogens. The AMPs are typically small (10 kDa), cationic and amphiphilic molecules, with low to potent activities against a variety of microbes—gram-positive and gram-negative bacteria, fungi and viruses (1). Alternatively termed as host-defense peptides, the functions of these peptides extend beyond antimicrobial function to a range of roles in innate and adaptive immunity, including immuno-modulation, chemotaxis, inflammatory response and wound repair (2).
There have been attempts to catalog known and newly-discovered AMPs, most notably in the form of three databases: Antimicrobial Sequence Database (AMSDb) [http://www.bbcm.univ.trieste.it/~tossi/amsdb.html, unpublished] with 880 AMP entries, Antimicrobial Peptide database (APD) [http://aps.unmc.edu/AP/main.php, (3)] with 559 records, and [http://research.i2r.a-star.edu.sg/Templar/DB/ANTIMIC/, (4)] (ANTIMIC) with 1788 entries, (as of October 4, 2006). These databases capture different levels of basic information on the constituent AMPs and reflect a burgeoning interest in the area.
One of the largest and most-studied families of AMPs is that of the defensins, represented across mammals, birds, invertebrates and plants (5). More than 300 defensins have been identified so far, including some recently discovered in black-cup fungus (6). The human defensins are potent against specific microbes, but also show a synergistic increase in antimicrobial range in combination. In addition, some human defensins have demonstrated roles in chemotaxis and in bridging the innate and adaptive immune responses (7).
Defensins are essentially ancient natural antibiotics with potent activity extending from lower organisms to humans; their mode of action makes them less likely to provoke microbial resistance in comparison to conventional antibiotics. Hence, knowledge of how these peptides work may provide us with insights to develop improved antibiotics.
Given the increasing research on defensins in diverse disciplines such as biochemistry, structural biology, microbiology, immunology, pathology and computational biology (8–10), it is useful to establish a single, integrated knowledgebase on defensins. This manually curated knowledgebase will serve as a centralized repository of all information pertaining to defensins. As the search for improved antimicrobial compounds continues, we trust that this defensins knowledgebase can be a valuable resource to accelerate understanding, avoid duplication of research, increase interdisciplinary collaborations and evolve new ideas in the realms of defensins and AMPs.
CONSTRUCTION AND FEATURES OF THE KNOWLEDGEBASE
Most of the data compiled in the knowledgebase was gathered from primary (bibliographic databases) and secondary (sequence databases) literature sources. For the primary source, a search query with the keyword ‘defensin’ performed in SciFinder Scholar 2006 [http://www.cas.org/SCIFINDER/SCHOLAR] retrieved 2583 articles (data correct as on October 4, 2006). The academic version of SciFinder Scholar 2006 was chosen mainly because of its broad coverage. It covers the two largest bibliographic databases: Chemical Abstracts Plus (CAplus) Database, which contains over 23 million references and U.S. National Library of Medicine (MEDLINE) database, which covers over 15 million references (11). For the secondary sources, we use the UniProt knowledgebase and GenBank to collate amino acid sequences of the individual defensin peptides.
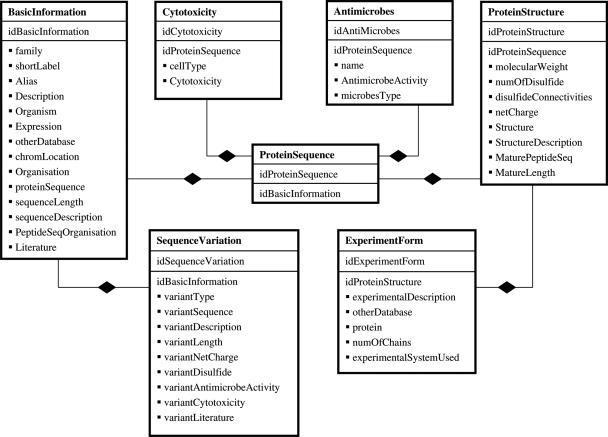
The defensins knowledgebase comprises a MySQL relational database management system that contains information on amino acid sequences, structures and activities of this family of AMP. The layout of the database is shown in Figure 1.
Figure 1.
Schematic representation of the MySQL relational database underlying defensins knowledgebase.
The basic information table contains elementary data such as aliases, family, expression sites and protein sequence for each characterized defensin peptide (including isoforms). This is linked to additional tables comprising information on sequence, structure and activity of the corresponding defensin. The current version of the database holds a comprehensive collection of over 350 individual defensin records, including 46 experimentally determined 3D structures from the Protein Data Bank (PDB) (12); structures can be viewed using either JMol [www.jmol.org] or the AstexViewer™ (13) structure viewing tools, both of which are implemented in the knowledgebase. Additionally, we are building and incorporating into the database, homology-built models of defensins whose 3-dimensional structures remain unresolved.
