Abstract
The circadian clock synchronizes the internal biology of an organism with the environment and has been shown to be widespread among organisms. Microarray experiments have shown that the circadian clock regulates mRNA abundance of about 10% of the transcriptome in plants, invertebrates, and mammals. In contrast, the circadian clock regulates the transcription of the virtually all cyanobacterial genes. To determine the extent to which the circadian clock controls transcription in Arabidopsis, we used in vivo enhancer trapping. We found that 36% of our enhancer trap lines display circadian-regulated transcription, which is much higher than estimates of circadian regulation based on analysis of steady-state mRNA abundance. Individual lines identified by enhancer trapping exhibit peak transcription rates at circadian phases spanning the complete circadian cycle. Flanking genomic sequence was identified for 23 enhancer trap lines to identify clock-controlled genes (CCG-ETs). Promoter analysis of CCG-ETs failed to predict new circadian clock response elements (CCREs), although previously defined CCREs, the CCA1-binding site, and the evening element were identified. However, many CCGs lack either the CCA1-binding site or the evening element; therefore, the presence of these CCREs is insufficient to confer circadian regulation, and it is clear that additional elements play critical roles.
Timing is everything, making biological clocks central to life in a rhythmic world (Young and Kay, 2001). One such biological clock is the circadian clock, an endogenous oscillator with a period of about 24 h. The circadian clock synchronizes internal biology with the external daily cycle of light and temperature. Biological activities are partitioned, or phased, to specific times of the day, and, therefore, occur in distinct relationships, or phase angles, with each other and the external environment. For example, humans are maximally active during the day, whereas melatonin synthetic rates and blood melatonin levels peak at night (Rutter et al., 2002). In unicellular nitrogen-fixing cyanobacteria, it is critical that oxygenic photosynthesis and anaerobic nitrogen assimilation occur at distinct times of day (Johnson and Golden, 1999). In higher plants, photosynthetic rates peak during the day, whereas hypocotyl and inflorescence stem elongation is maximal at night (McClung et al., 2002).
Rhythmic gene expression underlies many of these complex circadian rhythms. Recent advances in microarray technology have provided the ability in model organisms to ask how many mRNAs oscillate with a circadian period. Such microarray experiments have revealed in mammals, Drosophila melanogaster, Neurospora crassa, and Arabidopsis that between 2% and 10% of mRNAs are regulated by the circadian clock (Harmer et al., 2000; Claridge-Chang et al., 2001; McDonald and Rosbash, 2001; Schaffer et al., 2001; Akhtar et al., 2002; Ceriani et al., 2002; Duffield et al., 2002; Lin et al., 2002; Panda et al., 2002a; Storch et al., 2002; Ueda et al., 2002). These microarray experiments highlight the regulation by the circadian clock of many key biological pathways, including pathways involved in metabolism, detoxification, and stress response. It is possible that the circadian clock regulates a larger portion of biological processes by regulating key steps in pathways rather than entire pathways (Harmer et al., 2000). Although we are accustomed to thinking of “circadianregulated” genes as those whose transcripts oscillate with a period of 24 h, perhaps this definition should be broadened. Clearly oscillations in mRNA abundance cannot account for all circadian-mediated biology because both posttranscriptional and posttranslational regulation have been shown to play a major role in clock function (Lee et al., 2000, 2001). Moreover, there is potentially another class of genes for which the response to environmental or other stimuli is temporally gated by the circadian clock, and such genes would not be identified in the absence of the appropriate stimulus.
Efforts to understand the molecular mechanisms by which the circadian clock regulates transcription have focused on the identification and characterization of cis-acting elements necessary for circadian-regulated transcription and their cognate DNA-binding proteins. In mammals and D. melanogaster, both the enhancer box (E box; CACGTG) and the cAMP response element (TGACGTCA) have been shown to be functional circadian clock response elements (CCREs; Young and Kay, 2001). It is interesting that both the E box and the cAMP response element serve in output genes as the targets of central clock components but are also found in the promoters of genes encoding these same central clock components (Kyriacou and Rosato, 2000). In D. melanogaster, CCREs have been shown to be overrepresented in the predicted promoters of genes whose mRNA oscillates (Claridge-Chang et al., 2001). In contrast, in mice (Mus musculus), the E box was only found in the predicted promoters of a small proportion of cycling genes (Panda et al., 2002a). At least two CCREs have been defined in N. crassa. Some output genes are controlled by the circadian clock through the activating clock element (GTTGGGAT; Bell-Pedersen et al., 1996). In addition, the distal light response element confers light- and circadianregulated transcription upon the central clock component frequency (Froehlich et al., 2002).
Transcriptional/translational negative feedback loops underpin the molecular mechanisms of the circadian clock (Young and Kay, 2001). Although still incompletely defined, the Arabidopsis circadian clock includes reciprocal regulation between the pseudo response regulator TOC1 (TIMING OF CAB EXPRESSION 1) and two single MYB domain transcription factors, LHY (LATE ELONGATED HYPOCOTYL) and CCA1 (CIRCADIAN CLOCK ASSOCIATED 1; Alabadí et al., 2001, 2002; Matsushika et al., 2002b; Mizoguchi et al., 2002). In plants, functionally defined CCREs are distinct from those identified in mammals and flies. The CCA1-binding site (CBS; AAAAATCT) and evening element (EE; AAAATATCT) are targets of the central clock components CCA1 and LHY (Wang et al., 1997; Schaffer et al., 1998; Alabadí et al., 2001). In one study, the EE was found to be overrepresented in the predicted promoters of genes with mRNA abundance peaking in the evening (Harmer et al., 2000). Both CCA1 and LHY are induced by light through direct binding of PIF3 (PHYTOCHROME INTERACTING FACTOR 3) at the G box (CCACGTGG; Martínez-García et al., 2000). Although the plant G box shares the same core sequence as the animal E box, the factors that bind to these elements are distinct, and it is likely that this sequence similarity is coincidental.
Current efforts to define the subset of the transcriptome that is regulated by the circadian clock have largely relied on transcript profiling, in which large numbers of genes were assayed in parallel to identify transcripts that showed circadian oscillations in abundance. This approach, however, does not directly address circadian regulation of transcription. Transcript stability might obscure oscillations in transcriptional activity, as has been demonstrated for Arabidopsis CAB1 (Millar and Kay, 1991). Random insertion of a transposon-borne promoter-less luciferase reporter gene was used to establish widespread circadian regulation of transcription in cyanobacteria (Liu et al., 1995a). Gene or enhancer trapping (Sundarasan et al., 1995; Campisi et al., 1999; Springer, 2000), in which a reporter gene either lacking a promoter or possessing a minimal unregulated promoter is randomly inserted through T-DNA transformation, has proven effective in Arabidopsis. Therefore, utilizing an in vivo luciferase enhancer trapping technique, we set out to ask to what extent the circadian clock in Arabidopsis regulates transcription. In this study, we reveal that 36% of the Arabidopsis genome is under circadian regulation at the transcriptional level. We identify 23 clock-controlled genes (CCG-ETs) with peak luciferase activity at distinct circadian phases spanning the day. The CBS and EE were found in the predicted promoters of the CCG-ETs identified in the enhancer trap lines.
