Abstract
Our aim was to generate and prove the concept of “smart” plants to monitor plant phosphorus (P) status in Arabidopsis. Smart plants can be genetically engineered by transformation with a construct containing the promoter of a gene up-regulated specifically by P starvation in an accessible tissue upstream of a marker gene such as β-glucuronidase (GUS). First, using microarrays, we identified genes whose expression changed more than 2.5-fold in shoots of plants growing hydroponically when P, but not N or K, was withheld from the nutrient solution. The transient changes in gene expression occurring immediately (4 h) after P withdrawal were highly variable, and many nonspecific, shock-induced genes were up-regulated during this period. However, two common putative cis-regulatory elements (a PHO-like element and a TATA box-like element) were present significantly more often in the promoters of genes whose expression increased 4 h after the withdrawal of P compared with their general occurrence in the promoters of all genes represented on the microarray. Surprisingly, the expression of only four genes differed between shoots of P-starved and -replete plants 28 h after P was withdrawn. This lull in differential gene expression preceded the differential expression of a new group of 61 genes 100 h after withdrawing P. A literature survey indicated that the expression of many of these “late” genes responded specifically to P starvation. Shoots had reduced P after 100 h, but growth was unaffected. The expression of SQD1, a gene involved in the synthesis of sulfolipids, responded specifically to P starvation and was increased 100 h after withdrawing P. Leaves of Arabidopsis bearing a SQD1::GUS construct showed increased GUS activity after P withdrawal, which was detectable before P starvation limited growth. Hence, smart plants can monitor plant P status. Transferring this technology to crops would allow precision management of P fertilization, thereby maintaining yields while reducing costs, conserving natural resources, and preventing pollution.
Phosphorus (P), an essential mineral nutrient for plants, is required in large amounts to maintain growth (Raghothama, 1999; Abel et al., 2002). Plants acquire P in the form of phosphate from the soil solution via their roots. Phosphate exists in the soil solution as one of three orthophosphate species (H3PO4, H2PO4–, and HPO42–) depending on the pH (Bohn et al., 1979). The predominant form taken up by plants is thought to be H2PO4– (Ullrich-Eberius et al., 1984; Schachtman et al., 1998). Plants have evolved many morphological and enzymatic adaptations to tolerate low phosphate availability. Increased root to shoot ratios (Lynch, 1995; Williamson et al., 2001), production of roots in P-rich patches (Drew, 1975; Robinson, 1994), and the proliferation of long root hairs (Bates and Lynch, 2001; Jungk, 2001) are often observed when plants are starved of P. These adaptations enhance the exploitation of the soil volume. In addition, the associations of some plant species with symbiotic mycorrhizal fungi are stimulated by P starvation (Harrison, 1999). The transcription and activity of RNases (Bariola et al., 1994) and acid phosphatases (Goldstein, 1992; Duff et al., 1994; del Pozo et al., 1999; Haran et al., 2000; Baldwin et al., 2001; Miller et al., 2001; Li et al., 2002) are increased by P starvation. These enzymes release phosphate from both cellular (Bariola et al., 1994) and extracellular (Duff et al., 1994) organic compounds. The roots of P-deficient plants also exude organic acids into the soil solution to increase the availability of phosphate bound to soil particles and from phosphate salts (Lipton et al., 1987; Holford, 1997; Raghothama, 1999; López-Bucio et al., 2000). The transcripts and activity of phosphate transporters are increased to optimize uptake and remobilization of phosphate in P-deficient plants (Muchhal et al., 1996; Daram et al., 1999; Kai et al., 2002; Karthikeyan et al., 2002; Mudge et al., 2002; Versaw and Harrison, 2002). The internal P economy of P-starved plants is also improved by increasing the activity of enzymes that replace P in metabolites (Malboobi and Lefebvre, 1997; Plaxton and Carswell, 1999; Ciereszko et al., 2001) and structural compounds, for example the proteins involved in the replacement of chloroplast phospholipids with sulfolipids encoded by the SQD genes (Essigmann et al., 1998; Yu et al., 2002). It is thought that these morphological and enzymatic responses to P starvation are coordinated by both general stress-related and P-specific signaling cascades.
It has been suggested that manipulating the expression of genes to optimize root morphology for exploiting the soil volume, to maximize the uptake of phosphate, or to improve P use efficiency could reduce the phosphate fertilizer requirement of crops (López-Bucio et al., 2000; Vance, 2001; Brinch-Pedersen et al., 2002). This has stimulated research toward the identification of genes and signaling cascades involved in responses to P starvation through, for example: (a) homologies with genes to the phosphate-up-regulated (pho) regulon in yeast (Goldstein et al., 1988; Raghothama, 1999; Abel et al., 2002), (b) differential screens of P-starved and -replete plants (Martín et al., 2000; Kai et al., 2002; Wang et al., 2002), and (c) screening for mutants with aberrant root morphology (Bates and Lynch, 2000), uptake, or distribution of P within the plant (pho1 and pho2; Poirier et al., 1991; Delhaize and Randall, 1995; Hamburger et al., 2002), altered root exudate composition (López-Bucio et al., 2000), or acid phosphatase activity (pup1 and pho3; Trull and Deikman, 1998; Zakhleniuk et al., 2001). Recently, the mutagenesis of transgenic Arabidopsis, bearing a construct in which the promoter for a gene induced by P starvation (AtIPS1) controlled the expression of a marker gene (β-glucuronidase [GUS]), was used to identify genes in the signaling cascades initiated by P starvation (Rubio et al., 2001). These included PHR1, which encodes an MYB transcription factor (Rubio et al., 2001). Such global regulators coordinating pleiotropic responses to P starvation are likely to prove extremely useful in transgenic strategies to improve P efficiency in crops. The development of similar smart plants with contrasting promoters would enable further dissection of the signaling cascades by which plants sense and respond in a coordinated manner to P starvation.
In addition to their use in laboratory studies to elucidate the signaling cascades initiated by P starvation, smart plants also have a practical use. To realize the growth potential of crop plants, the phosphate concentration in the rhizosphere must be maximized, and to maintain crop yields and quality, the horticultural and agricultural industries in the United States and western Europe routinely apply excessive amounts of phosphate fertilizer (Goldstein, 1992). This leads to the leaching or erosion of phosphate bound to soil particles into watercourses (National Research Council, 1989; Sharpley, 1995; Withers et al., 2001), which results in nutrient imbalance and contributes to the process of eutrophication in aquatic environments (Raghothama, 1999). Worldwide, the application of phosphate fertilizers accounts for 90% of the total mineral phosphate used (Bieleski and Ferguson, 1983). Unfortunately, the nonrenewable nature of phosphate fertilizers means that cheap sources, such as phosphate rocks, will be exhausted within the next 60 to 90 years (Runge-Metzger, 1995). To prevent environmental problems and to reduce the consumption of a nonrenewable resource, phosphate fertilizers must be used sustainably. This might be achieved by various strategies including (a) the cultivation of crops that require less phosphate fertilizer, because they either acquire or use P more efficiently (Fageria and Baligar, 1999; Baligar et al., 2001), (b) the use of cultural techniques to maintain phosphate levels in the soil, such as crop rotation and intercropping (Frossard et al., 2000; Vance, 2001), and (c) the use of computer programs to inform the quantity and temporal application of phosphate fertilizers (Greenwood et al., 2001). Smart plants complement decision support systems by allowing farmers to monitor crop P status from the expression of a nontoxic, easily assayed marker gene in situ. It is important, therefore, that marker gene expression should increase specifically in response to P deficiency before P starvation restricts growth and reduces yield. The detection of marker gene activity would report a physiological P requirement and would indicate the need for remedial phosphate fertilization.
In this paper, we have identified Arabidopsis genes whose expression increases specifically in response to P starvation, when the P content of plant tissues begins to decline but before the lack of P affects growth. The promoters for these genes are suitable for the generation of smart plants. We have generated transgenic Arabidopsis bearing a construct containing a marker gene (GUS) under the control of the promoter sequence for one of these P-sensitive genes (SQD1), and we demonstrate that these smart plants can be used to monitor P deficiency in plants. Transferring this technology to crop plants will help to manage the application of phosphate fertilizers for sustainable agriculture.
RESULTS
We are interested in developing smart plant technology as both a practical tool to monitor crop P status and as a laboratory tool to discover the regulatory cascades that alter gene expression in response to P deficiency. Smart plants can be genetically engineered by transformation with a construct containing the promoter of a gene up-regulated by P starvation upstream of a gene encoding a visible marker. Detection of the visible marker in the leaves of smart plants indicates P stress and the need for fertilizer application. Because any period of P deficiency that reduces crop growth incurs a yield penalty (Broadley et al., 2002), it is important to detect P deficiencies in crops before they become acute and impact on growth. Thus, the P-sensitive promoter driving marker gene expression in the smart plant must become active early enough in response to phosphate depletion to enable remedial fertilizer application. Transcript profiling of Arabidopsis leaves using high-density oligonucleotide arrays was used to identify promoters active in the early stages of incipient P deficiency.
Phosphate Starvation Reduces Shoot P Concentration and Plant Growth
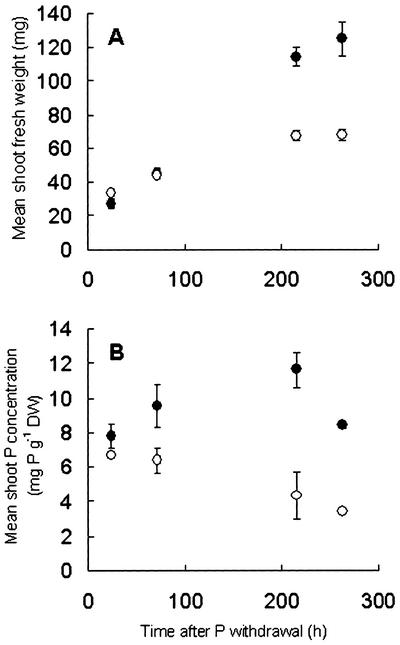
The effects of removing P supply on shoot P concentration and growth was determined on 28-d-old Arabidopsis growing hydroponically (Fig. 1). The shoot fresh weight of plants growing in a solution lacking P (–P solution) was comparable with that of control plants grown in a complete nutrient solution for at least 72 h after P was withdrawn. However, the shoot fresh weight of plants grown without P was significantly (P < 0.001) lower than plants grown in a complete nutrient solution 216 h after P was withdrawn. A decline in P concentration was observed in shoots of Arabidopsis growing in –P solutions. A significant decrease in shoot P concentration in plants lacking a P supply was observed before there was any significant difference in the shoot fresh weight of plants growing in complete and –P solutions. There was no significant difference in the shoot P concentrations of plants growing in complete and –P solutions 24 h after P was withdrawn. However, the shoot P concentrations of plants grown in a –P solution for 72 h were significantly less than those of plants grown in a complete nutrient solution. Thus, between 24 and 72 h after P withdrawal, Arabidopsis grown in –P solutions had a significantly lower shoot P concentration but retained comparable growth to plants grown in a complete nutrient solution. Hence, promoters from genes with up-regulated expression between 24 and about 72 h after P withdrawal could be used to drive the expression of a marker gene in smart plants because they would allow the plant to respond to P deficiency before growth was affected.
Figure 1.
The effect of P withdrawal on shoot fresh weight (A) and shoot P concentration expressed on a dry weight basis (B). Arabidopsis plants were grown hydroponically in a complete nutrient solution (•) or a solution lacking P (○). Plants were 28 d old at the beginning of the experiment. Data are expressed as mean ± SE (n = 3 experiments).
Shoot Gene Expression Changes during Phosphate Starvation
To study changes in the Arabidopsis shoot transcriptome in response to P withdrawal, total RNA samples from shoot material were used to challenge Affymetrix GeneChips representing 8,100 Arabidopsis genes. Total RNA was isolated from rosette leaves of Arabidopsis accession Columbia-5 (Col-5) at growth stage 3.90 (Boyes et al., 2001). Rosette leaves were harvested 4, 28, and 100 h after P withdrawal, and transcript levels were analyzed in two biological replicates at each time point.
Between 3,483 and 5,043 transcripts were declared present in the RNA samples used to challenge the Affymetrix GeneChips. Changes in transcript abundance after P starvation were calculated as their abundance in mRNA from shoots of plants grown in –P solutions divided by their abundance in control plants grown in complete nutrient solutions and harvested at the same growth stage. Differentially regulated transcripts were defined as those with a -fold change > 2.5 in two biological replicates. To reduce the number of transcripts falsely classified as differentially regulated, GeneChip signal values less than 20 were reset to 20 (see “Materials and Methods”).
