Abstract
The basic/helix-loop-helix (bHLH) proteins are a superfamily of transcription factors that bind as dimers to specific DNA target sites and that have been well characterized in nonplant eukaryotes as important regulatory components in diverse biological processes. Based on evidence that the bHLH protein PIF3 is a direct phytochrome reaction partner in the photoreceptor's signaling network, we have undertaken a comprehensive computational analysis of the Arabidopsis genome sequence databases to define the scope and features of the bHLH family. Using a set of criteria derived from a previously defined consensus motif, we identified 147 bHLH protein–encoding genes, making this one of the largest transcription factor families in Arabidopsis. Phylogenetic analysis of the bHLH domain sequences permits classification of these genes into 21 subfamilies. The evolutionary and potential functional relationships implied by this analysis are supported by other criteria, including the chromosomal distribution of these genes relative to duplicated genome segments, the conservation of variant exon/intron structural patterns, and the predicted DNA binding activities within subfamilies. Considerable diversity in DNA binding site specificity among family members is predicted, and marked divergence in protein sequence outside of the conserved bHLH domain is observed. Together with the established propensity of bHLH factors to engage in varying degrees of homodimerization and heterodimerization, these observations suggest that the Arabidopsis bHLH proteins have the potential to participate in an extensive set of combinatorial interactions, endowing them with the capacity to be involved in the regulation of a multiplicity of transcriptional programs. We provide evidence from yeast two-hybrid and in vitro binding assays that two related phytochrome-interacting members in the Arabidopsis family, PIF3 and PIF4, can form both homodimers and heterodimers and that all three dimeric configurations can bind specifically to the G-box DNA sequence motif CACGTG. These data are consistent, in principle, with the operation of this combinatorial mechanism in Arabidopsis.
INTRODUCTION
The basic/helix-loop-helix (bHLH) proteins are a superfamily of transcription factors that have been well characterized in nonplant eukaryotes, especially in mammalian systems, in which considerable structural, functional, and phylogenetic analyses have been performed (Atchley and Fitch, 1997; Littlewood and Evan, 1998; Ledent and Vervoort, 2001). The data indicate that bHLH proteins are important regulatory components in transcriptional networks in these systems, controlling a diversity of processes from cell proliferation to cell lineage establishment (Grandori et al., 2000; Massari and Murre, 2000).
This family is defined by the bHLH signature domain, which consists of ∼60 amino acids with two functionally distinct regions. The basic region, located at the N-terminal end of the domain, is involved in DNA binding and consists of ∼15 amino acids with a high number of basic residues. The HLH region, at the C-terminal end, functions as a dimerization domain (Murre et al., 1989; Ferre-D'Amare et al., 1994) and is constituted mainly of hydrophobic residues that form two amphipathic α-helices separated by a loop region of variable sequence and length (Nair and Burley, 2000). Outside of the conserved bHLH domain, these proteins exhibit considerable sequence divergence (Atchley et al., 1999). Cocrystal structural analysis has shown that the interaction between the HLH regions of two separate polypeptides leads to the formation of homodimers and/or heterodimers and that the basic region of each partner binds to half of the DNA recognition sequence (Ma et al., 1994; Shimizu et al., 1997). Some bHLH proteins form homodimers or restrict their heterodimerization activity to closely related members of the family. On the other hand, some can form heterodimers with one or several different partners (Littlewood and Evan, 1998).
The core DNA sequence motif recognized by the bHLH proteins is a consensus hexanucleotide sequence known as the E-box (5′-CANNTG-3′). There are different types of E-boxes, depending on the identity of the two central bases. One of the most common is the palindromic G-box (5′-CACGTG-3′). Certain conserved amino acids within the basic region of the protein provide recognition of the core consensus site, whereas other residues in the domain dictate specificity for a given type of E-box (Robinson et al., 2000). In addition, flanking nucleotides outside of the hexanucleotide core have been shown to play a role in binding specificity (Littlewood and Evan, 1998; Atchley et al., 1999; Massari and Murre, 2000), and there is evidence that a loop residue in the protein plays a role in DNA binding through elements that lie outside of the core recognition sequence (Nair and Burley, 2000).
In animal systems, bHLH proteins have been classified into six main groups (designated A to F) that reflect their evolutionary origin and sequence relatedness as well as the information available on their DNA binding specificities and functional activities (Dang et al., 1992; Atchley and Fitch, 1997; Ledent and Vervoort, 2001). In brief, group-A proteins bind to the E-box variant CAGCTG and include proteins such as MyoD, Twist, Acheate-Scute, Hen, Atonal, and Delilah. Group B includes a large number of functionally unrelated proteins such as Pho4 and R that bind to the G-box (CACGTG). A subclass in group B is represented by the bHLH-Leu zipper proteins exemplified by Myc, Mad, USF, and SREBP. Group C is formed by bHLH proteins that have a second protein–protein interaction domain, the PAS domain, and that bind to non-E-box (NACGTG or NGCGTG) core sequences. Examples of proteins included in this group are Per, Arnt, and Sim. Group D encompasses the HLH proteins (represented by Id, Emc, and Heira), which lack the basic DNA binding domain. Group E (previously considered part of group B by Atchley and Fitch [1997]) is formed by WRPW-bHLH proteins such as Hairy and Enhancer of Split (Ledent and Vervoort, 2001) that preferentially bind to N-boxes (CACGGC or CACGAC), have only low affinity for E-boxes, and possess a Pro instead of an Arg residue at a crucial position in the bHLH domain (Fisher and Caudy, 1998). Group F is formed by COE-bHLH proteins that have an additional domain involved in dimerization and DNA binding and that are divergent in sequence from the other groups described (Crozatier et al., 1996; Fisher and Caudy, 1998; Ledent and Vervoort, 2001).
Dimerization and the recognition of different E-boxes are believed to provide mechanisms by which bHLH proteins generate sufficient diversity to regulate a variety of different transcriptional programs (Fairman et al., 1993). In this context, the HLH proteins can function as negative regulators of bHLH proteins by forming non-DNA binding heterodimers with otherwise DNA binding bHLH proteins (Littlewood and Evan, 1998).
In plants, the R gene product Lc, which is involved in the control of anthocyanin synthesis in maize, was the first plant protein reported to possess a bHLH motif (Ludwig et al., 1989). However, only a few plant bHLH proteins have been studied to date, and the family remains largely uncharacterized in terms of the identification of its members and the biological processes they control. The relevance of bHLH proteins to our specific research interest, phytochrome-regulated light signaling pathways, was established with the identification of PHYTOCHROME INTERACTING FACTOR3 (PIF3). PIF3 is a bHLH protein identified in a yeast two-hybrid screen for potential phytochrome signaling partners (Ni et al., 1998). Molecular characterization of PIF3 demonstrated that it is a G-box binding bHLH protein that interacts preferentially with the active form of phytochrome and is involved in controlling the expression of light-regulated genes such as CCA1 and LHY (Martinez-Garcia et al., 2000). Given the potential for bHLH proteins to diversify the control of gene expression by the formation of a spectrum of different homodimer and heterodimer combinations, coupled with the recognition of a range of different types of DNA sequence motifs, it was of considerable interest to us to characterize the Arabidopsis bHLH (AtbHLH) protein family.
