Abstract
miRGen is an integrated database of (i) positional relationships between animal miRNAs and genomic annotation sets and (ii) animal miRNA targets according to combinations of widely used target prediction programs. A major goal of the database is the study of the relationship between miRNA genomic organization and miRNA function. This is made possible by three integrated and user friendly interfaces. The Genomics interface allows the user to explore where whole-genome collections of miRNAs are located with respect to UCSC genome browser annotation sets such as Known Genes, Refseq Genes, Genscan predicted genes, CpG islands and pseudogenes. These miRNAs are connected through the Targets interface to their experimentally supported target genes from TarBase, as well as computationally predicted target genes from optimized intersections and unions of several widely used mammalian target prediction programs. Finally, the Clusters interface provides predicted miRNA clusters at any given inter-miRNA distance and provides specific functional information on the targets of miRNAs within each cluster. All of these unique features of miRGen are designed to facilitate investigations into miRNA genomic organization, co-transcription and targeting. miRGen can be freely accessed at http://www.diana.pcbi.upenn.edu/miRGen.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs, 19–24 nt long, which play a crucial regulatory role by inhibiting the translation of protein-coding mRNAs in various eukaryotic organisms. In 1993, two of these tiny RNAs were fortuitously identified in a classical forward genetics screen (1). However, it was not until 2001 that miRNAs were realized to be widespread and abundant (2–5).
MiRNA biogenesis and function
A miRNAs is processed from a longer transcript, referred to as the primary transcript (pri-miRNA). miRNAs can be located within the introns of protein-coding genes, outside of protein-coding genes entirely (‘intergenic’) or more rarely in a coding exon, untranslated region (UTR) or exon of a non-coding transcript. There is evidence that miRNAs located within the introns of protein-coding genes are processed from the mRNAs of these ‘host genes’ (6).
The very few pri-miRNAs which have been experimentally characterized in animals have lengths up to ∼4 kb (8–10). Furthermore, these pri-miRNAs often contain several miRNAs (11). These co-transcribed miRNAs are referred to as a ‘cluster’. In cases where primary transcripts are unknown, miRNAs located close to each other on the genome are thought to be likely members of a ‘cluster’. The maximum length of a miRNA cluster is still unknown. Statistical evidence from microarray expression data has suggested that mammalian pri-miRNAs may be up to 50 kb in length (6). The distribution of pri-miRNA lengths in different species is likely to vary, just as protein-coding gene length distribution varies among animal organisms (12–16). After a pri-miRNA is transcribed, precursor miRNA (pre-miRNA) hairpin structures are cleaved from the transcript and exported to the cytoplasm where the final ‘mature’ miRNAs of 19–24 nt are produced (17,18).
These tiny miRNAs inhibit the translation of a mRNA into protein through imperfect base pairing to one or more target sequences in the mRNA. The identification of animal miRNA targets is a challenging assignment for both experimental and computational groups. The lack of a canonical high-throughput experimental method for miRNA target identification provides the motivation for the continued development of computational target prediction programs. Since the first computational programs in late 2003/early 2004 (19–23), ∼15 programs have been developed and published (24,25). Of these, ∼10 are available for download and/or public online use (24,25). Very recently the target predictions from two of these programs have been made accessible as UCSC Genome Browser tracks. In a recent comparative study of five widely used mammalian target prediction programs, TargetScan (20), DIANA-microT (19), miRanda (26), TargetScanS (27) and PicTar (28), we compared the output of these programs on the experimentally supported miRNA target interactions in TarBase (24). We find that no one program can be considered as consistently superior to the rest; however, in many cases it may be helpful to use the intersection of the predictions of a subset of these programs (29). This can yield improved specificity with only a marginal decrease in sensitivity relative to any individual program.
Existing databases and related tools
The discovery of novel miRNAs, the characterization of their biogenesis and the identification of their functions are areas of research that have been highly interdisciplinary in nature, bringing together a number of experimental and computational groups. There are several existing tools and resources that provide updated data regarding each of these areas of research.
Sanger Institute's miRBase serves as the central database for experimentally supported mature miRNA sequences (30).For each supported miRNA, miRBase provides the genomic coordinates of the predicted precursor sequence, the nucleotide sequences of both the precursor and mature miRNA sequences and predicted targets of the mature miRNA according to prediction programs miRanda, PicTar and TargetScanS. For each individual miRNA, miRBase reports Ensembl transcripts which contain the miRNA.
