Abstract
Alternative polyadenylation leads to mRNAs with variable 3′ ends. Since a 3′-untranslated region (3′-UTR) often contains cis elements that impact stability or localization of mRNA or translation, selection of poly(A) sites in a 3′-UTR is regulated in mammalian cells. However, the molecular basis for alternative poly(A) site selection within a 3′-UTR has been unclear. Here we show involvement of cleavage factor Im (CFIm) in poly(A) site selection within a 3′-UTR. CFIm is a heterodimeric 3′ end-processing complex, which functions to assemble other processing factors on pre-mRNA in vitro. We knocked down 25 kDa subunit of CFIm (CFIm25) in HeLa cells and analyzed alternative poly(A) site selection of TIMP-2, syndecan2, ERCC6 and DHFR genes by northern blotting. We observed changes in the distribution of mRNAs in CFIm25 depleted cells, suggesting a role for CFIm in alternative poly(A) site selection. Furthermore, tissue specific analysis demonstrated that the CFIm25 gene gave rise to 1.1, 2.0 and 4.6 kb mRNAs. The 4.6 kb mRNA was ubiquitously expressed, while the 1.1 and 2.0 kb mRNAs were expressed in a tissue specific manner. We found three likely poly(A) sites in the CFIm25 3′-UTR, suggesting alternative polyadenylation. Our results indicate that alternative poly(A) site selection is a well-regulated process in vivo.
INTRODUCTION
Numerous genes in mammalian species undergo alternative polyadenylation, which results in transcripts with variable 3′ ends (1,2). Alternative poly(A) sites can be located in the last or 3′-most exon, giving rise to mRNAs with variable 3′-untranslated region (3′-UTR), or in a different exon, resulting in protein products that vary at the C-terminus (1). In addition, pre-mRNA 3′ end processing contributes directly to transcription termination, pre-mRNA splicing and mRNA export (3,4). Many genes are reported to be subject to alternative 3′ end processing, and several lines of evidence suggest that poly(A) site selection may be modulated developmentally or in a tissue specific manner (5), however the fundamental mechanisms responsible for regulating alternative poly(A) site selection have not been elucidated.
Polyadenylation of eukaryotic mRNA occurs in a step-wise process, which includes a specific cleavage at the 3′ end of nascent mRNA followed by an addition of a poly(A) tail. Six factors, namely CPSF, CstF, CFIm, CFIIm (cleavage factor II), PAP (poly(A) polymerase) and PABII (poly(A) binding protein II) have been characterized to facilitate the 3′ end processing [reviewed in (6–8)]. The cleavage reaction requires recognition by CPSF of AAUAAA hexamer upstream of the cleavage site and by CstF of degenerate GU- and U-rich sequences downstream of the cleavage site. CFIm is an essential pre-mRNA 3′ end-processing factor unique to metazoans, which facilitates assembly of 3′ end processing factors on pre-mRNA in vitro (9). CFIm was first purified from HeLa cell nuclear extracts as four polypeptides of 25, 59, 68 and 72 kDa (CFIm25, CFIm59, CFIm68 and CFIm72, respectively). Subsequent studies revealed that CFIm59, CFIm68, and CFIm72 are structurally related and that CFIm can be reconstituted with CFIm68 and CFIm25 subunits, suggesting that CFIm is a heterodimer composed of the small CFIm25 subunit and any one of the three larger subunits (10). CFIm25 protein possesses a NUDIX domain and CFIm68 and CFIm59 proteins possess C-terminal RS-like alternating charge domains, similar to motifs seen in the SR protein family, which functions in basal and regulated pre-mRNA splicing. SELEX analysis has indicated that the CFIm68/25 heterodimer preferentially binds the sequence UGUAN (11). Sequence specific binding of CFIm to RNA can direct the recruitment of the CPSF on pre-mRNA through interaction with a CPSF subunit, hFip1 (12).
Here, we show that CFIm participates in alternative poly(A) site selection. We knocked down CFIm25 in HeLa cells, and analyzed, by northern blotting, alternative polyadenylation of several genes that have multiple poly(A) sites within the 3′-UTRs. We detected distributional changes in mRNAs following CFIm25 knock-down, suggesting that CFIm is involved in alternative polyadenylation. Furthermore, analysis of CFIm25 expression profiles in various tissues revealed 1.1, 2.0 and 4.6 kb species of CFIm25 mRNA, which were likely derived from multiple poly(A) signals within the CFIm25 3′-UTR. We found that the 4.6 kb mRNA was ubiquitously expressed in human tissues, while the 1.1 and 2.0 kb species were expressed in a tissue specific manner.
MATERIALS AND METHODS
Antibodies
The entire ORF of CF1m25 was cloned into the expression vector (pGEX-6P-1) and GST-CFIm25 protein was expressed in Escherichia coli and purified by affinity chromatography on glutathione Sepharose (GE healthcare). α-CFIm25 rabbit polyclonal antibody was raised against recombinant GST-CFIm25. α-CstF64 (H-300) was purchased from Santa Cruz and anti-actin (C4) was from Chemicon.
Cell culture and RNA interference
HeLa cells were cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin. For silencing experiments, HeLa cells (∼2.0 × 105/ml) were transfected with small interfering RNA duplexes (200 nM) using Oligofectamine (Invitrogen), according to the manufacturer's instructions, and harvested 84 h after transfection to obtain total RNA. Two siRNAs, GCAAUCGUCAAUGACCCAGUCUUGC (siRNA-408) and AAAUGAUGGGUCCAUAUCCUGGUGC (siRNA-613) targeting CF1m25, and control siRNA, CAGGAACGACUUGAUACGGCUACAG were selected and synthesized by Invitrogen (Stealth RNAi).