RESULTS
Enhancer Trapping Circadian-Regulated Elements in Arabidopsis
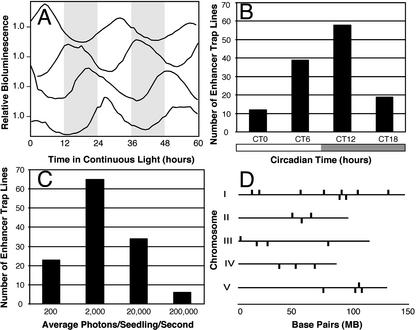
To identify circadian CCGs and their cognate CCREs and also to establish the extent of transcriptional regulation by the circadian clock in the Arabidopsis genome, we applied an in vivo luciferase-based enhancer trap strategy. Utilizing T-DNA-mediated transformation, a minimal promoter::LUC (LUCIFERASE) construct that was not clock regulated was randomly integrated into the Arabidopsis genome (Michael and McClung, 2002). Individual T1 plants resistant to gentamicin and, hence, carrying the transgene were identified and allowed to self-fertilize. Individual T2 plants resistant to gentamicin were entrained to a light/dark (12/12) cycle and then assayed under continuous light for clock-regulated luciferase activity in vivo using a Packard TopCount Luminometer (Packard, Meriden, CT; Michael and McClung, 2002). Each independent transgenic line exhibiting clock-regulated luciferase activity displayed a characteristic and consistent circadian phase and expression level (Fig. 1; see supplemental data Table III at http://www.plantphysiol.org). Of 335 lines assayed, 128 (36%) were scored as rhythmic by FFT-NLLS (Plautz et al., 1997). Rhythmic lines were defined as having a period of 20 to 30 h and a relative amplitude error (RAE), a measure of the strength of the rhythm, of <1.0. A perfect noise-free cosine wave would return an RAE = 0, whereas an RAE = 1 is the limit of statistical significance for any given rhythmic amplitude. We also generated lines bearing insertions of luciferase lacking a minimal promoter and found the same frequency of rhythmic lines, indicating that the minimal promoter sequences were not necessary for rhythmic transcription of the construct (data not shown). The range of periods (22.41–28.46 h) exhibited by the rhythmic lines is relatively broad. For example, Thain et al. (2002) observed a 2-h difference in the periods of CAB2 and CHALCONE SYNTHASE, which are thought to be driven by tissue-specific oscillators of distinct period. The broader range in periods observed among these lines may reflect inaccuracy arising from the relatively short (72 h) time series analyzed. However, it may be that we have trapped genes regulated by additional tissue- or cell-type-specific oscillators that collectively exhibit a broader range of periods, under the conditions tested, than has been previously observed. An additional and exciting possibility is that in some lines, the T-DNA insertion may have been mutagenic and may have disrupted the function of a gene that contributes to period specification. This is under investigation.
Figure 1.
Circadian enhancer trap using luciferase. Seedlings were grown 7 d in 12 h of light and 12 h of dark, and luminescence was measured in continuous light for 5 to 7 d. A, Circadian clock-controlled genes identified by enhancer trapping (CCG-ET) are phased to distinct times of day. Traces were normalized to the average expression over the experiment. All traces are the average of at least 12 seedlings. B, Number of CCG-ET lines with specific circadian phases: circadian time (CT) 0 (0–5.9 h), 6 (6–11.9 h), 12 (12–17.9 h), and 18 (18–23.9 h). All luminescence traces were analyzed for rhythmicity by fast Fourier transform-nonlinear least squares analysis (FFT-NLLS), and circadian phase was determined by normalizing for the period (phase = 24 h × phase/period). C, Expression level of the CCG-ET lines. Expression level was determined directly from the number of photons collected over the sampling interval: 200 (0–199 photons/s), 2,000 (200–1,999 photons/s), 20,000 (2,000–19,999 photons/s), and 200,000 (>20,000 photons/s). All data represent averages from at least two independent experiments. D, Map positions of CCG-ETs on the five Arabidopsis chromosomes.
Most of the clock-regulated enhancer trap lines display luciferase activity of approximately 2,000 photons s–1 seedling–1. We also identified lines with extremely high (>200,000 photons s–1 seedling–1) and low (<200 photons s–1 seedling–1) luciferase activity (Fig. 1C). One hundred photons per second per seedling is our standard low cutoff for background in the bioluminescence assay. We identified 22 lines with expression levels just above this cutoff level. Two factors contributed to our ability to identify with confidence lines expressing circadian transcription despite such low expression: Luciferase is an extremely sensitive assay, and our screening allowed us to test 12 to 24 genetically identical siblings over multiple (five–seven) circadian cycles. Thus, we were able to identify circadian regulation of transcription in lines with either low expression levels or with low amplitude rhythms. Such genes might not be identified by northern-blot or microarray analysis, in which relatively few cycles can be tested with lower levels of replication. The fact that we were able to identify lines with peak luciferase activity at circadian phases spanning the circadian day (Fig. 1B; see supplemental data Table III at http://www.plantphysiol.org) and over a broad range of expression levels (Fig. 1C; see supplemental data Table III at www.plantphysiol.org) leads us to believe that we were successful in enhancer trapping CCREs and identifying CCGs.
Identification of Enhancer-Trapped Sequences
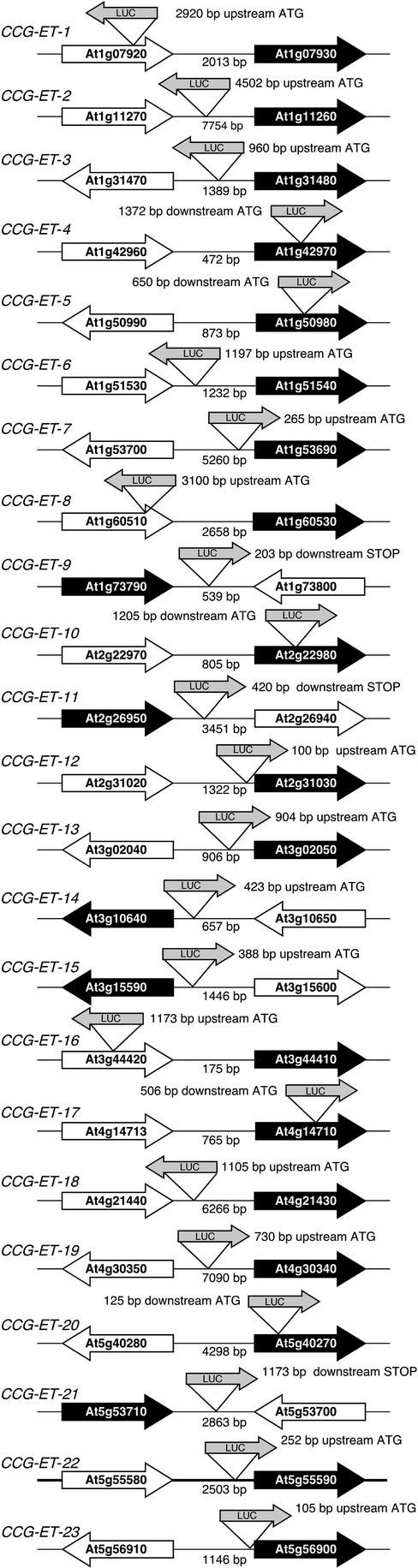
We defined the T-DNA insertion sites in a subset of 23 circadian-regulated enhancer trap lines by thermal asymmetric interlaced (TAIL)-PCR (Table I; Fig. 2). We chose the 23 enhancer trap lines randomly from the 128 rhythmic lines identified and found that they were evenly distributed over the Arabidopsis genome (Fig. 1D). All 23 of the recovered flanking sequences were aligned to the Arabidopsis sequence (www.arabidopsis.org) and corresponded to Arabidopsis genomic DNA. One additional CCG-ET corresponded to expressed cDNAs in the Arabidopsis database but failed to correspond to genomic, mitochondrial, or chloroplastic DNA, suggesting that it originates from a region of the Arabidopsis genome for which the sequence has not yet been determined (data not shown). Clock-controlled genes as determined by enhancer trapping (CCG-ET) were defined by the orientation of T-DNA insertion relative to predicted Arabidopsis genes: promoter, 5′-UTR, gene (intron or exon), 3′-UTR, and other (Table I; Fig. 2).
