Abstract
We have characterized the global gene expression patterns of Arabidopsis pollen using Serial Analysis of Gene Expression (SAGE). A total of 21,237 SAGE tags were sequenced and 4,211 unique tags were identified. Interestingly, the number of unique tags in pollen was low compared with the SAGE library of the leaf constructed on a similar scale. The transcript profiles of pollen reflect accurately the characteristics of pollen as a reproductive organ. Functional classification of the expressed genes reveals that those involved in cellular biogenesis such as polygalacturonase, pectate lyase, and pectin methylesterase make up more than 40% of the total transcripts. However, genes involved in energy and protein synthesis, which are prevalent in leaves, were expressed at a relatively low level. The expression level of the great majority of transcripts was unaffected by cold treatment at 0°C for 72 h, whereas pollen tube growth and seed production were substantially reduced. Interestingly, many genes thought to be responsible for cold acclimation such as COR, lipid transfer protein, and β-amylase, that are highly induced in Arabidopsis leaves, were only expressed at their normal level or weakly induced in the pollen. The expression patterns of the cold-responsive transcripts identified by SAGE were confirmed by microarray analysis. Our results strongly suggest that poor accumulation of proteins that play a role in stress tolerance may be why Arabidopsis pollen is cold sensitive.
The functional and biochemical features of specific cell types are determined by their particular gene expression profiles. Such global gene expression patterns can be represented by a “transcriptome”, which reveals the identity and the level of expression of each expressed gene in a defined population of cells (Velculescu et al., 1997). The transcriptome can be modulated by both external and internal factors, and thereby provide not only information useful for the understanding of the basic cellular biology but also a global view of biological responses over environmental stimuli.
Gene expression profiles can be obtained and compared by various methods, such as RNA-DNA hybridization measurements, subtractive hybridization, subtraction libraries, and differential display. However, these methods have been limited in providing overall gene expression patterns due to their technical shortcoming. The recent DNA microarray technique allows large-scale quantitative gene expression analysis. Especially, it has been possible to cover most of the genome in GeneChips for several model systems. However, in many experimental systems, it is still limited by the fact that it only analyzes arbitrarily chosen genes. Another technology, Serial Analysis of Gene Expression (SAGE), in part overcomes this limitation. SAGE allows simultaneous, comparative, and quantitative analysis of gene-specific, 9- to 10-bp sequence tags (Velculescu et al., 1995). The expression patterns of any population of transcripts can be evaluated qualitatively and quantitatively by identifying the gene corresponding to each tag and by determining the abundance of the individual tags. The comparison of gene expression patterns in different physiological states by SAGE can also provide unbiased and quantitative analysis of the genes that are differentially expressed in a variety of processes.
The male gametophyte of flowering plants, represented by the pollen grain, contains all the genetic information required to unite with the female gamete at fertilization and form a new sporophyte. Because of this crucial function in the plant's reproductive cycle, pollen has been the object of energetic cytological, biochemical, and molecular biological research. In recent years, insight into processes underlying pollen development and function has been extended by studying pollen-specific gene expression.
Male gametogenesis in flowering plants, from archesporal cells to mature pollen, involves a series of complicated events (Goldberg et al., 1993). A wide variety of specific gene products are involved. The differentiation and development of the male gametophyte of angiosperms depend on the expression of the haploid genome after meiosis. Mature pollen grains contain messenger RNAs whose protein products appear to function during the late maturation stages of pollen tube growth and germination (Mascarenhas, 1975). Mascarenhas (1990) separated pollen gene expression into two phases. Transcripts of the “early” genes are first detectable soon after meiosis and are reduced or undetectable in mature pollen. Transcripts of the “late” genes are first detected around the time of microspore mitosis and accumulate continuously as the pollen matures. The late genes include many pollen-specific sequences thought to function during pollen germination and tube growth.
Temperature is one of the most important environmental factors affecting plant development and crop productivity. Most plants, such as Arabidopsis, develop tolerance to freezing after being exposed to low nonfreezing temperatures. This adaptive process, known as cold acclimation, involves a number of biochemical and physiological changes. The most notable include reduction or cessation of growth, reduction of tissue water content (Levitt, 1980), transient increase in abscisic acid levels (Chen et al., 1983), changes in membrane lipid composition (Lynch and Steponkus, 1987; Uemura et al., 1995), accumulation of compatible osmolytes such as Pro, betaine, polyols, and soluble sugars, and increased levels of antioxidants (Nomura et al., 1991; Koster and Lynch, 1992; Kishitani et al., 1994; Murelli et al., 1995). Many of these changes have been shown to be regulated through changes in gene expression (Thomashow, 1998). Chilling sensitivity is manifested by certain plants whose germination, growth, development of reproductive organs, and postharvest longevity are affected in the range of chilling temperatures from 0°C (nonfreezing) to about 15°C (Lyons, 1973; Wang, 1990). Certain stages in the life cycle of a plant are more sensitive to chilling than others. For example, seedlings appear to be more susceptible than plants at advanced stages of development (Lyons, 1973), and the maturation of pollen is the most sensitive process in the entire life cycle of chilling-sensitive plants (Sataka and Koike, 1983; Patterson et al., 1987).
In recent years, several comprehensive genomic studies based on DNA microarray performed in the field of plant cold acclimation, identified large amount of cold-responsive genes, and discussed their signal pathways (Seki et al., 2001; Fowler and Thomashow, 2002; Kreps et al., 2002; Seki et al., 2002a). Our SAGE data on Arabidopsis leaves also identified many cold-inducible genes including various COR, lipid transfer protein, alcohol dehydrogenase, β-amylase, and many novel genes (Jung et al., 2003). Although cold stress has been extensively studied in whole plants, there has been little investigation into the effect of cold stress on plant organ development and function, especially that of pollen.
In this study, a comprehensive transcript profile of Arabidopsis pollen was analyzed by SAGE and compared with that of cold-treated pollen. Additionally, the transcript profiles of the Arabidopsis leaf, which is thought to be more cold resistant than pollen, were used as references to understand the molecular behavior of pollen. Information on gene expression profiles in pollen and gene regulation by cold stress provides valuable information for understanding why pollen cells are so sensitive to cold stress as well as yielding extensive knowledge of the characteristics of pollen itself. In addition, studies in these areas may reveal new strategies for improving the freezing tolerance of agronomic plants by genetic engineering.
RESULTS
SAGE Analysis of Arabidopsis Pollen
A total of 21,237 tags from Arabidopsis pollen, consisting of 4,211 unique tags, were obtained and analyzed (Table I). Of these, tags at an abundance level of 2 or greater represented 1,507 genes. Of 4,211 unique tags, only 45% of the tags matched the entries of the National Center for Biotechnology Information Arabidopsis SAGE tag-to-gene mapping database. Currently, the proportion of no match is high, probably because a large fraction of the low-abundance class of transcripts might not be reported yet. Even among the reported sequences in the UniGene DB, the 5′ expressed sequence tags (ESTs) are excluded because it is not proper target sequences considering that the tag sequences result from the 10 base sequences 3′ adjacent to the 3′-most NlaIII sites (CATG) of the transcripts.
Table I.
Sequence analysis of SAGE tags from Arabidopsis pollen
| Total No. of Clones | Total No. of Tags | No. of Unique Tags | No. of Tags Matched in the UniGene DBa |
|---|---|---|---|
| 769 | 21,237 | 4,211 | 1,877(45%) |
Indicates the number of genes that match a cDNA or EST entry in Arabidopsis UniGene DB in National Center for Biotechnology Information. No. in parentheses indicates the percentage of matched unique tags to the total number of unique tags.
It has been estimated that the total number of average-sized transcripts ranges from 100,000 to 500,000 per cell in higher plants (Kiper et al., 1979; Kamalay and Goldberg, 1980). Therefore, tag numbers can be converted to number of transcripts per cell on the assumption that 100,000 transcripts are present per cell in a plant with small genome such as Arabidopsis. On the basis of this assumption, a library of approximately 20,000 tag sequences provides 1-fold coverage for transcripts present at a minimum of 5 copies per cell. Our findings indicate that the copy number of highly expressed genes in Arabidopsis pollen is very high. Genes expressed at more than 10 copies per cell comprised 87% of the total number of transcripts (18,532 tags—the total number of tags observed twice or more—of the 21,237 tags) and even genes with more than 100 copies per cell made up 58% of the total number of transcripts (12,267 tags—the total number of tags observed 20 times or more—of the 21,237 tags). On the other hand, more than one-half of the mRNAs (64%) were present at ≤5 copies per cell, and in the aggregate, this low-abundance class represented only 13% of the mRNA mass, indicating that most of the expressed genes are present at a low basal level.