Northern blotting
Briefly, 2.5 μg of poly(A)+ RNA were selected on oligo-dT columns, glyoxylated, separated on a 1.0% agarose gel, and transferred to a membrane (Hybond-N+, Amersham). The membrane was hybridized with a radio-labeled probe (1.0–2.0 × 106 c.p.m./ml) for 2 h (or overnight) at 65°C in PerfectHyb solution (TOYOBO) and analyzed by autoradiography. Probes were radio-labeled by BcaBEST labeling (TAKARA). RNA size markers were purchased from Invitrogen (RNA ladder 0.24–9.5). For tissue specific analysis, a human MTN blot (BD Biosciences) was analyzed.
3′ RACE analysis and RT–PCR analysis
For 3′ RACE experiments, 1 μg of total RNA, from control HeLa cells or knocked-down cells, was reverse transcribed with an adapter linked oligo-dT primer to the final volume of 20 μl following the manufacture's procedure (3′-Full RACE Core Set, TAKARA). Then, 0.5 μl of the cDNA was subjected to PCR (the reaction volume of 20 μl) with a specific primer (4 pmol) and an adapter primer (1 pmol) in the cycle number of 33 (for TIMP-2 gene) or 26 (for GAPDH and CF1m25 genes).
For RT–PCR analysis of DHFR mRNA, 200 ng of poly(A)+ RNAs from control HeLa cells or CF1m25 knock-down cells were reverse transcribed by SuperScriptIII (Invitrogen) with oligo-dT primer, and then DHFR cDNA fragments were augmented using DHFR primers. The DHFR primers were designed in the first and the last exons, respectively.
The PCR products were resolved on a 1.0% agarose gel and stained with ethidium bromide.
Nucleotides
Oligonucleotides were synthesized to amplify the cDNA fragments of TIMP-2, syndecan2, ERCC6, DHFR and CF1m25 by PCR. The fragments were radio-labeled for probes in northern blotting. TIMP-2 (forward) oligonucleotide was also served as a specific primer (F1) in 3′ RACE experiments. DHFR oligonucleotides were also used in RT–PCR experiments. The sequences were as follows: TIMP-2 (forward) 5′-CGCAACAGGCGTTTTGCAAT-3′; TIMP-2 (reverse) 5′-TGGTGCCCGTTGATGTTCTT-3′; syndecan2 (forward) 5′-TGTACCTTGACAACAGCTCC-3′; syndecan2 (reverse) 5′-GCCAATAACTCCACCAGCAA-3′; ERCC6 (forward) 5′-AAATCAGTTGGCGTGCACAG-3′; ERCC6 (reverse) 5′-GCAGTATTCTGGCTTGAGTT-3′; DHFR (forward) 5′-GTTGGTTCGCTAAACTGCAT-3′; DHFR (reverse) 5′-TACTTAATGCCTTTCTCCTC-3′; CF1m25 (forward) 5′-TCACTCAGTTCGGCAACAAG-3′; CF1m25 (reverse) 5′-TGCAGCTACCAGCTTGTAAT-3′. For the 3′ RACE experiments, the specific oligonucleotide of TIMP-2 gene (F2) and an adapter primer were designed as follows: F2 (TIMP-2) 5′-CTGTTCGCTTCCTGTATGGT-3′; Adapter 5′-CTGATCTAGAGGTACCGGATCC-3′.
RESULTS
Knock-down of CFIm25 in HeLa cells
CFIm is an essential 3′ end processing factor facilitating assembly of other processing factors on pre-mRNA templates (10). Although precise studies in vitro have demonstrated a critical role for CFIm in pre-mRNA cleavage, in vivo function of CFIm has not been addressed. We knocked down CFIm25 in HeLa cells to evaluate potential effects on pre-mRNA cleavage.
First, we expressed a GST-fusion of CFIm25 protein in E.coli and raised a rabbit polyclonal antibody against purified GST-CFIm25. Figure 1A indicates that the antibody specifically recognized CFIm25.
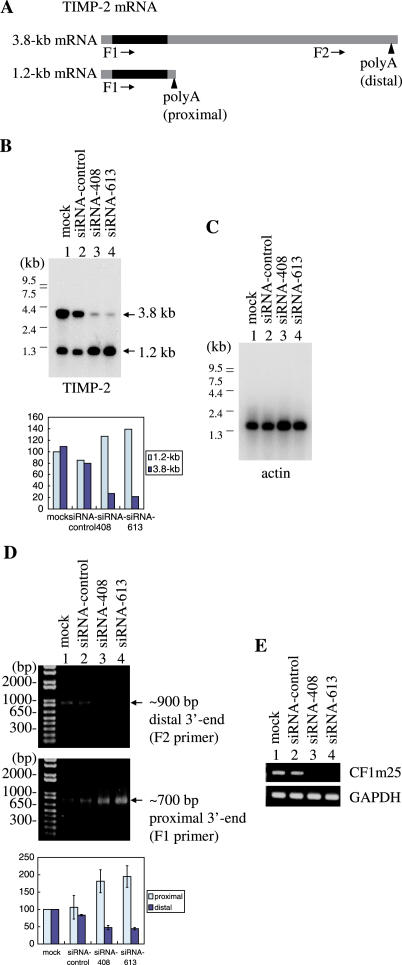
Figure 1.