Table I.
Clock-controlled genes as predicted by enhancer trapping (CCG-ET)
| Name | Putative Functiona | Gene Identification | Insb | Exprc | Phased | Microe | CBSf | EEf | G Boxf | HEXf |
|---|---|---|---|---|---|---|---|---|---|---|
| CCG-ET-1 | Elongation factor 1-α | At1g07930 | O | 200 | 18 | S | 1 (10) | 0 (8) | 0 (0) | 0 (2) |
| CCG-ET-2 | STP1, Glc transporter | At1g11260 | O | 2,000 | 8 | S, Hg | 0 (2) | 0 (4) | 1 (4) | 1 (3) |
| CCG-ET-3 | SGR2, phospholipase | At1g31480 | P | 200,000 | 6 | S | 0 (10) | 0 (7) | 0 (0) | 0 (1) |
| CCG-ET-4 | Glyceraldehyde-3-phosphate dehydrogenase | At1g42970 | G | 2,000 | 6 | H | 0 (11) | 0 (9) | 0 (0) | 0 (0) |
| CCG-ET-5 | Hypothetical, transcription factor/F box | At1g50980 | G | 200,000 | 12 | ND | 1 (11) | 1 (3) | 0 (0) | 0 (0) |
| CCG-ET-6 | Unknown, MYB, kelch domains | At1g51540 | P | 20,000 | 20 | ND | 1 (6) | 0 (6) | 0 (2) | 0 (2) |
| CCG-ET-7 | RBP10-α putative RNA polymerase | At1g53690 | P | 200 | 12 | ND | 1 (6) | 0 (4) | 0 (0) | 0 (1) |
| CCG-ET-8 | Putative, GTP-binding protein | At1g60530 | P | 200 | 0 | Hh | 0 (4) | 0 (8) | 0 (0) | 0 (0) |
| CCG-ET-9 | Hypothetical protein | At1g73790 | G | 2,000 | 8 | ND | 2 (9) | 0 (10) | 0 (2) | 0 (3) |
| CCG-ET-10 | Putative, Ser carboxypeptidase I | At2g22980 | G | 20,000 | 10 | Hh | 0 (6) | 1 (9) | 0 (4) | 0 (0) |
| CCG-ET-11 | Putative, MYB domain | At2g26950 | 3 | 2,000 | 3 | Hh | 1 (15) | 1 (11) | 0 (0) | 0 (1) |
| CCG-ET-12 | Putative oxysterol binding | At2g31030 | 5 | 2,000 | 6 | ND | 1 (7) | 0 (8) | 0 (0) | 0 (0) |
| CCG-ET-13 | AtKUP3, K+ transporter | At3g02050 | G | 2,000 | 0 | S, Hh | 0 (3) | 0 (6) | 0 (0) | 0 (0) |
| CCG-ET-14 | unknown protein | At3g10640 | P | 200 | 12 | S | 0 (13) | 1 (5) | 0 (0) | 0 (0) |
| CCG-ET-15 | Putative DNA binding | At3g15590 | P | 2,000 | 12 | ND | 1 (9) | 0 (4) | 0 (0) | 0 (0) |
| CCG-ET-16 | Putative protein | At3g44410 | P | 2,000 | 12 | S | 0 (1) | 0 (1) | 0 (0) | 0 (1) |
| CCG-ET-17 | Expressed protein | At4g14710 | G | 2,000 | 11 | S | 0 (6) | 1 (5) | 0 (0) | 0 (2) |
| CCG-ET-18 | Putative, chloroplast DNA binding | At4g21430 | P | 20,000 | 12 | ND | 0 (8) | 0 (8) | 0 (0) | 0 (0) |
| CCG-ET-19 | Putative, diacylglycerol kinase iota | At4g30340 | P | 20,000 | 12 | ND | 0 (7) | 0 (6) | 0 (0) | 0 (0) |
| CCG-ET-20 | Putative protein | At5g40270 | G | 2,000 | 12 | S | 0 (8) | 0 (5) | 0 (2) | 0 (2) |
| CCG-ET-21 | Unknown protein | At5g53710 | O | 20,000 | 11 | ND | 0 (7) | 0 (5) | 0 (0) | 0 (0) |
| CCG-ET-22 | AtPME3 like, pectin methylesterase | At5g55590 | P | 2,000 | 0 | ND | 0 (8) | 0 (3) | 0 (0) | 0 (0) |
| CCG-ET-23 | Putative, mRNA process (splicing) | At5g56900 | G | 200 | 4 | ND | 0 (7) | 1 (2) | 0 (0) | 0 (1) |
The Arabidopsis Information Resource and the Munich Information Center for Protein Sequences. b T-DNA insertion position in reference to predicted gene location. P, promoter (-2,000/-150); G, exon or intron; 5, 5′-UTR (-150/-1 bp); 3, 3′-UTR; O, other. c Average bioluminescence (photons per second per seedling). d Phase, Time of the peak luciferase activity in continuous conditions normalized to CT. e Micro, Microarray experiments; S, Schaffer et al. (2001); H, Harmer et al. (2000). f CBS (AAAAATCT), EE (AAATATCT), G box (CCACGTGG), and HEX (TGACGTGG); no. of exact matches in predicted promoters (one mismatch). g Rhythmically expressed. h ND, Not determined.
Figure 2.
Insertions of LUCIFERASE relative to predicted Arabidopsis genes in the enhancer trap lines. T-DNA carrying the LUCIFERASE gene (shaded arrow) inserted as indicated into Arabidopsis genomic sequence in relation to the predicted CCG-ET (black arrow) and the nearest adjacent gene (white arrow). The distance (base pairs) from the ATG (or STOP) of the predicted CCG-ET to the start of the LUCIFERASE gene is indicated. Also indicated, below the horizontal line of the chromosome, is the distance between the predicted CCG-ET and the nearest adjacent gene. Depictions are not to scale.
Recently, two groups have utilized microarray experiments to identify Arabidopsis mRNAs with a circadian pattern of expression. Six and eight of the CCG-ETs were present on the microarrays used by Harmer et al. (2000) and Schaffer et al. (2001), respectively. CCG-ET2, encoding the STP1 Glc transporter (At1g11260), was determined to be circadian regulated by Harmer et al. (2000), whereas Schaffer et al. (2001) characterized it as diurnally regulated, yet not circadian regulated (Table I). CCG-ET4, encoding glyceraldehyde-3-phosphate dehydrogenase (At1g42970), was not found to be rhythmic in continuous light by Harmer et al. (2000), yet is circadian regulated in other systems (Lillo, 1993; Shinohara et al., 1998; Fagan et al., 1999). The four remaining CCG-ETs represented on the microarray tested by Harmer et al. (2000) displayed no detectable signal (Table I; S. Harmer and S.A. Kay, personal communication), and so neither confirm nor refute their regulation by the circadian clock. Although CCG-ET22 (AtPME3-like, At5g55590; Table I) was not on either of the microarrays, its putative paralog, AtPME3, was characterized as rhythmic by Harmer et al. (2000).