The 50 most highly expressed genes are listed in Table II. An extended list is available as supplemental material (Supplemental Table I; supplementary data can be viewed at http://www.plantphysiol.org). As expected, a large proportion of these corresponded to well-characterized enzymes required for cell wall metabolism such as polygalacturonase, pectate lyase, and pectin methylesterase. The growth of the pollen tube is extremely rapid and is associated with the presence of numerous vesicles in the apical tip. The principle component of the vesicles is pectin (Drashek and Rosen, 1966; Rosen and Gawlick, 1966) and during pollen tube growth, vesicles fuse with the cell membrane in the growing tip to contribute cell wall precursor materials. Because pectin is a primary component of the cell wall, rapid growth of the pollen tube may involve rapid mobilization of pectin molecules from these vesicles to form the new wall. Pectin methylesterase is responsible for the demethylation of pectin before its degradation by the combined activities of polygalacturonase, pectate lyase, and pectin esterase. It is not surprising, therefore, that many genes involved in these functions are abundantly expressed in Arabidopsis pollen.
Table II.
Highly expressed genes in Arabidopsis pollen
| Tag_Sequence | Abundance | UniGene ID | Description |
|---|---|---|---|
| GCTGCTGAGG | 520 | At.24742 | Nuclear-encoded chloroplast stromal cyclophilin (ROC4) mRNA, complete cds |
| GCTGCTGAGG | 520 | At.34811 | Chromosome 3 CHR3v07142002 genomic sequence (pollen-specific protein BAN102-like protein) |
| CAGAGTGTCA | 457 | At.22350 | mRNA for exopolygalacturonase |
| CAGCGCTCCA | 393 | At.3 | mRNA for polygalacturonase |
| TAGGTACGTT | 369 | At.22342 | Putative pectinesterase (At2g47030; F14M4.14) mRNA, complete cds |
| TAGGTACGTT | 369 | AT.24875 | mRNA for pectin methylesterase (pme5 gene) |
| TTTCTCATTG | 318 | At.22223 | ESTs |
| TTTCTCATTG | 318 | At.36416 | Chromosome 1 CHR1v07142002 genomic sequence (expressed protein) |
| TGTGGACGGG | 264 | At.42166 | Chromosome 1 CHR1v07142002 genomic sequence (expressed protein) |
| TCGAGGCTCC | 238 | At.15900 | Chromosome 1 CHR1v07142002 genomic sequence (putative agp1) |
| TCCCTATTAA | 208 | No match | |
| GGGTAAAAAT | 197 | At.18310 | Chromosome 3 CHR3v07142002 genomic sequence (putative pectate lyase) |
| GCTGCAGAGG | 183 | At.44216 | ESTs, moderately similar to pollen-specific protein BAN102-like protein |
| GTTCAGACTC | 174 | At.27756 | Chromosome 5 CHR5v07142002 genomic sequence (Ole e I [main olive allergen]-like protein) |
| CAAAACGAAG | 173 | At.35593 | Chromosome 3 CHR3v07142002 genomic sequence |
| CTCAATCCGC | 166 | At.44875 | Clone 4426 mRNA, complete sequence |
| GAGTTTAGCT | 161 | At.28481 | Unknown protein (At3g01240) mRNA, complete cds |
| GGAGGCTGTG | 159 | At.29514 | Chromosome 5 CHR5v07142002 genomic sequence (expressed protein) |
| TATGGACCTC | 157 | At.12580 | Chromosome 2 CHR2v07142002 genomic sequence (putative pectinesterase) |
| TCCCCGTACA | 149 | No match | |
| GTTTCTGGCG | 148 | At.29756 | Chromosome 5 CHR5v07142002 genomic sequence (putative protein) |
| CTTATAAACC | 143 | At.42723 | Chromosome 3 CHR3v07142002 genomic sequence (hypothetical protein) |
| GAGGAGTTGA | 138 | At.20152 | Chromosome 5 CHR5v07142002 genomic sequence (pectin methyl-esterase-like protein) |
| TCAGAACGTT | 134 | At.7075 | Chromosome 5 CHR5v07142002 genomic sequence (polygalacturonase) |
| CTGTACATTG | 133 | At.36415 | Chromosome 1 CHR1v07142002 genomic sequence (expressed protein) |
| GATGCAGGAG | 129 | At.27687 | Chromosome 1 CHR1v07142002 genomic sequence (protein Ser/Thr phosphatase-α, putative) |
| GATGCAGGAG | 129 | At.41974 | Chromosome 1 CHR1v07142002 genomic sequence (pectate lyase, putative) |
| TCTTGTGACA | 128 | No match | |
| TGTTTGACTT | 127 | At.24426 | Peroxiredoxin TPx2 mRNA, complete cds |
| CAAGGAAATG | 127 | At.172 | mRNA for shaggy-like kinase beta |
| TGAGTGGTGG | 126 | At.38803 | Chromosome 2 CHR2v07142002 genomic sequence (expressed protein) |
| TAAAAGGTGA | 123 | At.24875 | mRNA for pectin methylesterase (pme5 gene) |
| CTCCGGCTCC | 113 | At.43891 | Chromosome 5 CHR5v07142002 genomic sequence (putative protein) |
| AGAGTGTTAT | 111 | No match | |
| CAATGCAACG | 111 | No match | |
| TCCACCCACG | 110 | At.30848 | Chromosome 5 CHR5v07142002 genomic sequence (nectarin-like protein) |
| GATCCTGCTA | 98 | No match | |
| GCACGTGGTG | 94 | At.45253 | Chromosome 1 CHR1v07142002 genomic sequence (putative transposon protein) |
| GCTACTTCCT | 91 | At.27768 | Chromosome 5 CHR5v07142002 genomic sequence (pectin methyl-esterase-like protein) |
| CAATCCTTCC | 90 | At.8258 | AT3g13400/MRP15_3 mRNA, complete cds |
| CGAGAGCACA | 89 | At.34199 | Chromosome 4 CHR4v07142002 genomic sequence (putative calmodulin) |
| TGTTTGCTCT | 89 | No match | |
| AAGACGAGGG | 87 | No match | |
| TTCTGTTATG | 86 | At.40793 | Chromosome 3 CHR3v07142002 genomic sequence (hypothetical protein) |
| TTTTCTTAAA | 83 | No match | |
| TGAAGTTTGT | 82 | At.24426 | Peroxiredoxin TPx2 mRNA, complete cds |
| TGAAGTTTGT | 82 | At.20543 | Peroxiredoxin TPx1 mRNA, complete cds |
| TTGGCTGGTG | 79 | At.35248 | Chromosome 3 CHR3v07142002 genomic sequence (hypothetical protein) |
| TTGGCTGGTG | 79 | At.45117 | Chromosome 3 CHR3v07142002 genomic sequence |
| GTTCAAACTC | 78 | At.32885 | Chromosome 4 CHR4v07142002 genomic sequence (predicted protein) |
| CGCCTTCGCG | 77 | At.2545 | AppB protein (AppB1) mRNA, complete cds |
| CACACCCACA | 74 | At.43123 | Chromosome 3 CHR3v07142002 genomic sequence (unknown protein) |
| ATGCATATAC | 74 | At.45700 | Chromosome 2 CHR2v07142002 genomic sequence (hypothetical protein) |
| GCGTTTCCCT | 74 | No match | |
| TCAAAATCAA | 74 | No match | |
| ATGTTGATGA | 69 | At.35281 | Chromosome 1 CHR1v07142002 genomic sequence (unknown protein) |
Furthermore, several of the genes highly represented in Arabidopsis pollen reflect the characteristics of pollen itself. For example, Agp1 is supposed to be related to Bcp1, an anther-specific cDNA isolated from Chinese cabbage (Brassica campestris; Theerakulpisut et al., 1991). It is active in both diploid tapetum and haploid microspores and is known to be required for pollen fertility (Xu et al., 1995). Other genes expressed at high levels in pollen include those that encode pollen-specific proteins similar to Bnm1 and BAN102-like protein whose function have not yet been characterized. Bnm1, a cDNA isolated from canola (Brassica napus cv Topas; Treacy et al., 1997), is specifically expressed in the bicellular and tricellular stages of pollen development but has no significant sequence similarity to cloned genes of known function. Ole e I (main olive allergen)-like protein is also known to be related structurally to the tomato (Lycopersicon esculentum) anther-specific protein LAT52 (Twell et al., 1989) and maize (Zea mays) pollen-specific protein ZmC13 (Hanson et al., 1989). The LAT52 and Zm13 proteins both show sequence similarity to Kunitz trypsin inhibitor sequences (McCormick et al., 1991), but neither is likely to encode a functional proteinase inhibitor because they lack essential active site residues. The antisense inhibition approach applied to LAT52 (Muschietti et al., 1994) showed a direct correlation between reduced expression of LAT52 protein and abnormal pollen function in transgenic plants, and suggested that the LAT52 protein plays a role in pollen hydration and/or germination.