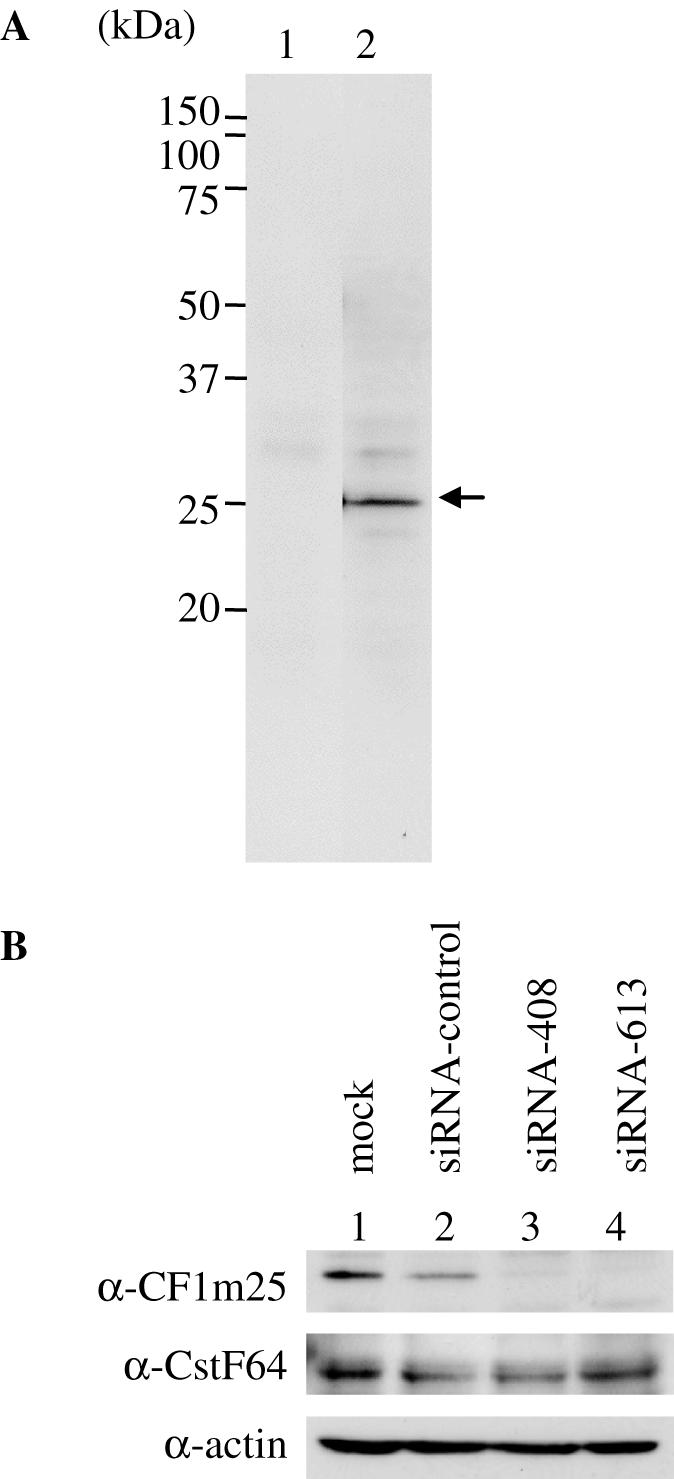
(A) A rabbit polyclonal antibody was raised against GST-CFIm25. A glutathione S-transferase (GST)-fusion protein of CFIm25 was expressed in E.coli and purified by affinity chromatography on glutathione Sepharose (GE Healthcare). HeLa whole cell extracts (20 μg) were separated on 12% SDS–polyacrylamide gels and analyzed by western blotting with pre-immune serum (lane 1) or anti-serum (lane 2). The arrowhead at right indicates CFIm25. (B) Knock-down of CFIm25. Two siRNAs (siRNA-408 and siRNA-613) were designed to target CFIm25. Whole cell extracts (20 μg) from mock-transfected (lane 1), siRNA transfected (control siRNA for lane 2, siRNA-408 for lane 3, siRNA-613 for lane 4) HeLa cells were subjected to western blot analysis with α-CFIm25, α-CstF-64 and α-actin antibodies. Actin was served as a loading control.
Next, we designed siRNAs (designated siRNA-408 and siRNA-613) targeting CFIm25 to sequester CFIm function. As CFIm is a heterodimer composed of CFIm25 and one other larger subunit, we reasoned that depleting cells of CFIm25 protein by RNAi would block CFIm function. HeLa cells were mock-transfected or independently transfected with control siRNA, siRNA-408 and or siRNA-613 siRNAs, 84 h later cells were harvested, and then whole cell extracts were subjected to western blotting. Preliminary time course analysis showed a ∼50% reduction in CFIm25 expression detected after 60 h and a ∼90% reduction after 84 h (data not shown). Expression levels recovered to 50% after 132 h and to 80% after 156 h (data not shown). Knocked-down cells appeared healthy and grew normally. As shown in Figure 1B, either siRNA significantly reduced CFIm25 protein levels. Actin levels, analyzed as a loading control, were not altered by CFIm25 knock-down, indicating that each siRNA specifically knocked down CFIm25.
We also analyzed CstF-64 expression in control cells and knocked-down cells, since it was previously shown that expression levels of CstF-64 influence alternative poly(A) site selection (13–16). As shown in Figure 1B, CstF-64 expression levels were not changed by CFIm25 knock-down.