Two additional databases, ARGONAUTE (31) and miRNAMap (32), offer enhanced interfaces to the data contained in miRBase for human, mouse, rat and dog. miRNAMap also reports computationally predicted miRNAs and their predicted targets according to programs miRanda and RNAhybrid (33). Finally, it provides cross-links to other biological databases in order to provide tissue expression and cross-species sequence conservation data for each supported and predicted miRNA. ARGONAUTE, published simultaneously with miRNAMap, provides much of the sameinformation with perhaps a larger miRNA tissue expression dataset—collected from various miRNA expression studies.
TarBase offers a manually curated and comprehensive set of experimentally supported targets in eight different species (24). It contains over 550 target genes and over 750 individual target sites. For each miRNA:target interaction that has gained experimental support, TarBase reports on the sufficiency of the interaction to independently induce translational silencing, the type of translational silencing that is induced (repression versus immediate cleavage), the location of the target site along the 3′-UTR, the nature of the base pairing between the miRNA and target sequence according to the minimum free energy hybridization and the types of experimental methods used for verification.
MOTIVATION FOR miRGen
The aforementioned resources have been useful as centralized sources of basic miRNA genomic and target information, however there are a number of limitations that make it difficult to conduct systematic analyses of the relationship between miRNA genomic organization and miRNA function. For example, one limitation eliminated by miRGen is that it provides direct queries for miRNAs that are located only in introns, exons, overlapping exon boundaries or in UTRs for several popular gene sets including UCSC Known Genes, Refseq Genes and Genscan Genes. It also provides direct queries for miRNAs located in other genomic entities such as pseudogenes and CpG islands. This facility is particularly useful when studying the targets of miRNAs because it provides a straight-forward method of determining whether genomic location or relationship to a particular genomic entity affects function.
Furthermore, although a few of the existing databases, such as ARGONAUTE, do provide the option of searching for predicted miRNA targets from several widely used computational programs, none of them allow the user to determine which predictions are shared by one or more of the programs. Computational programs for target prediction use different gene id systems (Ensembl gene id, Refseq id, Gene symbol, etc) to represent the predicted targets genes. miRGen performs gene id integration so that users can search for and compare target predictions of several programs.
ARGONAUTE and miRNAMap contain miRNA data for up to four genomes (human, mouse, rat and dog). Except for predicted target information, miRGen currently provides miRNA data for 11 animal genomes. Additionally, while surveys with respect to miRBase miRNAs and Ensembl annotation have been published for a few genomes in the past (34), a constant influx of new data makes it very difficult to obtain timely information from these sources. miRGen is updated quarterly for all genomes and all of the programs used to generate the miRGen database are also made available for public use with user-provided datasets.
Finally, the current state of the art for miRNA databases still does not afford the user a rapid and undemanding method of studying the positional relationship among miRNAs. Only a few poly-cistronic miRNA clusters have been experimentally characterized. Therefore, at the moment, it is necessary to computationally predict likely miRNA clusters for those wishing to study the properties and utilities of miRNA co-transcription. miRGen provides a database of pre-computed miRNA clusters, at various inter-miRNA distances. It also affords a user interface for dynamic computation of miRNA clusters at any user-specified inter-miRNA distance. This facility is useful for the study of the phylogenetic conservation of miRNA clusters as well as the study of miRNA cluster utilities—for example, are clusters generated at various points in evolution in order to consolidate similarly functioning miRNAs?
DATABASE STATISTICS
These unique features of miRGen make rapid and up-to-date customized analyses of miRNA genomic organization and function accessible to every laboratory, regardless of previous bioinformatics experience. The following statistics provide a brief snapshot of miRGen's capabilities as well as several insights on the genomic organization and function of miRNAs.
We first computed the percentages of miRNAs that are located within the introns of UCSC Genome Browser's Known Genes, Refseq Genes and Genscan Genes (Table 1). Genscan gene predictions are available for nearly all animal organisms and we note that the average percentage of miRNAs located within introns of Genscan genes across all of the organisms in Table 1 is ∼42%. miRGen revealed that a surprising number of mammalian miRNA precursors overlap exonic and UTR regions of Known Genes. In human, mouse and rat together there are four cases of precursors contained in exons, 11 cases overlapping exons and 90 cases inside UTRs.
Table 1.