During the sequencing of the Arabidopsis genome, it became apparent from sequence similarity searches of the growing databases that this genome contains a large number of bHLH-encoding genes. An estimate published at the time of the completion of the genome sequence indicated the existence of 139 such genes (Riechmann et al., 2000). To more precisely determine the extent of the bHLH family in Arabidopsis, we have systematically analyzed candidate genes in the fully sequenced genome using a set of minimal criteria to define the signature bHLH domain. By this process, we have identified 147 bHLH-encoding genes. A recent report published since the completion of our analysis has identified 133 Arabidopsis bHLH-encoding genes (Heim et al., 2003). Here, we explore the phylogenetic relationships among the encoded proteins and those from other organisms, examine the chromosomal distribution and diversity in gene structure in the bHLH domain of these genes, and consider the structural and functional activities predicted from the encoded sequences. We also experimentally test the predicted DNA binding activity and heterodimerization potential of two related members of the family, PIF3 and PIF4 (Huq and Quail, 2002), that are involved in phytochrome signaling to test the hypothesis that heterodimeric interactions between the members of the family may provide a combinatorial mechanism for the control of multiple transcriptional pathways in plants, similar to that proposed for other organisms (Grandori et al., 2000; Quail, 2000; Levens, 2003).
RESULTS
The AtbHLH Protein Family Consists of at Least 147 Members
To provide criteria for defining a bHLH protein, we referred to the studies of the amino acid sequence distribution within the bHLH domain performed by Atchley et al. (1999). In brief, these authors analyzed the occurrence of amino acids at individual positions in the bHLH domain for 392 bHLH proteins. Based on patterns of sequence conservation, a hypothetical consensus motif that includes 19 amino acids dispersed across the bHLH domain was generated: 18 amino acids from the basic and helix regions and 1 from the loop (Table 1).
Table 1.
bHLH Domain Consensus Motif
| Position in the Alignment
|
Region | Consensus Motif Amino Acid Frequency within the bHLH Domaina (Atchley et al., 1999) |
Amino Acid Frequency within the Arabidopsis bHLH Domainsb (This Study) |
|
|---|---|---|---|---|
| Atchley et al. (1999) |
This study |
|||
| 1 | 1 | Basic | K (27%), R (61%) | K (22%), R (24%), other (53%) |
| 2 | 2 | Basic | K (16%), R (77%) | K (7%), R (35%), other (58%) |
| 9 | 13 | Basic | E (93%) | E (76%), A (10%), other (14%) |
| 10 | 14 | Basic | R (81%), K (14%) | R (74%), K (14%), other (12%) |
| 12 | 16 | Basic | R (91%) | R (91%), other (9%) |
| 16 | 20 | Helix 1 | I (35%), L (33%), V (23%) | I (52%), L (27%), M (12%), V (3%), other (6%) |
| 17 | 21 | Helix 1 | N (74%) | N (51%), S (19%), other (30%) |
| 20 | 24 | Helix 1 | F (72%), I (9%), L (14%) | F (26%), I (14%), L (26%), M (20%), other (15%) |
| 23 | 27 | Helix 1 | L (98%) | L (100%) |
| 24 | 28 | Helix 1 | K (35%), R (44%) | K (4%), R (35%), Q (42%), G (4%), other (15%) |
| 47 | 39 | Loop | K (58%), R (24%) | K (66%), R (7%), other (27%) |
| 50 | 42 | Helix 2 | K (93%) | K (45%), T (13%), other (42%) |
| 53 | 45 | Helix 2 | I (74%), T (15%), V(7%) | I (27%), T (4%), V (16%), L (14%), M (33%), other (6%) |
| 54 | 46 | Helix 2 | L (98%) | L (76%), V (14%), other (12%) |
| 57 | 49 | Helix 2 | A (76%) | A (60%), I (16%), V (12%), T (9%), other (3%) |
| 58 | 50 | Helix 2 | I (31%), T (23%), V (27%) | I (63%), T (2%), V (22%), other (13%) |
| 60 | 52 | Helix 2 | Y (77%) | Y (78%), other (22%) |
| 61 | 53 | Helix 2 | I (69%), L (16%), V (8%) | I (40%), L (13%), V (33%), other (14%) |
| 64 | 56 | Helix 2 | L (80%), M (7%) | L (93%), M (1%), other (6%) |
Amino acids (one-letter code) and positions within the bHLH domain used to define the members of the AtbHLH protein family. The conserved amino acids that define the motif are those reported by Atchley et al. (1999). The original position for each conserved residue based on sequence alignments by Atchley et al. (1999) and the corresponding position for our multiple sequence alignments are indicated in the first two columns, respectively. In the last two columns, the frequencies of the residues at each position reported by Atchley et al. (1999) and those found in this study for the AtbHLH proteins are compared. Boldface letters indicate residues in the AtbHLH proteins that differ from the consensus motif but that have a representation in the group of at least 10%.
Percentages refer to the 392 bHLH proteins analyzed by Atchley et al. (1999).
Percentages refer to the 147 Arabidopsis bHLH proteins identified in this study.
Initially, we performed multiple BLAST (Basic Local Alignment Search Tool) searches of the Arabidopsis databases using the bHLH domain (58 amino acids) of PIF3 as our query sequence and obtained a large number of protein hits (see Methods). We identified the unique hits and removed duplications from our data set caused by the multiple identification numbers frequently assigned to the same DNA or protein sequence in the databases. The procedure was repeated several times as the genome sequence was being completed and updated versions became available. The last database search to confirm the data included in this work used the August 2002 version of the genome sequence.
Frequent apparent misannotations were encountered, often because of the presence of multiple introns in the bHLH domain. The gene structure for each bHLH domain was assessed using the program NetGene2 and by comparison with transcript sequence where available. The protein sequences were corrected, when appropriate, and used in this analysis.
Having identified nonredundant and verified protein sequences, we developed a set of criteria to objectively define those sequences to be considered bona fide bHLH proteins as follows. To select from the initial hits obtained, we used the Atchley et al. (1999) bHLH consensus motif, representing the most conserved amino acids in the bHLH region (Table 1). This motif allows some sequence divergence, represented as mismatches from the consensus. The most divergent class defined by Atchley et al. (1999) had up to 7 mismatches from the motif, including an average of 3.4 mismatches in the basic region alone and 3.9 mismatches in the rest of the motif.
To define the conserved amino acids and select the putative bHLH proteins in Arabidopsis, we conducted multiple protein sequence alignments using Multalin (Corpet, 1988) (see Methods). We calculated manually the number of matches and mismatches from the predicted motif for each protein. A match was scored if the residue present in the Arabidopsis sequence was the same as any of those at that position listed by Atchley et al. (1999). The frequencies of the consensus amino acids within the bHLH domains are shown in Table 1. We defined bHLH proteins here as those that had up to 9 mismatches compared with the conserved 19 amino acids constituting the motif described by Atchley et al. (1999). This criterion was used because of the inherent divergence of the consensus motif and the fact that, in our case, proteins with 8 and 9 mismatches had an average of 3.4 and 4.1 mismatches, respectively, in the basic region, and the mismatches within the HLH part of the protein corresponded either to conservative changes within the AtbHLH proteins or were in positions with higher variation within the motif. Proteins with more than nine mismatches had many of the mismatches in the more conserved HLH region and were not included in our analysis.