CFIm knock-down alters poly(A) site usage in TIMP-2 gene
Based on in vitro observations, we predicted that RNAi-mediated depletion of CFIm might reduce the efficient cleavage of pre-mRNA. To evaluate a role of CFIm for pre-mRNA cleavage in vivo, we undertook northern blotting and 3′ RACE PCR to analyze mRNAs transcribed from genes with multiple poly(A) sites in their 3′-UTR.
We first analyzed TIMP-2 gene, which is reported to have two poly(A) sites in the 3′-UTR (17), resulting in 1.2 and 3.8 kb mRNAs (Figure 2A). Total RNA was, 84 h after siRNA transfection, were extracted from mock-transfected, control siRNA transfected or knocked-down HeLa cells, knocked-down or mock-transfected HeLa cells, poly(A)+ RNA were selected on oligo-dT columns and the RNA were analyzed by hybridization. As shown in Figure 2B, significant changes in TIMP-2 mRNA patterns were detected in knock-down cells compared to control cells. In knock-down cells, 1.2 kb mRNA levels were increased, while 3.8 kb decreased relative to controls (Figure 2B, lower panel). Actin was analyzed as a loading control and also a specific control (Figure 2C), indicating that the same amounts of RNA were analyzed in control and knocked-down cells and that polyadenylation of actin mRNA was not affected by CF1mCFIm25 knock-down. The two siRNAs were independently transfected to knock down CFIm25 and the same results were obtained (Figure 2B, lanes 23 and 34), suggesting that the results were reproducible. Moreover we reproduced the results at least three times. Because it is a distributional change of mRNAs but not the appearance of a novel mRNA that was observed by CFIm25 knock-down, we prefer the idea suggest that CFIm is involved in alternative poly(A) site selection.
Figure 2.
Analysis of CFIm25 knock-down on TIMP-2 mRNAs in HeLa cells by northern blotting and 3′ RACE PCR. (A) The TIMP-2 mRNA structures are diagramed. Black boxes indicate open reading frame. The arrowheads indicate proximal and distal poly(A) sites (1077 and 3650). Shaded boxes indicate UTRs and arrows indicate the F1 primer (400–419) and F2 primer (2777–2796) in 3′ RACE experiments. (B) Northern blotting analysis on TIMP-2 mRNAs. Poly(A)+ RNA from mock-transfected (lane 1), siRNA transfected (siRNA-control for lane 2, siRNA-408 for lane 3 and siRNA-613 for lane 4) HeLa cells were analyzed by northern blotting with a TIMP-2 probe. RNA size markers are indicated on the left. Quantification is shown in the lower panel, relative to the 1.2 kb signal in lane1. (C) Northern analysis with a β-actin cDNA probe, serving as a loading control. (D) 3′ RACE PCR experiments. Specific primers F1 and F2 were used for the amplification of the 3.8 kb mRNA 3′ ends (upper panel) and the 1.2 kb mRNA 3′ end (middle panel), respectively. Lane 1 mock-transfected, lane 2 control siRNA, lane 3 siRNA-408 and lane 4 siRNA-613 transfected. Quantification is shown in the lower panel, relative to the signals in lane 1. (E) RT–PCR experiments. CF1m25 (upper panel) was analyzed to show the knock-down efficiency and GAPDH (lower panel) was analyzed for the loading control.
We further performed 3′ RACE PCR experiments on TIMP-2 gene. Total RNA were reverse-transcribed with adapter linked oligo-dT primer and then 3′ end was amplified with an adapter primer and a specific primer by PCR. The specific primers were designed (designated as F1 and F2) as diagramed in Figure 2A. As shown in Figure 2E, F1 primer and F2 primer yielded bands of about 700 and 900 bp, corresponding to the proximal poly(A) site and the distal poly(A) site, respectively. We found that the relative amounts of the amplified bands of 700 bp increased in knocked-down cells compared to control cells, and bands of 900 bp decreased in knocked-down cells compared to control cells (compare lane1,2 and 3,4), suggesting that the proximal poly(A) site usage increased and the distal poly(A) site usage decreased in CFIm25 knocked-down cells. We could not obtain a longer band with F1 primer, which would represent the distal poly(A) site. With specific primer sets, GAPDH mRNA was analyzed as a loading control and CFIm25 mRNA was analyzed to show that the knock-down was successful (Figure 2E).
Taken together, it was suggested that CFIm may be involved in alternative polyadenylation.
CFIm affects alternative poly(A) site selection in syndecan2, ERCC6 and DHFR genes
To determine whether or not participation of CFIm in alternative poly(A) site selection is restricted to TIMP-2 gene, we also analyzed by northern blotting syndecan2 (18), ERCC6 (19) and DHFR genes (20–22), all of which possess multiple poly(A) sites in their 3′-UTRs.