In addition to pollen-specific genes, nectarin-like protein is abundantly expressed in this library. Nectarin I, a protein that accumulates in the nectar of Nicotiana sp. (Carter et al., 1999), is a germin-like protein and has been shown to have manganese superoxide dismutase activity (Carter and Thornburg, 2000). Germin was first identified in germinating wheat (Triticum aestivum) embryos and has subsequently been identified in all species examined to date, from mosses to gymnosperms and from monocots to dicots (Carter et al., 1998; Neutelings et al., 1998; Yamahara et al., 1999). At least 12 germin-like proteins were detected in the Arabidopsis genome (Carter et al., 1998).
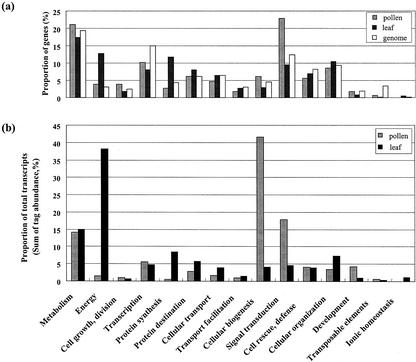
Predicted or known genes were classified according to function, which was assigned essentially by applying the Munich Information Center for Protein Sequences (http://mips.gsf.de) analysis system as well as additional publications. The proportion of genes in each category is shown in Figure 1. Of the 437 genes analyzed, the most highly enriched functional categories are those of cellular communication/signal transduction (23%), metabolism (21%), and transcription (10%). There is no great difference in the overall pattern of gene distribution between pollen and total genome (Fig. 1A). However, if the tag abundance of all transcripts corresponding to each category is considered, genes involved in cellular biogenesis are the most abundant class, indicating that the average copy number per gene of this category is significantly higher than that of any other group of genes (Fig. 1B). This finding suggests that transcripts encoding proteins required for cellular biogenesis represent high copy genes, even if there are not many such genes. Compared with leaves, genes involved in energy and protein synthesis were expressed at lower levels.
Figure 1.
Functional classification of expressed genes in Arabidopsis pollen and leaves. a, The number of genes in each category is presented as a percentage. Functional category is based on the criteria of the Munich Information Center for Protein Sequences (MIPS) Arabidopsis Database (http://mips.gsf.de/proj/thal/). Functional analysis of the genes in Arabidopsis leaves is based on the SAGE data from this laboratory (Jung et al., 2003). The results for the total genome are cited from a recent paper (Seki et al., 2002b). b, The tag abundance of all transcripts corresponding to each transcript is summed and plotted. Functional analysis of the genes in Arabidopsis leaves is based on the SAGE data from this laboratory (Jung et al., 2003).
The Effect of Cold Stress on Pollen Function
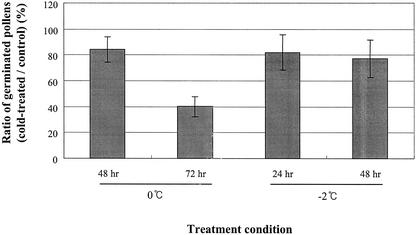
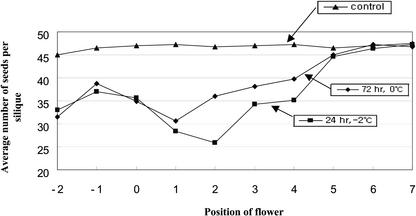
To examine the physiological changes caused by cold stress, its effects on pollen tube growth in vitro and on seed production were measured. The germination rate of pollen was reduced to a greater or lesser extent in all conditions tested (Fig. 2). Exposure of pollen grains to 0°C for 72 h had an especially marked effect, leading to a 60% reduction. Although the effect on pollen germination at –2°C and 0°C was similar, the effect on other parts of plants was quite different. Plants undergoing 0°C treatment re-initiate normal growth after transfer to normal conditions, whereas plants treated at –2°C for long periods tend to wilt due to freezing injury. Plants maintained at 0°C for 72 h and –2°C for 24 h were chosen for seed production assay. As shown in Figure 3, the seed number per silique in unstressed plants is about 47, and this number is reduced in cold-treated plants. Worst of all, some siliques produced abnormal withered seed. Reduced seed production is likely to depend on the stage of flower development. The floral stage of the flower primordia at the time of cold treatment varied from 9 to 16 depending on their location in the plant. Generally, two different stages of flower primordia, that is, stages 11 to 12 and stage 16, suffered severe effects of cold treatment. At stage 16, petals and sepals are starting to wither, and siliques form subsequently. Therefore, the effect on stage 16 is unlikely to be related to pollen development but may instead affect silique formation. However, floral stage 9 to 12 is an important phase in the formation of mature pollen grains (Bedinger, 1992; Goldberg et al., 1993; Scott et al., 2001). Thus, it is likely that flower primordia undergoing pollen development are highly vulnerable to cold stress and that this results in loss of productivity. Stage-specific influences of other types of stress on pollen development have also been reported in barley (Hordeum vulgare) and Arabidopsis (Sakata et al., 2000; Kim et al., 2001).
Figure 2.
Effect of cold treatment on pollen tube growth. Flowers were excised from cold-treated plants and submerged in Tris-EDTA solution (10 mm Tris and 1 mm EDTA, pH 8.0), shaken twice for 15 min, and filtered through a Nitex filter. The pollen in the filtrate was incubated in 20% (w/v) Suc medium containing 2 mm calcium chloride and 1.65 mm boric acid. Total numbers of germinated pollen after 4 h were counted and plotted relative to the control. Approximately 500 pollen grains were used in each experiment, and all experiments were repeated four times. In each bin, the value of the sd is presented as an error bar.
Figure 3.
Effect of cold treatment on seed production. Cold-treated plants were transferred to normal conditions and grown until seeds had been produced. All experiments were repeated twice, and at least eight individual plants were included in each experiment. The average number of seeds was calculated from all the plants. Flower primordia at stage 14 at the time of cold treatment are numbered “0” and earlier blown flowers are given negative numbers.
Gene Expression in Response to Cold Stress
To generate a profile of relative gene expression patterns in cold-treated pollen, RNA was extracted from pollen cells obtained from plants exposed to 0°C for 72 h, where pollen tube growth and seed production were greatly affected, and used for constructing a SAGE library. With 21,075 tags, similar to the number of tags analyzed from untreated pollen, 4,927 different genes were obtained. The 50 most highly expressed genes are shown in Table III. An extended list is available as supplemental material (Supplemental Table II). The expression levels of 57 genes were not much altered, and these ranked among the top hundred most highly expressed genes in both experimental conditions. The finding that the most abundantly expressed genes in cold-treated pollen were also highly expressed in untreated pollen suggests that the highly abundant genes responsible for pollen function are not greatly affected by cold stress.
Table III.