CCREs
Enhancer trapping takes an unbiased approach to the identification of genes whose transcript is clock-regulated and, therefore, may reveal novel CCREs. To identify possible CCREs, we searched the predicted promoters (defined as –2,000/–1 upstream of the predicted translational start site) of all the CCGETs for overrepresented elements using the Webbased regulatory sequence analysis tools (RSAs; http://embnet.cifn.unam.mx/rsa-tools/). Only one element between 5 and 8 bp in length, GATATAC, was found to be significantly overrepresented. It was only present in 39% (9 of 23) of the CCG-ET promoters. The promoter region of CCG-ET15 (At3g15590) contained 17 of these motifs, accounting for 56% (17 of 30) of the motifs found in this set of 23 promoters. When CCG-ET15 is removed from the analysis, there are no significantly overrepresented elements in the promoters of the CCG-ETs. Repeating the analysis on subsets of the CCG-ETs grouped according to the circadian phase of peak expression did not reveal overrepresented elements.
A similar promoter alignment tool (AlignAce; http://arep.med.harvard.edu/) identified the EE (AAATATCT) in the promoters of the evening-specific (CT 8 and CT 12, where CT 0 is subjective dawn) CCGs identified by Harmer et al. (2000). Utilizing RSA, we also identified the EE (AAAATATCTCA and AAAATATCTT) in promoters of rhythmic genes with evening-specific phases. In genes with morning-specific phases (CT 20 and CT 0), the G-box core (CACGTG) was overrepresented with the consensus (GACACGTGG). G boxes located in the CCA1 and LHY promoters serve as the site of activation by the light-transducing transcription factor PIF3 (Martínez-García et al., 2000). Therefore, like the light response element in N. crassa that confers both light and circadian regulation (Froehlich et al., 2002), the G box may also play dual roles as has been hypothesized (Menkens et al., 1995).
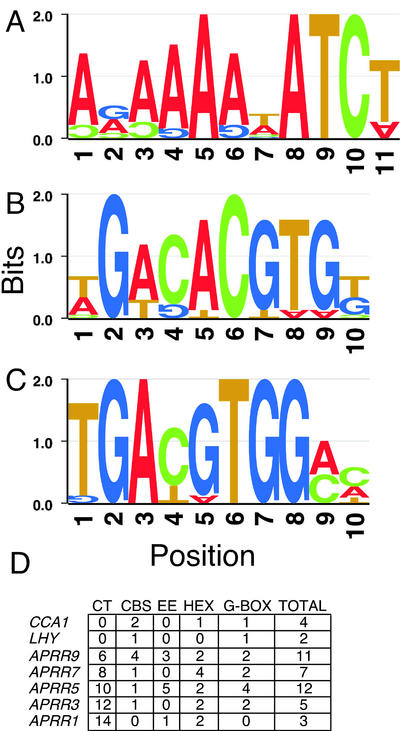
We also used RSA to search the predicted promoters of genes thought to be intimately involved in the circadian clock. When the promoters of CCA1, LHY, TOC1 (APRR1), APRR3, APRR5, APRR7, and APRR9 (Matsushika et al., 2002a; McClung et al., 2002; Sato et al., 2002) were searched together, three oligos were overrepresented: EE (AAATATCT), G box (CACGTGTC) and the Hex element (TGACGTGG; Schindler et al., 1992; Fig. 3). The Hex element is related to the G box and is bound by G-box-binding factors (Schindler et al., 1992). The Hex element is overrepresented specifically in the promoters of the family of Arabidopsis APRRs (PSEUDO-RESPONSE REGULATORS), of which the central clock component TOC1 is the founding member (Fig. 3D; Matsushika et al., 2000; Strayer et al., 2000).
Figure 3.
CCREs in the promoters of genes encoding clock components. Predicted promoters (–2,000/–1 upstream of the ATG) of CCA1, LHY, APRR1/TOC1, APRR3, APRR5, APRR7, and APRR9 were aligned using AlignACE (http://arep.med.harvard.edu/; Hughes et al., 2000) and RSA (http://embnet.cifn.unam.mx/rsa-tools/). Motif models were created as sequence logos (Schneider and Stephens, 1990) using the program GENIO/logo (http://genio.informatik.unistuttgart.de/GENIO/logo.cgi). At each position, the height of each residue is proportional to its frequency in that position. The total height of all the residues in the position is proportional to the degree of conservation (information content) of the position, presented in bits. Two bits are sufficient to specify a single nucleotide as always present at a given position and, therefore, are the maximum information content possible at any position. Upside-down letters appear if the probability of that nucleotide occupying the position is less than the equally distributed a priori probability of all letters. A, CBS/EE (AAAa/tATCT); B, G box (CACGTG); C, HEX element (TGACGTGG); D, Number of each element and total number of elements in each predicted promoter.
Promoter Analysis of the Arabidopsis Genome
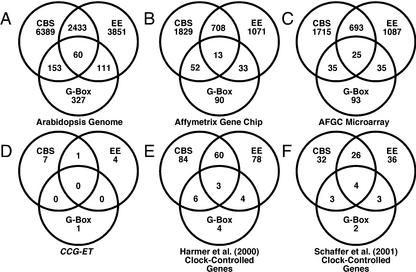
To place the CBS, EE, G box, and Hex element results from the CCG-ETs in the context of the whole Arabidopsis genome, we searched for these motifs in promoters (–2,000/–1 from the ATG) from most (25,545) of the predicted Arabidopsis genes. To determine the “expected” frequency of these motifs, we also randomly generated 25,545 “promoters,” each 2,000 bp, which were derived with the same base pair composition as actual Arabidopsis promoters (see “Materials and Methods”). We also analyzed the predicted promoters from 8,200 genes on the Affymetrix GeneChip, the Harmer et al. (2000) clock-regulated genes, the Arabidopsis Functional Genomics Consortium (AFGC) microarray genes, and the genes described by Schaffer et al. (2001) as clock regulated, diurnally regulated, light induced, or dark induced. Figure 4 illustrates the occurrence of the CBS, EE, and G box in the predicted promoters of these sets of genes, and Table II summarizes our findings.
Figure 4.
Occurrence of CBS, EE, and G box in predicted Arabidopsis promoters. Predicted promoters (–2,000/–1 upstream of the ATG) for most Arabidopsis genes (25,545) were searched for the following motifs: CBS (AAAAATCT), EE (AAATATCT), G box (CCACGTGG), and G box core (CACGTG). Venn diagrams illustrate the occurrences of the CBS (upper left), EE (upper right), and G box (bottom). A, Genes from the Arabidopsis genome; B, genes on the Affymetrix Gene Chip (AffyGC); C, genes on the AFGC microarray; D, CCG-ETs; E, clock-controlled genes detected by Harmer et al. (2000); F, clock-controlled genes detected by Schaffer et al. (2001).
Table II.