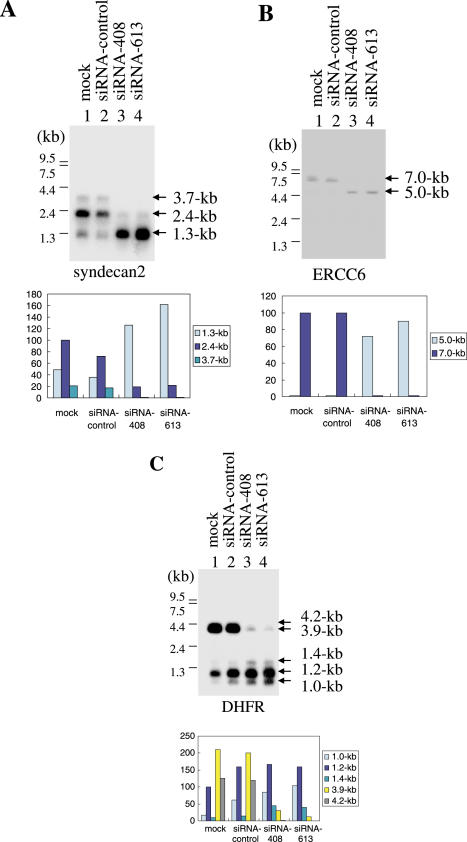
In HeLa cells, there are three syndecan2 transcripts of 1.3, 2.4 and 3.7 kb derived from three poly(A) sites (18). We found that syndecan2 mRNA patterns were drastically altered following RNAi-mediated depletion of CFIm25. As shown in Figure 3A, the 2.4 kb mRNA was most abundant in control HeLa cells, while the 1.3 and the 3.7 kb mRNAs were minor. However, when CFIm was knocked down, the 1.3 kb mRNA became predominant and the 2.4 and 3.7 kb mRNAs were greatly reduced, suggesting that alternative poly(A) site selection was affected.
Figure 3.
Northern blotting analysis on syndecan2 (A), ERCC6 (B) and DHFR (C) transcripts. Poly(A)+ RNA from control (lanes 1 and 2) or from CFIm25 knocked-down (lanes 3 and 4) cells was immobilized on a membrane and analyzed using following cDNA probes: syndecan2 (A), ERCC6 (B) and DHFR (C). Lane 1 mock-transfected, lane 2 control siRNA transfected, lane 3 siRNA-408 transfected, lane 4 siRNA-613 transfected. Quantifications are shown in the lower panels, relative to the 2.4 kb signal in lane 1 (syndecan2), the 7.0 kb signal in lane 1 (ERCC6) and the 1.2 kb signal in lane 1 (DHFR). RNA size markers are indicated at left.
ERCC6 is known to possess two poly(A) sites in the 3′-UTR, resulting in 7.0 and 5.0 kb mRNAs (19). Northern blotting analysis showed that the 7.0 kb mRNA was detectable in control HeLa cells and the 5.0 kb was undetectable (Figure 3B, lane 1), indicating that the downstream site was preferentially used and the upstream site was rarely used in HeLa cells. However, when CFIm25 was knocked down, the 5.0 kb mRNA turned to be detectable, while the 7.0 kb mRNA became a minor species (Figure 3B), suggesting that the potential poly(A) site was used when CFIm25 was knocked down in HeLa cells. These observations indicate that CFIm may regulate poly(A) site selection but not mRNA stability.
Remarkable changes in DHFR mRNA patterns were also detected in knock-down cells. DHFR gene is reported to possess multiple poly(A) sites in its 3′-UTR (20–22), and we detected five mRNA species of 1.0, 1.2, 1.4, 3.9 and 4.2 kb in control HeLa cells by northern blotting (Figure 3C). In contrast, in knock-down cells the 1.0, 1.2 and 1.4 kb mRNA species increased while the 3.9 and 4.2 kb species decreased relative to control cells, indicating that alternative poly(A) site selection was affected (Figure 3C).
We have reproduced the results of northern blotting experiments that knockdown of CFIm25 altered poly(A) site selection at least three times for each gene. Taken together, these observations of the three mRNAs suggest that CFIm may function in alternative poly(A) site selection in the 3′-UTRs.
CFIm does not alter splicing of DHFR gene transcripts in HeLa cells
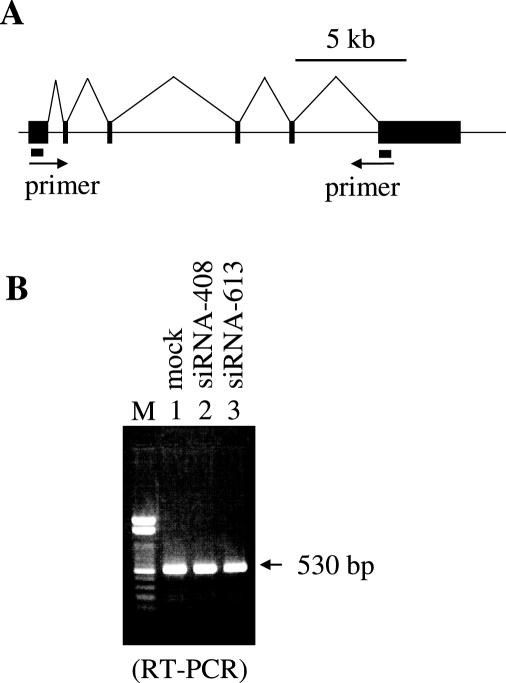
The fact that CFIm68 and CFIm59 are SR proteins suggests a possible participation of CFIm in splicing (23–25). To exclude this possibility, we performed RT–PCR analysis of CFIm25 knockdown cells and control HeLa cells using DHFR primers. Poly(A)+ RNA isolated from CFIm25 knock-down or control cells were reverse transcribed using an oligo-dT primer and amplified using DHFR primers shown in Figure 4A. As shown in Figure 4B, a single band was amplified in control and knock-down cells. Furthermore, western blotting analysis with a DHFR antibody of whole cell extracts derived from control and CFIm25 knock-down cells detected a single band (data not shown), suggesting that CFIm dose not participate in alternative splicing of DHFR transcripts in HeLa cells.
Figure 4.