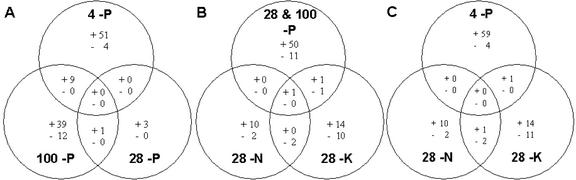
For the majority of genes represented on the GeneChip, expression levels appeared unchanged after phosphate withdrawal (Fig. 2). The expression of no gene was differentially regulated at all time points after P withdrawal, and only 10 genes overlapped between two time points (Fig. 2; Tables I and II). The expression of 64 genes was altered >2.5-fold in two biological replicates 4 h after withdrawing P (Table I). This group of genes was termed “early” P response genes. Sixty genes increased their expression (between 2.5- and 59.6-fold), and four genes decreased their expression. There was great variability in the relative expression of some genes at this time point. This can be attributed to biological variability and may be a consequence of the heterogeneous or asynchronous responses of individual plants to P withdrawal. The differences in gene expression between biological replicates are unlikely to arise from experimental error, because the microarray data were normalized and neither biological replicate showed consistently greater changes in expression. Also, variability in expression was observed for genes that were highly expressed. Twenty-eight h after P was withdrawn, the expression of only four genes differed between shoots of plants grown in –P solutions and shoots of plants grown in complete nutrient solutions (Table II). This lull in differential gene expression has also been observed several hours after other environmental challenges (Reymond et al., 2000; Kreps et al., 2002; Ramonell et al., 2002). One hundred hours after P was withdrawn, the expression of 61 genes differed between the shoots of plants grown in –P solutions and the shoots of plants grown in complete nutrient solutions (Table II). Forty-nine of these genes were up-regulated in plants grown without P, and 12 genes were down-regulated. Because the expression of so few genes differed between shoots of plants grown in –P solutions and shoots of plants grown in complete nutrient solutions at the 28-h time point, these genes were grouped with genes whose expression was altered 100 h after P withdrawal. This combined group was termed the late P response genes. There was only one gene (At2g04160; AIR3) that was up-regulated at both 28 and 100 h after P withdrawal (Table II).
Figure 2.
Venn diagrams showing the numbers of genes differentially expressed in shoots of Arabidopsis plants in response to 4, 28, and 100 h of P starvation (A); 28 + 4 h of P starvation (late P-responsive genes), 28 h of K starvation, and 28 h N starvation (B); 4 h of P starvation (early P-responsive genes), 28 h of K starvation, and 28 h of N starvation (C). Differentially expressed genes were defined as those with a -fold difference in expression in two biological replicates of 2.5 between shoots of plants grown in solutions lacking specific elements and control plants grown in complete nutrient solutions and harvested at the same growth stage. The identities of genes that were differentially expressed in response to P, K, or N starvation are given in Tables I and II.
Table I.
Genes whose expression was altered 4 h after the withdrawal of P from the nutrient solution
Elements (P, K, or N) were withdrawn from the nutrient solutions supplied to 28-d-old Arabidopsis plants growing hydroponically. Gene expression profiles were determined in shoots 4 h after the withdrawal of P and 28 h after the withdrawal of K or N. Gene expression was calculated as the ratio of transcript abundance in shoots of plants starved of P, K, or N divided by transcript abundance in shoots of plants supplied with full nutrient solution.
| Gene Expression Relative to Nutrient Replete Plants
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GeneChip IDa | Expression Levelb | P Starved for 4 h
|
K Starved for 28 h
|
N Starved for 28 h
|
Specificityc | Other Stimulid | Arabidopsis Genome Initiative (AGI) IDe | Functional Categoryf | ||||
| 1 | 2 | 1 | 2 | 1 | 2 | |||||||
| 12097_g_at | M | 0.23 | 0.03 | 1.57 | 1.17 | 15.26 | 1.43 | -P | At2g20870 | Expressed protein | Unclassified | |
| 14633_at | M | 0.34 | 0.14 | 0.70 | 3.17 | 1.03 | 2.30 | -P | At5g54190 | NADPH:protochlorophyllide oxidoreductase A | Control of cellular organisation | |
| 15178_s_at | L | 0.23 | 0.18 | 0.52 | 0.75 | 0.71 | 1.01 | -P | t, hs, c, a, b | At4g14130 | Xyloglucan endotransglycosylase (XTR7) | Metabolism |
| 20518_at | L | 0.39 | 0.30 | 1.18 | 2.11 | 0.89 | 3.20 | -P | At1g10060 | LEA76 homolog type 1 | Metabolism | |
| 12115_at | M | 5.84 | 4.50 | 3.15 | 0.63 | 0.90 | 0.49 | +P | ms, c, cc | At4g22470 | Extensin-like protein | Unclassified |
| 12332_s_at | H | 18.10 | 4.27 | 0.88 | 0.20 | 5.30 | 0.37 | +P | At3g12500 | Basic chitinase | Cell rescue, defense, and virulence | |
| 12642_at | M | 2.54 | 2.90 | 1.64 | 0.71 | 2.96 | 0.72 | +P | At2g15390 | Xyloglucan fucosyltransferase, putative | Unclassified | |
| 12764_f_at | H | 2.80 | 4.20 | 0.32 | 1.04 | 2.38 | 0.84 | +P | cc | At2g02930 | Putative glutathione S-transferase | Metabolism |
| 13154_s_at | H | 4.48 | 11.29 | 2.07 | 0.13 | 2.35 | 0.09 | +P | cc | At2g43590 | Putative endochitinase | Cell rescue, defense, and virulence |
| 13244_s_at | H | 4.91 | 4.17 | 3.25 | 0.47 | 2.47 | 0.46 | +P | w | At4g37990 | Cinnamyl-alcohol dehydrogenase ELI3-2 | Metabolism |
| 13273_at | H | 3.17 | 6.96 | 2.99 | 1.11 | 2.42 | 0.90 | +P | hs, ms, cc | At4g36990 | Heat shock transcription factor HSF4 | Transcription |
| 13467_at | H | 2.94 | 2.74 | 1.08 | 0.53 | 1.08 | 0.76 | +P | At3g47420 | Putative protein | Transport facilitation | |
| 13842_at | M | 4.04 | 3.71 | 0.79 | 0.73 | 0.75 | 0.76 | +P | At1g24140 | Putative metalloproteinase | Protein fate | |
| 14016_s_at | M | 6.19 | 3.38 | 0.70 | 0.67 | 2.58 | 1.99 | +P | g, c | At1g30700 | FAD-linked oxidoreductase family | Metabolism |
| 14032_at | M | 2.59 | 3.47 | 1.78 | 0.43 | 3.59 | 0.58 | +P | lo, o, s, c | At4g37370 | Cytochrome P450-like protein | Metabolism |
| 14041_at | L | 3.45 | 2.59 | 0.95 | 0.79 | 2.14 | 0.90 | +P | At1g09480 | Putative cinnamyl alcohol dehydrogenase | Metabolism | |
| 14635_s_at | M | 3.16 | 7.84 | 0.83 | 1.23 | 2.22 | 0.72 | +P | c, d | At2g14610 | Pathogenesis-related PR-1-like protein | Cell rescue, defense, and virulence |
| 14636_s_at | H | 6.48 | 3.96 | 2.97 | 3.05 | 1.96 | 1.39 | +PK | w, c, d | At1g75040 | Thaumatin-like protein | Cellular communication |
| 14638_at | H | 2.99 | 2.57 | 2.13 | 0.42 | 2.76 | 0.35 | +P | Al, Oz, cc | At3g49120 | Putative peroxidase | Cell rescue, defense, and virulence |
| 14964_at | H | 2.91 | 6.73 | 1.03 | 0.45 | 0.61 | 0.20 | +P | At1g65500 | Expressed protein | Unclassified | |
| 15137_s_at | H | 2.86 | 5.70 | 0.57 | 0.30 | 0.89 | 0.37 | +P | cc | At2g44790 | Phytocyanin | Unclassified |
| 15162_at | H | 9.69 | 8.20 | 0.78 | 0.39 | 1.85 | 0.35 | +P | w, ms | At3g04720 | Hevein-like protein precursor (PR-4) | Unclassified |
| 15216_at | H | 2.99 | 2.89 | 1.44 | 1.47 | 1.38 | 0.98 | +P | cc | At1g09560 | Germin-like protein | Metabolism |
| 15415_at | L | 4.55 | 3.29 | 1.77 | 0.38 | 1.68 | 0.17 | +P | At1g22900 | Disease resistance response protein-related | Cell rescue, defense, and virulence | |
| 15665_at | M | 2.62 | 5.66 | 0.20 | 0.25 | 2.35 | 0.50 | +P | g, c, o | At5g04340 | Putative c2h2 zinc finger transcription factor | Transcription |
| 15672_s_at | H | 3.83 | 4.23 | 2.33 | 0.77 | 0.79 | 0.47 | +P | g, cc | At2g22470 | Arabinogalactan-protein (AGP2) | Unclassified |
| 15866_s_at | H | 2.99 | 3.36 | 1.76 | 0.95 | 2.50 | 0.85 | +P | At2g38860 | Expressed protein | Unclassified | |
| 15985_at | M | 4.03 | 3.53 | 0.43 | 0.61 | 0.65 | 0.37 | +P | Sulf | At5g64100 | Putative peroxidase | Cell rescue, defense, and virulence |
| 16014_at | H | 2.89 | 3.28 | 0.87 | 0.43 | 0.60 | 0.40 | +P | g, cc | At1g75750 | Expressed protein | Unclassified |
| 16053_i_at | H | 4.10 | 4.63 | 2.23 | 0.51 | 3.29 | 0.34 | +P | g, lo, w | At1g02920 | Glutathione S-transferase | Metabolism |
| 16150_at | H | 5.56 | 9.53 | 5.84 | 0.32 | 11.13 | 0.19 | +P | Al, c | At4g12480 | pEARLI 1 | Unclassified |
| 16198_at | M | 2.59 | 3.93 | 0.63 | 0.46 | 1.55 | 0.36 | +P | At2g28710 | Putative C2H2-type zinc finger protein | Transcription | |
| 16888_at | M | 4.64 | 4.12 | 2.24 | 0.31 | 2.07 | 0.30 | +P | At2g37770 | Aldo/keto reductase family | Metabolism | |
| 16914_s_at | H | 19.47 | 4.44 | 0.57 | 0.28 | 4.14 | 0.49 | +P | lo | At4g11650 | Osmotin precursor | Cell rescue, defense, and virulence |
| 17014_s_at | H | 3.92 | 59.55 | 3.87 | 0.66 | 0.57 | 0.22 | +P | P, d | At2g02990 | Ribonuclease, RNS1 | Metabolism |
| 17413_s_at | H | 14.85 | 43.39 | 0.12 | 0.48 | 5.73 | 0.18 | +P | lo | At5g64120 | Putative peroxidase | Cell rescue, defense, and virulence |
| 17485_s_at | H | 44.30 | 4.66 | 1.09 | 0.17 | 6.02 | 0.20 | +P | ms, cc | At4g16260 | Glycosyl hydrolase family 17 | Metabolism |
| 17533_s_at | M | 4.62 | 3.69 | 5.08 | 0.19 | 4.26 | 0.15 | +P | t, hs, c, a, b | At4g25810 | Xyloglucan endotransglycosylase (XTR-6) | Metabolism |
| 17840_at | H | 2.94 | 5.31 | 0.86 | 0.67 | 2.41 | 0.28 | +P | cc | At2g43570 | Endochitinase isolog | Cell rescue, defense, and virulence |
| 17894_at | H | 2.53 | 5.96 | 2.28 | 0.58 | 1.84 | 0.40 | +P | w | At2g18690 | Expressed protein | Unclassified |
| 17899_at | H | 4.69 | 11.87 | 1.68 | 0.21 | 1.35 | 0.23 | +P | o | At4g15610 | Expressed protein | Unclassified |
| 17907_s_at | M | 5.13 | 3.55 | 0.68 | 0.34 | 0.32 | 0.36 | +P | lo | At2g37750 | Expressed protein | Unclassified |
| 17930_s_at | H | 9.13 | 2.77 | 0.92 | 0.41 | 4.49 | 0.46 | +P | ms, lo | At4g37520 | Peroxidase, prxr2 | Cell rescue, defense, and virulence |
| 17963_at | H | 2.52 | 25.79 | 1.06 | 0.49 | 0.79 | 0.27 | +P | c | At4g12470 | pEARLI 1-like protein | Unclassified |
| 18217_g_at | H | 3.16 | 6.07 | 1.30 | 0.65 | 2.50 | 0.68 | +P | s, c | At1g27730 | Salt-tolerance zinc finger protein | Transcription |
| 18228_at | H | 5.83 | 13.73 | 3.40 | 0.65 | 4.14 | 0.59 | +P | ms | At3g15356 | Lectin-like protein | Unclassified |
| 18591_at | M | 2.83 | 3.32 | 2.16 | 0.73 | 2.22 | 0.38 | +P | o | At5g08790 | Expressed protein | Unclassified |