The data stored in the database can be accessed through a web-based interface which allows for text-based searching on the data fields and filtering of results by organism, family, antimicrobial target and other attributes. Each individual record contains detailed information on the sequence, structure and activity of the peptide (e.g. a limited screen capture is shown in Figure 2). Since the database is intended to supplement existing biological databases, all records are cross-linked to their corresponding entries, where available, on these databases.
Figure 2.
Defensins knowledgebase sample record (protein HBD-3). Only data pertaining to expression sites, cytotoxicity and structure are displayed in this screen-capture.
Other features of the knowledgebase include a glossary that defines jargon terms and two integrated analysis tools for alignment of sequences. A Basic Local Alignment Search Tool (BLAST) (14) has been incorporated to allow users to query any sequence against the defensin sequences stored in the database. Both blastp and blastx versions are included, to support protein and nucleotide sequence queries respectively. Multiple sequence alignment of user-selected defensin sequences from the database can be carried out using the built-in ClustalW (15) tool. Users can select or limit sequences by family or organism. The resulting alignment can be displayed graphically using the embedded JalView tool (16).
The knowledgebase also captures information on defensin-related publications, coupled with a browse-and-search system that allows searching and filtering of records by keywords, journal name, year and type of publication. Additionally, the website features four sub-sections of tabulated information: (i) laboratories and group leaders in the field of defensin research (ii) awarded grants (iii) patents and (iv) commercial entities, all related to defensins. These pages will be updated on a regular basis, as well as ad hoc based on communications within the defensin research community. Additional static pages are devoted to reviews on defensins, culled by manual curation of the literature, and these sections will evolve and expand, in conjunction with research developments and the expansion of research cross-talk and collaborations arising from use of the knowledgebase.
The knowledgebase will be continually updated with data harvested from bibliographic, gene and protein databases. In addition, we plan to include mini-reviews on topics such as mechanism of action, membrane interaction and emerging roles of defensins will be added in the near future. Given the mounting interest in defensins, we anticipate that the knowledgebase will prove to be a useful resource in the field of AMP research. Researchers in the field are invited to use this knowledgebase, suggest improvements, and submit their defensin-related sequences, 3D models and data for inclusion by contacting the authors. The defensins knowledgebase is available at http://defensins.bii.a-star.edu.sg/.
Acknowledgments
Development of the knowledgebase was generously supported by NMRC CPG 007/2004 (RWB) to the Singapore Consortium for Antimicrobial Peptides (SCAMP) and the Biomedical Research Council of A*STAR Singapore. We thank the referees for useful suggestions. Funding to pay the Open Access publication charges for this article was provided by A*STAR (Singapore).
Conflict of interest statement. None declared.
REFERENCES
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Jenssen H., Hamill P., Hancock R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z., Wang G. APD: the Antimicrobial Peptide Database. Nucleic Acids Res. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahmachary M., Krishnan S.P., Koh J.L., Khan A.M., Seah S.H., Tan T.W., Brusic V., Bajic V.B. ANTIMIC: a database of antimicrobial sequences. Nucleic Acids Res. 2004;32:D586–D589. doi: 10.1093/nar/gkh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 6.Mygind P.H., Fischer R.L., Schnorr K.M., Hansen M.T., Sonksen C.P., Ludvigsen S., Raventos D., Buskov S., Christensen B., De Maria L., et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437:975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- 7.Yang D., Chertov O., Bykovskaia S.N., Chen Q., Buffo M.J., Shogan J., Anderson M., Schroder J.M., Wang J.M., Howard O.M., et al. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 8.Pazgiera M., Hoover D.M., Yang D., Lu W., Lubkowski J. Human beta-defensins. Cell Mol. Life Sci. 2006;63:1294–1313. doi: 10.1007/s00018-005-5540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kluver E., Adermann K., Schulz A. Synthesis and structure-activity relationship of beta-defensins, multi-functional peptides of the immune system. J. Pept. Sci. 2006;12:243–257. doi: 10.1002/psc.749. [DOI] [PubMed] [Google Scholar]
- 10.Selsted M.E., Ouellette A.J. Mammalian defensins in the antimicrobial immune response. Nature Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 11.Wagner A.B. SciFinder Scholar 2006: an empirical analysis of research topic query processing. J. Chem. Inf. Model. 2006;46:767–774. doi: 10.1021/ci050481b. [DOI] [PubMed] [Google Scholar]
- 12.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartshorn M.J. AstexViewer™: An aid for structure-based drug design. J. Comput. Aided Mol. Des. 2002;16:871–881. doi: 10.1023/a:1023813504011. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clamp M., Cuff J., Searle S.M., Barton G.J. The Jalview Java Alignment Editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]