RT–PCR analysis. (A) The DHFR gene structure is diagramed. Black boxes indicate exons. Primer sets used for PCR are shown. (B) Poly(A)+ RNA from control mock-transfected (lane 1) or from siRNA-408 and siRNA-613 transfected CFIm25 knocked-down (lanes 2 and 3) HeLa cells were reverse transcribed using an oligo-dT primer, and then DHFR cDNA fragments were amplified by PCR. A 100 bp DNA ladder was loaded in lane M.
CFIm25 has tandem multiple poly(A) signals in its 3′-UTR
It is known that alternative poly(A) site selection sometimes occurs in a tissue specific fashion (see Supplementary Figure) (5), which may be mediated in part by tissue specific expression of 3′ end processing factors. To determine if CFIm25 functions in such a tissue specific manner, we examined its expression profile in human tissues.
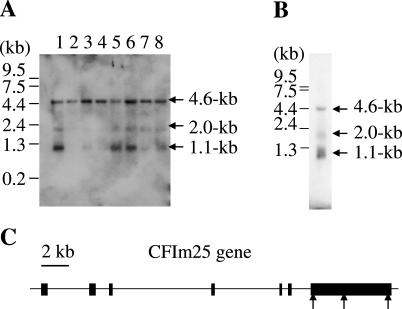
Northern blotting analysis of eight different tissues revealed 1.1, 2.0 and 4.6 kb CFIm25 transcripts. In those human tissues examined, the 4.6 kb mRNA was ubiquitously expressed, but the 1.1 and 2.0 kb mRNAs were expressed in a tissue specific fashion (Figure 5A). Specifically the 1.1 kb mRNA was highly expressed in heart, liver and skeletal muscle, and the 2.0 kb mRNA species was detectable in heart, liver, skeletal muscle, kidney and pancreas. The three mRNAs were also detected in HeLa cells (Figure 5B). To characterize the three mRNAs, we analyzed EST databases and found that there are at least three different poly(A) sites, with canonical poly(A) signals, in CFIm25 gene (NCBI: BG753954, BI260021 and DB558396). The three mRNAs were further analyzed computationally, and we found that the three cleavage sites are located in the 3′-UTR, 82, 1040 and 3579 nt downstream from the stop codon, respectively. Moreover, we found that the calculated mRNA sizes exactly matched the bands detected by northern blotting analysis, suggesting that the three mRNAs represent the three poly(A) sites in the CFIm25 3′-UTR.
Figure 5.
(A) Tissue specific expression of CFIm25. A human MTN blot membrane containing approximately 2.0 μg of poly(A)+ RNA per lane from eight different human tissues was analyzed by northern blotting (lane 1, heart; lane 2, brain; lane 3, placenta; lane 4, lung; lane 5, liver; lane 6, skeletal muscle; lane 7, kidney and lane 8, pancreas). Three transcripts of 1.1, 2.0 and 4.6 kb were detected. (B) Approximately 2.5 μg of poly(A)+ RNA from HeLa cells were analyzed using the CFIm25 probe. Three transcripts of 1.1, 2.0 and 4.6 kb were detected. (C) The gene structure of CFIm25 is diagramed. Black boxes indicate exons. Arrows indicate likely poly(A) sites.
Taken together, these results indicate that CFIm25 is ubiquitously expressed in human tissues and there are three likely poly(A) sites in its 3′-UTR.
DISCUSSION
Here, we present in vivo evidence that CFIm regulates alternative poly(A) site selection of genes with tandem poly(A) signals in their 3′-UTRs.
Alternative poly(A) site selection is widely employed in mammalian cells to regulate gene expression (1,2,26). Alternative poly(A) sites can be located in the last or 3′-most exon, usually giving rise to mRNAs with variable 3′-UTRs, or in a different exon leading to distinct protein products. In terms of alternative polyadenylation, one of the best studied examples is the IgM heavy chain gene (13,14,27–32). During B lymphocyte activation, the IgM heavy chain gene switches from using one poly(A) site to another, resulting in a shift in protein production from a membrane-bound to a secreted form following the deletion of a C-terminal membrane interaction region. An increase in CstF-64 levels during the transition of resting B cells to proliferating B lymphoblasts causes preferential use of an upstream signal (13). Although many genes that possess multiple poly(A) sites within the 3′-UTR have been reported and characterized (1), the molecular basis for alternative polyadenylation in a 3′-UTR has remained unknown. We show here that CFIm is involved in alternative selection among tandem arrays of poly(A) signals within a single 3′-UTR.
In this article, we analyzed TIMP-2, syndecan2, ERCC6 and DHFR genes that have multiple poly(A) signals in their 3′-UTRs (17–22). In addition to these genes, we also investigated for alpha-tropomyosin and vesl genes (33–35) in HeLa cells whose alternative poly(A) sites are located in a body of genes. In these genes, we did not observe alternative poly(A) site selection following CFIm knock-down (data not shown). At this point we cannot determine whether CFIm serves a role for selection of poly(A) sites located within a body of genes.
What is the meaning of alternative poly(A) site selection in a single 3′-UTR? In our experiments, we observed that CFIm depletion in HeLa cells did not affect the cell viability. This may be explained by the fact that HeLa cells can often overcome some defects that would otherwise be critical. We speculate that alternative polyadenylation may most impact in cooperation with some regulatory factors. Recently it was demonstrated that there are many regulatory elements in a 3′-UTR (36,37). For example, mRNA stability is controlled by AU rich elements in the 3′-UTR (38) and microRNAs target the 3′-UTR to inhibit translation (39,40). The regulation by microRNA occurs frequently in development and in cellular differentiation. We took an advantage of knocking down zCFIm25 (NCBI: AAH53172. 97% identical to hCFIm25) in fertilized zebrafish eggs by morpholino oligonucleotide (MO) and observed some morphological defects, suggesting that CFIm may function in embryogenesis (T. Kubo and T. Wada, unpublished data).