| 18888_at | M | 3.56 | 2.55 | 0.33 | 0.51 | 1.12 | 0.85 | +P | At1g15380 | Expressed protein | Unclassified | |
| 18966_at | M | 2.56 | 4.93 | 0.72 | 0.57 | 1.25 | 0.61 | +P | cc | At2g29420 | Glutathione transferase, putative | Metabolism |
| 19171_at | H | 4.46 | 7.62 | 2.89 | 0.34 | 2.48 | 0.31 | +P | ms, cc | At2g43510 | Putative trypsin inhibitor | Unclassified |
| 19178_at | H | 3.32 | 15.50 | 1.00 | 0.46 | 1.07 | 0.35 | +P | hl, c, o | At5g20230 | Blue copper-binding protein | Unclassified |
| 19284_at | M | 6.02 | 4.08 | 1.63 | 0.77 | 2.24 | 0.56 | +P | ms | At2g38240 | Putative anthocyanidin synthase | Metabolism |
| 19640_at | H | 2.54 | 11.96 | 3.39 | 0.54 | 3.39 | 0.29 | +P | ms | At2g29460 | Putative glutathione S-transferase | Metabolism |
| 19840_s_at | M | 3.54 | 6.47 | 3.72 | 0.67 | 1.93 | 0.29 | +P | cc | At1g30720 | FAD-linked oxidoreductase family | Metabolism |
| 19892_at | H | 2.84 | 5.76 | 2.19 | 0.64 | 1.40 | 0.36 | +P | cc | At2g38870 | Putative protease inhibitor | Protein fate |
| 19991_at | H | 2.51 | 7.87 | 0.50 | 0.58 | 1.77 | 0.47 | +P | g | At2g35980 | Similar to harpin-induced protein hin1 from tobacco | Cell rescue, defense, and virulence |
| 20194_at | H | 2.94 | 3.95 | 6.40 | 0.53 | 2.85 | 0.57 | +P | lo, cc | At2g17500 | Expressed protein | Unclassified |
| 20238_at | H | 3.09 | 4.09 | 2.49 | 1.10 | 1.95 | 0.37 | +P | At3g13790 | Glycosyl hydrolase family 32 | Metabolism | |
| 20269_at | M | 4.33 | 4.13 | 1.19 | 0.56 | 2.34 | 0.52 | +P | ms | At2g45220 | Pectinesterase family | Metabolism |
| 20287_at | M | 3.74 | 6.65 | 2.12 | 0.66 | 1.34 | 0.53 | +P | w | At3g54420 | Glycosyl hydrolase family 19 (class IV chitinase) | Cell rescue, defense, and virulence |
| 20420_at | M | 4.63 | 3.12 | 1.52 | 0.14 | 1.13 | 0.11 | +P | g,c | At4g19810 | Glycosyl hydrolase family 18/putative chitinase | Cell rescue, defense, and virulence |
| 20491_at | H | 8.86 | 4.00 | 2.85 | 0.27 | 2.22 | 0.28 | +P | w | At2g29350 | Putative tropinone reductase | Unclassified |
| 20499_at | M | 2.96 | 4.81 | 2.09 | 2.60 | 1.94 | 2.12 | +P | At2g33480 | Putative NAM (no apical meristem)-like protein | Development | |
| 20685_at | M | 2.55 | 4.72 | 2.00 | 0.85 | 1.18 | 0.36 | +P | o | At4g13180 | Short-chain alcohol dehydrogenase like protein | Metabolism |
Affymetrix probe set number. b Qualitative estimates of absolute gene expression, classified as low (L, Affymetrix signal values < 150), medium (M) and high (H, Affymetrix signal values > 800). c Treatments for which genes were differentially expressed 2.5-fold in two biological replicates. (+), Genes whose expression has increased; (-), Genes whose expression has decreased. d Genes whose expression responds to auxin (a); aluminum (Al); brassinosteroids (b); cold (c); cell-cycle regulated (cc); drought (d); gravity (g); high light (hl); heat shock (hs); low oxygen (lo); mechanical stimulus (ms); oxidative stress (o); ozone (Oz); phosphate stress (P); salinity (s); sulfur deficiency (sulf); touch (t); and wounding and/or pathogen attack (w); see text for references. e AGI numbers and brief descriptions of the transcript identified by BLAST searching and from the descriptions provided by Ghassemian et al. (2002). f Functional categories for genes were identified using the AGI number to search the MIPS database (http://mips.gsf.de/proj/thal/db/; 28/11/02), where no category had been experimentally determined the category with the highest probability score was used.
Table II.
Genes whose expression was altered 28 and 100 h after the withdrawal of P from the nutrient solution
Elements (P, K, or N) were withdrawn from the nutrient solutions supplied to 28-d-old Arabidopsis plants growing hydroponically. Gene expression profiles were determined in shoots 28 and 100 h after the withdrawal of P and 28 h after the withdrawal of K or N. Gene expression was calculated as the ratio of transcript abundance in shoots of plants starved of P, K, or N divided by transcript abundance in shoots of plants supplied with full nutrient solution.
| Gene Expression Relative to Nutrient Replete Plants
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GeneChip IDa | Time Point | Expression Levelb | P Starved for 28 h
|
P Starved for 100 h
|
K Starved for 28 h
|
N Starved for 28 h
|
Specificityc | Other Stimulid | AGI IDe | Functional Categoryf | |||||
| 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | ||||||||
| 17012_at | 28 | M | 2.53 | 2.55 | 0.85 | 1.12 | 2.92 | 1.00 | 0.95 | 0.52 | +P | w, d | At1g72260 | Thionin | Cell rescue, defense, and virulence |
| 19186_s_at | 28 | H | 3.36 | 2.91 | 10.08 | 2.27 | 8.81 | 1.20 | 1.53 | 0.47 | +P | c, w, d, ab, s | At3g50970 | Dehydrin Xero2 | Cell rescue, defense, and virulence |
| 20641_at | 28 | H | 2.77 | 3.67 | 3.31 | 1.07 | 2.50 | 0.47 | 1.00 | 0.35 | +P | d | At1g52690 | LEA76 homolog type1 | Unclassified |
| 14025_s_at | 28/100 | M | 2.54 | 3.84 | 3.05 | 2.84 | 2.39 | 2.49 | 3.56 | 2.36 | +P | At2g04160 | Subtilisin-like Ser protease AIR3 | Protein fate | |
| 12608_i_at | 100 | M | 0.94 | 0.60 | 0.19 | 0.32 | 0.92 | 1.50 | 0.43 | 1.05 | -P | a, b | At4g38850 | Small auxin up RNA (SAUR-AC1) | Unclassified |
| 13322_at | 100 | H | 0.61 | 0.68 | 0.28 | 0.40 | 0.42 | 0.60 | 0.25 | 0.54 | -P | b | At4g38860 | Putative auxin-regulated protein | Unclassified |
| 13660_i_at | 100 | L | 0.76 | 0.98 | 0.27 | 0.37 | 1.39 | 1.66 | 0.23 | 0.90 | -P | a, w | At1g15580 | IAA5 | Transcription |
| 13972_at | 100 | M | 0.72 | 0.66 | 0.36 | 0.35 | 0.43 | 0.71 | 0.30 | 0.69 | -P | At4g17810 | SUPERMAN-like protein | Transcription | |
| 14048_at | 100 | M | 0.87 | 0.77 | 0.28 | 0.24 | 0.47 | 0.81 | 0.50 | 1.53 | -P | c | At2g18890 | Putative protein kinase | Cellular communication |
| 14946_at | 100 | H | 0.45 | 0.52 | 0.36 | 0.39 | 0.37 | 0.22 | 0.47 | 0.44 | -PK | At4g21620 | Putative protein | Unclassified | |
| 15084_at | 100 | M | 0.75 | 0.93 | 0.35 | 0.25 | 0.67 | 0.81 | 0.31 | 0.91 | -P | c | At4g35320 | Putative protein | Unclassified |
| 15671_s_at | 100 | M | 0.82 | 1.19 | 0.40 | 0.31 | 0.61 | 1.11 | 0.51 | 1.45 | -P | w | At1g75840 | Rac-like GTP binding protein (ARAC5) | Cellular communication |
| 15817_at | 100 | M | 0.53 | 0.63 | 0.40 | 0.34 | 0.42 | 0.76 | 0.33 | 0.69 | -P | At4g37240 | Putative protein | Unclassified | |
| 16312_at | 100 | M | 0.51 | 0.53 | 0.39 | 0.15 | 0.47 | 1.10 | 0.18 | 1.05 | -P | At4g12970 | Putative protein | Unclassified | |
| 18755_at | 100 | L | 1.05 | 0.79 | 0.33 | 0.40 | 0.68 | 1.38 | 1.02 | 1.90 | -P | At4g25780 | Putative pathogenesis-related protein | Cell rescue, defense, and virulence | |
| 20575_at | 100 | L | 0.33 | 0.92 | 0.35 | 0.26 | 0.32 | 0.76 | 0.37 | 1.52 | -P | At4g32890 | Putative protein | Transcription | |
| 12341_s_at | 100 | M | 1.00 | 1.53 | 2.61 | 4.42 | 1.76 | 0.71 | 3.08 | 0.76 | +P | ms | At4g20110 | Vacuolar sorting receptor-like protein | Protein fate |
| 12500_s_at | 100 | M | 2.86 | 1.56 | 5.06 | 4.87 | 5.62 | 1.00 | 5.48 | 0.65 | +P | d | At1g51760 | Indole acetic acid-Ala hydrolase (IAR3) | Unclassified |
| 12597_at | 100 | M | 0.83 | 0.61 | 5.36 | 2.63 | 0.58 | 1.46 | 2.17 | 1.28 | +P | At2g22780 | Putative glyoxysomal malate dehydrogenase precursor | Energy | |
| 12880_at | 100 | H | 2.18 | 1.33 | 2.80 | 5.33 | 2.02 | 1.05 | 2.96 | 0.63 | +P | w, d, g | At3g28930 | AIG2-like protein | Unclassified |
| 13004_at | 100 | M | 1.39 | 2.16 | 6.39 | 3.20 | 4.12 | 9.54 | 2.90 | 4.71 | +PKN | c, s | At2g17840 | Putative senescence-associated protein 12 | Unclassified |
| 13230_at | 100 | H | 2.00 | 0.76 | 2.89 | 3.11 | 0.03 | 0.95 | 3.81 | 1.14 | +P | At4g16190 | Cys proteinase | Energy | |
| 13666_s_at | 100 | M | 2.10 | 1.03 | 2.85 | 2.88 | 5.28 | 1.04 | 4.35 | 1.02 | +P | At2g04400 | Putative Indole-3-glycerol phosphate synthase | Metabolism | |
| 13695_at | 100 | M | 1.37 | 0.94 | 3.72 | 2.76 | 0.83 | 1.34 | 1.70 | 1.87 | +P | w | At3g20600 | Non-race-specific disease resistance protein (NDR1) | Cell rescue, defense, and virulence |
| 13934_g_at | 100 | M | 2.58 | 1.81 | 2.53 | 3.43 | 3.03 | 2.38 | 6.32 | 1.71 | +P | At4g17230 | Scarecrow-like 13 (SCL 13) | Transcription | |
| 14016_s_at | 100 | M | 2.33 | 1.37 | 2.80 | 2.54 | 0.70 | 0.67 | 2.58 | 1.99 | +P | g, c | At1g30700 | FAD-linked oxidoreductase family | Metabolism |
| 14032_at | 100 | M | 1.83 | 1.37 | 6.30 | 5.18 | 1.78 | 0.43 | 3.59 | 0.58 | +P | o, c, lo, s | At4g37370 | Cytochrome P450-like protein | Metabolism |
| 14116_at | 100 | H | 2.52 | 1.17 | 6.06 | 2.55 | 1.88 | 1.00 | 4.72 | 0.92 | +P | cc | At5g26340 | Hexose transporter-like protein | Transport facilitation |
| 14614_at | 100 | M | 1.83 | 1.49 | 3.62 | 3.41 | 1.40 | 1.22 | 2.14 | 0.80 | +P | cc | At2g30140 | Putative glucosyltransferase | Metabolism |
| 14672_at | 100 | H | 2.38 | 1.30 | 5.56 | 2.80 | 0.98 | 1.50 | 3.88 | 1.44 | +P | cc, d, g | At3g54640 | Tryptophan synthase α-chain | Metabolism |
| 14978_at | 100 | M | 2.66 | 2.07 | 2.69 | 6.34 | 1.00 | 0.94 | 3.56 | 1.27 | +P | At2g43820 | Putative glucosyltransferase | Metabolism | |
| 15124_at | 100 | H | 2.61 | 1.24 | 2.60 | 8.41 | 1.96 | 0.79 | 2.75 | 0.36 | +P | g, c | At3g30775 | Osmotic stress-induced Pro dehydrogenase | Metabolism |
| 15156_at | 100 | H | 1.68 | 1.10 | 3.14 | 5.18 | 1.01 | 2.05 | 3.69 | 1.84 | +P | At5g50850 | Pyruvate dehydrogenase E1 component β-subunit | Metabolism | |
| 15195_s_at | 100 | H | 2.82 | 1.29 | 3.87 | 2.86 | 4.52 | 0.82 | 3.09 | 0.64 | +P | cc | At2g30490 | Cinnamate-4-hydroxylase | Metabolism |
| 15616_s_at | 100 | H | 1.06 | 2.24 | 2.85 | 7.28 | 2.69 | 4.35 | 5.65 | 2.11 | +PK | At1g21250 | Ser-Thr kinase | Cellular communication | |
| 15629_s_at | 100 | H | 2.52 | 1.93 | 5.08 | 3.85 | 4.70 | 1.07 | 4.88 | 0.66 | +P | At1g17740 | Phosphoglycerate dehydrogenase | Energy | |
| 15656_at | 100 | M | 1.00 | 0.76 | 2.72 | 3.61 | 1.41 | 0.68 | 1.93 | 1.15 | +P | At5g54080 | Homogentisate 1,2-dioxygenase | Unclassified | |
| 15866_s_at | 100 | H | 1.20 | 1.03 | 2.79 | 2.47 | 1.76 | 0.95 | 2.50 | 0.85 | +P | At2g38860 | Expressed protein | Unclassified | |
| 15976_at | 100 | M | 0.68 | 1.10 | 3.09 | 5.52 | 1.44 | 2.21 | 2.15 | 1.05 | +P | cc, sulf | At1g21750 | Putative protein disulfide isomerase precursor | Protein fate |