Highly expressed genes in cold-treated pollen
| Tag_Sequence | Abundance | Unigene ID | Description |
|---|---|---|---|
| TTTCTCATTG | 542 | At.22223 | ESTs |
| TTTCTCATTG | 542 | At.36416 | Chromosome 1 CHR1v07142002 genomic sequence (expressed protein) |
| TAGGTACGTT | 366 | At.22342 | Putative pectinesterase (At2g47030; F14M4.14) mRNA, complete cds |
| TAGGTACGTT | 366 | At.24875 | mRNA for pectin methylesterase (pme5 gene) |
| TAAAAGGTGA | 339 | At.24875 | mRNA for pectin methylesterase (pme5 gene) |
| CAGCGCTCCA | 314 | At.3 | mRNA for polygalacturonase |
| GAGGAGTTGA | 302 | At.20152 | Chromosome 5 CHR5v07142002 genomic sequence (pectin methyl-esterase-like protein) |
| TATGGACCTC | 274 | At.12580 | Chromosome 2 CHR2v07142002 genomic sequence (putative pectinesterase) |
| GCTACTTCCT | 242 | At.27768 | Chromosome 5 CHR5v07142002 genomic sequence (pectin methyl-esterase-like protein) |
| GATCCTGCTA | 237 | No match | |
| GAGTTTAGCT | 229 | At.28481 | Unknown protein (At3g01240) mRNA, complete cds |
| GGGTAAAAAT | 210 | At.18310 | Chromosome 3 CHR3v07142002 genomic sequence (putative pectate lyase) |
| TGTTTGACTT | 206 | At.24426 | Peroxiredoxin TPx2 mRNA, complete cds |
| GCTGCTGAGG | 190 | At.24742 | Nuclear-encoded chloroplast stromal cyclophilin (ROC4) mRNA, complete cds |
| GCTGCTGAGG | 190 | At.34811 | Chromosome 3 CHR3v07142002 genomic sequence (pollen-specific protein BAN102-like protein) |
| CTGTACATTG | 176 | At.36415 | Chromosome 1 CHR1v07142002 genomic sequence (expressed protein) |
| AACGGAAAAG | 166 | No match | |
| TGTGGACGGG | 138 | At.42166 | Chromosome 1 CHR1v07142002 genomic sequence (expressed protein) |
| GCTGCAGAGG | 138 | At.44216 | ESTs, moderately similar to pollen-specific protein BAN102-like protein |
| TTTTTATGTA | 135 | At.44216 | ESTs, moderately similar to pollen-specific protein BAN102-like protein |
| TATTGTGTTT | 135 | At.706 | mRNA for putative β -galactosidase (BGAL11 gene) |
| TACGGACGGA | 134 | No match | |
| AGAGATTTTC | 129 | At.27756 | Chromosome 5 CHR5v07142002 genomic sequence (Ole e I [main olive allergen]-like protein) |
| TAAACCTTTT | 121 | At.19923 | Chromosome 5 CHR5v07142002 genomic sequence (putative protein) |
| CAGAGTGTCA | 116 | At.22350 | mRNA for exopolygalacturonase |
| GCGAATGATA | 116 | No match | |
| CAATCCTTCC | 114 | At.8258 | AT3g13400/MRP15_3 mRNA, complete cds |
| GTTCAGACTC | 110 | At.27756 | Chromosome 5 CHR5v07142002 genomic sequence (Ole e I [main olive allergen]-like protein) |
| CGAGAGCACA | 103 | At.34199 | Chromosome 4 CHR4v07142002 genomic sequence (putative calmodulin) |
| TACATTCTGT | 102 | At.26829 | Chromosome 4 CHR4v07142002 genomic sequence (hypothetical protein) |
| TCGAGGCTCC | 101 | At.15900 | Chromosome 1 CHR1v07142002 genomic sequence (putative agp1) |
| ATGCATATAA | 97 | At.41351 | Chromosome 1 CHR1v07142002 genomic sequence (expressed protein) |
| AAAATTCAAA | 92 | At.29916 | Chromosome 5 CHR5v07142002 genomic sequence (peroxidase) |
| GAGTTTGGCT | 90 | At.41311 | Chromosome 3 CHR3v07142002 genomic sequence (hypothetical protein) |
| AGGATGCTCA | 85 | At.19923 | Chromosome 5 CHR5v07142002 genomic sequence (putative protein) |
| TACTACTTTA | 83 | No match | |
| AGAGTGTTAT | 82 | No match | |
| CTTATAAACC | 79 | At.42723 | Chromosome 3 CHR3v07142002 genomic sequence (hypothetical protein) |
| ATGCATATAC | 76 | At.45700 | Chromosome 2 CHR2v07142002 genomic sequence (hypothetical protein) |
| CTCCGGCTCC | 75 | At.43891 | Chromosome 5 CHR5v07142002 genomic sequence (putative protein) |
| TTCTTCAGTA | 75 | At.20183 | AT5g55930/MYN21_4 mRNA, complete cds |
| TGTTTGCTCT | 72 | No match | |
| CTCAATCCGC | 71 | At.44875 | Clone 4426 mRNA, complete sequence |
| TTAATGACGC | 71 | At.14786 | At1g78460/F3F9_3 mRNA, complete cds |
| AAATTGAAAT | 70 | At.39426 | Chromosome 3 CHR3v07142002 genomic sequence (L-ascorbate oxidase precursor, putative) |
| CGCCTTCGCG | 65 | At.2545 | AppB protein (AppB1) mRNA, complete cds |
| GCGTTTCCCT | 64 | No match | |
| CCGGAGACAA | 63 | At.19124 | At1g55570/T5A14_1 mRNA, complete cds |
| TTTTGTGCTT | 61 | At.29641 | Chromosome 5 CHR5v07142002 genomic sequence (unknown protein) |
| TTTTGTGCTT | 61 | At.37415 | Chromosome 3 CHR3v07142002 genomic sequence (expressed protein) |
| TCAGAACGTT | 59 | At.7075 | Chromosome 5 CHR5v07142002 genomic sequence (polygalacturonase) |
| TGTTCAGATG | 59 | At.32125 | Chromosome 5 CHR5v07142002 genomic sequence (hypothetical protein) |
| TATTCCAATA | 58 | At.43992 | Chromosome 5 CHR5v07142002 genomic sequence (carbonate dehydratase-like protein) |
| GAGGATTAGT | 57 | No match |
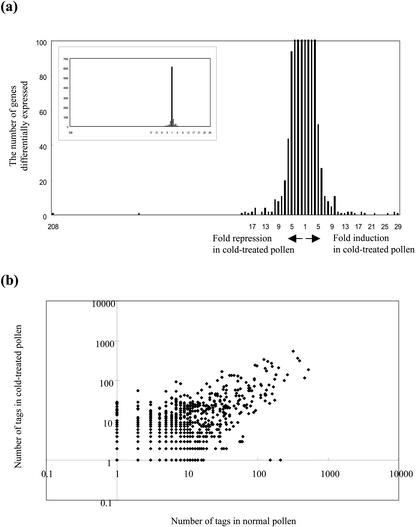
The distribution of fold-changes in tag number between normal and cold-treated cells is shown in Figure 4A. The great majority of transcripts were expressed at similar levels in cold-treated and normal pollen: approximately 92% of the genes showed less than a 3-fold difference in expression. The similarities in the global expression profiles of the two samples can be readily visualized using a scatterplot (Fig. 4B). Twenty-six tags were expressed at least 10-fold more in the cold-treated than in the normal cells, and 28 tags were expressed at least 10-fold more in normal pollen (Table IV). Interestingly, most of the known genes whose products are thought to be responsible for cold acclimation were not cold-induced. Even of the genes whose expression was increased more than 8-fold, only two are known to be involved in stress responses or defense: aluminum tolerance-associated protein and glutathione S-conjugate transporting ATPase (AtMRP1), a multidrug-resistance-associated protein homolog. However there is no direct evidence that the function of these genes is related to the cold stress.
Figure 4.
Changes of copy number of SAGE tags in cold-treated versus normal pollens. a, The ratio of tags was determined by dividing the number of tags in cold-treated and normal pollen. To avoid division by zero, we used a tag value of 1 for tags that were not detected in the tag list. The y axis indicates the number of tags expressed at the ratio indicated on the x axis. b, The number of times each unique SAGE tag was observed is plotted on a logarithmic scale, using a total of 4,927 tags from cold-treated (y axis) versus 4,211 tags from normal pollen (x axis). Tags with no expression in one of the two groups are set to a value of one. The line of slope of 1 through the center corresponds to equal expression in the two tissues.
Table IV.