Displays the proportion of miRNAs in UCSC known genes, refseq genes and genscan genes for each species
| Organism | UCSC known genes | Refseq genes | Genscan genes |
|---|---|---|---|
| A.gambiae | 16/37 (43.2%) | ||
| C.elegans | 20/116 (17.2%) | ||
| C.familiaris | 1/6 (16.7%) | 2/6 (33.3%) | |
| D.melanogaster | 21/78 (26.9%) | 26/78 (33.3%) | |
| G.gallus | 10/147 (6.8%) | 81/147 (55.1%) | |
| H.sapiens | 166/466 (35.6%) | 157/466 (33.7%) | 237/466 (50.9%) |
| M.musculus | 129/367 (35.1%) | 117/367 (31.9%) | 203/367 (55.3%) |
| P.troglodytes | 21/65 (32.3%) | 35/65 (53.8%) | |
| R.norvegicus | 27/228 (11.8%) | 33/228 (14.5%) | 117/228 (51.3%) |
| T.nigroviridis | 52/143 (36.4%) |
For the human genome, we also computed the percentage of miRNAs that are located in regions that overlap other UCSC Genome Browser genomic entities. This analysis reveals that only ∼10% of human miRNAs are located in regions that overlap with CpG Islands. Furthermore, corroborating a recent investigation (35) we report that there may not be a widespread organizational link between miRNAs and pseudogenes. Only four of the 466 human miRNAs are contained within a pseudogene from the Yale and Vega Pseudogene sets. Finally, six miRNAs are located within regions undergoing genomic analysis by the ENCODE Project. While this number is currently small, these miRNAs are particularly useful case studies due to the extensive amount of experimental data (i.e. regulatory elements, gene expression, etc.) which has been reported for ENCODE regions.
We then investigated which miRNAs are highly likely to be processed as co-transcribed units in human, mouse, rat and chicken (Table 2). For each of these organisms, we note that even at a very conservative maximum inter-miRNA distance of 1000 nt, over 30% of all miRNAs are organized into clusters. Furthermore, we analyzed the conservation of cluster composition at this distance. We found that for 48 of the 50 clusters in mouse, if the cluster contained more than one miRNA with a like-named ortholog in human then all of these orthologs were found together in a human miRNA cluster. This same phenomenon was observed for 18 out of the 20 clusters in chicken. miRGen provides the opportunity to perform additional customized analyses by accessing GO functional annotation for all miRNA targets of each cluster for any given inter-miRNA distance.
Table 2.
Displays the proportion of miRNAs falling into clusters of size two or more for a sample collection of species
| Organism | MID* 500 nt | MID* 1 kb | MID* 5 kb | MID* 50 kb |
|---|---|---|---|---|
| H.sapiens | 99/466 (21.2%) | 142/466 (30.5%) | 204/466 (43.8%) | 230/466 (49.4%) |
| M.musculus | 107/367 (29.2%) | 133/367 (36.2%) | 169/367 (46.0%) | 200/367 (54.5%) |
| R.norvegicus | 73/228 (32.0%) | 91/228 (39.9%) | 112/228 (49.1%) | 127/228 (55.7%) |
| G.gallus | 39/147 (26.5%) | 50/147 (34.0%) | 65/147 (44.2%) | 79/147 (53.7%) |
Cluster distance is the maximum distance between any two miRNAs considered to be in the same cluster.
MID (Maximum Inter-miRNA Distance) is the maximum distance between any two miRNAs considered to be in the same cluster.
miRGen uniquely provides the predicted target sets of optimized combinations of the widely used target prediction programs. In a recent analysis we demonstrate how the use of different combinations of programs can affect sensitivity and specificity when evaluated on experimentally supported target sets (29). We refer the reader to this analysis for a direct comparison of predicted target sets when using an individual program, the intersection of the two most recent programs, the intersection of the 3 second generation programs and the union of the five most widely used programs. We make each of these predicted target sets available to the user through miRGen.