Occurrence of CBS, EE, G-Box, and Hex elements in predicted Arabidopsis promoters
Expected values were determined by establishing the frequency of the oligonucleotide in the 25,545 random Arabidopsis promoters. Gene sets are defined as follows: Harmer Circadian, genes defined as circadian regulated in continuous light by Harmer et al. (2000); Schaffer Circadian, genes defined as circadian regulated in continuous light by Schaffer et al. (2001); Schaffer Light Induced, the subset of circadian genes defined Schaffer et al. (2001) in which expression was higher in the day than in the night; Schaffer Dark Induced, the subset of circadian genes defined Schaffer et al. (2001) in which expression was higher in the night than in the day; Schaffer Diurnal, genes shown to oscillate in a light-dark cycle by Schaffer et al. (2001).
| Arabidopsis Genome
|
Affymetrix Gene Chip
|
AFCG Chip
|
||||||||
| Motif | 8mer | Observed % (#) | Expected % (#) | Obs/exp | Observed % (#) | Expected % (#) | Obs/exp | Observed % (#) | Expected % (#) | Obs/exp |
| CBS | AAAAATCT | 35 (9035) | 53 (13641) | 0.66 | 36 (2602) | 53 (3798) | 0.69 | 35 (2468) | 53 (3692) | 0.67 |
| EE | AAATATCT | 25 (6455) | 35 (9042) | 0.71 | 25 (1825) | 35 (2508) | 0.73 | 26 (1840) | 35 (2508) | 0.73 |
| G-Box | CCACGTGG | 3 (651) | 1 (332) | 1.96 | 3 (188) | 1 (71) | 2.64 | 3 (188) | 1 (71) | 2.64 |
| Hex | TGACGTGG | 5 (1274) | 3 (817) | 1.56 | 6 (406) | 3 (214) | 1.89 | 6 (389) | 3 (214) | 1.80 |
| Total Genes | 25,545 | Total Genes | 7,166 | Total Genes | 6,966 | |||||
|
CCG-ET
|
Harmer Circadian
|
Schaffer Circadian
|
||||||||
| Motif | 8mer | Observed % (#) | Expected % (#) | Obs/exp | Observed % (#) | Expected % (#) | Obs/exp | Observed % (#) | Expected % (#) | Obs/exp |
| CBS | AAAAATCT | 35 (8) | 53 (12) | 0.66 | 39 (153) | 53 (204) | 0.75 | 40 (64) | 53 (85) | 0.75 |
| EE | AAATATCT | 22 (5) | 35 (8) | 0.62 | 37 (145) | 35 (135) | 1.07 | 43 (69) | 35 (56) | 1.23 |
| G-Box | CCACGTGG | 4 (1) | 1 (0.23) | 4.35 | 4 (17) | 1 (4) | 4.25 | 7 (11) | 1 (2) | 5.50 |
| Hex | TGACGTGG | 4 (1) | 3 (0.69) | 1.45 | 8 (30) | 3 (12) | 2.50 | 10 (10) | 3 (5) | 2.00 |
| Total Genes | 23 | Total Genes | 386 | Total Genes | 161 | |||||
| Schaffer Light Induced
|
Schaffer Dark Induced
|
Schaffer Diurnal
|
||||||||
| Motif | 8mer | Observed % (#) | Expected % (#) | Obs/exp | Observed % (#) | Expected % (#) | Obs/exp | Observed % (#) | Expected % (#) | Obs/exp |
| CBS | AAAAATCT | 34 (32) | 53 (50) | 0.64 | 38 (199) | 53 (280) | 0.71 | 41 (276) | 53 (354) | 0.80 |
| EE | AAATATCT | 27 (25) | 35 (33) | 0.75 | 37 (198) | 35 (185) | 1.07 | 38 (254) | 35 (234) | 1.08 |
| G-Box | CCACGTGG | 9 (8) | 1 (0.93) | 8.60 | 5 (27) | 10 (53) | 0.50 | 4 (27) | 1 (7) | 3.85 |
| Hex | TGACGTGG | 8 (7) | 3 (3) | 2.33 | 8 (41) | 3 (16) | 2.56 | 9 (57) | 3 (20) | 2.85 |
| Total Genes | 93 | Total Genes | 530 | Total Genes | 668 | |||||
| Harmer % (rhythmic with motif/total genes with motif)
|
Schaffer % (rhythmic with motif/total genes with motif)
|
|||||||||
| Motif
|
8mer
|
1 copy
|
2 copies
|
3 or more
|
1 copy
|
2 copies
|
3 or more
|
|||
| CBS | AAAAATCT | 6 (119/1991) | 6 (28/483) | 5 (6/128) | 3 (47/1897) | 4 (15/437) | 2 (2/134) | |||
| EE | AAATATCT | 6 (96/1493) | 12 (32/278) | 32 (17/54) | 3 (44/1493) | 6 (17/278) | 13 (8/62) | |||
Of the four motifs sampled, the two known CCREs are found slightly less frequently (approximately 0.7-fold) in the Arabidopsis promoter database than predicted by chance, whereas the G-box and Hex elements are found more frequently (1.96- and 1.56-fold, respectively). The frequencies of these elements in the promoters of genes on both microarrays are representative of their frequency in the whole genome, except for a modest enrichment of the G box (Table II). A number of observations can be made. First, the G-Box is overrepresented in the predicted promoters of three sets of clock-regulated genes (Table II; CCGET, Harmer circadian and Schaffer circadian) and of genes that are diurnally and light regulated (Table II; Schaffer light induced and Schaffer diurnal). Consistent with the G box playing a role in light signaling, it is overrepresented among light-induced genes and underrepresented among dark-induced genes (Table II) (Schaffer et al., 2001). Second, the EE is slightly overrepresented in the Harmer circadian, Schaffer circadian, Schaffer dark-induced, and Schaffer diurnal sets of genes (Table II). Third, the Hex element is overrepresented in the Schaffer light-induced, dark-induced, and diurnal gene sets, and among the Harmer circadian genes (Table II). Finally, the occurrence of the CBS does not vary among the sets of genes (Table II).
The CBS and EE are found in the promoters of 35% and 25%, respectively, of Arabidopsis genes, which is consistent with our estimation that 36% of the Arabidopsis genome is circadian regulated. Is the presence of a CBS or EE sufficient to predict clock regulation? Apparently not at the level of mRNA abundance, because the CBS and EE are found in 35% and 25%, respectively, of the promoters on both the Affymetrix and AFGC microarrays, yet only 6% and 2%, respectively, of all the genes on these microarrays were found to be clock regulated. In fact, 93% (3,236 of 3,471) and 97% (3,384 of 3,487) of the genes on the Affymetrix and AFGC, respectively, whose predicted promoters have the CBS or EE, were not identified as rhythmic. Clearly, the presence of a CBS or EE in insufficient to confer circadian-regulated mRNA abundance. This is consistent with the accumulating evidence that additional elements are required to provide a context in which CCREs can properly function (Michael and McClung, 2002; Muñoz et al., 2002).
Combinatorial interaction of light-responsive elements confers light-regulated expression in Arabidopsis (Puente et al., 1996; Chattopadhyay et al., 1998). Clock regulation may require multiple copies or combinations of the CBS, EE, and/or G box. Consistent with this idea, 76% and 83% of the G boxes identified in the promoters of Harmer et al. (2000) and Schaffer et al. (2001) CCGs, respectively, occurred in combination with either the CBS or EE (Fig. 4, E and F), which is an enrichment relative to the whole-genome frequency of co-occurrence of approximately 50% (Fig. 4A, B, C). Among the CCGs identified by Harmer et al. (2000), the presence of three or more EEs in a promoter is a stronger predictor of rhythmicity than the presence of a single element, but only one-third of the genes with three or more copies of the EE are rhythmic, whereas two-thirds are not. In contrast, the presence of three or more copies of the CBS did not correlate with an increased frequency of rhythmicity (Table II).