Differentially expressed genes in cold-treated pollen
| Tag_Sequence | Normal | Cold | Ratio | Unigene ID | Description |
|---|---|---|---|---|---|
| Cold-induced genes | |||||
| TAGAATGATG | 1 | 29 | 29.0a | At.42700 | Chromosome 3 CHR3v07142002 genomic sequence (cellulase-like protein) |
| GACAACAACA | 1 | 28 | 28.0a | At.16827 | FRIGIDA (FRI) mRNA, complete cds |
| GACAACAACA | 1 | 28 | 28.0a | At.35897 | Chromosome 1 CHR1v07142002 genomic sequence (MADS-box transcription factor, putative) |
| TGAAAACTGG | 2 | 55 | 27.5a | No match | |
| AGTTCATCTC | 1 | 26 | 26.0a | No match | |
| TACATACCAT | 1 | 22 | 22.0a | No match | |
| CCGGATGGAT | 1 | 19 | 19.0a | At.22234 | mRNA for exopolygalacturonase |
| AAAATTTGGT | 1 | 18 | 18.0a | No match | |
| TTTTCTCATT | 1 | 18 | 18.0a | At.24853 | mRNA for serine glyoxylate aminotransferase, complete cds |
| TTTGAGAATC | 1 | 16 | 16.0a | No match | |
| AGAAAAAGAC | 1 | 15 | 15.0a | No match | |
| ATTCCAAAAA | 2 | 29 | 14.5a | At.14365 | At2g42580/F14N22.15 mRNA, complete cds |
| AAAATAAAGA | 1 | 14 | 14.0a | No match | |
| AGTATAGAGG | 1 | 14 | 14.0a | No match | |
| AAAATTCAAA | 7 | 92 | 13.1a | At.29916 | Chromosome 5 CHR5v07142002 genomic sequence (peroxidase) |
| ATATTTAAAG | 2 | 23 | 11.5a | No match | |
| TTTAAGAGTC | 5 | 54 | 10.8a | No match | |
| AGGATGCTCA | 8 | 85 | 10.6a | At.19923 | Chromosome 5 CHR5v07142002 genomic sequence (putative protein) |
| TATCTCATTG | 2 | 20 | 10.0a | No match | |
| TAAAGATTAA | 1 | 10 | 10.0a | At.5375 | mRNA for JR3 protein |
| TAAAGATTAA | 1 | 10 | 10.0a | At.32958 | Chromosome 4 CHR4v07142002 genomic sequence (putative protein) |
| ACATCAATTA | 1 | 10 | 10.0a | No match | |
| TATCTGTTCA | 1 | 10 | 10.0a | No match | |
| TCTTAACATT | 1 | 10 | 10.0a | No match | |
| TATATATTCT | 1 | 10 | 10.0a | At.44082 | EST, Moderately similar to glyoxal oxidase (glx1), putative |
| ATAATAAAAT | 1 | 10 | 10.0a | At.20152 | Chromosome 5 CHR5v07142002 genomic sequence (pectin methyl-esterase-like protein) |
| ATAATAAAAT | 1 | 10 | 10.0a | At.32723 | Chromosome 5 CHR5v07142002 genomic sequence (pectin methyl-esterase-like protein) |
| AAATTAAAAT | 1 | 10 | 10.0a | At.6411 | AT3g25770/K13N2_9 mRNA, complete cds |
| TGTATTTAAC | 1 | 10 | 10.0a | No match | |
| TGTATTTAAC | 1 | 10 | 10.0a | No match | |
| Cold-repressed genes | |||||
| TCCCTATTAA | 208 | 1 | 208.0b | No match | |
| TCCCCGTACA | 149 | 1 | 149.0b | No match | |
| TTGGATTGGC | 60 | 3 | 20.0b | At.31475 | Chromosome 4 CHR4v07142002 genomic sequence (putative protein) |
| ACGTCGTAGT | 58 | 3 | 19.3b | No match | |
| GAGACGTTAT | 56 | 3 | 18.7b | No match | |
| AAGACAGGAG | 36 | 2 | 18.0b | At.21711 | Zinc finger protein 3 (ZFN3) mRNA, complete cds |
| GCTGCAAATA | 17 | 1 | 17.0b | At.37952 | Chromosome 2 CHR2v07142002 genomic sequence (putative thioredoxin) |
| GCCATTATGA | 33 | 2 | 16.5b | At.26579 | C7A10_870/C7A10_870 mRNA, complete cds |
| TGGATTATTG | 16 | 1 | 16.0b | At.2344 | Unknown protein (At4g27350) mRNA, complete cds |
| GGTCAGCTCA | 16 | 1 | 16.0b | At.20411 | AT5g01350/T10O8_60 mRNA, complete cds |
| GCAGGTTTGG | 62 | 4 | 15.5b | At.41998 | Chromosome 1 CHR1v07142002 genomic sequence (hypothetical protein) |
| GACCCTTACC | 31 | 2 | 15.5b | At.45700 | Chromosome 2 CHR2v07142002 genomic sequence (hypothetical protein) |
| GACAAGTTCG | 14 | 1 | 14.0b | At.27187 | Unknown protein (At3g52360) mRNA, complete cds |
| GCCCTCGCCG | 14 | 1 | 14.0b | At.23312 | Germin-like protein (GLP8) mRNA, complete cds |
| GTTCTCTTTT | 13 | 1 | 13.0b | At.7295 | Unknown protein (At5g43930; MRH10.2) mRNA, complete cds |
| GTTCTCTTTT | 13 | 1 | 13.0b | At.21104 | Unknown protein (At1g04140) mRNA, complete cds |
| TCCCTATAAG | 13 | 1 | 13.0b | No match | |
| GGAACATACG | 25 | 2 | 12.5b | At.36357 | Chromosome 1 CHR1v07142002 genomic sequence (protein kinase, putative) |
| TGGAGCAGCA | 12 | 1 | 12.0b | At.19998 | AT5g10270/F18D22_40 mRNA, complete cds |
| GGACGGGCTG | 12 | 1 | 12.0b | At.23506 | AT5g54810/MBG8_7 mRNA, complete cds |
| GGCTGATTGG | 11 | 1 | 11.0b | No match | |
| CAACGAGAAC | 11 | 1 | 11.0b | No match | |
| CGGTTTCTGA | 10 | 1 | 10.0b | At.37419 | Chromosome 2 CHR2v07142002 genomic sequence (putative histone H2B) |
| GATGGAGTTT | 10 | 1 | 10.0b | At.20144 | ESTs |
| GTGGCAAGAA | 10 | 1 | 10.0b | At.31838 | Chromosome 5 CHR5v07142002 genomic sequence (probable aldose 1-epimerase-like protein) |
| AACTAAGGGA | 10 | 1 | 10.0b | At.33661 | Chromosome 4 CHR4v07142002 genomic sequence (AIG1-like protein) |
| ACTCCAATCT | 10 | 1 | 10.0b | At.9578 | CBL-interacting protein kinase 10 (CIPK10) mRNA, complete cds |
| GCCCCTGCGC | 10 | 1 | 10.0b | No match |
Cold:Normal. b Normal:Cold.
Confirmation of Gene Expression Patterns by Microarray Analysis
According to our SAGE data, most known genes thought to be responsible for cold acclimation were not induced in Arabidopsis pollen. To confirm this result, a specialized cDNA microarray, constructed in this laboratory to study cold stress, was used. Total RNA from normal and cold-treated pollen was hybridized to the cDNA microarray to examine the expression of hundreds of cold-responsive genes. Among them, the expression patterns of 20 well-known cold-induced genes (Seki et al., 2002a; Fowler and Thomashow, 2002) and three housekeeping genes as revealed by the SAGE and DNA microarray approaches are compared in Table V. Although all of the selected cold-responsive genes were highly induced in the Arabidopsis leaf under cold stress, the same set of genes was not induced or was only slightly induced in Arabidopsis pollen. Such differences in gene expression may contribute to the difference in ability to resist cold stress.
Table V.