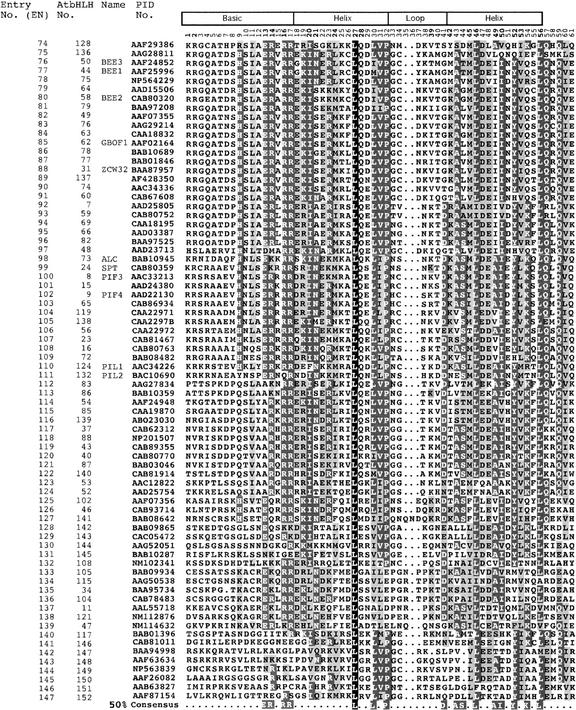
Based on these criteria, we identified 147 proteins as members of the bHLH family in Arabidopsis. Compared with the recent report by Heim et al. (2003), we identified an additional 19 bHLH protein–encoding genes in the present study. Therefore, the combined total number of Arabidopsis bHLH genes from the two studies should be 152. However, the three sequences designated AtbHLH127 (At4g28815), AtbHLH131 (At4g38071), and AtbHLH133(At1g20095) by Heim et al. (2003) do not appear to correspond to bHLH proteins, and the two sequences designated AtbHLH109 (At1g68240) and AtbHLH84 (At2g14760) contain more than nine mismatches and therefore do not conform to our minimal criteria for inclusion. Therefore, these five sequences are not included in our total estimate or in the various analyses performed here. The complete multiple sequence alignment of the bHLH domains of these 147 proteins is shown in Figure 1.
Figure 1.
Multiple Sequence Alignment of the bHLH Domains of the 147 Members of the AtbHLH Protein Family.
Each protein is identified by its PID number and AtbHLH number (Heim et al., 2003). The EN assigned in this study is based on the order in which the proteins are shown in this alignment. The scheme at top depicts the locations and boundaries of the basic, helix, and loop regions within the bHLH domain. The numbers below the scheme (1 to 61) indicate the position within the bHLH motif as defined in this study. For those proteins for which a name has been given, the name is provided after the PID number. The shading of the alignment presents identical residues in black, conserved residues in dark gray, and similar residues in light gray. Dots denote gaps. The Arabidopsis consensus motif at bottom is based on the residues with 50% conservation among the 147 proteins shown.
An information summary including Atg number, protein identification (PID) number, name given (for those bHLHs that have been identified by various researchers), GenBank accession number, chromosome location, and BAC/clone coordinates for each of these proteins is provided in Table 2. For convenience, we have assigned each bHLH protein an “entry number” (EN) in the various tables and figures, representing the order of these sequences from top to bottom in the multiple sequence alignment shown in Figure 1. In accord with the report by Heim et al. (2003), we also included the AtbHLH numbers assigned by those authors. Because we have identified 19 additional members of the family beyond those reported by Heim et al. (2003), we assigned AtbHLH numbers to these new members (AtbHLH 134 to 152) in the order of increasing EN in Figure 1. For the purpose of the analysis here, we refer to each protein by its EN (to aid ease of location in the tables and figures) as well as by PID and/or AtbHLH number, all of which are provided in Figure 1 and Table 2.
Table 2.
Summary of Information on the Arabidopsis bHLH Proteins
| EN | AtbHLH Numbera |
Atg Number | PID Number Used |
Most Recent PID Number |
Name | Chromosome | Map Position (Mb) |
GenBank Accession Number |
BAC/Clone Coordinates |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 55 | At1g12540 | AAF79643 | AAF88076 | 1 | 4.2 | AC025416 | F5O11.28/102605-103305 | |
| 2 | 125 | At1g62975 | AAF75809 | NP_683462 | 1 | 22.8 | AC011000 | F16P17/55268-56889 | |
| 3 | 126 | At4g25410 | NM118673 | 4 | 11.9 | AL079350 | T30C3/35130-36041 | ||
| 4 | 120 | At5g51790b | NP199992 | 5 | 20.6 | AB010074 | MI024/43665-44497 | ||
| 5 | 118 | At4g25400 | NM118672 | 4 | 11.9 | AL161563 | T30C3/0653-31693 | ||
| 6 | 36 | At5g51780 | NM124557 | 5 | 20.6 | AB010074 | MI024.9/38782-40349 | ||
| 7 | 100 | At2g41240 | AAC78547 | NP_181657 | 2 | 17.1 | AC005662 | T3K9/2154-26143 | |
| 8 | 38 | At3g56970 | CAB72167 | AAM10940 | 3 | 21 | AL138655 | F24I3.60/12214-13104 | |
| 9 | 39 | At3g56980 | CAB72168 | AAM10941 | 3 | 20.9 | AL138635 | T47758/F24I3.60/14622-15504 | |
| 10 | 101 | At5g04150 | NM120497 | 5 | 1.1 | AL391716 | F21E1/42106-43023 | ||
| 11 | 67 | At3g61950 | CAB71902 | NP_567121 | 3 | 22.9 | AL138642 | F21F14/102333-104004 | |
| 12 | 57 | At4g01460 | CAB77716 | NP_192055 | 4 | 0.6 | AL161492 | F11O4.13/43899-45262 | |
| 13 | 70 | At2g46810b | AAC33499 | NP_182204 | 2 | 19.2 | AC005310 | F19D11/39254-41933 | |
| 14 | 97 | At3g24140 | BAB01355 | NP_189056 | 3 | 8.7 | AB028621 | MUJ8/13023-14413 | |
| 15 | 96 | At1g72210 | AAG51804 | NP_177366 | 1 | 26.8 | AC067754 | T9N14.4/F14J22/10438-12278 | |
| 16 | 94 | At1g22490 | NM102098 | 1 | 7.9 | AC006551 | F12K8/67443-69736 | ||
| 17 | 71 | At5g46690 | NM124039 | 5 | 18.5 | AB016882 | MZA15/27590-29492 | ||
| 18 | 99 | At5g65320 | BAB11554 | NP_201335 | 5 | 25.8 | AB011479 | MNA5/19267-20539 | |
| 19 | 98 | At5g53210 | BAB09783 | NP_200133 | 5 | 21.3 | AB013388 | K19E1.1/795-3130 | |
| 20 | 45 | At3g06120 | AAF30305 | NP_187263 | 3 | 1.8 | AC018907 | F28L1.6/12214-13104 | |