Interestingly, the northern blotting analysis of CFIm25 expression in human tissues demonstrated that in addition to the predominant mRNA species, the CFIm25 gene was transcribed as several minor mRNAs, likely resulting from use of multiple poly(A) sites in the 3′-UTR. The CFIm25 3′-UTR structure is conserved between humans and mice (data not shown), suggesting that the alternative polyadenylation may be similarly regulated. Thus we attempted to investigate for an auto-regulation mechanism of poly(A) site selection (41,42). Since siRNA destabilizes mRNA itself, we took an approach of synthesizing MO against CFIm25, which blocks translation but not decays mRNA. So far, low efficiency of delivering MO to HeLa cells prevented us from succeeding in blocking translation of CFIm25.
In the four genes analyzed, CFIm knockdown influenced preferential use of polyadenylation sites upstream of those favored in control cells within 3′-UTR. Since CFIm is an essential 3′ end processing factor, our initial prediction was that CFIm depletion by RNAi would reduce efficient pre-mRNA cleavage, resulting in an appearance of longer transcripts. However, in all genes tested, we observed increases in upstream poly(A) site usage following CFIm25 knock-down, leading us to hypothesize that CFIm suppresses pre-mRNA cleavage. This idea has also been proposed by Gilmartin and colleagues (11). In that work, increasing levels of CFIm68/25 suppressed pre-mRNA cleavage in vitro (11). Given that CPSF and CstF are present throughout the transcription unit, there should be a mechanism to restrict access of 3′ end processing factors to nascent RNA (12,43,44). We favor the idea that CFIm may play a role in regulating RNA binding of CPSF and CstF. Moreover, our results in TIMP-2, syndecan2 and ERCC6 indicate that in the presence of CFIm the most upstream poly(A) sites in the terminal exons are not primarily used in HeLa cells, while in the absence of CFIm the most upstream poly(A) sites predominate. In this context, we speculate that CFIm may serve a role for recognizing the most upstream poly(A) site in a terminal exon. The terminal exon definition has been predicted to occur by communication between 3′ splice site and poly(A) signal (45–47), which are recognized by splicing factors and 3′ end processing factors, respectively. Interaction of CFIm with splicing factors may contribute to selection of poly(A) sites in the terminal exon (11,23–25). CFIm may inhibit pre-mRNA cleavage, recognizing the most upstream poly(A) site within a terminal exon through physical interactions with splicing factors and other processing factors.
Acknowledgments
The authors thank Chikanori Kuramori and Wenyan Zhu for assistance and Hirofumi Furuhashi for valuable discussions. This study was supported by a Grant-in Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology and a Tokyo Tech. Award for Challenging Research (T.W.), by Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology, the Japanese government (T.W. and H.H.) and also supported in part by a Grant from the 21st Century COE Program from the Ministry of Education, Culture, Sports, Science and Technology and a grant for Research (H.H.). T.K. was supported by the Japan Society for the Promotion of Science. Funding to pay the Open Access publication charges for this article was provided by the Ministry of Education, Culture, Sports, Science and Technology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Edwalds-Gilbert G., Veraldi K.L., Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian B., Hu J., Zhang H., Lutz C.S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;12:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosonina E., Kaneko S., Manley J.L. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- 4.Proudfoot N. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 2000;25:290–293. doi: 10.1016/s0968-0004(00)01591-7. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H., Lee J.Y., Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colgan D.F., Manley J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 7.Wahle E., Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruegsegger U., Beyer K., Keller W. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 1996;271:6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- 10.Ruegsegger U., Blank D., Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 11.Brown K.M., Gilmartin G.M. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol. Cell. 2003;12:1467–1476. doi: 10.1016/s1097-2765(03)00453-2. [DOI] [PubMed] [Google Scholar]
- 12.Venkataraman K., Brown K.M., Gilmartin G.M. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagaki Y., Seipelt R.L., Peterson M.L., Manley J.L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 14.Takagaki Y., Manley J.L. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 15.Martincic K., Campbell R., Edwalds-Gilbert G., Souan L., Lotze M.T., Milcarek C. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc. Natl Acad. Sci. USA. 1998;95:11095–11100. doi: 10.1073/pnas.95.19.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shell S.A., Hesse C., Morris S.M., Jr, Milcarek C. Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J. Biol. Chem. 2005;280:39950–39961. doi: 10.1074/jbc.M508848200. [DOI] [PubMed] [Google Scholar]
- 17.Hammani K., Blakis A., Morsette D., Bowcock A.M., Schmutte C., Henriet P., DeClerck Y.A. Structure and characterization of the human tissue inhibitor of metalloproteinases-2 gene. J. Biol. Chem. 1996;271:25498–25505. doi: 10.1074/jbc.271.41.25498. [DOI] [PubMed] [Google Scholar]
- 18.Marynen P., Zhang J., Cassiman J.J., Van den Berghe H., David G. Partial primary structure of the 48- and 90-kilodalton core proteins of cell surface-associated heparan sulfate proteoglycans of lung fibroblasts. Prediction of an integral membrane domain and evidence for multiple distinct core proteins at the cell surface of human lung fibroblasts. J. Biol. Chem. 1989;264:7017–7024. [PubMed] [Google Scholar]