Comparison of expression patterns of 20 well-known cold responsive genes between pollen and leaf
| SAGE (Tag Counts)
|
Microarray (Ratio)
|
||||||
|---|---|---|---|---|---|---|---|
| Gene Products | Tag_Sequence | Pollen
|
Leafa
|
Pollen C/N
|
Leaf C/N
|
||
| N | C | N | C | ||||
| Cold-responsive genes | |||||||
| Chalcone synthase | TCGAGCGCGT | 0 | 0 | 0 | 80 | 1.17 | 2.80 |
| pyrroline-5-carboxylate synthetase A | CTCGTGGTCC | 0 | 0 | 1 | 21 | 0.91 | 13.72 |
| β -Amylase enzyme (ct-bmy gene) | GGAGGAGACT | 0 | 0 | 0 | 144 | 1.33 | 4.68 |
| Alcohol dehydrogenase | GGTGCTTGAA | 0 | 0 | 0 | 28 | 1.37 | 19.33 |
| Lipid transfer protein 3 | TATTTGTTTT | 0 | 0 | 1 | 38 | 1.40 | 4.50 |
| Lipid transfer protein 4 | TTTAAGATAT | 0 | 0 | 0 | 17 | 1.09 | 52.22 |
| Fatty acid desaturase/cytochrome b5 fusion protein | GAAGAGATGT | 6 | 36 | 0 | 19 | 0.87 | 8.21 |
| Cold-regulated protein cor15a precursor | TTTAATAGTA | 0 | 0 | 0 | 389 | 5.46 | 133.04 |
| Early light-induced protein | TGTACTAAGT | 0 | 0 | 0 | 203 | 1.20 | 19.46 |
| Iti30 | AAAATAAAAG | 0 | 0 | 0 | 104 | 2.09 | 305.80 |
| Early light-inducable protein | ACTTCAGACG | 0 | 0 | 0 | 80 | 1.52 | 149.94 |
| cDNA 5′ similar to dehydrin erd10 (low-temperature-induced protein Iti45) | CCAGCACCAC | 0 | 0 | 3 | 105 | 1.53 | 8.95 |
| Gly-rich protein (RAB18) | AGTGTGTAAT | 0 | 0 | 0 | 30 | 1.24 | 25.76 |
| Cold acclimation protein WCOR413-like protein α -form | TAAGAGTGAT | 0 | 0 | 0 | 42 | 1.42 | 12.18 |
| cor15b | TTCAATAGTT | 0 | 0 | 0 | 19 | 1.22 | 8.37 |
| rd22 | TTGTGTGGTT | 0 | 0 | 4 | 75 | 0.98 | 8.12 |
| Senescence-associated protein (SAG29) | AGGATCTGTC | 0 | 0 | 0 | 13 | 1.23 | 23.86 |
| Putative dessication-related protein LEA1 | ACGGCTCTTG | 0 | 0 | 3 | 63 | 1.52 | 10.98 |
| LEA D113 type1 protein | CAATGTAACA | 0 | 0 | 0 | 16 | 0.81 | 38.50 |
| FL3-5A3 mRNA for cold acclimation protein homolog | AGGTGTTAGT | 0 | 0 | 3 | 27 | 0.78 | 8.90 |
| Housekeeping genes | |||||||
| α -Tubulin | TATGCGAAGA | 0 | 0 | 3 | 2 | 0.97 | 0.84 |
| Actin8 | AAGATTAAGG | 3 | 2 | 1 | 2 | 1.03 | 0.92 |
| Ubiquitin-conjugating enzymeE2 | GGCTGACACA | 19 | 6 | 3 | 11 | 0.54 | 1.25 |
Data from Jung et al. (2003); N, normal; C, cold.
DISCUSSION
No comprehensive study of gene expression in pollen cells has been reported previously. In this study, in an effort to better understand the development and function of the pollen cell at the molecular level, SAGE analysis was performed, and a comprehensive gene expression profile of Arabidopsis pollen was obtained. SAGE data from Arabidopsis pollen shed light on the transcriptional profile of pollen as a reproductive tissue and hence reveal the classes of proteins and metabolic pathways that are probably used during pollen development. It has been surmised that the early genes might encode cytoskeletal proteins and proteins needed for wall synthesis or starch deposition, whereas the late genes whose mRNAs accumulates throughout microsporogenesis might be required during maturation or pollen tube growth. The primary synthetic events that occur during pollen tube growth are concerned with pollen tube wall synthesis and the synthesis of the cell membrane of the elongating tube. Accordingly, one might expect that many of the pre-existing mRNAs and proteins in the mature pollen grain code for enzymes required in these two processes. By analyzing the SAGE cDNA tag library, many genes involved in cell wall metabolism and other pollen-specific genes were identified. In addition, numerous novel genes not identified previously were also detected.
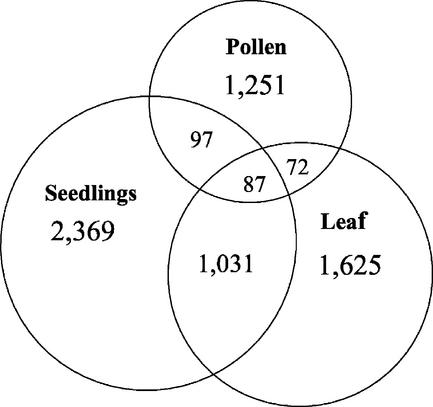
Comparison of the tags present in the pollen cells with those identified in other tissues throws light on the similarities and differences in gene expression among different tissue types. Two SAGE tag sets, one from Arabidopsis leaves constructed in this laboratory (Jung et al., 2003), the other from Arabidopsis seedlings (from SAGEmap; http://www.ncbi.nlm.nih.gov/), were used for this purpose. Because these three libraries were of similar size, comparison between them directly reflects the characteristics of each tissue type. First, the number of unique tag sequences in pollen was only 42% and 54% of that in seedling and leaves, respectively. This observation correlates with previous evidence for low complexity of the mRNA sequences in the mature pollen grains of both Tradescantia sp. and corn (Willing and Mascarenhas, 1984; Willing et al., 1988). Among the 1,507 tags observed twice or more in pollen, only 159 (10%) were common to pollen and leaves. The proportion of tags shared by pollen and seedlings was also approximately 12% of the pollen tags. On the other hand, leaves and seedlings have 1,031 tags in common, which represents approximately 40% of the unique tags in leaves (Fig. 5). This may imply that the overall gene expression regulatory system in pollen is quite different from that in the other two tissues. The identification of 1,251 tags—the number of unique tags observed in pollen but not in leaves and seedlings of the 1,507 tags—may help to account for the characteristics of pollen. Thus 46 of the 50 most highly expressed genes, including well-known pollen-specific genes such as SF21, BAN102-like, and Bnm1-like, are expressed only in pollen not in the two other tissues analyzed.
Figure 5.
Comparison of the pollen SAGE library with other Arabidopsis libraries. Only tags observed more than once are included in this analysis. A total of 1,507 tags in pollen, 2,815 tags in leaves, and 3,583 tags in seedlings were compared. Tag sequences of Arabidopsis seedlings were retrieved from http://www.ncbi.nlm.nih.gov/SAGEmap. Tag sequences of Arabidopsis leaves are from this laboratory (Jung et al., 2003).
Pollen is a specialized organ for accomplishing reproduction. Therefore, its gene expression pattern differs from that of normal vegetative growing leaves. Generally, genes involved in energy production were strongly expressed in leaves (Fig. 1). On the other hand, in pollen, a high level of expression of genes responsible for cellular biogenesis was reveled. The gene expression patterns identified by SAGE reflect well the characteristics of leaves as photoautotrophic organs and pollen as reproductive organs. Thus, the specific composition of a SAGE tag set from a tissue can provide a useful fingerprint representing the tissue-specific pattern of gene expression.
One aim of functional genome analysis is to understand the temporal and spatial expression patterns of all genes with roles in the developmental processes of an organism, and to understand how they function in response to biotic and abiotic cues such as pathological conditions and environmental stresses. In the present work, SAGE was applied to analyze the gene regulation pattern in response to cold stress in Arabidopsis pollen. Because pollen is an important reproductive organ, it is highly likely that the reduced productivity induced by stress originates from malfunction of the pollen. Although cold stress in plants has been extensively studied, there has been little investigation into the effect of stress on pollen development and function. In this study, as a step in unraveling the complexities of gene expression in pollen cells, the SAGE profile of transcripts derived from pollen cells before and after cold treatment was compared. In addition, comparison of transcriptomes between leaf and pollen has yielded interesting information on the dynamics of total genome expression attributable to a change in environmental conditions, and provides clues to the function of those genes whose contribution to cold acclimation is still unknown.
Approximately 20,000 transcripts were analyzed, and numerous genes showing differential expression between cold-treated and untreated cells were identified. The availability of the transcriptome of the cold-treated leaf (Jung et al., 2003) made it possible to compare the differences in gene expression patterns under cold stress between leaf and pollen. Previous SAGE studies have shown that overall gene expression patterns were greatly altered in the leaf, and many cold-related genes known to be responsible for cold acclimation were identified. On the other hand, among the genes differentially induced (P < 0.01) in leaf, only one gene, fatty acid desaturase/cytochrome b5 fusion protein, was also induced (6-fold) in pollen.
Expression of several genes thought to be cold inducible, such as nonspecific lipid transfer protein and δ-8-spingolipid desaturase, was detected in pollen. However, their expression was not significantly increased by cold stress. Although it is possible that additional genes may be expressed at levels below the present detection threshold, it seems unlikely that such genes would contribute to cold tolerance. Microarray-based analysis of several genes showing strong induction in cold-treated Arabidopsis leaves also supports the observations that these genes are not strongly induced in cold-treated pollen. It seems probable that the genes induced or repressed by cold stress in pollen merely reflect general cellular changes unrelated to cold acclimation, and it is possible that the inability to induce expression of genes important in cold acclimation may be responsible for the cold sensitivity of pollen.