| 21 | 95 | At1g49770 | AAG13058 | NP_175399 | 1 | 18 | AC011807 | F14J22.2/10438-12278 | |
| 22 | 92 | At5g43650 | BAB11628 | NP_199178 | 5 | 17.2 | AB016875 | K9D7.15/57218-58666 | |
| 23 | 10 | At2g31220 | AAD20667 | NP_180680 | 2 | 13.2 | AC006593 | F16D14/21932-23451 | |
| 24 | 89 | At1g06170 | AAF80214 | NP_172107 | 1 | 1.9 | AC025290 | F9P14.3/10177-11595 | |
| 25 | 91 | At2g31210 | AAD20666 | NP_180679 | 2 | 13.2 | AC006593 | F16D14.5/15445-16739 | |
| 26 | 19 | At2g22760 | AAC63587 | NP_179861 | 2 | 9.6 | AC005617 | T30L20/9335-10327 | |
| 27 | 20 | At2g22770 | AAC63588 | NP_179862 | 2 | 9.6 | AC005617 | T30L20/16181-17536 | |
| 28 | 18 | At2g22750 | AAC63586 | NP_179860 | 2 | 9.6 | AC005617 | T30L20/3468-4711 | |
| 29 | 25 | At4g37850 | CAB38933 | NP_195498 | 4 | 16.8 | AL035709 | T28I19/95156-96198 | |
| 30 | 2 | At1g63650 | AAB72192 | NP_176552 | EGL1 | 1 | 23.2 | AF013465 | F24D7/160-2994 |
| 31 | 1 | At5g41315 | BAB08503 | NP_680372 | GL3 | 5 | 15.8 | AB006707 | MYC6/10271-13680 |
| 32 | 42 | At4g09820 | CAC14865 | NP_192720 | TT8 | 4 | 5.1 | AL049482 | F17A8.170/45-1601 |
| 33 | 14 | At4g00870 | AAB62853 | NP_56719 | 4 | 0.3 | AF013294 | TM018A10.7/64238-65647 | |
| 34 | 3 | At4g16430 | CAB78685 | NP_193376 | 4 | 8.2 | AL161544 | ATAFAC6/10029-11432 | |
| 35 | 17 | At2g46510 | AAD20162 | NP_566078 | 2 | 19 | AC006418 | MHK10/14876-17530 | |
| 36 | 5 | At5g46760 | BAB08920 | NP_199488 | ATR2 | 5 | 18.6 | AB016882 | MZA15.18/56206-57984 |
| 37 | 4 | At4g17880 | CAA17131 | NP_193522 | AtMYC4 | 4 | 9.3 | AL021889 | T6K21.60/17670-19439 |
| 38 | 6 | At1g32640 | BAA25078 | NP_174541 | AtMYC2 | 1 | 11.8 | AB000875 | F6N18.4/31-1902 |
| 39 | 13 | At1g01260 | AAF97322 | NP_171634 | Myc7E | 1 | 0.1 | AC023628 | F6F3.7/50503-52275 |
| 40 | 28 | At5g46830 | BAA97217 | NP_199495 | 5 | 18.7 | AB022221 | MSD23/4699-6234 | |
| 41 | 35 | At5g57150 | BAA97365 | NP_568850 | 5 | 22.8 | AB023042 | MUL3.10/58409-59341 | |
| 42 | 27 | At4g29930 | CAB43668 | NP_194722 | 4 | 13.6 | AL050352 | F27B13.170/68686-70083 | |
| 43 | 29 | At2g28160 | AAC98450 | NP_180383 | 2 | 11.9 | AC005851 | F24D13/72083-73201 | |
| 44c | 33 | At1g12860 | AAF78492 | NP_172746 | 1 | 4.3 | AC012187 | F13K23.12/31576-33210 | |
| 45c | 116 | At3g26744 | NM113586 | ICE1 | 3 | 9.8 | AP000602 | NMDJ14/1-1257 | |
| 46 | 61 | At5g10570 | CAB89386 | NP_179283 | 5 | 3.3 | AL353995 | F12B17/21984-23504 | |
| 47 | 93 | At5g65640 | NM125962 | NP_569014 | 5 | 26 | AB026639 | K21L13.6/53440-55207 | |
| 48 | 21 | At2g16910 | AAC64222 | AMS | 2 | 7.2 | AC005167 | F12A24.3/39374-41285 | |
| 49 | 22 | At4g21330 | CAB79132 | 4 | 10.3 | AL161554 | T6K22.60/161501-162273 | ||
| 50 | 90 | At1g10610 | AAD39586 | 1 | 3.5 | AC007067 | T10O24.26/87549-89690 | ||
| 51 | 41 | At5g56960 | BAA97026 | NP_200506 | 5 | 22.7 | AB024035 | F24I3/25154-26143 | |
| 52 | 134 | At4g38070 | CAB80472 | NP_195520 | 4 | 16.8 | AL161592 | F20D10.19/178968-185002 | |
| 53 | 30 | At1g68810 | AAF07352 | NP_564944 | 1 | 25.5 | AC011665 | F14K14.8/2929-4522 | |
| 54 | 32 | At3g25710 | BAA95758 | NP_189199 | 3 | 9.3 | AB028607 | K13N2/6294-7792 | |
| 55 | 107 | At3g56770 | CAC00740 | NP_191236 | 3 | 21 | AL390921 | T51265/45125-46541 | |
| 56 | 106 | At2g41130 | AAD11998 | NP_181646 | 2 | 17.4 | AC004261 | T3K9/21984-23504 | |
| 57 | 51 | At2g40200 | AAD25935 | NP_181549 | 2 | 16.7 | AC018721 | T7M7.8/36136-37065 | |
| 58 | 12 | At4g00480 | BAA11933 | NP_191957 | AtMYC1 | 4 | 0.2 | AL161472 | F6N23/850-3428 |
| 59 | 110 | At1g27660 | AAF24944 | NP_174087 | 1 | 9.6 | AC012375 | T22C5.11/33684-37649 | |
| 60 | 68 | At4g29100 | CAB79668 | NP_194639 | 4 | 13.3 | AL161574 | F19B15/69220-72655 | |
| 61 | 113 | At3g19500 | BAA99700 | NP_566639 | 3 | 6.7 | AB025624 | MLD14.24/72515-74119 | |
| 62 | 103 | At4g21340 | CAA20199 | NP_193865 | 4 | 10.3 | AL031187 | T24B6/164643-165806 | |
| 63 | 123 | At3g20640 | BAB02240 | NP_188700 | 3 | 7.2 | AP002034 | F3H11.2/4659-7561 | |
| 64 | 112 | At1g61660 | AAD21412 | NP_564782 | 1 | 22.4 | AC005882 | T13M11.1/5898-7895 | |
| 65 | 114 | At4g05170 | CAB81059 | NP_192426 | 4 | 2.6 | AL161503 | NM_116756/6443-7820 | |
| 66 | 111 | At1g31050 | AAF98179 | 1 | 11 | AC000107 | F17F8.26/11084-13337 | ||
| 67 | 135 | At1g74500b | AAF15922 | NP_177590 | 1 | 27.7 | AC011765 | F1M20/20273-21661? | |
| 68 | 26 | At1g02340 | AAK15282 | NP_563650 | HFR1 | 1 | 0.45 | AF324245 | T6A9.34/469818-470693 |
| 69 | 130 | At2g42280 | AAB88652 | NP-181757 | 2 | 17.5 | AC002561 | T24P15.19/52239-53974 | |
| 70 | 122 | At1g51140 | AAG50543 | NP_564583 | 1 | 18.5 | AC079828 | F23H24.3/9727-1155 | |
| 71 | 80 | At1g35460 | AAG12608 | NP_174776 | 1 | 13.2 | AC023064 | F12A4/26892-28707 | |
| 72 | 81 | At4g09180 | CAB78042 | NP_192657 | 4 | 4.8 | AL161514 | T8A17/14666-16092 | |
| 73 | 129 | At2g43140 | AAC64303 | NP_181843 | 2 | 17.8 | AC004450 | F14B2/22038-25301 | |
| 74 | 128 | At1g05805 | AAF29386 | NP_563749 | 1 | 1.7 | AC009999 | T20M3/23344-25799 | |