DISCUSSION
Our observation that 36% of the Arabidopsis genome is potentially under transcriptional control by the circadian clock suggests a much more pervasive role for the clock in the regulation of gene expression than do estimates generated through microarray analysis of mRNA oscillations, which suggest that 2% to 6% of the transcriptome is clock regulated (Harmer et al., 2000; Schaffer et al., 2001). In D. melanogaster, microarray analysis and enhancer trapping provide similar estimates of the degree (approximately 10%) of clock control of gene expression (Claridge-Chang et al., 2001; McDonald and Rosbash, 2001; Stempfl et al., 2002; Ueda et al., 2002), and these estimates are largely congruent with estimates from microarray analyses conducted in mammals (Grundschober et al., 2001; Akhtar et al., 2002; Duffield et al., 2002; Panda et al., 2002a; Storch et al., 2002). However, promoter trap experiments in cyanobacteria have revealed that transcription of most genes is regulated by the circadian clock (Liu et al., 1995a; Aoki et al., 2002).
Possibly the most parsimonious way to explain the difference between the transcriptional and mRNA abundance estimates of circadian regulation is to recall that mRNA abundance does not strictly reflect transcriptional activity. For oscillations in transcription to yield oscillations in mRNA abundance requires that the transcript be sufficiently unstable to turn over within a circadian cycle. For example, it has been demonstrated that both oscillations in de novo transcription rates and in mRNA stability contribute to oscillations in mRNA abundance for chloroplast genes in Chlamydomonas reinhardtii (Salvador et al., 1993; Hwang et al., 1996). A recent estimate is that the majority of Arabidopsis transcripts (>95%) have half-lives >2 h (Gutierrez et al., 2002), suggesting that there may be a subset of genes for which the transcript stability is too great to allow clock-regulated changes in de novo transcription to be apparent as mRNA abundance oscillations. For instance, circadian oscillations in transcriptional activity of the CATALASE 3 (CAT3) promoter in light-dark cycles or in continuous light result in robust oscillations in CAT3 transcript abundance (Zhong and McClung, 1996; Michael and McClung, 2002). However, in continuous dark, the robust circadian oscillations in transcriptional activity persist, but the CAT3 transcript accumulates to a high and nonoscillating level, presumably due to an increase in transcript stability (Zhong et al., 1997; Michael and McClung, 2002). Similarly, although the majority of genes of Synechococcus elongatus PCC7942 exhibit robust circadian oscillations in transcription, as measured by activity of luciferase gene fusions (Liu et al., 1995a), parallel oscillations are not always detected at the level of mRNA abundance by northern-blot analysis (S.S. Golden, personal communication).
Other factors may prevent the detection of circadian oscillations in transcription at the level of microarray analysis. Sampling of total mRNA may mask tissue-specific oscillations of genes that only cycle in a subset of the tissues in which they are expressed. Enhancer trapping does not bias against tissue-specific expression, although it should be noted that our activity measure is primarily from the leaves (Thain et al., 2002). The luciferase assay is extremely sensitive, allowing the reproducible detection of oscillations at or below the limits of detection in a standard mRNA-based assay. This sensitivity is enhanced by our ability to monitor rhythms of multiple seedlings for each insertion for 5 to 7 d, taking measurements every hour. In addition, we can retest specific lines in multiple experiments. Thus, our assay allows us to reliably identify low-amplitude rhythms or weakly expressed CCGs (Fig. 1C). Such genes are very likely filtered out by the stringent statistical criteria used in microarray analysis to reduce the identification of false positives.
Our prediction of the enhancer-trapped CCG-ETs is based on proximity and orientation of the inserted LUC gene relative to the start site of the putative CCG-ET. In each case, we chose the gene whose start is closest to the LUC start except when LUC had inserted downstream of the start and in the antisense orientation. These criteria may have allowed the misidentification of some CCG-ETs. Targeted transcriptional LUC fusions to each of the 46 genes adjacent to the sites of LUC insertion in the 23 characterized lines would allow unambiguous resolution of this issue. It is also possible that we have identified “orphan” CCREs that fail to confer circadian regulation on adjacent genes but are capable of conferring circadian regulation on our LUC construct. Although plant regulatory regions tend to be compact, it is also possible that CCREs may be able to act at a distance (Bertolino and Singh, 2002). Therefore, circadian regulation may have been incorrectly attributed to a gene that is not regulated itself but that resides adjacent to a gene that is regulated by the clock. In any case, we do not think that the incorrect prediction of some of the CCG-ETs undermines the conclusion that there may be more circadian regulation of transcription than is indicated by analysis of mRNA abundance.
Although our CCG-ETs are distributed evenly over the chromosomes, T-DNA integration is not completely random. Two studies have suggested a slight bias toward integration into intergenic regions of high A + T base composition (Brunaud et al., 2002; Krysan et al., 2002). This should serve to increase insertion near promoter elements and, hence, overestimate the frequency of regulatory elements capable of conferring circadian regulation. However, in both studies, this bias was modest and insufficient to account for the much higher estimate of clock-controlled transcription (36%) versus mRNA abundance oscillations (6%–10%). Rhythmic histone acetylation recently has been correlated with circadian rhythms in mRNA abundance in mice, consistent with a key role of chromatin remodeling in circadian transcription (Etchegaray et al., 2003). It has also been suggested that clock-mediated condensation and/or supercoiling of the cyanobacterial chromosome is responsible for global circadian oscillations of transcript abundance (Mori and Johnson, 2001; Mori et al., 2002), similar to that found in the chloroplast of C. reinhardtii (Salvador et al., 1998). Possibly, T-DNA insertions could be biased toward chromosomal regions whose chromatin structure is under circadian control. Clustering of clock-regulated genes has been observed in D. melanogaster (Ueda et al., 2002). In Arabidopsis, the identification of TEJ has implicated polyADP-ribosylation in circadian regulation, possibly via chromatin modification (Panda et al., 2002b). DET1 (DEETIOLATED 1) associates with the core histone H2B in the context of the nucleosome, suggesting that chromatin remodeling plays a role in photomorphogenesis (Benvenuto et al., 2002). The det1 mutation also results in circadian defects (Millar et al., 1995), consistent with a role of chromatin structure in circadian regulation of transcription.
Our study suggests that transcriptional control of the Arabidopsis circadian clock may be more widespread then previously demonstrated through microarray experiments. The discrepancies between estimates of clock regulation based on transcription versus mRNA abundance, together with the observation that clock-regulated transcripts are enriched in the set of relatively unstable transcripts (Gutierrez et al., 2002), suggests that mRNA stability may be an important component of circadian regulation of gene expression. High levels of mRNA stability may reduce the effective number of cycling gene products by obscuring oscillations in abundance of transcripts whose transcription is circadian regulated. Conversely, circadian regulation of transcript stability, either periodically stabilizing or periodically destabilizing a transcript, could confer oscillations in mRNA abundance on transcripts whose transcription rates do not vary over the circadian cycle.