| 16053_i_at | 100 | H | 1.84 | 0.94 | 4.71 | 3.70 | 2.23 | 0.51 | 3.29 | 0.34 | +P | g, lo, w | At1g02920 | Glutathione S-transferase | Metabolism |
| 16077_s_at | 100 | M | 1.22 | 1.33 | 3.36 | 3.04 | 1.39 | 2.73 | 3.87 | 1.64 | +P | hm | At5g44070 | Phytochelatin synthase | Unclassified |
| 16440_at | 100 | M | 1.33 | 1.08 | 2.53 | 4.46 | 2.20 | 1.02 | 2.14 | 0.47 | +P | lo, c, o | At2g40000 | Putative nemalode-resistance protein | Cell rescue, defense, and virulence |
| 16493_at | 100 | H | 2.35 | 1.89 | 2.59 | 3.71 | 8.84 | 0.63 | 2.53 | 0.71 | +P | s, c | At1g54010 | Myrosinase-associated protein | Metabolism |
| 16609_at | 100 | M | 1.74 | 0.72 | 4.12 | 2.64 | 1.32 | 0.76 | 5.50 | 1.09 | +P | w, e, cc | At5g47220 | Ethylene-responsive element-binding factor 2 (EREB2) | Transcription |
| 17104_s_at | 100 | H | 1.86 | 1.36 | 2.72 | 2.98 | 4.47 | 0.62 | 3.62 | 0.38 | +P | At4g35630 | Phospho-Ser aminotransferase | Metabolism | |
| 17105_at | 100 | M | 2.94 | 1.07 | 2.95 | 3.44 | 3.28 | 1.10 | 7.11 | 1.32 | +P | w, lo | At5g47910 | Respiratory burst oxidase protein | Cell rescue, defense, and virulence |
| 17207_at | 100 | M | 1.83 | 0.82 | 2.70 | 3.82 | 1.43 | 0.67 | 4.05 | 0.72 | +P | At4g36670 | Sugar transporter like protein | Metabolism | |
| 17413_s_at | 100 | H | 6.29 | 0.36 | 8.64 | 5.51 | 0.12 | 0.48 | 5.73 | 0.18 | +P | lo | At5g64120 | Putative peroxidase | Cell rescue, defense, and virulence |
| 17484_at | 100 | M | 0.52 | 2.95 | 3.37 | 3.17 | 1.30 | 0.70 | 0.54 | 0.45 | +P | At1g17020 | SRG1-like protein | Metabolism | |
| 17775_at | 100 | M | 1.42 | 0.84 | 4.14 | 2.70 | 0.77 | 0.48 | 3.08 | 0.47 | +P | w, ms | At1g61800 | Glc-6-phosphate/phosphate-translocator precursor | Transport facilitation |
| 17877_g_at | 100 | H | 1.54 | 1.73 | 2.77 | 2.66 | 2.36 | 0.71 | 1.92 | 0.49 | +P | lo | At4g15760 | Monooxygenase | Metabolism |
| 17917_s_at | 100 | H | 1.07 | 1.53 | 4.56 | 3.68 | 1.73 | 2.54 | 2.49 | 1.94 | +P | At2g41090 | Putative calcium binding protein CaBP-22 | Cellular communication | |
| 17930_s_at | 100 | H | 3.07 | 1.68 | 7.35 | 3.27 | 0.92 | 0.41 | 4.49 | 0.46 | +P | ms, lo | At4g37520 | Peroxidase, prxr2 | Cell rescue, defense, and virulence |
| 18228_at | 100 | H | 2.29 | 0.96 | 5.23 | 3.21 | 3.40 | 0.65 | 4.14 | 0.59 | +P | ms | At3g15356 | Lectin-like protein | Unclassified |
| 18946_at | 100 | M | 3.05 | 0.48 | 3.46 | 2.88 | 5.39 | 0.31 | 4.19 | 0.29 | +P | ms | At5g39580 | Peroxidase ATP24a | Cell rescue, defense, and virulence |
| 19373_at | 100 | M | 0.77 | 1.67 | 2.71 | 3.46 | 0.80 | 1.09 | 2.55 | 2.16 | +P | At2g29670 | Expressed protein | Unclassified | |
| 19614_at | 100 | H | 2.78 | 2.05 | 3.28 | 3.77 | 3.62 | 0.15 | 0.68 | 0.23 | +P | At1g09500 | Putative cinnamyl alcohol dehydrogenase | Metabolism | |
| 19640_at | 100 | H | 2.97 | 1.60 | 4.31 | 3.81 | 3.39 | 0.54 | 3.39 | 0.29 | +P | ms | At2g29460 | Putative glutathione S-transferase | Metabolism |
| 19704_i_at | 100 | H | 4.82 | 1.23 | 3.98 | 4.42 | 1.77 | 1.09 | 4.94 | 1.49 | +P | ms | At5g24160 | Squalene monooxygenase 1,2 (squalene epoxidase 1,2) | Metabolism |
| 19843_at | 100 | M | 2.84 | 0.92 | 3.11 | 3.19 | 2.16 | 1.16 | 3.69 | 2.14 | +P | At1g61890 | Expressed protein | Transport facilitation | |
| 19844_at | 100 | M | 1.13 | 0.69 | 2.57 | 3.60 | 1.79 | 0.72 | 2.52 | 0.63 | +P | w | At4g38540 | Monooxygenase 2 (MO2) | Metabolism |
| 19991_at | 100 | H | 1.00 | 0.95 | 6.61 | 2.66 | 0.50 | 0.58 | 1.77 | 0.47 | +P | g | At2g35980 | Similar to harpin-induced protein hin1 from tobacco | Cell rescue, defense, and virulence |
| 20344_at | 100 | M | 0.93 | 1.72 | 2.88 | 2.79 | 1.20 | 2.02 | 1.02 | 1.74 | +P | At2g15090 | Putative fatty acid elongase | Metabolism | |
| 20442_i_at | 100 | H | 0.97 | 0.75 | 2.63 | 2.87 | 1.49 | 0.32 | 1.52 | 0.74 | +P | At1g16410 | Putative cytochrome P450 protein | Metabolism | |
Affymetrix probe set number. b Qualitative estimates of absolute gene expression, classified as low (L, Affymetrix signal values < 150), medium (M), and high (H, Affymetrix signal values > 800). c Treatments for which genes were differentially expressed 2.5-fold in two biological replicates. (+), Genes whose expression has increased; (-), Genes whose expression has decreased. d Genes whose expression responds to auxin (a); abscisic acid (ab); brassinosteroids (b); cold (c); cell-cycle regulated (cc); drought (d); ethylene (e); gravity (g); high light (hl); heat shock (hs); low oxygen (lo); mechanical stimuli (ms); salinity (s); sulfur deficiency (sulf); and wounding and/or pathogen attack (w); see text for references. e AGI numbers and brief descriptions of the transcript identified by BLAST searching and from the descriptions provided by Ghassemian et al. (2002). f Functional categories for genes were identified using the AGI number to search the MIPS database (http://mips.gsf.de/proj/thal/db/; 28/11/02), where no category had been experimentally determined the category with the highest probability score was used.
Genes Regulated Specifically by P Starvation
Smart plant technology to monitor crop P status requires promoters that increase the expression of a marker gene specifically in response to P starvation. It was therefore necessary to confirm that the genes up-regulated by P starvation in our experiments were unaffected by the withdrawal of other mineral elements (K and N). The shoot K concentration of plants grown in solutions lacking K for 28 h (73.2 μmol g–1 fresh weight) was lower than the shoot K concentration of plants grown in full nutrient solution (83.6 μmol g–1 fresh weight). However, the shoot N concentration of plants grown in solutions lacking N for 28 h was not significantly lower than the shoot N concentration of plants grown in full nutrient solution (3.93% dry weight), although it declined to 3.71% dry weight 100 h after N was withdrawn. The P concentrations in shoots of plants starved of K or N for 28 h did not decline (data not shown). Thus, the changes in gene expression 28 h after the withdrawal of K or N will be responses to a decline in the shoot concentration of K or N specifically. Only one of the early genes (At1g75040; Table I) and three of the late genes (At4g21620, At1g21250, and At2g17840; Table II) responding to P withdrawal responded also to K or N withdrawal (Fig. 2). Furthermore, only two genes responding to P withdrawal (At5g64100 and At1g21750) were found to respond to S starvation (Nikiforova et al., 2003).
A literature survey revealed that the expression of several genes that responded to P withdrawal was also affected by other environmental challenges (Tables I and II). Many genes responding to P withdrawal also increased after wounding or pathogen attack (Uknes et al., 1992; Potter et al., 1993; Epple et al., 1995; Reuber and Ausubel, 1996; Rouse et al., 1996; Somssich et al., 1996; Century et al., 1997; de A Gerhardt et al., 1997; Winge et al., 1997; Torres et al., 1998; Fujimoto et al., 2000; Chen et al., 2002; Mahalingam et al., 2003). Several genes responding to P withdrawal also responded to anoxia, heat shock, cold, drought, salinity, or plant growth regulators (Gil et al., 1994; Abel et al., 1995; Nover et al., 1996; Rouse et al., 1996; Xu et al., 1996; Fujimoto et al., 2000; Reymond et al., 2000; Fowler and Thomashow, 2002; Goda et al., 2002; Klok et al., 2002; Kreps et al., 2002). The expression of a phytochelatin gene increased after exposure of plants to heavy metals (Vatamaniuk et al., 1999) and several genes that responded to P withdrawal also responded to oxidative stress (Sharma and Davis, 1994; Desikan et al., 2001). With the exception of At2g40000, the expression of these genes were found to respond early to P withdrawal. The gene for the blue copper-binding protein (At5g20230) was previously shown to be induced in high-light environments (Rossel et al., 2002), and the expression of a putative peroxidase gene (At3g49120) increased in response to ozone and aluminum stress (Sharma and Davis, 1994; Richards et al., 1998). Several genes whose expression changed in response to P withdrawal were up-regulated by mechanical or gravitational stimulation (Moseyko et al., 2002) or showed significant fluctuation during the cell cycle in suspension cells (Menges et al., 2002). Interestingly, with the exception of At2g17500, At2g38870, At4g16260, and At4g22470, all of the latter genes had maximal expression during the S phase. The majority of genes that were up-regulated during the cell cycle and changed their expression early in response to P withdrawal were involved in cell defense and detoxification. By contrast, those genes that were up-regulated during the S phase and changed in expression late in response to P withdrawal were predominantly involved in cellular metabolism. Promoters from none of these genes would be suitable for use in smart plant technologies to monitor crop P status.
Promoters from genes whose expression is increased by P withdrawal but is unaffected by other environmental or developmental challenges can be used in smart plant technologies. The expression of 13 early genes increased specifically in response to P withdrawal (Table I). These encoded proteins with diverse cellular functions, including transcriptional regulation (At2g28710). However, the increase in their expression was transient. Consequently, smart plants based on their promoters would not be suitable for the periodical monitoring of plant P status. The expression of 18 genes was increased 100 h after P withdrawal and appeared unaffected by other stimuli (Table II). One of these genes, a subtilisin-like Ser protease (At2g04160; AIR3), was up-regulated both 28 and 100 h after P withdrawal. Twelve late genes encoded proteins involved in aspects of cellular metabolism (At1g09500, At1g16410, At1g17020, At1g17740, At2g04400, At2g15090, At2g22780, At2g43820, At4g16190, At4g35630, At4g36670, and At5g50850). A further late gene, whose expression responded solely to P withdrawal, encoded a scarecrow-like transcription factor (At4g17230; SCL13). Because increased expression of late genes was sustained and their up-regulation occurred as tissue P declined but before growth was compromised, their promoters would be suitable for smart plant technologies.