| 75 | 136 | AAG28811 | AAF48607 | 1 | 8.8 | AC079374 | F4F7.28/24886-26134 | ||
| 76 | 50 | At1g73830 | AAF24852 | NP_177524 | BEE3 | 1 | 27.4 | AC012679 | F25P22.25/87971-89290 |
| 77 | 44 | At1g18400 | AAF25996 | AAL38882 | BEE1 | 1 | 6.3 | AC013354 | F4F7.28/24886-26134 |
| 78 | 75 | At1g25330 | NP_564229 | NP_563839 | 1 | 8.8 | AC084785 | F4F7.28/81-752 | |
| 79 | 64 | At2g18300 | AAD15506 | NP_565434 | 2 | 7.9 | AC006439 | T30D6/30391-31792 | |
| 80 | 58 | At4g36540 | CAB80320 | AAK96779 | BEE2 | 4 | 16.2 | AL161589 | ATAP22/124723-126274 |
| 81 | 79 | At5g62610 | BAA97208 | NP_201067 | 5 | 24.4 | AB020751 | MRG21.2/2904-4387 | |
| 82 | 49 | At1g68920 | AAF07355 | NP_177058 | 1 | 25.5 | AC011665 | T6L1.10/57260-59315 | |
| 83 | 76 | At1g26260 | AAG29214 | NP_173950 | 1 | 9.04 | AC079829 | F28B23/28697-30518 | |
| 84 | 63 | At4g34530 | CAA18832 | NP_195179 | 4 | 15.4 | AL023094 | T4L20/34476-35872 | |
| 85 | 62 | At3g07340 | AAF02164 | NP_187390 | GBOF1 | 3 | 2.3 | AC009853 | F21O3/BAC T8C13 15938-18038 |
| 86 | 78 | At5g48560 | BAB10689 | NP_199667 | 5 | 18.9 | AB015468 | K15N18.2/5281-7992 | |
| 87 | 77 | At3g23690 | BAB01846 | NP_189011 | 3 | 8.5 | AP000377 | MYM9.3/2955-4677 | |
| 88 | 31 | At1g59640 | BAA87957 | NP_683448 | ZCW32 | 1 | 21.5 | AB028232 | ZCW32/RNA/T30E16.21 |
| 89 | 137 | At5g50915 | AF428350 | NP_568745 | 5 | 20 | AB017063 | K3K7/14232-13838 | |
| 90 | 74 | At1g10120 | AAC34336 | NP_172483 | 1 | 3.3 | AC004122 | T27I1.15/49647-51298 | |
| 91 | 60 | At3g57800 | CAB67608 | AAM10949 | 3 | 21.4 | AL132977 | T10K17.10/3113-6400 | |
| 92 | 7 | At1g03040 | AAD25805 | NP_563672 | 1 | 0.7 | AC006550 | F10O3/59042-61220 | |
| 93 | 59 | At4g02590 | CAB80752 | NP_567245 | 4 | 1.1 | AL161494 | T10P11.13/173619-175957 | |
| 94 | 69 | At4g30980 | CAA18195 | NP_194827 | 4 | 14 | AL022198 | F6I18/40330-42618 | |
| 95 | 66 | At2g24260 | AAD03387 | NP_180003 | 2 | 10.2 | AC005967 | F27D4 (cDNA)/75522-78053 | |
| 96 | 82 | At5g58010 | BAA97525 | NP_200609 | 5 | 23.1 | AB026635 | F2C19.2/757-1976 | |
| 97 | 48 | At2g42300 | AAD23713 | NP_181759 | 2 | 17.5 | AC005956 | MHK10/6734-8744 | |
| 98 | 73 | At5g67110 | BAB10945 | NP_201512 | ALCATRAZ | 5 | 26.5 | AB020742 | K21H1.7/26828-27834 |
| 99 | 24 | At4g36930 | CAB80359 | NP_568010 | SPATULA | 4 | 16.4 | AL161590 | AP22.25/23-1144 |
| 100 | 8 | At1g09530 | AAC33213 | NP_172424 | PIF3 | 1 | 3 | AAC95156 | F14J9/58196-61045 |
| 101 | 15 | At2g20180 | AAD24380 | NP_179608 | 2 | 8.6 | AC006224 | T2G17.2/88309-90528 | |
| 102 | 9 | At2g43010 | AAD22130 | AAL55716 | PIF4 | 2 | 17.8 | AC006224 | MLF.18/T18B20 (cDNA) |
| 103 | 65 | At3g59060 | CAB86934 | AAM10954 | 3 | 21.8 | AL163527 | F17J16/(cDNA)/41420-43126 | |
| 104 | 119 | CAA22971-1 | –d | 4 | 13.2 | AL035353 | F16A16.80/26792-30927 | ||
| 105 | 138 | CAA22971-2 | – | 4 | 13.2 | F16A16.90/32379-34363 | |||
| 106 | 56 | At4g28800b | CAA22972 | – | 4 | 13.2 | AL161573 | F16A16.90/32379-34363 | |
| 107 | 23 | At4g28790 | CAB81467 | NP_194608 | 4 | 13.2 | AL161573 | F16A16.100/139063-140907 | |
| 108 | 16 | At4g00050 | CAB80763 | AAM20933 | 4 | 0.006 | AL161471 | F6N15.11/16894-18848 | |
| 109 | 72 | At5g61270 | BAB08482 | – | 5 | 24.3 | AB010073 | MFB13.4 (cDNA)/9431-10589 | |
| 110 | 124 | At2g46970 | AAC34226 | NP_182220 | PIL1 | 2 | 19.5 | AC004411 | F14M4.20/45222-47283 |
| 111 | 132 | At3g62090 | BAC10690 | PIL2 | 3 | 23 | AB090874 | T17J13/16742-18191 | |
| 112 | 83 | At1g66470 | AAG27834 | NP_176820 | 1 | 24.4 | AC013288 | F28G11.9/39417-40689 | |
| 113 | 86 | At5g37800 | BAB10359 | NP_198596 | 5 | 14.7 | AB016873 | K22F20.8/38153-39326 | |
| 114 | 54 | At1g27740 | AAF24948 | NP_564293 | 1 | 9.6 | AC012375 | T22C5.19/66736-67789 | |
| 115 | 85 | At4g33880 | CAA19870 | NP_195114 | 4 | 15.2 | AL031032 | F17I5.70/24099-25576 | |
| 116 | 139 | At5g43175 | NP_680385 | 5 | 17 | AB023030 | K24F5.1/3318-4709 | ||
| 117e | 37 | At3g50330 | CAB62312 | NP_190602 | 3 | 18.6 | AL132976 | F11C1.170/57803-58498 | |
| 118e | 88 | At5g67060 | NP201507 | 5 | 26.3 | AB026640 | K8A10/38904-39629 | ||
| 119 | 43 | At5g09750 | CAB89355 | NP_196537 | 5 | 3 | AL353994 | F17I14.60/21646-22320 | |
| 120 | 40 | At4g00120 | CAB80770 | NP_191923 | 4 | 0.003 | AL161471 | F6N15.18/41601-42197 | |
| 121 | 87 | At3g21330 | BAB03046 | NP_188770 | 3 | 7.5 | AP001305 | MHC9.1/3842-5050 | |
| 122 | 140 | At5g01310 | CAB81914 | NP_195751 | 5 | 0.1 | AL161746 | T10O8.20/13362-17018 | |
| 123 | 53 | At2g34820 | AAC12822 | NP_181028 | 2 | 14.6 | AC004238 | F19I3/19911-20798 | |
| 124 | 52 | At1g30670 | AAD25754 | NP_174355 | 1 | 10.8 | AC007060 | T5I8.12/54562-55439 | |
| 125 | 102 | At1g69010 | AAF07356 | NP_177064 | 1 | 25.5 | AC011665 | T6L10/83444-85239 | |
| 126 | 46 | At5g08130 | CAB93714 | NP_196430 | 5 | 2.6 | AL357612 | T22D6.70/33324-36240 | |
| 127 | 141 | At5g38860 | BAB08642 | NP_198702 | 5 | 15.2 | AB009048 | K15E6.7/21572-23400 | |
| 128 | 142 | At5g64340 | BAB09865 | NP_201239 | 5 | 25.4 | AB008268 | MSJ1.18/55674-56720 | |