The expression levels of many genes responsible for pollen germination and tube growth are not influenced by cold stress. In particular, genes that are strongly expressed under normal condition retain their level of expression in cold-treated samples (Table II). This result may imply that the physiological changes induced by cold stress are not caused by transcriptional repression of genes responsible for pollen function but rather by the inhibition of protein function at the translational or posttranslational level. It is possible that absence of proteins involved in protein stabilization, such as molecular chaperones and heat shock proteins, causes the failure of normal cellular homeostasis. In addition, the fact that many cold-inducible genes that promote membrane stability such as COR15a (Steponkus et al., 1998) and fatty acid desaturase (Kodama et al., 1994, 1995) are not induced by cold stress may be of particular significance in accounting for the cold sensitivity of pollen.
In conclusion, SAGE was performed on normal and cold-treated Arabidopsis pollen, and permitted the identification of their characteristic gene expression profiles. The databases of over 20,000 SAGE tags may be useful resources for investigators interested in the relative expression level of transcripts in the pollen of Arabidopsis. In addition, such SAGE libraries offer important data sets for analyzing comparative gene expression patterns among different tissue types. The present study suggests that an inability to induce certain genes that are important for cold acclimation is responsible the cold sensitivity of pollen and is directly linked to pollen function.
MATERIALS AND METHODS
Growth Conditions and Preparation of Plant Materials
Arabidopsis cv Columbia plants were grown for 4 weeks in an environmentally controlled growth chamber at 23°C under constant light (∼100 μmol m–2 s–1). Flower stages from 9 to 14 were excised and collected in 50-mL tubes. Approximately 70% of the samples were at stage 12 to 14 and the rest were at stage 9 to 11. Pollen was harvested by shaking a tube full of flowers with 25 mL of Tris-EDTA solution (10 mm Tris and 1 mm EDTA, pH 8.0) for 15 min. Two such suspensions were filtered through a Nitex filter (40-μm pore size; Tetko, Elmsford, NY), and the pollen was pelleted at 4,000 rpm for 10 min with a clinical centrifuge. It was then resuspended in 100 μL of RNAlater (Ambion, Austin, TX) and stored at –80°C until use. For cold treatment of the plants, plants grown as above were moved to a growth chamber at the desired temperature under the same light condition and grown for the time period as specified time.
RNA Isolation and cDNA Synthesis
The pollen from 20 mL of flowers was repelleted by centrifugation in an Eppendorf microcentrifuge at 12,000g for 2 min. The supernatant was removed, and the pellet was resuspended in 200 μL of Tri-reagent (Molecular Research Center, Cincinnati). The pollen cells were disrupted with a pellet pestle motor (Thomas Scientific, Swedesboro, NJ) and 200 μL of Tri-reagent was again added. After homogenizing twice, total RNA was prepared following the manufacturer's protocol. mRNA was purified using the Oligotex mRNA mini kit (Qiagen USA, Valencia, CA) according to the manufacturer's instructions. Poly(A+) RNA was converted to double-stranded cDNA as directed by the SuperScript Choice System for cDNA synthesis (catalog series 18090, Invitrogen, Carlsbad, CA).
SAGE Library Construction and Analysis
For construction of the SAGE tag library from normal pollen, SAGE was performed as previously described (SAGE protocol v1.0c) with minor modifications. In brief, double-stranded cDNA was digested with NlaIII, and biotin-containing 3′ cDNA fragments were collected with Streptavidin beads (Roche Diagnostic, Mannheim, Germany). After ligation of oligonucleotides containing recognition sites for BsmFI, the linked cDNA was released from the beads by digestion with BsmFI. The released tags were ligated to one another, blunt-ended, amplified, and redigested with NlaIII. The ditag pellets were resuspended in 3 mm Tris-HCl, pH 7.5, and 0.2 mm EDTA, pH 8.0 containing 25 mm NaCl to prevent denaturation. The ditags were then concatemerized at their NlaIII overhangs using DNA ligase. The ligation mixture was heated at 65°C for 15 min and separated on an 8% (w/v) polyacrylamide gel. DNA fragments between 500 and 1,000 bp in size were isolated and cloned into the SphI site of the pZero-1 vector (Invitrogen). Sequencing was carried out at GENE Inc. (Suwon, Korea). In the case of cold-treated pollen, a modified SAGE method for microscale RNA sample preparation (Lee et al., 2001) was applied for construction of the tag library due to difficulty in obtaining the large amounts of mRNA required for standard SAGE. In brief, mRNA from 25 μg of total RNA was converted to double-stranded cDNA using 5′-biotinylated 3′-anchored oligo(dT) primers (biotin-ATCTAGAGCGGCCGCdT16[A/G/CA/CG/CC]). After digestion with NlaIII and ligation with SAGE linker 1 or 2, the 3′ cDNAs were PCR amplified with the sense primer of SAGE primer 1 (GGATTTGCTGGTGCAGTACA) or SAGE primer 2 (CTGCTCGAATTCAAGCTTCT) and the antisense primer (5′-ACTATCTAGAGCGGCCGCTT-3′), which was located at the 3′ end of all cDNAs generated from the anchored oligo(dT) primers. The BsmFI-released fragments containing the SAGE tags were gel purified before being used for ditag formation and concatenation to provide high-quality tags for SAGE. Sequence files were analyzed using SAGE 2000 software (kindly provided by Dr. Kenneth Kinzler's laboratory, Johns Hopkins University, Baltimore), which automatically detects and counts tags from sequence files. The identities of SAGE tags were analyzed with the National Center for Biotechnology Information Arabidopsis SAGE tag-to-gene mapping database (ftp://ftp.ncbi.nih.gov/pub/sage/map/At/NlaIII/), which matches possible 14-mer tags with known Arabidopsis genes and ESTs.
In Vitro Germination Assay and Seed Production Assay
For germination assays, flowers were submerged in Tris-EDTA solution (10 mm Tris and 1 mm EDTA, pH 8.0), shaken twice for 15 min, and filtered through a Nitex filter. The pollen in the filtrate was plated on in vitro pollen germination medium (20% [w/v] Suc, 2 mm calcium chloride, and 1.65 mm boric acid) and incubated at 25°C for 4 h. Pollen tube growth was observed with a light microscope. Four different conditions (temperature of 0°C or –2°C, and growth period of 24 or 72 h) were used for this analysis. Total numbers of germinated pollen were monitored at times up to 4 h. Approximately 500 pollen grains harvested from flowers in the B4 phase (corresponding to stage 14 [Smyth et al., 1990]) were used in each experiment. Pollen grains at this developmental stage are known to be showing the greatest vitality (Pickert, 1988). For the seed production assay, cold-treated plants were transferred to normal conditions and grown for about 3 weeks until seed production was complete. All the seeds in each silique were counted.
Microarray Analysis
A specialized cDNA microarray generated in this laboratory for cold stress research was used. This microarray contains 712 cDNAs selected on the basis of SAGE and SSH (suppression subtractive hybridization) data, as well as additional reference genes. Pollen RNA was labeled by an indirect method with the 3DNA Array 50 Expression Array Detection Kit (Genisphere, Hatfield, PA). In brief, 25 μg of total RNA was reverse transcribed using reverse transcription primers tagged with either Cy3- or Cy5-specific 3DNA capture sequences. The synthesized tagged cDNAs were then fluorescently labeled by Cy3–3DNA or Cy5–3DNA based on the complementarity of capture sequences with 3DNA capture reagents. Hybridized microarrays were scanned using an ArrayWoRx (Applied Precision, Issaquah, WA). Intensity values were quantified from the resultant pairs of TIFF files using ImaGene image analysis software (BioDiscovery, Los Angeles) and analyzed using the GeneSight software package (BioDiscovery, Los Angeles). Analyses were performed using mean signal intensity values for each spot. Local background was subtracted from the signal intensity and minimum intensity was raised to 20 using “floor” function. The ratio of mean hybridization intensity for each element was normalized by dividing them by the mean of the selected subset (α-tubulin and actin-2). The mean of the two elements was used to combine duplicate spots on one slide.
Supplementary Material
Acknowledgments
We thank Dr. Kenneth W. Kinzler (Johns Hopkins University, Baltimore) for the SAGE protocol and analysis program. We are also grateful to Dr. Sanggyu Lee (University of Chicago) for providing the modified SAGE protocol and technical support.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020511.