The Functions of Genes Regulated by P Starvation
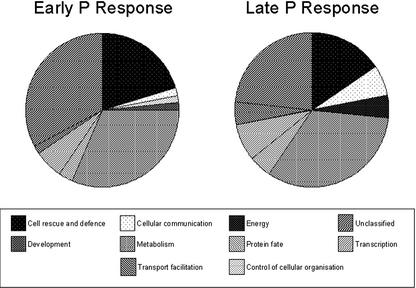
The functional categories of the gene products whose expression was altered by P starvation were obtained from the Munich Information on Protein Sequences (MIPS) database using their AGI identifier (Tables I and II). When no category was assigned by functional characterization, the most probable category based on sequence homology was assigned. Approximately 20% to 30% of both the early and late P response gene products were assigned no known function (Fig. 3), which is comparable with the 38% of the 8,100 gene products represented on the Affymetrix GeneChip whose function is unknown.
Figure 3.
The MIPS functional categories of genes whose expression had changed early (4 h; Table I) or late (28 or 100 h; Table II) after the withdrawal of P from 28-d-old hydroponically grown Arabidopsis. The AGI number for each gene was used to identify its functional category from the MIPS database (http://mips.gsf.de/proj/thal/db/tables/tables_func_frame.html). When no functional category was given by MIPS, the first predicted category identified in PENDANT was used.
The early gene products (Table I) were dominated by those involved in metabolism (31%) and in cell rescue and defense (20%). Of the 20 metabolism genes, 13 were involved in primary metabolic functions, such as amino acid metabolism and carbohydrate metabolism, and seven were involved in the production of secondary metabolites involved in plant defense, such as alkaloids and phenylpropanoids. The gene products involved in cell rescue and defense included five chitinases, four peroxidases, a PR-1-like protein, and a tobacco (Nicotiana tabacum) hin-1 homolog. Other significant functions of the early gene products included transcription (6%), protein fate (3%), cellular communication (2%), and transport facilitation (2%).
There were nine gene products common to both early and late responses. These included a cytochrome P450 (At4g37370), two glutathione-S-transferases (At1g02920, At2g29460), a FAD-linked oxidoreductase (At1g30700), two peroxidases (At5g64120 and At4g37520), the tobacco hin-1 homolog (At2g35980), and a lectin (At3g15356). Curiously, the expression of no chitinase or endochitinase genes remained elevated 100 h after P withdrawal. In a way similar to the early gene products, roles in metabolism (33%) and cell rescue and defense (16%) dominated the late gene products (Table II). At this time, however, the number of gene products involved in secondary metabolism (11) was greater than those involved in primary metabolism (10). The gene products involved in cell rescue and defense included three peroxidases, a non-race-specific disease resistance protein (NDR1), a putative nematode-resistance protein, thionin, and a respiratory burst oxidase protein. A greater proportion of the late gene products was involved in transcription (8%), protein fate (5%), cellular communication (6%), and transport facilitation (5%). Interestingly, the identity of these genes differed between 4 and 100 h after P withdrawal (Tables I and II). For example, the expression of five transcription factors (three zinc finger proteins, a heat shock transcription factor, and a blue copper-binding protein) was up-regulated 4 h after P withdrawal, but the expression of two other transcription factors (for SCL13 and EREB2) was up-regulated, and three other transcription factors (IAA5, a SUPERMAN-like protein, and a putative transcription factor protein) were down-regulated 100 h after P withdrawal. Because auxin orchestrates plant morphology, it is noteworthy that four of the 12 genes whose expression was down-regulated after 100 h of P starvation were potentially auxin regulated.
Verifying the Temporal Expression Patterns of Target Genes
To confirm the gene expression patterns observed using microarray technology, quantitative PCR was performed. The expression of several genes with potentially contrasting magnitude and relative changes in expression during P starvation was monitored (Table III). Genes with contrasting levels of expression that responded to the withdrawal of P but not K or N were chosen, which were identified as up-regulated (At4g12470) or down-regulated (At1g10060) solely at the 4-h time point, up-regulated (At2g22780; At5g26340; At1g17740; At4g35630; At1g61800) or down-regulated (At2g18890) solely at the 100-h time point, or up-regulated at both the 4- and 28-h time points (At2g02990) in the microarray analyses. The expression of two genes previously identified as being up-regulated immediately (IPS1; Martín et al., 2000) or slowly (SQD1, At4g33030; Essigmann et al., 1998) and specifically to P starvation were also included as positive controls in the quantitative PCR assays. The rRNA 18S gene was used to quantitate transcript abundance.
Table III.
Genes selected for quantitative PCR to confirm Affymetrix GeneChip results
Genes were selected to have contrasting magnitudes (low, L; medium, M; or high, H) and change in gene expression when assayed using Affymetrix GeneChips in response to the withdrawal of P from hydroponcally grown 28-d-old Arabidopsis for 4, 28, and 100 h. The primers used for quantitative PCR are indicated. -, not determined.
| Expression Level
|
GeneChip Analysis
|
GeneChip ID
|
AGI ID
|
Forward Primer
|
Reverse Primer
|
Mean-Fold Change on GeneChip
|
Fold Change for Quantitative PCR
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P Starved for 4 h | P Starved for 28 h | P Starved for 100 h | P Starved for 4 h | P Starved for 28 h | P Starved for 100 h | ||||||
| L | Down at 4 h | 20518_at | At1g10060 | AATAGAGGGGATGAAAGC | ATGAACAGAAGGAGAATGC | 0.34 | 1.23 | 0.44 | 1.00 | 0.78 | 0.20 |
| H | Up at 4 h | 17963_at | AT4g12470 | CCTCTCTTGCTCTTTTCTTTG | GGGACTGGCTTTGGTTTAG | 14.15 | 1.43 | 0.96 | 0.14 | 1.78 | 0.01 |
| H | Up at 4 + 28 h | 17014_s_at | At2g02990 | CTTGCCTTCTGTCTTCTCTG | GGATAACAACACTTCTTCTGTG | 31.73 | 4.77 | 1.88 | 3.33 | 11.41 | 8.45 |
| M | Down at 100 h | 14048_at | At2g18890 | GGATAACAAGAGGAGGAAGAGATG | CAACAACCAAGAAGAGACAAGAC | 0.58 | 0.82 | 0.26 | 0.97 | 1.50 | 0.42 |
| M | Up at 100 h | 12597_at | At2g22780 | GCTTCCCTTCTTCGCATC | GCCTTCTCTAATCCCATCCTC | 1.57 | 0.72 | 4.00 | 1.52 | 1.79 | 1.54 |
| H | Up at 100 h | 14116_at | At5g26340 | GCCGTTCCGTTGTTCTTG | CTTGGCGGTCCCATAGTTG | 2.35 | 1.85 | 4.31 | 0.32 | 4.45 | 0.77 |
| H | Up at 100 h | 15629_s_at | At1g17740 | GAAGGGGAGGTTGAAAGTGG | CACCGTATTAGCCGTTGGAG | 3.74 | 2.22 | 4.47 | 0.29 | 4.79 | 3.23 |
| H | Up at 100 h | 17104_s_at | At4g35630 | ACACGCCTCCTTGCTTTG | GCTTTCCTCTGGTTCTTCTTCTC | 2.01 | 1.61 | 2.85 | 0.60 | 5.45 | 3.08 |
| M | Up at 100 h | 17775_at | At1g61800 | AAGAAAGGGATGAAAGGGAAG | CACGGCAATAGAAAATGGAG | 2.74 | 1.13 | 3.42 | 1.11 | 19.76 | 5.14 |
| - | - | Not present | 18s rRNA | CATAAACGATGCCGACCAG | AGCCTTGCGACCATACTCC | 1.13 | 1.25 | 1.15 | |||
| - | - | Not Present | IPS1 | AGGGGATGGCCTAAATACAAAATG | GGGAGATAAACAAAACTCGCAGTC | 0.89 | 14.68 | 1143.05 | |||
| - | - | Not Present | SQD1 | CACCACCCGAAACATCTACC | ACCGCAATAACCATCTCCAC | 0.68 | 2.1 | 4.5 | |||
In general, the results from the quantitative PCR experiments confirmed the changes in gene expression after the withdrawal of P observed using microarray technology (Table III). The temporal changes in gene expression after P withdrawal followed similar patterns of change in experiments using microarrays and quantitative PCR, but the magnitude and kinetics of these changes apparently differed. This difference may be a consequence of using different biological samples, contrasting primers, or normalizing data to different genes. Most of the discrepancies between experiments performed using microarrays and quantitative PCR occurred at the 4-h time point. This probably reflects biological variability and may be a consequence of the general nature of any initial shock response to an environmental perturbation (Desikan et al., 2001; Fowler and Thomashow, 2002; Kreps et al., 2002). Data were more consistent at later time points, which was presumably because plant physiology responded specifically to P starvation. Both techniques indicated a decrease in the expression of LEA76 (At1g10060) and the pEARLI 1-like (At4g12470) gene as shoot P concentration declined and indicated an up-regulation of RNS1 (At2g02990) after P withdrawal. The same temporal changes in the expression of the protein kinase (At2g18890) gene were observed using both microarrays and quantitative PCR techniques. With the exception of glycosomal malate dehydrogenase (At2g22780), whose expression was apparently reduced at the 28-h time point according to the microarray data, and the hexose transporter gene (At5g26340), whose expression was apparently reduced at the 100-h time point according to quantitative PCR, the expression of all up-regulated late genes showed increased expression both 28 and 100 h after P withdrawal when assayed by either technique.
Proving the Smart Plant Concept
Promoters for genes that are expressed early enough in response to P starvation to enable remedial fertilizer application (< 100 h after withdrawing P), but whose expression is not affected by transient vagaries in environmental conditions (< 4 h after withdrawing P) or by other stimuli are suitable for the generation of smart plants. One such gene is SQD1 (Table III), a gene involved in sulfolipid biosynthesis, originally isolated from a cDNA library with probes homologous to the bacterial sqdB genes by Essigmann et al. (1998). Expression of this gene is increased when Arabidopsis are grown under P-limiting conditions (Essigmann et al., 1998) and also when P is withdrawn from Arabidopsis growing hydroponically (Table III). To generate a smart plant, we isolated the promoter for SQD1 through inverse PCR of genomic DNA using nested primers to the published cDNA sequence (AF022082).
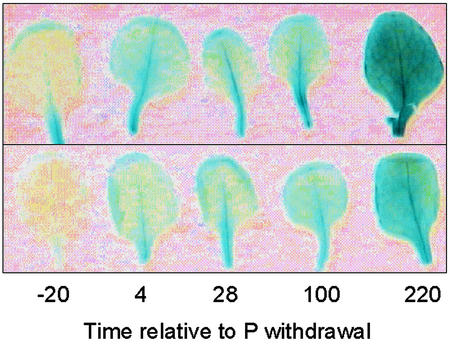
Transgenic Arabidopsis harboring constructs containing the promoter for SQD1 driving the expression of a GUS marker gene showed a gradual increase in leaf GUS activity after P withdrawal (Fig. 4). No GUS activity was observed 20 h before the withdrawal of P. Four hours after P was withdrawn, GUS activity had increased noticeably and appeared to remain constant as plants approached incipient P deficiency (28–100 h). A further marked increase in GUS activity was observed 220 h after the withdrawal of P. These observations demonstrate the potential for smart plant technology based on the SQD1::GUS construct to monitor plant P status.
Figure 4.
The activity of GUS in the leaves of two independent transgenic Arabidopsis lines bearing constructs containing the GUS marker gene under the control of the promoter sequence for the P-sensitive gene, SQD1. Plants were grown hydroponically and fully expanded leaves were harvested 20 h before and 4, 28, 100, and 220 h after withdrawing P. Excised leaves were vacuum infiltrated with staining solution containing 520 mg L–1 5-bromo-4-chloro-3-indolyl β-d-glucopyranoside and incubated overnight at 37°C. To assist the visualization of the blue product, chlorophyll was removed from leaves by stepwise replacement of the staining solution with ethanol and mounted in a solution containing 50% (v/v) glycerol. The transgenic lines were GUS13/4 (top) and GUS22/1 (bottom).