| 129 | 143 | At5g09460 | CAC05472 | NP_196508 | 5 | 2.9 | AL391712 | 152E12T/85222-86202 | |
| 130 | 144 | At1g29950 | AAG52051 | NP_564342 | 1 | 10.5 | AC022455 | T1P2/14666-16092 | |
| 131 | 145 | At5g50010 | BAB10287 | NP_199812 | 5 | 8.2 | AB006707 | MOP9.3/13425-15115 | |
| 132 | 108 | At1g25310 | NM102341 | 1 | 8.8 | AC079374 | F4F7/31815-32525 | ||
| 133 | 105 | At5g54680 | BAB09934 | NP_567195 | 5 | 21.9 | AB022214 | K5F14.2/7451-9174 | |
| 134 | 115 | At1g51070 | AAG50538 | NP_175518 | 1 | 18.5 | AC079828 | F24H24.8/25827-27322 | |
| 135 | 34 | At3g23210 | BAA95734 | NP_188962 | 3 | 8.2 | AB025608 | K14B15.12/42016-43676 | |
| 136 | 104 | At4g14410 | CAB78483 | NP_567431 | 4 | 7.2 | AL161538 | 197859-198944 | |
| 137 | 11 | At4g36060 | AAL55718 | NP_195330 | 4 | 16 | AY090362 | T19K4.190/201-1007 | |
| 138 | 121 | At3g19860 | NM112876 | 3 | 6.9 | AB025631 | MPN9/36652-38887 | ||
| 139 | 47 | At3g47640 | NM114632 | 3 | 17.5 | AL132955 | F1P2.190/80945-82655 | ||
| 140 | 117 | At3g22100 | BAB01396 | NP_188848 | 3 | 7.7 | AB028622 | MZN24.31/74616-75749 | |
| 141 | 146 | At4g30180 | CAB81011 | NP_194747 | 4 | 13.7 | AL109796 | F9N11.30/108679-109155 | |
| 142 | 147 | At3g17100 | BAA94988 | NP_566567 | 3 | 5.8 | AB026636 | K14F17.17/63352-64044 | |
| 143 | 148 | At3g06590 | AAF63634 | AAG51338 | 3 | 2 | AC020580 | F5E6.8/31055-31720 | |
| 144 | 149 | At1g09250 | NP565839 | 1 | 2.9 | AC003114 | T12M4.4/11899-12872 | ||
| 145 | 150 | At3g05800 | AAF26082 | NP_566260 | 3 | 1.7 | AC012393 | F10A16.9/32345-32980 | |
| 146 | 151 | At2g47270 | AAB63827 | NP_566098 | 2 | 19.3 | AC002337 | T8I13/46017-46325 | |
| 147 | 152 | At1g22380 | AAF87154 | 1 | 7.9 | AC002423 | T23E23.14/53107-58309 |
The AtbHLH proteins identified in the present study are listed according to their EN determined by the multiple sequence alignment in Figure 1. Proteins are designated according to their TAIR Atg numbers, protein accession numbers (PID), other reference numbers (AtbHLH numbers), names, chromosomal locations, map positions within the chromosome, and clone information (GenBank accession numbers and coordinates within BAC/clone).
AtbHLH numbers correspond to those assigned by Heim et al. (2003). Numbers in boldface indicate AtbHLH proteins for which no number was assigned previously.
Sequence designated by Atg number does not coincide 100% with predicted bHLH gene sequence.
Identical bHLH domain sequence in these two different proteins.
–, The database (NCBI) annotation differs from that determined in this study.
Identical bHLH domain sequence in these two different proteins.
Eight other Arabidopsis proteins described as bHLH proteins in the databases also are not included in this study because they do not comply with the criteria we have established (up to nine mismatches from the predicted motif) and do not have a high similarity to any of the proteins reported here. The accession numbers of these proteins (PID numbers) are AAG50594, AAG50694, AAF79358, CAC08333, AAC26786, CAB72153, CAB61988, and CAB93708. None of them has been shown experimentally to possess the properties of bHLH proteins. However, the possibility that divergent members of the family that did not match our criteria and were not included in our analysis do exist (including those listed above) cannot be excluded.
The AtbHLH proteins fit well the consensus motif used to select the set of proteins reported in this study, because 77% (113) of these proteins had fewer than seven mismatches from the consensus motif, and even the most divergent of them retained at least a 52% conservation of the 19 residues that constitute the motif. Two AtbHLH proteins matched the consensus motif perfectly (EN44 and EN45), and five proteins had only two mismatches (see supplemental Figure 1 online). On average, the AtbHLH proteins had 5.3 mismatches from the consensus motif and 1.6 mismatches in the basic region. However, there are certain positions that are less conserved in the AtbHLH proteins than in the consensus described by Atchley et al. (1999). Those differences are indicated in boldface letters in the summary of the conserved amino acids that form the motif (Table 1) and are observable in the multiple sequence alignment shown in supplemental Figure 1 online, where the fit of each of the proteins to the conserved motif is analyzed. Most of the changes are conservative in terms of the type of residue. To illustrate some of the differences in residue conservation, at position 1 (all position numbers referred to are those defined in this study), the residues are conserved in 88% of the animal proteins analyzed, whereas in Arabidopsis, the conservation decreases to 46%. We found similar results for the residues at positions 2 (93% conservation in animals versus 42% in Arabidopsis), 13 (93% in animals versus 76% in Arabidopsis), 28 (79% in animals versus 39% in Arabidopsis), and 42 (93% in animals versus 45% in Arabidopsis).