IMPLEMENTATION AND DESIGN
Data sources
miRGen computes and stores (1) positional relationships between miRNAs and genomic annotation sets and (2) miRNA targets according to combinations of widely used target prediction programs. miRGen builds these primary contents from six underlying types of data: genomic coordinates for miRNA precursors, UCSC genome annotation files, genome browser views, experimentally supported miRNA target data, predicted miRNA target data and Gene Ontology (GO) data. Genomic coordinates for miRNA precursors are downloaded from the Sanger Institute's miRBase (30) ftp site. All genome annotation files are downloaded from the UCSC genome annotation database (36) and all genome browser views are through the UCSC Genome Browser. miRGen interfaces whole-genome collections of Sanger miRBase miRNAs with UCSC genomic data for the following genomes: Anopheles gambiae, Caenorhabditis elegans, Canis familiaris, Danio rerio, Drosophila melanogaster, Gallus gallus, Homo sapiens, Mus musculus, Rattus norvegicus, Tetraodon nigroviridis and Pan troglodytes. Experimentally supported miRNA targets are accessed from the DIANA laboratory's TarBase (24) and predicted miRNA targets are accessed from results of the PicTar, TargetScanS, miRanda and DIANA-microT programs (19,26–28). Each experimentally supported or predicted gene is associated with its GO function as defined by the Gene Ontology Consortium (37). Regular data updates are facilitated by an automated system for retrieving and integrating the most recent miRBase and UCSC annotation track files.
Data integration
The proper function of the genomic locations component requires that miRNA coordinates used for each genome be compatible with all annotation coordinates. miRGen includes every eukaryotic organism handled by both miRBase and UCSC for which genome builds are either identical or can be rendered compatible via UCSC's genome LiftOver facility. The Clusters interface generates cluster links to UCSC Genome Browser views and to experimentally supported and computationally predicted miRNA targets. miRGen links miRNA genomic locations to computationally predicted targets by querying four prominent and publicly available programs, PicTar, TargetScanS, Miranda and DIANA-microT. Currently the predicted targets query facility is focused on mammalian targets, though it is designed to be expanded to other species as predictions become available through the prediction program websites. Experimentally supported targets are loaded from TarBase, which currently handles: H.sapiens, M.musculus, D.melanogaster, C.elegans and D.rerio. A critical data integration task performed by the underlying targets and cluster facilities is to collate target gene sets across multiple prediction programs, while these programs use many different gene identifier systems (Refseq, Ensembl Gene ID, Ensembl Transcript ID, Gene Symbol).
Database generation
The primary goals of the underlying miRNA locations, targets and clusters database generation and query programs are speed and efficiency. The miRNA genomic locations program performs a binary search into each UCSC genome annotation file to find items which overlap the precursor and then the sub-structure of each overlapping item is examined. The purpose of the binary search is to efficiently extract these overlapping items from large annotation files. This program provides the ability to scan 466 miRNAs over a gene annotation file of 10 Mb in 7 s on a Pentium IV PC. The resulting set of miRNA relationships to the sub-structure of each gene is stored in the database. A similar procedure is performed for all other annotation sets such as CpG islands. The Targets interface facilities of miRGen rely on SQL database queries to TarBase, as well as predicted miRNA target sets. Optimized intersections and unions of the predicted target sets are stored separately for query by the Targets interface and for public download as flat files. The Clusters interface facility initially performs a binary sort of the miRNA locations by chromosome and strand, then for each gene set invokes a version of the genomic locations program to efficiently determine which miRNAs are co-located within the same gene and which inter-miRNA regions contain a gene and finally stores this pre-computed information in the database. When the Clusters interface is invoked by the user, it then ‘walks’ each chromosome strand using this pre-stored information to determine which adjacent miRNAs are added to a cluster given a specific user-defined inter-miRNA distance. All programs which generate the miRNA-gene relationship data used by miRGen can be freely downloaded and expanded under the GNU public license.
USER INTERFACE
miRNA genomics
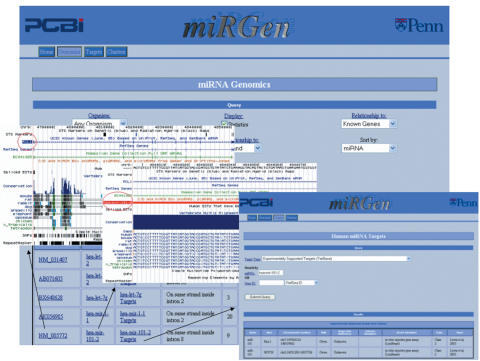
For each organism, the user can select and sort subgroups of miRNAs according to their relationship to genes—for example, one can select all miRNAs located on the same strand inside the introns of UCSC Known Genes. The results of each selection are displayed in a table, where each row contains either an intergenic miRNA or a unique miRNA-gene relationship pair. If predicted or experimentally supported targets are available for the selected organism, a miRNA Targets interface query link is provided for each miRNA. Within all selected subsets, links to the UCSC genome browser for each miRNA and gene also allow a graphical view of any individual entity of interest, shown in Figure 1.