DISCUSSION
Changes in Gene Expression after P Withdrawal
Changes in gene expression in Arabidopsis shoots during P starvation assayed using microarray technology (Tables I and II) were confirmed using quantitative PCR techniques (Table III). When P was with-held from Arabidopsis plants growing hydroponically, changes in gene expression occurred that were either specific to P starvation or common to a variety of environmental perturbations (Tables I and II). The expression of 60 genes was transiently up-regulated 4 h after withdrawing P. Many of these genes are ubiquitous “shock” response genes up-regulated by various pathogens and environmental perturbations (Table I; Desikan et al., 2001; Fowler and Thomashow, 2002; Kreps et al., 2002). Several of the genes transiently up-regulated 4 h after withdrawing P are involved in cell rescue and defense (Table I; Fig. 3), which is consistent with the common horticultural observation that plants subject to a mild P stress are less susceptible to pathogens. These included five chitinases, four peroxidases, a PR-1-like protein, which may be induced by peroxidase activity (Mackerness et al., 2001), and a homolog of a harpin-induced gene (hin-1) in tobacco. Harpin has been shown to induce peroxidase activity (Wang and Liu, 1999). Other genes that are up-regulated by oxidative stress were also transiently up-regulated 4 h after withdrawing P (Table I; Desikan et al., 2001). These included a cytochrome P450 (At4g37370), a C2H2-type zinc finger protein (Att5g04340), and a blue copper-binding protein (At5g20230; Table I). It is noteworthy that phosphate deficiency has been shown to induce the production of reactive oxygen species (Gonzàlez-Meler et al., 2001). Interestingly, the expression of the alternative oxidase 1a precursor (At3g22370) was up-regulated 100 h after P withdrawal, with 2.2- and 3.2-fold changes in the two replicate experiments. This may reduce the production of harmful reactive oxygen species by increasing electron transport through the alternative oxidase pathway.
Surprisingly, the expression of only four genes differed between nutrient-replete plants and plants starved of P for 28 h (Table II). This lull in differential gene expression has also been observed in the time course of changes in gene expression after other environmental perturbations (Reymond et al., 2000; Kreps et al., 2002; Ramonell et al., 2002) and may demarcate the progression from an initial common shock response to a more specific response associated with each perturbation. One hundred hours after withdrawing P, the expression of 49 genes was greater in plants starved of P than in nutrient-replete plants. Many of these genes appeared to be specific for P starvation, and several are involved in P metabolism (Table II). Curiously, the expression of none of the five genes present on the Affymetrix GeneChip previously found to be up-regulated by P starvation in Arabidopsis (At2g02990, RNS1; At2g32830, Pht1;5; At3g26570, PHT2;1; At5g43350, Pht1;1; and At2g27190, PAP12) was classified as up-regulated 100 h after withdrawing P by the criteria employed here. However, the expression of RNS1 (At2g02990), which is up-regulated during P starvation in Arabidopsis leaves (Bariola et al., 1994), was observed to be significantly up-regulated after 4 h (Table I) and 28 h (7.27- and 2.27-fold increases in the two replicate experiments) of P starvation and marginally up-regulated after 100 h of P starvation (2.69- and 1.07-fold increases in the two replicate experiments). Genes for the phosphate transporters Pht1;5 (At2g32830) and PHT2;1 (At3g26570) are also expressed in the shoots of Arabidopsis and show increased expression in P-starved plants (Daram et al., 1999; Mudge et al., 2002; Versaw and Harrison, 2002). Unfortunately, transcripts of Pht1;5 could not be detected in our study, but the expression of PHT2;1 was increased in one of the replicate experiments 100 h after the withdrawing P (0.76- and 2.87-fold changes in the two replicate experiments). The gene encoding the phosphate transporter Pht1;1 (At5g43350) appears to be only expressed in Arabidopsis roots (Muchhal et al., 1996; Mudge et al., 2002), and consequently, no transcripts for this gene were detected in our study. The expression of PAP12 (At2g27190), which encodes a purple acid phosphatase, has been shown to increase in Arabidopsis suspension cells starved of P (Li et al., 2002). In our experiments the expression of PAP12 was marginally increased 100 h after withdrawing P in shoots of Arabidopsis (2.76- and 1.76-fold increases in the two replicate experiments), which is in agreement with the data presented by Li et al. (2002).
Plant P Status Can Be Monitored Using Smart Plants
Smart plants can be used to assay crop P status. They can be genetically engineered in transgenic plants by harnessing a phosphate-sensitive promoter to the expression of a gene encoding a visible marker. Typical marker genes that could be used are GUS or green fluorescent protein. Detection of the visible marker in the leaves of a smart plant would indicate a physiological P stress and the need for fertilizer application. It is important that the expression of the marker gene responds solely to P and increases as tissue P concentration declines but before P starvation affects plants growth. In the experiments described here, the shoot P concentration of Arabidopsis plants had decreased without an effect on growth between 24 and 72 h after P withdrawal (Fig. 1). Promoters for genes whose expression is increased during this time period are suitable for smart plant technology. One such gene is SQD1 (Table III; Essigmann et al., 1998).
The smart plant concept was tested using the promoter from the gene SQD1 to control the expression of the GUS marker gene. It was observed that GUS expression in leaves of transgenic Arabidopsis bearing the SQD1::GUS construct increased after the withdrawal of P (Fig. 4) and that an increase could be detected before P starvation affected plant growth. However, because the GUS activity continued to increase with acute P starvation, smart plants based on the SQD1::GUS construct would require careful calibration. In practice, a grower may also need to know whether P stress had been relieved after remedial P fertilization. Therefore, it would be necessary for the expression of any marker gene in a smart plant to return rapidly to basal levels after the application of P. This was not investigated in the present study, but the responsiveness of SQD1 promoter activity to remedial P fertilization is under investigation. In addition, the marker gene product itself would need to have a rapid turnover time that would allow the grower to determine quickly whether remedial P fertilization had been successful. A maximal half-life on the order of a few days is probably necessary.
The Identification of cis-Regulatory Elements in the Promoters of P-Responsive Genes
It is possible that promoters for other P-responsive genes might be used for smart plant technologies. Because P stress can be induced by a variety of factors and because many different signaling and metabolic pathways are implicated, it might be wise to identify a suite of promoters responding differentially to P starvation to allow plant P stress to be monitored in all environments. These promoters could be stacked in a single construct in which each controlled the expression of a different marker gene, for example other colored or fluorescent proteins, such as yellow fluorescent protein, cyan fluorescent protein, and red fluorescent proteins (Lansford et al., 2001; Visser et al., 2002), in a single smart plant. To identify cis-regulatory elements in the promoters of P-responsive genes that may be involved in the up-regulation of gene expression in P-starved plants, we searched for common sequences contained within the promoters of the genes whose expression changed either early or late in response to P withdrawal (Table IV) using the GeneSpring program (Silicon Genetics, Redwood City, CA). These sequences may act as cis-regulatory elements binding a common transcription factor. Two novel sequences were present significantly (P < 0.05) more frequently in the promoters of genes up-regulated early in response to P withdrawal than in the promoters of all genes represented on the Affymetrix GeneChip (Table IV). The first promoter consensus sequence (CGCGTGGG) had similarities to a PHO element (Mukatira et al., 2001), and the second promoter consensus sequence (TATAAATA) had similarities to TATA box elements (Rossel et al., 2002). The promoters of three genes (At2g02930, At4g25810, and At1g30720) that were up-regulated early in response to P withdrawal contained both putative cis-regulatory elements. The promoter regions of several genes whose expression changed in P-starved plants in our experiments also contained cis-regulatory elements previously identified as common to genes whose expression responds to P starvation (Mukatira et al., 2001; Rubio et al., 2001; Table IV). However, only the PHO element CACGT(G/C) was present slightly more often in the promoters of genes up-regulated in response to P withdrawal than in the promoters of all genes represented on the Affymetrix GeneChip. The four other cis-regulatory elements we surveyed occurred at a similar frequency in the genes we identified as responding to P withdrawal as in the regulatory sequences of all genes on the Affymetrix GeneChip (Table IV). Other cis-regulatory elements in promoters that respond to P starvation might be identified as the targets of transcription factors whose expression is increased by P stress (Tables I and II; Rubio et al., 2001).
Table IV.
The occurrence of novel cis-regulatory elements in the promoters of genes whose expression changed early or late in response to P withdrawal in the experiments reported here (Tables I and II) and cis-regulatory elements previously identified as common to genes whose expression responded to P starvation
The occurrence of these cis regulatory elements in the promoters of all genes on the Affymetrix GeneChip are given for comparison. —, not determined.
| No. of Genes with Element Upstream of Start Codon
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Element Name | Element Sequence | Early P-Responsive Genes
|
Late P-Responsive Genes
|
Affymetrix GeneChip
|
Reference | |||
| No. | % | No. | % | No. | % | |||
| Novel elements | ||||||||
| PHO-like | C(G/T/A)(C/T/A)GTGG | 18 | 32.1 | — | — | 588 | 7.1 | — |
| TATA box-like | TATAAATA | 20 | 35.7 | — | — | 753 | 9.1 | — |
| P-responsive elements | ||||||||
| PHR1-binding site | GNATATNC | 10 | 16.7 | 9 | 17.6 | 1498 | 18.2 | Rubio et al. (2001) |
| PHO element | CACGT(G/C) | 17 | 28.3 | 16 | 31.4 | 1574 | 19.1 | Mukatira et al. (2001) |
| ATGCCAT | 1 | 1.7 | 2 | 3.9 | 349 | 4.2 | Mukatira et al. (2001) | |
| Helix-loop-helix | CA(T/G)(A/C)TG | 32 | 53.3 | 23 | 45.1 | 4431 | 53.7 | Mukatira et al. (2001) |
| NIT 2 | TATC(A/T)(A/T) | 42 | 70.0 | 36 | 70.6 | 6296 | 76.3 | Mukatira et al. (2001) |
Potential for Smart Plants
Smart plants can be used as a basic research tool as well as for more applied ends. In the laboratory, smart plants could be used (a) after mutation of the transgenic lines to identify components of signaling cascades impacting on nutrient-specific gene regulation (Martín et al., 2000), (b) in conjunction with a screen of mutants or ecotypes, to identify plants with enhanced nutrient use efficiency, which could be used in breeding programs, (c) to investigate the role of root and/or soil structure in the acquisition of P from soils with low P availability, and (d) as experimental material to assist and ratify computer predictions for the application of P fertilizers on crop P status. Transferring smart plant technology to crops would provide a rapid bioassay for P availability to plants in situations where existing chemical tests provide unclear results (Smethurst, 2000). Thus, sentinel plants monitoring the P status of crops will allow efficient temporal and spatial applications of P fertilizer and further the development of decision-making systems for precision farming. Ultimately, these will lower costs, reduce pollution, and enhance biodiversity.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis accession Col-5 were obtained from the Nottingham Arabidopsis Stock Centre (N1644; Nottingham, UK). Seeds were washed in 70% (v/v) ethanol-water, rinsed in distilled water, and surface sterilized using 50% (v/v) domestic bleach-water. Seeds were then rinsed and imbibed for 3 to 5 d at 4°C in sterile distilled water to break dormancy.
After imbibition, seeds were sown in un-vented, polycarbonate culture boxes (Broadley et al., 2001). Seedlings were grown aseptically for 21 d on perforated black polycarbonate discs (diameter 91 mm) placed on 75 mL of 0.8% (w/v) agar containing 1% (w/v) Suc and a basal salt mix (Murashige and Skoog, 1962). Roots grew into the agar, but shoots remained on the opposite side of the disc. Seedlings were grown with a 16-h photoperiod at a constant temperature of 24°C. Illumination was provided by a bank of 100-W 84 fluorescent tubes (Philips, Eindhoven, Netherlands) giving a photon flux density between 400 and 700 nm of 50 to 80 μmol photons m–2 s–1 at plant height.
After 21 d, plants were transferred, still on polycarbonate discs, to a hydroponics system situated in a Saxcil growth cabinet (S.K. Saxton Ltd., ARC Works, Cheshire, UK). Plants were grown with a 16-h photoperiod at a temperature of 24°C during the day and 16°C at night. Illumination was provided by a bank of OSRAM L 58 W/23 fluorescent tubes (OSRAM, Langley, UK) giving a photon flux density between 400 and 700 nm of 75 μmol photons m–2 s–1 at plant height. Relative humidity was approximately 80%. Each polycarbonate disc containing plants was placed on a light-proof 500-mL beaker over 450 mL of aerated complete nutrient solution (pH 5.6) containing: 8.0 mm NO3–, 4.025 mm Ca2+, 0.764 mm SO42–, 0.75 mm K+, 0.75 mm Mg2+, 0.25 mm H2PO42–, 0.1mm FeNaEDTA, 0.05 mm Cl–, 0.03 mm H2BO3–, 0.01 mm Mn2+, 0.001 mm Na+, 0.001 mm Zn2+, 0.003 mm Cu2+, and 0.0005 mm MoO42–. Nutrient solution was recirculated using a peristaltic pump at a flow rate of 30 mL min–1 (M045 OEM peristaltic pump, Autoclude, Essex, UK) through four beakers and a central reservoir of 6 L. Four hydroponic units could be operated simultaneously. Nutrient solutions were replaced twice a week. Plants were grown hydroponically for 7 d in complete nutrient solution before experimentation.
Analysis of Plant Growth and Mineral Content
To determine the effects of removing the P, K, or N supply on shoot growth and mineral content, the complete nutrient solution was replaced with nutrient solutions lacking these elements. These are referred to as –P, –K, and –N solutions. In solutions lacking P, KH2PO4 was replaced with KH2SO4 to maintain potassium levels. In solutions lacking K, KH2PO4 and KOH were replaced with Ca(H2PO4)2 to maintain phosphate levels. In solutions lacking N, Ca(NO3)2 was replaced with CaSO4 to maintain calcium levels.