Multiple Sequence Alignments
Although the signature bHLH domain of the AtbHLH proteins is well conserved, the regions outside of this domain in the remainder of the protein generally are poorly conserved (data not shown). Therefore, our analysis here is restricted primarily to consideration of the bHLH domain, as has been the case for previous studies of this kind (Atchley and Fitch, 1997; Atchley et al., 1999; Morgenstern and Atchley, 1999; Ledent and Vervoort, 2001). On average, the AtbHLH proteins have 5.8 basic residues in the first 17 positions that correspond to the basic region. We identified a subset of proteins that have fewer basic residues than others and that are discussed below. The loop is the most divergent region of the domain in terms of size and amino acid composition, as has been observed for bHLH proteins from other organisms (Massari and Murre, 2000).
An alignment of the 147 members selected (Figure 1) shows that the extremes, represented by EN1 and EN147, have a low sequence similarity (12.5%) with each other and that the common residues are restricted mainly to those in the consensus predictive motif. As revealed by database searches, the closest homologs for the identified Arabidopsis proteins are always plant proteins. Animal bHLHs have reduced sequence similarity to the AtbHLHs, often being restricted to the signature amino acids that constitute the bHLH consensus motif (data not shown).
Conversely, some AtbHLH proteins have high amino acid conservation not only in the generally more conserved helices but also in the basic region. Indeed, two pairs of AtbHLH proteins are identical to their respective counterparts across the entire bHLH domain (EN44/AtbHLH33, which is identical to EN45/AtbHLH116, and EN118/AtbHLH88, which is identical to EN117/AtbHLH37). Otherwise, among subsets of the family, conservation can be up to 79%. In supplemental Figure 2 online, we show closeups of some of the bHLH proteins that have the highest conservation of amino acid sequence among themselves.
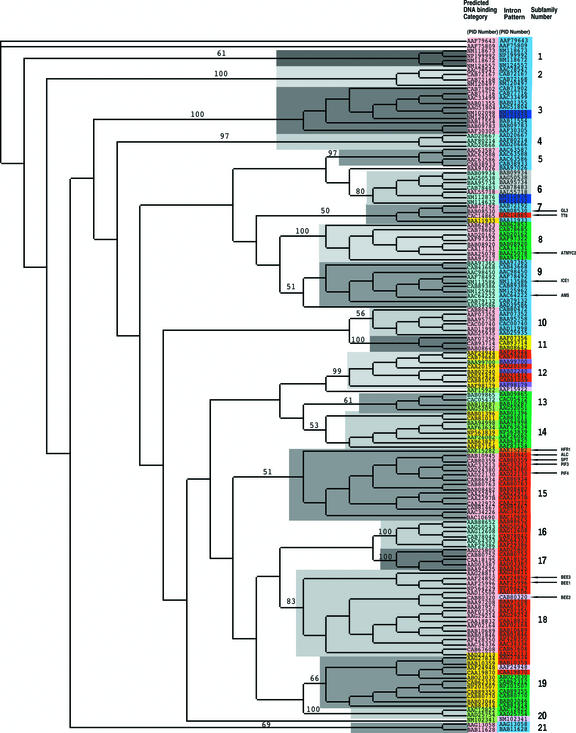
Figure 2.
Neighbor-Joining Phylogenetic Tree of the AtbHLH Domains Indicating the Predicted DNA Binding Activities and the Intron Distribution Pattern within the Domain.
The unrooted tree, constructed using PAUP 4.0, summarizes the evolutionary relationships among the 147 members of the AtbHLH protein family. The proteins are named according to their PID numbers (see Figure 1 and Table 2). The tree was constructed using the amino acid sequence of the bHLH domain for each protein. The tree shows the 21 phylogenetic subfamilies (right column, numbered 1 to 21 and marked with different alternating tones of a gray background to make subfamily identification easier) with high predictive value (bootstrap support of 50 or greater). The internal nodes are not supported by the sampling method and do not necessarily give a true indication of the phylogenetic relationships between the different subfamilies of bHLH proteins. Functionally characterized AtbHLH proteins are indicated with arrows and their names (Table 2; see also supplemental Table 4 online). The tree shown has branch lengths that are not proportional to the distance between sequences. The alignment on which the tree is based is shown in Figure 1. The color code in the central column (Intron Pattern) indicates the numbers and positions of the introns localized in the bHLH domain of each protein. The colors correspond to the intron patterns shown in Figure 3. The color code in the left column (Predicted DNA binding Category) indicates the predicted DNA binding activity of each protein. Pink indicates putative G-box binders; blue indicates putative non-G-box binders; green indicates putative non-E-box binders (i.e., possible DNA binding capacity but no predicted recognition of an E-box); and yellow indicates putative non-DNA binders (see Table 3 for categories).
In the alignment, we identified 17 residues with at least 50% conservation across all members (shaded in black/dark gray and indicated at the bottom of the alignment shown in Figure 1). Ten of the 17 correspond to residues included in the consensus domain used to select the family members (Glu-13, Arg-14, Arg-16, Leu-27, Lys-39, Leu-46, Ala-49, Ile-50, Tyr-52, and Leu-56 in our alignment). Some of these residues are reported from studies in animals to play a specific functional role (Winston and Gottesfeld, 2000) (see below). Some general differences between the AtbHLHs and the animal bHLHs were observable (data not shown). These include the previously mentioned differences in the percentages of the bHLH consensus motif amino acid conservation summarized in Table 1. Another difference involves the location of the start of the basic region. In our alignments, we considered the basic region to be 17 amino acids long, which is 4 amino acids longer than that described by Atchley et al. (1999). The reason for this difference is that few AtbHLHs (entry numbers 1 to 19, 22 to 25, 40, 43, 49, 50, 62, 112, 113, 117, 118, 125 to 127, 138, and 143 to 147) have at least one of the conserved basic amino acids in position 8 or 9 upstream of the conserved Glu (Glu-9 for Atchley et al., 1999). Based on our reference bHLH protein, PIF3, we observed that these basic residues are present but are located 11 or 12 amino acids upstream of the conserved Glu. In the set of proteins reported here, 68 and 63 proteins have the first and second conserved residue in the same position as does PIF3. Therefore, we adjusted the numbering of the amino acid positions in the bHLH consensus motif, as shown in Table 1, and concluded that the basic region of the AtbHLH proteins is 17 amino acids long.