Figure 1.
miRGen Genomics interface links to the UCSC genome browser and to the miRGen Targets interface Links to the UCSC genome browser for each miRNA and gene provide graphical views. Links to the miRGen Targets interface provide tabular views of predicted or experimentally supported targets.
Sorting by miRNA allows the user to easily see which miRNAs are contained in more than one gene listing—for example, many splice forms of a particular gene may contain a single miRNA, but the miRNA may potentially be in a different intron of each. Sorting by Gene allows the user to quickly see which genes contain more than one miRNA. If the ‘Statistics’ box is checked, a summary of gene and miRNA totals for the selected category is displayed below the table, along with a breakdown of gene–miRNA relationship pairs contained in each subcategory—the user can examine the number of gene–miRNA pairs where a miRNA sits inside a gene 3′-UTR for example.
When ‘All UCSC Tracks’ is selected from the ‘Relationship’ menu, a table of all miRNAs for the selected organism is displayed. In addition to Known, Refseq or Genscan gene tracks, this table shows which other UCSC track data entities—Vega Pseudogenes, for example—contain these miRNAs. A statistics table summarizes the total number and percentage of miRNAs overlapping each entity. Since all available annotation sets are included, this summary table is the best place to gain a quick overview for a particular genome.
miRNA targets
The miRGen Targets interface is functional for the following organisms: human, mouse, rat, worm, fruit fly and zebrafish. For any of these organisms, the user can search either for targets of a particular miRNA or miRNAs that target a particular gene. The user also can choose the type of targets to be reported in the search. For human, the type of targets can be chosen from the following options: (i) predicted targets from the union of first and second generation mammalian target prediction programs (PicTar, TargetScanS, miRanda, DIANA-microT), (ii) predicted targets from the intersection of second generation mammalian programs (PicTar, TargetScanS, miRanda), (iii) predicted targets from the optimal intersection of programs (PicTar, TargetScanS) and (iv) experimentally supported targets from TarBase (24). For mouse, rat, worm, and fruit fly, predictions from several individual programs are available. For zebrafish, only the TarBase option is currently available. For each target search type, the user is not limited to a search on one miRNA or family of miRNAs—any desired group of miRNAs may be entered so that target information can be analysed collectively. All major target gene ID types (RefSeq, Ensembl Gene, Entrez Gene, Gene Symbol) along with functional annotation and GO terms are provided for each predicted target gene, regardless of the gene's target prediction program of origin. Figure 1 provides a snapshot of the miRGen Targets interface and how it relates to the miRGen Genomics interface.
miRNA clusters
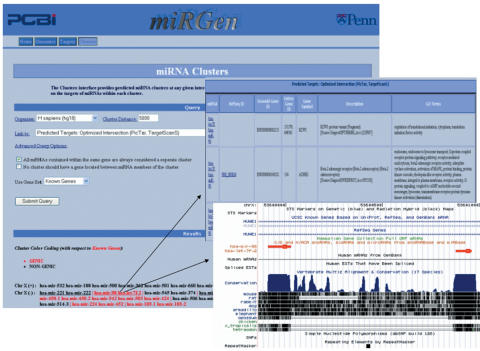
miRGen allows the user to define a cluster distance—the maximum distance between any two miRNAs considered to be in the same cluster—and view the resulting clusters of miRNAs on each chromosome. miRGen gives careful consideration to the issue of how clusters should be defined in relationship to protein-coding genes. The Clusters interface provides a set of advanced options which allow the user to choose whether all miRNAs within the same gene are automatically defined to constitute a separate cluster, whether a cluster should be split by a gene which is located between miRNA members of the cluster and which (if any) available gene sets for the chosen organism should be used for these tasks. Non-genic and ‘genic’ clusters are color-coded according to the selected gene set. The user can then choose to link the resulting clusters to a UCSC browser view, to experimentally supported targets provided by TarBase or predicted miRNA targets provided by PicTar, TargetScanS, Miranda and DIANA-microT. Selection of appropriate tracks in the UCSC browser can help the user to produce customized views which are immediately informative—for example, selection of the PhastCons conservation track produces a view such that the user can quickly examine the appearance of conservation patterns within each cluster. Predicted target tables for each cluster include a column for comparison of GO function ontology terms among target genes. Figure 2 provides a snapshot of the miRGen Clusters interface.
Figure 2.
miRGen Clusters Interface Users can choose which interface that the clusters should link to. The figure above shows links to the ‘Predicted Targets Optimized Intersection (PicTar, TargetScanS)’ target set and to the UCSC view for a particular cluster.