In the first experiment, which was repeated three times, plants were harvested 1, 3, 8, and 11 d after P was removed from the nutrient solution. At each harvest, the fresh weights of the shoots of 25 plants were determined. The shoot material was then dried at 80°C for 48 h, and the dry weights of individual shoots were determined. Shoot P, K, and N contents were determined from bulked samples of 25 plants. To determine P and K, samples were ashed overnight at 490°C, and the ash was dissolved in 1 mL of nitric acid and 5 mL of water. Solutions were filtered through No. 5 paper (Whatman, Kent, UK) and analyzed for P and K by inductively coupled plasma optical emission spectrophotometry (J Y Horiba Ultima 2 ICP-OES, Jobin Yvon, Middlesex, UK). To determine N, an aliquot of the bulked sample was loaded directly into a combustion analyzer (CN 2000, LECO UK, Stockport, Cheshire, UK) and analyzed for percentage of N via an internal thermal conductivity detector.
In the second experiment, which was also repeated three times, plant material was harvested 1, 3, 8, and 11 d after removing N or K from the nutrient solution and also in nutrient-replete plants. Shoot fresh and dry weights and shoot P, K, and N contents were determined as described above.
Profiling Gene Expression under Nutrient Starvation
To determine changes in gene expression in response to phosphate starvation, the complete nutrient solution was replaced by –P nutrient solution 4 h before the midpoint of the light-period 28 d after sowing. Shoot material was harvested 4, 28, and 100 h after the phosphate supply was removed, mid-way through the light period. Shoot material from control plants supplied with complete nutrient solution was also harvested at these time points and also 20 h before P starvation.
To identify genes up-regulated specifically in response to P starvation, rather than as a nonspecific response to mineral deficiency, the complete nutrient solution was replaced by –K or –N nutrient solutions. Again, the nutrient solutions were changed 4 h before the midpoint of the light period, and shoot material was harvested 28 h after K or N was removed, mid-way through the light period.
At each harvest, shoot material from eight to 12 plants from each treatment was bulked into 1.5-mL colorless, sterile, screw-cap polypropylene tubes and was snap-frozen in liquid nitrogen. Tissue samples were stored at –70°C before the extraction of total RNA. The entire experiment was performed twice.
Extraction of Total RNA
Tissue samples, previously stored at –70°C, were placed in liquid nitrogen before grinding. To each sample, 1 mL of TRIzol reagent was added, and total RNA was subsequently extracted according to the manufacturer's instructions (Invitrogen Life Technologies, Paisley, UK). Due to the high content of proteoglycans and polysaccharides in plant material, the following modifications were made: (a) After homogenization with the TRIzol reagent, the samples were centrifuged to remove any remaining plant material. The supernatant was then transferred to a clean Eppendorf tube. (b) To aid precipitation of RNA from the aqueous phase, 0.25 mL of isopropanol and 0.25 mL of 1.2 m NaCl solution containing 0.8 m sodium citrate were added. This procedure precipitates the RNA while maintaining the proteoglycans and polysaccharides in a soluble form. Samples of total RNA were then sent to AROS Applied Biotechnology (Aarhus, Denmark) for labeling and Arabidopsis GeneChip analysis (Affymetrix Inc., Santa Clara, CA).
Microarray Data Analysis
All data manipulations were performed using Microsoft Excel (Microsoft, Redmond, WA). Signal values (indicating the relative abundance of a particular transcript) and detection call values (indicating the probability that a particular transcript is present) were generated by Microarray Analysis Suite 5.0 software (Affymetrix Inc., Santa Clara, CA). Transcripts called “Absent” in both experiments were removed from subsequent analyses. Differences in transcript abundance were expressed as -fold changes, which were calculated as the ratio of signal values in different experiments or treatments. A difference in transcript abundance of 2.5-fold was taken as indicating a difference in gene expression. It has been estimated by challenging Arabidopsis GeneChips with identical RNA samples that the frequency of false calls (differences in transcript abundance 2.5-fold) due to technical reasons approximates 0.04% to 0.2% (Fowler and Thomashow, 2002; Zhu et al., 2001). A preliminary comparison of the expression data from identical experiments undertaken here indicated that the frequency of false calls due to biological/experimental variation was higher than this and was greatest at low signal values. For this reason, and following the precedent of Zhu et al. (2001), all signal values < 20 were floored to 20. The frequency of false calls in the two replicate experiments reported here was estimated to be 3% (n = 8 comparisons) per experiment.
The AGI numbers for transcripts present on the Affymetrix Arabidopsis GeneChip were cross-referenced against probe sets on the GeneChip to remove duplicates. The annotation of transcripts was confirmed using the AGI and GenBank numbers given by Affymetrix. The target nucleotide sequences used by Affymetrix to design probes for the GeneChip (Available from http://www.affymetrix.com) were used to BLAST the GenBank sequence database to confirm the identity of the assayed transcript. A similar confirmation of probe fidelity has been undertaken independently by Ghassemian et al. (2001).
The identification of cis-regulatory elements (five to 10 bases, with one or no single nucleotide discrepancies) that occurred with a greater frequency within the 10 to 1,000 bases upstream of genes whose expression increased early or late after the withdrawal of P in our study, compared with the regulatory sequences contained upstream of all genes on the Affymetrix GeneChip (P value of 0.05), was performed by the GeneSpring subprogram “Find potential regulatory sequences” (Silicon Genetics, Redwood City, CA). The occurrence of cis-regulatory elements previously identified as common to genes whose expression responds to P starvation (Mukatira et al., 2001; Rubio et al., 2001) was also determined using GeneSpring in the 10 to 1,000 bases upstream of genes whose expression increased early or late after the withdrawal of P in our study and of all genes present on the Affymetrix GeneChip.
Generating Smart Plants
Primers were designed to SQD1 based on its published cDNA sequence (GenBank accession no. AF022082). These consisted of an initial pair of primers (CCCTTCAGTTATCTCACTTAGCAG and AAGCTGAAAGTAGATGCGCCAT) to perform inverse PCR (IPCR) on circularized genomic DNA (Collins and Weissman, 1984) followed by a second “nested” pair of primers (ACTAAGCTTCAGCAGCAGCA and GCGCCATTTTAAAGCTTTGTTC). The second pair was adjusted to incorporate the underlined HindIII sites. Promoter fragments for SQD1 of approximately 0.8 kb (with DNA cut using PstI) and 3.5 kb (with DNA cut using HindIII) were obtained by IPCR techniques. On the first pass IPCR, no apparent major bands were present, but on a re-PCR with the nested primers (from the purified products of the first-pass IPCR) strong individual bands were obtained. Their sequences corresponded to both the published cDNA sequences for SQD1 and the bacterial artificial chromosome containing SQD1 (BAC F26P21/AL031804).
A 1.6-kb region upstream of the SQD1 gene was inserted into the pMOG24 plasmid (Mogen International, Leiden, The Netherlands) upstream of the GUS marker gene. The pMOG24 plasmid was then transformed into Agrobacterium tumefaciens C58 pGV3850 via electroporation (Wei-jun and Forde, 1989) and then into Arabidopsis ecotype Col-5 by vacuum infiltration (Bechtold et al., 1993). Seeds from transformed plants were imbibed and surface sterilized before being sown on 0.8% (w/v) agar, containing 1% (w/v) Suc, 50 mg L–1 kanamycin, and a basal salt mix (Murashige and Skoog, 1962). Twenty-four transformed lines were driven to homozygosity over three generations through selection on agar containing kanamycin.
To determine changes in GUS activity in untransformed and transgenic plants bearing the SQD1::GUS construct in response to phosphate starvation, fully expanded leaves of hydroponically growing plants were removed 20 h before and 4, 28, 100, and 220 h after complete nutrient solution was replaced by –P nutrient solution 28 d after sowing. Excised leaves were vacuum infiltrated with staining solution containing 520 mg L–1 5-bromo-4-chloro-3-indolyl β-d-glucopyranoside (Melford Laboratories, Ipswich, Suffolk, UK), 1.32 g L–1 potassium ferricyanide, 100 mg L–1 chloramphenicol, 0.1% (v/v) Triton X-100, and 50 mm sodium phosphate buffer (pH 7.0) and were incubated overnight at 37°C. To assist the visualization of the blue product, chlorophyll was removed from leaves by stepwise replacement of the staining solution with 10%, 30%, 50%, and finally 100% (v/v) ethanol. Leaves were kept in solutions containing 10%, 30%, and 50% (v/v) ethanol for 2 h and in 100% ethanol overnight. Leaves were transferred to a solution containing 50% (v/v) glycerol for mounting.
Quantitative PCR
Reverse transcription was performed on 1 μg of total RNA, isolated from plants grown under the same conditions as those grown for microarray analysis, using the ThermoScript RT-PCR system (Invitrogen Life Technologies). The cDNA synthesis reaction was carried out using random hexamers (50 ng μL–1) according to the manufacturer's instructions. Primers for quantitative PCR were designed to the cDNA sequences of genes identified by the microarray screening process and 18S rRNA gene control using the Primer Express Software (v2.0, Applied Biosystems, Cheshire, UK; Table IV). Analysis of transcript levels of each gene was achieved by quantitative PCR using an ABI Prism 7900 HT sequence detector and SYBR Green fluorescent dye. Triplicate reactions (15 μL volume) were performed for each gene and sample (control and –P plants, 4, 28, and 100 h after the withdrawal of P) using 384-well plates consisting of 20-ng cDNA sample, 1 μm forward and reverse primer, and 7.5 μL of 2× SYBR Green PCR master mix (Applied Biosystems). The quantitative PCR reaction conditions were 50°C (2 min) followed by 95°C (10 min) for 1 cycle, then 95°C (15 s) followed by 60°C (1 min) for 40 cycles. This was followed by a dissociation step of 95°C for 15 s, 60°C for 15 s and 95°C for 15 s. The dissociation step was included to generate data for melting curve analysis so that primer dimers or nonspecific products could be detected in the reaction. A control reaction was carried out on each sample using 2 ng of total RNA, to confirm that there was no contaminating chromosomal DNA in the original sample. The expression pattern of individual genes, as suggested by microarray data, dictated which cDNA samples were used to generate standard curves, i.e. the standard curves for genes whose expression increased 100 h after P withdrawal were generated from cDNA samples isolated from plants 100 h after the withdrawal of P. Samples were diluted to give a cDNA concentration range from 10 ng μL–1 to 0.01 ng μL–1. The cycle threshold and normalized fluorescence (ΔRn) values were determined for each sample during the quantitative PCR cycling reaction using the ABI Prism sequence detector software (v2.0). The cycle threshold value was calculated using a threshold value set at 0.2. Transcript levels of each gene were normalized to the 18S rRNA control gene.
Novel Materials
All novel materials described in this publication will be made available upon request in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank Andy Bradshaw, Martin Holdsworth, and Steve Coggins (Horticulture Research International [HRI]) for constructing the Arabidopsis hydroponics system; Simon Elliott and Mark Powell (HRI) for mineral analyses; Thomas Thykjaer (AROS Applied Biotechnology, Aarhus, Denmark) for microarray analyses; Martin Sergeant (HRI) for assistance with quantitative PCR; and Andrew Thompson and Ken Manning (HRI) for their comments on the original manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020941.
This work was supported by the Biotechnology and Biological Sciences Research Council (UK), by the Department for Environment, Food and Rural Affairs (UK; project nos. HH0915SFV, HH3502SFV, and HH3501SFV), and by a Horticulture Research International Gordon Browning Studentship to J.P.H.