MATERIALS AND METHODS
Circadian Enhancer Trap Lines
A modified pPZP plant transformation vector containing a minimal CAT3 promoter (80 bp, insufficient to drive circadian transcription [Michael and McClung, 2002]), an omega translational enhancer, the luciferase coding sequence (LUC+), and the nopaline synthase terminator was used for circadian enhancer trapping. Enhancer trap lines were created using floral dip transformation (Bent and Clough, 1998) of different Arabidopsis ecotypes (Columbia, CS933; Rschew, CS913; Wassilewskija, CS915; and Landsberg erecta, CS20). Agrobacterium tumefaciens strain GV3101 was used in all transformations. T1 seeds were collected, and resistant seeds were selected on 1% (w/v) agar Murashige and Skoog (Murashige and Skoog, 1962) plates with 70 μL mL–1 gentamicin and 150 μL mL–1 carbenicillin. T1 seedlings were collected, allowed to self, and T2 seeds were collected and analyzed for luciferase activity.
All enhancer trap lines were screened for circadian regulated luciferase activity using the Packard TopCount luminometer as described by Salomé et al. (2002). Seeds were vapor phase sterilized (Bent and Clough, 1998) and plated on 1% (w/v) agar Murashige and Skoog media containing 70 μL mL–1 gentamicin. Seeds were stratified for 3 d in the dark at 4°C and then transferred into continuous 12 h of white light (70 μmol m–2 s–1)/12 h of dark cycle for 7 d at 22°C. Seedlings were transferred to black microtiter plates (Dynex Technologies, Inc., Chantilly, VA), containing 200 μL of 0.8% (w/v) Murashige and Skoog agar plus 2% (w/v) Suc and 35 μL of 0.5 mm luciferin (Biosynth, Switzerland) per well. Microtiter plates were covered with clear plastic TopSeal (Packard) in which holes were placed above each well for seedling gas exchange. Plates were moved to the Packard TopCount and interleaved with four clear plates to allow light to reach the seedlings. Seedlings were entrained in white light (15–35 μmol m–2 s–1) for 3 d with 12-h-light/12-h-dark cycles. Luciferase activity was measured every 1 h by integrating photons emitted by seedlings during a 10-s sampling period. Seedlings were imaged for a minimum of 4 d and a maximum of 8 d in continuous light conditions.
Data Analysis
For each seedling, 3 d of data were analyzed by FFT-NLLS (Plautz et al., 1997). All data were normalized to the average luciferase activity of the individual seedling and are presented as relative bioluminescence. Seedlings were determined to be rhythmic if their period was between 20 and 30 h, the peak signal strength exceeded 100 photons s–1 seedling–1 and the RAE, a measure of the strength of the rhythm, was <1.0, the limit of statistical significance for any given rhythmic amplitude (Michael and McClung, 2002). For all experiments, between 12 and 24 independent T2 lines were tested in a minimum of two independent experiments. Phase is given in CT (phase = 24 h × phase/period) to facilitate comparison of phase between rhythmic traces with different periods.
Characterization of Enhancer Trap Lines
To characterize possible enhancer-trapped promoters/genes, TAIL-PCR (Liu et al., 1995b) was used to clone flanking genomic Arabidopsis DNA from enhancer trap lines. Genomic DNA was isolated using the Nucleon Phytopure DNA extraction system (Amersham Biosciences, Piscataway, NJ). Vector-specific (pPZP) primers to both the right and left borders were utilized (Hajdukiewicz et al., 1994). Primary TAIL reactions were subjected to secondary PCR and directly sequenced. The flanking sequence was aligned to the Arabidopsis genome using BLAST.
CCRE Mapping in Arabidopsis Promoters
RSAs (van Helden et al., 2000) were utilized for extracting Arabidopsis promoters, searching for overrepresented oligonucleotides, and searching for CCREs. Arabidopsis promoters were defined as between –2,000 and –1 bp relative to the predicted translational start. Random Arabidopsis promoters (25,545, each 2,000 bp) were generated using the Markov chain process based on nucleotide frequencies of the complete set of Arabidopsis intergenic sequences with a oligonucleotide size of six (http://rsat.ulb.ac.be/rsat/). Expected values were determined by establishing the frequency of the oligonucleotide in the 25,545 random Arabidopsis promoters. Gene sets are defined as follows: Harmer circadian, genes defined as circadian regulated in continuous light by Harmer et al. (2000); Schaffer circadian, genes defined as circadian regulated in continuous light by Schaffer et al. (2001); Schaffer light induced, the subset of circadian genes defined by Schaffer et al. (2001) in which expression was higher in the day than in the night; dark induced, the subset of circadian genes defined by Schaffer et al. (2001) in which expression was higher in the night than in the day; and Schaffer diurnal, genes shown to oscillate in a light-dark cycle by Schaffer et al. (2001). Genes and motifs were mapped to Arabidopsis chromosomes and visualized using GeneSpring (Silicon Genetics, Redwood City, CA).
Supplementary Material
Acknowledgments
We thank Chris Langmead and Bob Gross for Arabidopsis promoter databases and Francisca Cerda for technical assistance with TAIL-PCR. We thank Mary Lou Guerinot and Patrice Salomé for helpful discussions and two anonymous reviewers for useful suggestions. We thank the Arabidopsis Biological Resource Center for all Arabidopsis accessions used in this study.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.021006.
This work was supported by the National Science Foundation (grant nos. IBN 9817603 and MCB 0091008) and by the U.S. Department of Agriculture (National Research Initiative Competitive Grants Program grant no. 9602632 to C.R.M.).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP (2002) Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12: 540–550 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Yanovsky MJ, Más P, Harmer SL, Kay SA (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr Biol 12: 757–761 [DOI] [PubMed] [Google Scholar]
- Aoki S, Kondo T, Ishiura M (2002) A promoter-trap vector for clock-controlled genes in the cyanobacterium Synechocystis sp. PCC 6803. J Microbiol Methods 49: 265–274 [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Shinohara ML, Dunlap JC, Loros JJ (1996) Circadian clock-controlled genes isolated from Neurospora crassa are late night- to early morning-specific. Proc Natl Acad Sci USA 93: 13096–13101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Clough SJ (1998) Agrobacterium germ-line transformation: transformation of Arabidopsis without tissue culture. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp B7, 1–14
- Benvenuto G, Formiggini F, Laflamme P, Malakhov M, Bowler C (2002) The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr Biol 12: 1529–1534 [DOI] [PubMed] [Google Scholar]
- Bertolino E, Singh H (2002) POU/TBP cooperativity: a mechanism for enhancer action from a distance. Cell 10: 397–407 [DOI] [PubMed] [Google Scholar]
- Brunaud V, Balzergue S, Dubreucq B, Aubourg S, Samson F, Chauvin S, Bechtold N, Cruaud C, DeRose R, Pelletier G (2002) T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep 3: 1152–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi L, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T (1999) Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J 17: 699–707 [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA (2002) Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci 22: 9305–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Puente P, Deng X-W, Wei N (1998) Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J 15: 69–77 [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW (2001) Circadian regulation of gene expression systems in the Drosophila head. Neuron 32: 657–671 [DOI] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC (2002) Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol 12: 551–557 [DOI] [PubMed] [Google Scholar]
- Etchegaray J-P, Lee C, Wade PA, Reppert SM (2003) Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421: 177–182 [DOI] [PubMed] [Google Scholar]
- Fagan T, Morse D, Hastings JW (1999) Circadian synthesis of a nuclear-encoded chloroplast glyceraldehyde-3-phosphate dehydrogenase in the dinoflagellate Gonyaulax polyedra is translationally controlled. Biochemistry 38: 7689–7695 [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Liu Y, Loros JJ, Dunlap JC (2002) White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297: 815–819 [DOI] [PubMed] [Google Scholar]