CONCLUSIONS
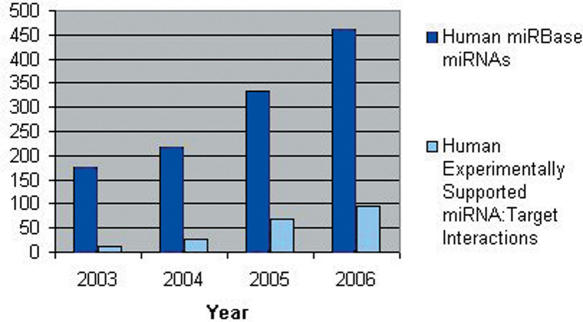
miRGen is designed to study the relationship between miRNA genomic organization and miRNA function in a rapidly evolving field with a constant flow of new data. The number of known miRNAs, as well as the number of experimentally supported targets for these miRNAs, has exploded within recent years (Figure 3), while at the same time target prediction programs are also rapidly changing. miRGen's three connected interfaces: miRNA Genomics, miRNA Clusters and miRNA Targets, provide a user friendly context for performing rapid up-to-date analyses. miRGen provides a particular advantage for efficiently examining large collections of miRNAs using UCSC genome annotation data where one-by-one look-up is not practical. Clusters can be analysed en masse at any given inter-miRNA distance, while at the same time specific functional information on the targets of individual miRNAs within each cluster is also accessible. Using the targets facility, any group of miRNAs may be analysed together as a collection using functional annotation data. miRGen's integration of target gene ID systems enables the user to quickly perform inquiries using unions or intersections of multiple target prediction programs, as well as TarBase.
Figure 3.
Growth in Number of miRNAs and Experimentally Supported Targets This chart tracks the number of Human miRNAs in miRBase and the number of experimentally supported target instances, published through recent years.
AVAILABILITY
miRGen is freely available at http://www.diana.pcbi.upenn.edu/miRGen. miRGen's data processing programs along with an FTP tool for retrieving all underlying miRNA and protein-coding gene data files can be freely downloaded according to the GNU Public License. miRNA targets predicted by individual programs as well as the union and intersections discussed above are also provided as flat files. All data sources used by miRGen are reviewed and updated quarterly.
Acknowledgments
We thank the reviewers and Shane Jensen for helpful commentary on this work. We also thank Julien Hirel for his contribution to the web interface style sheets. This material is based upon work supported by the National Science Foundation under Grant No. 0238295. P.S. is supported by a predoctoral NIH training grant (5T32GM008216). Funding to pay the Open Access publication charges for this article was provided by xxxxx.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lee R.C., Feinbaum R.L., Ambros V. The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 3.Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 5.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 6.Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 8.Bracht J., Hunter S., Eachus R., Weeks P., Pasquinelli A.E. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanzer A., Stadler P.F. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 12.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 13.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 14.Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 15.Hillier L.W., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 16.C.elegans Sequencing Consortium. Genome sequence of the nematode C.elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 17.Kim V.N., Nam J.W. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Seitz H., Zamore P.D. Rethinking the microprocessor. Cell. 2006;125:827–829. doi: 10.1016/j.cell.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Kiriakidou M., Nelson P.T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 21.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark A., Brennecke J., Russell R.B., Cohen S.M. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajewsky N., Socci N.D. Computational identification of microRNA targets. Dev. Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Sethupathy P., Corda B., Hatzigeorgiou A.G. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajewsky N. microRNA target predictions in animals. Nature Genet. 2006;38(Suppl. 1):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 26.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 28.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nature Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 29.Sethupathy P., Megraw M., Hatzigeorgiou A.G. A guide through present computational approaches for the identification of mammalian microRNA targets. Nature Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shahi P., Loukianiouk S., Bohne-Lang A., Kenzelmann M., Kuffer S., Maertens S., Eils R., Grone H.J., Gretz N., Brors B. Argonaute—a database for gene regulation by mammalian microRNAs. Nucleic Acids Res. 2006;34:D115–D118. doi: 10.1093/nar/gkj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu P.W., Huang H.D., Hsu S.D., Lin L.Z., Tsou A.P., Tseng C.P., Stadler P.F., Washietl S., Hofacker I.L. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Devor E.J. Primate microRNAs miR-220 and miR-492 lie within processed pseudogenes. J. Hered. 2006;97:186–190. doi: 10.1093/jhered/esj022. [DOI] [PubMed] [Google Scholar]
- 36.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]