References
- Abel S, Nguyen MD, Theologis A (1995) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Abel S, Ticconi CC, Delatorre CA (2002) Phosphate sensing in higher plants. Physiol Plant 115: 1–8 [DOI] [PubMed] [Google Scholar]
- Baldwin JC, Karthikeyan AS, Raghothama KG (2001) LEPS2, a phosphorus starvation-induced novel acid phosphatase from tomato. Plant Physiol 125: 728–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baligar VC, Fageria NK, He ZL (2001) Nutrient use efficiency in plants. Commun Soil Sci Plant Anal 32: 921–950 [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ (1994) The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J 6: 673–685 [DOI] [PubMed] [Google Scholar]
- Bates TR, Lynch JP (2000) Plant growth and phosphorus accumulation of wild type and two root hair mutants of Arabidopsis thaliana (Brassicaceae). Am J Bot 87: 958–963 [PubMed] [Google Scholar]
- Bates TR, Lynch JP (2001) Root hairs confer a competitive advantage under low phosphorus availability. Plant Soil 236: 243–250 [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris 316: 1194–1199 [Google Scholar]
- Bieleski RL, Ferguson IB (1983) Physiology and metabolism of phosphate and its compounds. In A Läuchli, RL Bieleski, eds, Encyclopedia of Plant Physiology, New Series, Vol 15A. Springer-Verlag, Berlin, pp 422–449 [Google Scholar]
- Bohn HL, McNeal BL, O'Connor GA (1979) Soil Chemistry. John Wiley & Sons, New York
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley MR, Burns A, Burns IG (2002) Relationships between phosphorus forms and plant growth. J Plant Nutr 25: 1075–1088 [Google Scholar]
- Broadley MR, Escobar-Gutiérrez AJ, Bowen HC, Willey NJ, White PJ (2001) Influx and accumulation of Cs+ by the akt1 mutant of Arabidopsis thaliana (L.) Heynh. lacking a dominant K+ transport system. J Exp Bot 52: 839–844 [DOI] [PubMed] [Google Scholar]
- Brinch-Pedersen H, Sørensen LD, Holm PB (2002) Engineering crop plants: getting a handle on phosphate. Trends Plant Sci 7: 118–125 [DOI] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ (1997) NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA et al. (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciereszko I, Johansson H, Hurry V, Kleczkowski LA (2001) Phosphate status affects the gene expression, protein content and enzymatic activity of UDP-glucose pyrophosphorylase in wild-type and pho mutants of Arabidopsis. Planta 212: 598–605 [DOI] [PubMed] [Google Scholar]
- Collins FS, Weissman SM (1984) Directional cloning of DNA fragments at a large distance from an initial probe: a circularization method. Proc Natl Acad Sci USA 81: 6812–6816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daram P, Brunner S, Rausch C, Steiner C, Amrhein N, Bucher M (1999) Pht2;1 encodes a low affinity phosphate transporter from Arabidopsis. Plant Cell 11: 2153–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de A Gerhardt LB, Sachetto-Martins G, Contarini MG, Sandroni M, de P Ferreira R, de Lima VM, Cordeiro MC, de Oliveira DE, Margis-Pinheiro M (1997) Arabidopsis thaliana class IV chitinase is early induced during the interaction with Xanthomonas campestris. FEBS Lett 419: 69–75 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ (1995) Characterisation of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Desikan R, Mackerness SAH, Hancock JT, Neill SJ (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC (1975) Comparison of the effects of a localised supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and the shoot, in barley. New Phytol 75: 479–490 [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC (1994) The role of acid phosphatases in plant phosphorus metabolism. Physiol Plant 90: 791–800 [Google Scholar]
- Epple P, Apel K, Bohlmann H (1995) An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol 109: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C (1998) Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 1950–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fageria NK, Baligar VC (1999) Phosphorus-use efficiency in wheat geno-types. J Plant Nutr 22: 331–340 [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frossard E, Condron LM, Oberson A, Sinaj S, Fardeau JC (2000) Processes governing phosphorus availability in temperate soils. J Environ Qual 29: 15–23 [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Waner D, Tchieu J, Gribskov M, Schroeder J (2001) An integrated Arabidopsis annotation database for Affymetrix Genechip data analysis, and tools for regulatory motif searches. Trends Plant Sci 6: 448–449 [DOI] [PubMed] [Google Scholar]
- Gil P, Liu Y, Orbovic V, Verkamp E, Poff KL, Green PJ (1994) Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol 104: 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AH (1992) Phosphate starvation inducible enzymes and proteins in higher plants. In JL Wray, ed, Society for Experimental Biology Seminar Series 49: Inducible Plant Proteins. Cambridge University Press, Cambridge, UK, pp 25–44
- Goldstein AH, Beartlein DA, McDaniel RG (1988) Phosphate starvation inducible metabolism in Lycopersicon esculentum: I. Excretion of acid phosphatase by tomato plants and suspension cultured cells. Plant Physiol 87: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzàlez-Meler MA, Giles L, Thomas RB, Siedow JN (2001) Metabolic regulation of leaf respiration and alternative pathway activity in response to phosphate supply. Plant Cell Environ 24: 205–215 [Google Scholar]
- Greenwood DJ, Karpinets TV, Stone DA (2001) Dynamic model for the effects of soil P and fertilizer P on crop growth, P uptake and soil P in arable cropping: model description. Ann Bot 88: 279–291 [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y (2002) Identification and characterisation of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran S, Logendra S, Seskar M, Bratanova M, Raskin I (2000) Characterization of Arabidopsis acid phosphatase promoter and regulation of acid phosphatase expression. Plant Physiol 124: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ (1999) Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu Rev Plant Physiol Plant Mol Biol 50: 219–243 [DOI] [PubMed] [Google Scholar]
- Holford ICR (1997) Soil phosphorus: its measurement, and its uptake by plants. Aust J Soil Res 35: 227–239 [Google Scholar]
- Jungk A (2001) Root hairs and the acquisition of plant nutrients from soil. J Plant Nutr Soil Sci 164: 121–129 [Google Scholar]
- Kai M, Takazumi K, Adachi H, Wasaki J, Shinano T, Osaki M (2002) Cloning and characterisation of four phosphate transporter cDNAs in tobacco. Plant Sci 163: 837–846 [Google Scholar]
- Karthikeyan AS, Varadarajan DK, Mukatira UT, Panio D'Urzo M, Damsz B, Raghothama KG (2002) Regulated expression of Arabidopsis transporters. Plant Physiol 130: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok EJ, Wilson IW, Wilson D, Chapman SC, Ewing RM, Somerville SC, Peacock WJ, Dolferus R, Dennis ES (2002) Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 14: 2481–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansford R, Bearman G, Fraser SE (2001) Resolution of multiple green fluorescent protein color variants and dyes using two-photon microscopy and imaging spectroscopy. J Biomed Optics 6: 311–318 [DOI] [PubMed] [Google Scholar]
- Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D (2002) Purple acid phosphatases of Arabidopsis thaliana. J Biol Chem 277: 27772–27781 [DOI] [PubMed] [Google Scholar]
- Lipton DS, Blancher RW, Blevins DG (1987) Citrate, malate and succinate concentrations in exudates from P-sufficient and P-starved Medicago sativa L. seedlings. Plant Physiol 85: 315–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Nieto-Jacobo MF, Ramírez-Rodríguez V, Herrera-Estrella L (2000) Organic acid metabolism in plants: from adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci 160: 1–13 [DOI] [PubMed] [Google Scholar]
- Lynch PJ (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerness SAH, John CF, Jordan B, Thomas B (2001) Early signalling components in ultraviolet-B responses: distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett 489: 237–242 [DOI] [PubMed] [Google Scholar]
- Mahalingam R, Gomez-Buitrago AM, Eckardt N, Shah N, Guevara-Garcia A, Day P, Raina R, Fedoroff NV (2003) Characterizing the stress/defense transcriptome of Arabidopsis. Genome Biol 4: R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malboobi MA, Lefebvre DD (1997) A phosphate-starvation inducible β-glucosidase (psr3.2) isolated from Arabidopsis thaliana is a member of a distinct subfamily of the BGA family. Plant Mol Biol 34: 57–68 [DOI] [PubMed] [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de la Peña A, Leyva A, Paz-Ares J (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24: 559–567 [DOI] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JAH (2002) Cell cycle-regulated gene expression in Arabidopsis. J Biol Chem 277: 41987–42002 [DOI] [PubMed] [Google Scholar]
- Miller SS, Liu J, Allan DL, Menzhuber CJ, Fedorova M, Vance CP (2001) Molecular control of acid phosphatase secretion into the rhizosphere of proteoid roots from phosphorus-stressed white lupin. Plant Physiol 127: 594–606 [PMC free article] [PubMed] [Google Scholar]
- Moseyko N, Zhu T, Chang HS, Wang X, Feldman LJ (2002) Transcriptional profiling of the early gravitropic response in Arabidopsis using high-density oligonucleotide probe microarrays. Plant Physiol 130: 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Mukatira UT, Liu C, Varadarajan DK, Raghothama KG (2001) Negative regulation of phosphate starvation-inducible genes. Plant Physiol 127: 1854–1862 [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- National Research Council (1989) Alternative Agriculture. National Academic Press, Washington, DC
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley WB (1996) The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones 1: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Carswell MC (1999) Metabolic aspects of phosphate starvation in plants. In HR Lerner, ed, Plant Responses to Environmental Stresses: From Phytohormones to Genome Reorganisation. Marcel Dekker, New York, pp 349–372
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S, Uknes S, Lawton K, Winter AM, Chandler D, DiMaio J, Novitzky R, Ward E, Ryals J (1993) Regulation of a hevein-like gene in Arabidopsis. Mol Plant-Microbe Interact 6: 680–685 [DOI] [PubMed] [Google Scholar]
- Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Ramonell KM, Zhang B, Ewing RM, Chen Y, Xu D, Stacey G, Somerville S (2002) Microarray analysis of chitin elicitation in Arabidopsis thaliana. Mol Plant Pathol 3: 301–311 [DOI] [PubMed] [Google Scholar]
- Reuber TL, Ausubel FM (1996) Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 8: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KD, Schott EJ, Sharma YK, Davis KR, Gardner RC (1998) Aluminium induces oxidative stress genes in Arabidopsis thaliana. Plant Physiol 116: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D (1994) The responses of plants to non-uniform supplies of nutrients. New Phytol 127: 635–674 [DOI] [PubMed] [Google Scholar]
- Rouse DT, Marotta R, Parish RW (1996) Promoter and expression studies on an Arabidopsis thaliana dehydrin gene. FEBS Lett 381: 252–256 [DOI] [PubMed] [Google Scholar]
- Rossel JB, Wilson IW, Pogson BJ (2002) Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol 130: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor in phosphate starvation signalling both in vascular plants and in unicellular algae. Gene Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge-Metzger A (1995) Closing the cycle: obstacles to efficient P management for improved global security. In H Tiessen, ed, Phosphorus in the Global Environment: Transfers, Cycles and Management. John Wiley & Sons, New York, pp 27–42
- Schachtman DP, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116: 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma YK, Davis KR (1994) Ozone-induced expression of stress-related genes in Arabidopsis thaliana. Plant Physiol 105: 1089–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpley A (1995) Identifying sites vulnerable to phosphorus loss in agricultural runoff. J Environ Qual 24: 947–951 [Google Scholar]
- Smethurst PJ (2000) Soil solution and other soil analyses as indicators of nutrient supply: a review. Forest Ecol Man 138: 397–411 [Google Scholar]
- Somssich IE, Wernert P, Kiedrowski S, Hahlbrock K (1996) Arabidopsis thaliana defence-related protein ELI3 is an aromatic alcohol:NADP+ oxidoreductase. Proc Natl Acad Sci USA 93: 14199–14203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Onouchi H, Hamada S, Machida C, Hammond-Kosack KE, Jones JD (1998) Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J 14: 365–370 [DOI] [PubMed] [Google Scholar]
- Trull MC, Deikman J (1998) An Arabidopsis mutant missing one acid phosphatase isoform. Planta 206: 544–550 [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich-Eberius CI, Novacky A, van Bel AJE (1984) Phosphate uptake in Lemna gibba G1: energetics and kinetics. Planta 161: 46–52 [DOI] [PubMed] [Google Scholar]
- Vance CP (2001) Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol 127: 390–397 [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk OK, Mari S, Lu YP, Rea PA (1999) AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proc Natl Acad Sci USA 96: 7110–7115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versaw WK, Harrison MJ (2002) A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14: 1751–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser NV, Hink MA, Borst JW, van der Krogt GNM, Visser AJWG (2002) Circular dichroism spectroscopy of fluorescent proteins. FEBS Lett 521: 31–35 [DOI] [PubMed] [Google Scholar]
- Wang WC, Liu ZH (1999) HarpinPSS-induced peroxidase and lignin accumulation in tobacco during the hypersensitive response. Aust J Plant Physiol 26: 265–272 [Google Scholar]
- Wang YH, Garvin DF, Kochian LV (2002) Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots: evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol 130: 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei-jun S, Forde BG (1989) Efficient transformation of Agrobacterium ssp. by high voltage electroporation. Nucleic Acids Res 17: 8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SPCP, Fitter AH, Leyser HMO (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge P, Brembu T, Bones AM (1997) Cloning and characterization of rac-like cDNAs from Arabidopsis thaliana. Plant Mol Biol 35: 483–495 [DOI] [PubMed] [Google Scholar]
- Withers PJA, Edwards AC, Foy RH (2001) Phosphorus cycling in UK agriculture and implications for phosphorus loss from soil. Soil Use Manag 17: 139–149 [Google Scholar]
- Xu W, Campbell P, Vargheese AK, Braam J (1996) The Arabidopsis XET-related gene family: environmental and hormonal regulation of expression. Plant J 9: 879–889 [DOI] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhleniuk OV, Raines CA, Lloyd JC (2001) pho3: a phosphorus-deficient mutant of Arabidopsis thaliana (L.) Heynh. Planta 212: 529–534 [DOI] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Han B, Brown D, Chang HS, Zou G, Wang X (2001) Toward elucidating the global gene expression patterns of developing Arabidopsis: parallel analysis of 8300 gene by a high-density oligonucleotide probe array. Plant Physiol Biochem 39: 221–242 [Google Scholar]