- Grundschober C, Delaunay F, Pühlhofer A, Triqueneaux G, Laudet V, Bartfai T, Nef P (2001) Circadian regulation of diverse gene products revealed by mRNA expression profiling of synchronized fibroblasts. J Biol Chem 276: 46751–46758 [DOI] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing R, M, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol 296: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Hwang S, Kawazoe R, Herrin DL (1996) Transcription of tufA and other chloroplast-encoded genes is controlled by a circadian clock in Chlamydomonas. Proc Natl Acad Sci USA 93: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Golden SS (1999) Circadian programs in cyanobacteria: adaptiveness and mechanism. Annu Rev Microbiol 53: 389–409 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Jester PJ, Monson S, Copenhaver G, Preuss D, Sussman MR (2002) Characterization of T-DNA insertion sites in Arabidopsis thaliana and the implications for saturation mutagenesis. Omics 6: 163–174 [DOI] [PubMed] [Google Scholar]
- Kyriacou CP, Rosato E (2000) Squaring up the E-box. J Biol Rhythms 15: 483–490 [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray J-P, Cagampang FRA, Loudon ASI, Reppert SM (2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107: 855–867 [DOI] [PubMed] [Google Scholar]
- Lee K, Loros JJ, Dunlap JC (2000) Interconnected feedback loops in the Neurospora circadian system. Science 289: 107–110 [DOI] [PubMed] [Google Scholar]
- Lillo C (1993) Light-induced circadian rhythms in NADP+-glyceraldehyde-3-phosphate dehydrogenase mRNA in corn seedlings. J Interdiscipl Cycle Res 24: 65–71 [Google Scholar]
- Lin Y, Han M, Shimada B, Wang L, Gibler TM, Amarakone A, Awad TA, Stormo GD, Van Gelder RN, Taghert PH (2002) Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA 99: 9562–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tsinoremas NF, Johnson CH, Golden SS, Ishiura M, Kondo T (1995a) Circadian orchestration of gene expression in cyanobacteria. Genes Dev 9: 1469–1478 [DOI] [PubMed] [Google Scholar]
- Liu Y-G, Mitsukawa N, Oosumi T, Whittier RF (1995b) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Matsushika A, Imamura A, Yamashino T, Mizuno T (2002a) Aberrant expression of the light-inducible and circadian-regulated APRR9 gene belonging to the circadian-associated APRR1/TOC1 quintet results in the phenotype of early flowering in Arabidopsis thaliana. Plant Cell Physiol 43: 833–843 [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Yamashino T, Mizuno T (2002b) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: II. Characterization with CCA1-overexpressing plants. Plant Cell Physiol 43: 118–122 [DOI] [PubMed] [Google Scholar]
- McClung CR, Salomé PA, Michael TP (2002) The Arabidopsis circadian system. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, pp 1–25 http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- McDonald MJ, Rosbash M (2001) Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107: 567–578 [DOI] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR (1995) The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci 20: 506–510 [DOI] [PubMed] [Google Scholar]
- Michael TP, McClung CR (2002) Phase-specific circadian clock regulatory elements in Arabidopsis thaliana. Plant Physiol 130: 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Kay SA (1991) Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis. Plant Cell 3: 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua N-H, Kay SA (1995) The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267: 1163–1166 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song H-R, Carré IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Mori T, Johnson CH (2001) Circadian programming in cyanobacteria. Semin Cell Dev Biol 12: 271–278 [DOI] [PubMed] [Google Scholar]
- Mori T, Saveliev SV, Xu Y, Stafford WF, Cox MM, Inman RB, Johnson CH (2002) Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci USA 99: 17203–17208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz E, Brewer M, Baler R (2002) Circadian transcription: thinking outside the E-box. J Biol Chem 277: 36009–36017 [DOI] [PubMed] [Google Scholar]
- Murashige TR, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JG (2002a) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320 [DOI] [PubMed] [Google Scholar]
- Panda S, Poirier GC, Kay SA (2002b) tej defines a role for poly(ADP Ribosyl) ation in establishing period length of the Arabidopsis circadian oscillator. Dev Cell 3: 51–61 [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW (1996) Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J 15: 3732–3743 [PMC free article] [PubMed] [Google Scholar]
- Rutter J, Reick M, McKnight SL (2002) Metabolism and the control of circadian rhythms. Annu Rev Biochem 71: 307–332 [DOI] [PubMed] [Google Scholar]
- Salomé PA, Michael TP, Kearns EV, Fett-Neto AG, Sharrock RA, McClung CR (2002) The out of phase 1 (oop1) mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol 129: 1674–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador ML, Klein U, Bogorad L (1993) Light-regulated and endogenous fluctuations of chloroplast transcript levels in Chlamydomonas: regulation by transcription and RNA degradation. Plant J 3: 213–219 [DOI] [PubMed] [Google Scholar]
- Salvador ML, Klein U, Bogorad L (1998) Endogenous fluctuations of DNA topology in the chloroplast of Chlamydomonas reinhardtii. Mol Cell Biol 18: 7235–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato E, Nakamichi N, Yamashino T, Mizuno T (2002) Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol 43: 1374–1385 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré IA, Coupland G (1998) LATE ELONGATED HYPOCOTYL, an Arabidopsis gene encoding a MYB transcription factor, regulates circadian rhythmicity and photoperiodic responses. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schindler U, Beckmann H, Cashmore AR (1992) TGA1 and G-box binding factors: two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell 4: 1309–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara ML, Loros JJ, Dunlap JC (1998) Glyceraldehyde-3-phosphate dehydrogenase is regulated on a daily basis by the circadian clock. J Biol Chem 273: 446–452 [DOI] [PubMed] [Google Scholar]
- Springer PS (2000) Gene traps: tools for plant development and genomics. Plant Cell 12: 1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stempfl T, Vogel M, Szabo G, Wülbeck C, Liu J, Hall JC, Stanewsky R (2002) Identification of circadian-clock-regulated enhancers and genes of Drosophila melanogaster by transposon mobilization and luciferase reporting of cyclical gene expression. Genetics 160: 571–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ (2002) Extensive and divergent circadian gene expression in liver and heart. Nature 417: 78–82 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Sundarasan VA, Springer P, Volpe T, Haward S, Jones JDG, Dean C, Martienssen R (1995) Patterns of gene action revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Thain SC, Murtas G, Lynn JR, McGrath RB, Millar AJ (2002) The circadian clock that controls gene expression in Arabidopsis is tissue specific. Plant Physiol 130: 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S (2002) Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem 277: 14048–14052 [DOI] [PubMed] [Google Scholar]
- van Helden J, Andre B, Collado-Vides J (2000) A web site for the computational analysis of yeast regulatory sequences. Yeast 16: 177–187 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW, Kay SA (2001) Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2: 702–715 [DOI] [PubMed] [Google Scholar]
- Zhong HH, McClung CR (1996) The circadian clock gates expression of two Arabidopsis catalase genes to distinct and opposite circadian phases. Mol Gen Genet 251: 196–203 [DOI] [PubMed] [Google Scholar]
- Zhong HH, Resnick AS, Straume M, McClung CR (1997) Effects of synergistic signaling by phytochrome A and cryptochrome 1 on circadian clock-regulated catalase expression. Plant Cell 9: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.