This research was supported in part by Korea Science and Engineering Foundation (grant no. 98–0401–06–01–3), by the Brain Korea 21 Project in 2001, and by the 21st Frontier R&D Program (grant no. CG1122).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Bedinger PA (1992) The remarkable biology of pollen. Plant Cell 4: 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Graham RA, Thornburg RW (1998) Arabidopsis thaliana contains a large family of germin-like proteins: characterization of cDNA and genomic sequences encoding 12 unique family members. Plant Mol Biol 38: 929–943 [DOI] [PubMed] [Google Scholar]
- Carter C, Graham RA, Thornburg RW (1999) Nectarin I is a novel, soluble germin-like protein expressed in the nectar of Nicotiana sp. Plant Mol Biol 41: 207–216 [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW (2000) Tobacco nectarin: I. Purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. J Biol Chem 275: 36726–36733 [DOI] [PubMed] [Google Scholar]
- Chen HH, Brenner ML, Li PH (1983) Involvement of abscisic acid in potato cold acclimation. Plant Physiol 71: 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drashek WV, Rosen WG (1966) Electron-microscopical localization of chemical components in the growth zone of lily pollen tubes. Protoplasma 61: 192–204 [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic principles and practical applications. Plant Cell 5: 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson DD, Hamilton DA, Travis JL, Bashe DM, Mascarenhas JP (1989) Characterization of a pollen-specific cDNA from Zea mays and its expression. Plant Cell 1: 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SH, Lee JY, Lee DH (2003) Use of SAGE technology to reveal changes in gene expression in Arabidopsis leaves undergoing cold stress. Plant Mol Biol (in press) [DOI] [PubMed]
- Kamalay JC, Goldberg RB (1980) Regulation of structural gene expression in tobacco. Cell 19: 935–946 [DOI] [PubMed] [Google Scholar]
- Kim SY, Hong CB, Lee I (2001) Heat shock stress causes stage-specific male sterility in Arabidopsis thaliana. J Plant Res 114: 301–307 [Google Scholar]
- Kiper M, Bartels D, Herzfeld F, Richter G (1979) The expression of a plant genome in hnRNA and mRNA. Nucleic Acids Res 6: 1961–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishitani S, Watanabe K, Yasuda S, Arakawa K, Takabe T (1994) Accumulation of glycinebetaine during cold acclimation and freezing tolerance in leaves of winter and spring barley plants. Plant Cell Environ 17: 89–95 [Google Scholar]
- Kodama H, Hamada T, Horiguchi G, Nishimura M, Iba K (1994) Genetic enhancement of cold tolerance by expression of a gene for chloroplast ω-3 fatty acid desaturase in transgenic tobacco. Plant Physiol 105: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama H, Horiguchi G, Nishiuchi T, Nishimura M, Iba K (1995) Fatty acid desaturation during chilling acclimation is one of the factors involved in conferring low-temperature tolerance to young tobacco leaves. Plant Physiol 107: 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster KL, Lynch DV (1992) Solute accumulation and compartmentation during the cold acclimation of Puma rye. Plant Physiol 98: 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chen J, Zhou G, Wang SM (2001) Generation of high-quality and quantity tag/ditag cDNAs for SAGE analysis. Biotechniques 31: 348–354 [DOI] [PubMed] [Google Scholar]
- Levitt J (1980) Responses of Plants to Environmental Stresses. New York, Academic Press
- Lynch DV, Steponkus PL (1987) Plasma membrane lipid alterations associated with cold acclimation of winter rye seedlings (Secale cereale L. cv Puma). Plant Physiol 83: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons JM (1973) Chilling injury in plants. Annu Rev Plant Physiol 24: 445–466 [Google Scholar]
- Mascarenhas JP (1975) The biochemistry of angiosperm pollen development. Bot Rev 41: 259–314 [Google Scholar]
- Mascarenhas JP (1990) Gene activity during pollen development. Annu Rev Plant Physiol Plant Mol Biol 41: 317–338 [Google Scholar]
- McCormick S, Twell D, Vancanneyt G, Yamaguchi J (1991) Molecular analysis of gene regulation and function during male gametophyte development. In GI Jenkins, W Schuch, eds, Molecular Biology of Plant Development. Company of Biologists, London, pp 229–244 [PubMed]
- Murelli C, Rizza F, Albini FM, Dulio A, Terzi V, Cattivelli L (1995) Metabolic changes associated with cold-acclimation in contrasting cultivars of barley. Physiol Plant 94: 87–93 [Google Scholar]
- Muschietti J, Dircks L, Vancanneyt G, McCormick S (1994) LAT52 protein is essential for tomato pollen development: pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J 6: 321–338 [DOI] [PubMed] [Google Scholar]
- Neutelings G, Domon JM, Membre N, Bernier F, Meyer Y, David A, David H (1998) Characterization of a germin-like protein gene expressed in somatic and zygotic embryos of pine (Pinus caribaea Morelet). Plant Mol Biol 38: 1179–1190 [DOI] [PubMed] [Google Scholar]
- Nomura K, Heino P, Tapio Palva E (1991) Separate signal pathways regulate the expression of a low-temperature-induced gene in Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 16: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Patterson BD, Mutton L, Paull RE, Nguyen VQ (1987) Tomato pollen development: stages sensitive to chilling and a natural environment for the selection of resistant genotypes. Plant Cell Environ 10: 363–368 [Google Scholar]
- Pickert M (1988) In vitro germination and storage of trinucleate Arabidopsis thaliana (L.) pollen grains. http://www.arabidopsis.org/ais/1988/picke-1988-aadeg.html
- Rosen WG, Gawlick SR (1966) Fine structure of lily pollen tubes following various fixation and staining procedures. Protoplasma 61: 181–191 [DOI] [PubMed] [Google Scholar]
- Sakata T, Takahashi H, Nishiyama I, Higashitani A (2000) Effects of high temperature on the development of pollen mother cells and microspores in barley Hordeum vulgare L. J Plant Res 113: 395–402 [Google Scholar]
- Sataka T, Koike S (1983) Sterility caused by cooling treatment at the flowering stage in rice plants. Jpn J Crop Sci 52: 207–213 [Google Scholar]
- Scott R, Hodge R, Draper J (2001) The molecular biology of anther differentiation. Plant Sci 80: 167–191 [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression patterns of 1,300 Arabidopsis genes under drought and cold stresses using full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T et al. (2002a) Monitoring the expression profiles of 7,000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y et al. (2002b) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF (1998) Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerakulpisut P, Xu H, Singh MB, Pettitt JM, Knox RB (1991) Isolation and developmental expression of Bcp1, an anther-specific cDNA clone in Brassica campestris. Plant Cell 3: 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1998) Role of cold-responsive genes in plant freezing tolerance. Plant Physiol 118: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treacy BK, Hattori J, Prud'homme I, Barbour E, Boutilier K, Baszczynski CL, Huang B, Johnson DA, Miki BL (1997) Bnm1, a Brassica pollen-specific gene. Plant Mol Biol 34: 603–611 [DOI] [PubMed] [Google Scholar]
- Twell D, Wing R, Yamaguchi J, McCormick S (1989) Isolation and expression of an anther-specific gene from tomato. Mol Gen Genet 217: 240–245 [DOI] [PubMed] [Google Scholar]
- Uemura M, Joseph RA, Steponkus PL (1995) Cold acclimation of Arabidopsis thaliana: effect on plasma membrane lipid composition and freeze-induced lesions. Plant Physiol 109: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Volgelstein B, Kinzler KW (1995) Serial analysis of gene expression. Science 5235: 484–487 [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE Jr, Hieter P, Vogestein B, Kinzler KW (1997) Characterization of the yeast transcriptome. Cell 88: 243–251 [DOI] [PubMed] [Google Scholar]
- Wang CY (1990) Chilling Injury of Horticultural Crops. CRC Press, Boca Raton, FL
- Willing RP, Bashe D, Mascarenhas JP (1988) An analysis of the quantity and diversity of messenger RNAs from pollen and shoots of Zea mays. Theor Appl Genet 75: 751–753 [Google Scholar]
- Willing RP, Mascarenhas JP (1984) Analysis of the complexity and diversity of mRNAs from pollen and shoots of Tradescantia. Plant Physiol 75: 865–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Knox RB, Taylor PE, Singh MB (1995) Bcp1, a gene required for male fertility in Arabidopsis. Proc Natl Acad Sci USA 92: 2106–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamahara T, Shiono T, Suzuki T, Tanaka K, Takio S, Sato K, Yamazaki S, Satoh T (1999) Isolation of a germin-like protein with manganese superoxide dismutase activity from cells of a moss, Barbula unguiculata. J Biol Chem 274: 33274–33278 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.