- 19.Troelstra C., Hesen W., Bootsma D., Hoeijmakers J.H. Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne's syndrome group B. Nucleic Acids Res. 1993;21:419–426. doi: 10.1093/nar/21.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Will C.L., Dolnick B.J. 5-Fluorouracil inhibits dihydrofolate reductase precursor mRNA processing and/or nuclear mRNA stability in methotrexate-resistant KB cells. J. Biol. Chem. 1989;264:21413–21421. [PubMed] [Google Scholar]
- 21.Feder J.N., Assaraf Y.G., Seamer L.C., Schimke R.T. The pattern of dihydrofolate reductase expression through the cell cycle in rodent and human cultured cells. J. Biol. Chem. 1989;264:20583–20590. [PubMed] [Google Scholar]
- 22.Lu Y., Han J., Scanlon K.J. Biochemical and molecular properties of cisplatin-resistant A2780 cells grown in folinic acid. J. Biol. Chem. 1988;263:4891–4894. [PubMed] [Google Scholar]
- 23.Zhou Z., Sim J., Griffith J., Reed R. Purification and electron microscopic visualization of functional human spliceosomes. Proc. Natl Acad. Sci. USA. 2002;99:12203–12207. doi: 10.1073/pnas.182427099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rappsilber J., Ryder U., Lamond A.I., Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dettwiler S., Aringhieri C., Cardinale S., Keller W., Barabino S.M. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 26.Iseli C., Stevenson B.J., de Souza S.J., Samaia H.B., Camargo A.A., Buetow K.H., Strausberg R.L., Simpson A.J., Bucher P., Jongeneel C.V. Long-range heterogeneity at the 3′ ends of human mRNAs. Genome Res. 2002;12:1068–1074. doi: 10.1101/gr.62002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980;20:323–329. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- 28.Peterson M.L., Gimmi E.R., Perry R.P. The developmentally regulated shift from membrane to secreted mu mRNA production is accompanied by an increase in cleavage-polyadenylation efficiency but no measurable change in splicing efficiency. Mol. Cell. Biol. 1991;11:2324–2327. doi: 10.1128/mcb.11.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwalds-Gilbert G., Milcarek C. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol. Cell. Biol. 1995;15:6420–6429. doi: 10.1128/mcb.15.11.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips C., Pachikara N., Gunderson S.I. U1A inhibits cleavage at the immunoglobulin M heavy-chain secretory poly(A) site by binding between the two downstream GU-rich regions. Mol. Cell. Biol. 2004;24:6162–6171. doi: 10.1128/MCB.24.14.6162-6171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips C., Jung S., Gunderson S.I. Regulation of nuclear poly(A) addition controls the expression of immunoglobulin M secretory mRNA. EMBO J. 2001;20:6443–6452. doi: 10.1093/emboj/20.22.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veraldi K.L., Arhin G.K., Martincic K., Chung-Ganster L.H., Wilusz J., Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lees-Miller J.P., Goodwin L.O., Helfman D.M. Three novel brain tropomyosin isoforms are expressed from the rat alpha-tropomyosin gene through the use of alternative promoters and alternative RNA processing. Mol. Cell. Biol. 1990;10:1729–1742. doi: 10.1128/mcb.10.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandona D., Tibaldo E., Volpe P. Evidence for the presence of two homer 1 transcripts in skeletal and cardiac muscles. Biochem. Biophys. Res. Commun. 2000;279:348–353. doi: 10.1006/bbrc.2000.3948. [DOI] [PubMed] [Google Scholar]
- 35.Kato A., Ozawa F., Saitoh Y., Hirai K., Inokuchi K. vesl, a gene encoding VASP/Ena family related protein, is upregulated during seizure, long-term potentiation and synaptogenesis. FEBS Lett. 1997;412:183–189. doi: 10.1016/s0014-5793(97)00775-8. [DOI] [PubMed] [Google Scholar]
- 36.Lewis J.D., Gunderson S.I., Mattaj I.W. The influence of 5′ and 3′ end structures on pre-mRNA metabolism. J. Cell. Sci. Suppl. 1995;19:13–19. doi: 10.1242/jcs.1995.supplement_19.2. [DOI] [PubMed] [Google Scholar]
- 37.Hughes T.A. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Barreau C., Paillard L., Osborne H.B. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2006;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carthew R.W. Gene regulation by microRNAs. Curr. Opin. Genet. Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 40.Alvarez-Garcia I., Miska E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 41.Audibert A., Simonelig M. Autoregulation at the level of mRNA 3′ end formation of the suppressor of forked gene of Drosophila melanogaster is conserved in Drosophila virilis. Proc. Natl Acad. Sci. USA. 1998;95:14302–14307. doi: 10.1073/pnas.95.24.14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juge F., Audibert A., Benoit B., Simonelig M. Tissue-specific autoregulation of Drosophila suppressor of forked by alternative poly(A) site utilization leads to accumulation of the suppressor of forked protein in mitotically active cells. RNA. 2000;6:1529–1538. doi: 10.1017/s1355838200001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dantonel J.C., Murthy K.G., Manley J.L., Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 44.Calvo O., Manley J.L. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- 45.Berget S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 46.Proudfoot N.J., Furger A., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 47.Fortes P., Cuevas Y., Guan F., Liu P., Pentlicky S., Jung S.P., Martinez-Chantar M.L., Prieto J., Rowe D., Gunderson S.I. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc. Natl Acad. Sci. USA. 2003;100:8264–8269. doi: 10.1073/pnas.1332669100. [DOI] [PMC free article] [PubMed] [Google Scholar]