Phylogenetic Analysis of the Arabidopsis bHLH Proteins
Using the bHLH domains from the alignments shown in Figure 1, a neighbor-joining phylogenetic tree was generated (Figure 2). In supplemental Figure 3 online, we provide the branch lengths for this tree. For statistical reliability, we conducted bootstrap analysis with 1000 replicates (see supplemental data Figure 4 online). From the values obtained in the bootstrap analysis, it was apparent that the deep nodes of the tree have low statistical support. This observation also is true for the phylogeny of bHLH proteins from other organisms, which has been attributed to the small size of the bHLH motif and the existence of numerous ancient paralogs (Atchley and Fitch, 1997). Nevertheless, in the outer clades, the bHLH domain has better resolution, permitting subfamilies of proteins to be delimited. Based on the statistical support of each branch, we selected those with a bootstrap value of >50 to divide the AtbHLH protein family into 21 subfamilies, numbered 1 to 21 (Figure 2, right). In supplemental Figure 5 online, we show the amino acid sequence of each bHLH domain and the phylogenetic subfamily to which it belongs. We could not infer evolutionary relationships between the different subfamilies of bHLH proteins because the internal nodes do not show high support. By contrast, within each subfamily, the strong amino acid sequence conservation is evident from the short branch lengths at the tips of the tree, suggestive of strong evolutionary relationships among subfamily members. The fact that plant bHLH proteins do not seem to have close animal homologs was demonstrated when we attempted to include classic examples of animal bHLH proteins in our tree. No monophyletic clades that included AtbHLH proteins were formed (data not shown). This phenomenon also was observed by Ledent and Vervoort (2001) in their study of Caenorhabditis elegans and Drosophila bHLHs.
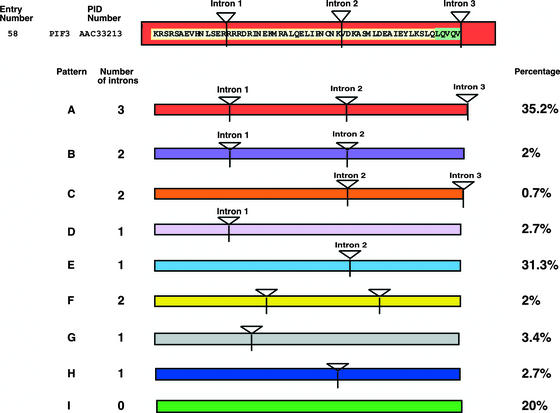
Figure 3.
Intron Distribution within the bHLH Domains of the AtbHLH Proteins.
Scheme of the intron distribution patterns (color coded and designated A to I) within the bHLH domains of the AtbHLH proteins. Introns are indicated by triangles and numbered (1 to 3) based on those present in the bHLH region of PIF3, which is shown at top. When the position of the intron coincides with that found in PIF3, the intron number is given above the triangle. For patterns F, G, and H, no intron number above the triangle indicates that the location of the intron within the bHLH domain is different from that found in PIF3. The percentage of proteins with each pattern is given at right. The correlation of intron distribution patterns and phylogenetic subfamilies is provided in Figure 2 (central column, color coded), and the chromosomal distribution of intron patterns is provided in Figure 4 (colored ovals adjacent to each entry number).
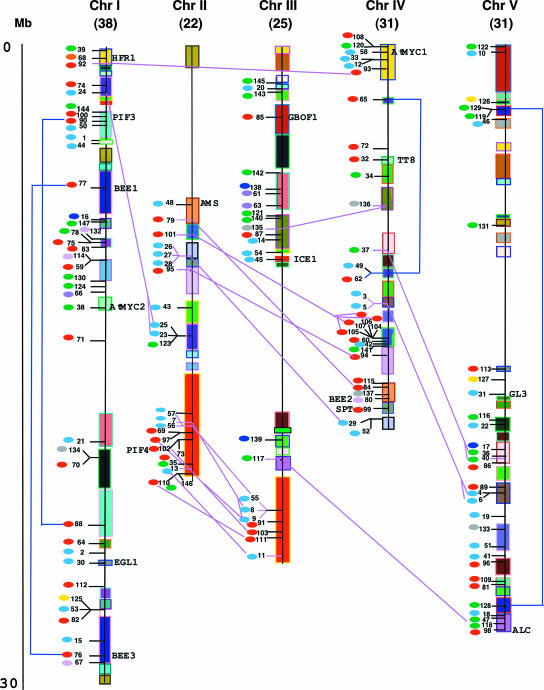
Figure 4.
Chromosomal Locations, Intron Distribution Patterns, and Duplication Events for AtbHLH Genes.
Deduced chromosomal positions of the AtbHLH genes are indicated by EN (assigned in Figure 1). Segmentally duplicated regions in the chromosomes (Chr I to V) are indicated by boxes of the same color (adapted from TIGR). The total number of bHLH genes per chromosome is indicated at the top of each chromosome in parentheses. The scale is in megabases (Mb) and is adapted from the scale available on the TIGR database (see Methods). The small colored ovals at left of the ENs indicate the intron distribution patterns within each gene. The color code corresponds to the intron patterns shown in Figure 3. Connecting lines (blue and pink) mark the specific cases in which there is a strong correlation between duplicated genomic regions and the presence of bHLH genes with both closely related predicted amino acid sequence (close ENs) and the same intron pattern. The blue lines link cases associated with apparent intrachromosomal duplications (see supplemental Figure 7B online), and the pink lines link cases associated with apparent interchromosomal duplications (for more details, see supplemental Figure 7C online).
Figure 5.
PIF4 Heterodimerizes with PIF3.
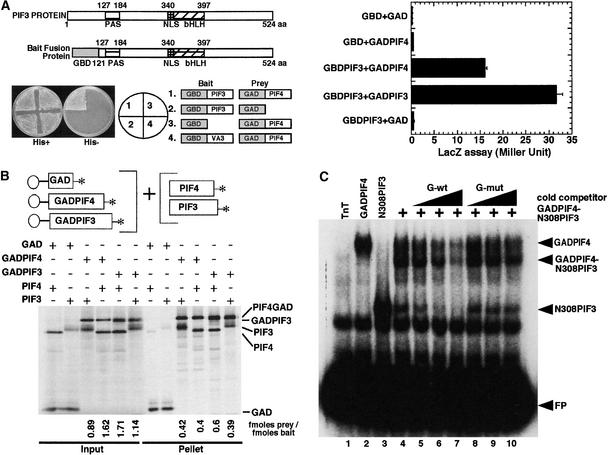
(A) PIF3 and PIF4 interact in a yeast two-hybrid assay. The left panel shows interaction in a plate growth assay. The combination of constructs used in each section is indicated in the circle (middle) and at right. The right panel shows Miller units in a quantitative liquid β-galactosidase assay. GBD and GAD denote GAL4 DNA binding and activation domains, respectively. GAD:PIF4 denotes the GAL4 activation domain:PIF4 fusion protein, and GBD:PIF3 denotes the GAL4 DNA binding domain:PIF3 fusion protein. aa, amino acids; NLS, nuclear localization signal.
(B) PIF3 and PIF4 interact in vitro. Full-length PIF3 or PIF4 cDNAs either alone or fused to GAD were used as templates for synthesizing the proteins for this coimmunoprecipitation assay. All proteins were synthesized as 35S-Met–labeled products in a TnT reaction. PIF4:GAD, PIF4 fused at its C terminus to the GAL4 activation domain; GAD:PIF3, PIF3 fused at its C terminus to the GAL4 activation domain.
(C) PIF3 and PIF4 bind to the G-box both as homodimers and as a PIF3:PIF4 heterodimer. PIF4:GAD and a truncated N308PIF3 clone were coexpressed in a TnT reaction, and 1 μL of this TnT mix was used for DNA binding. PIF4:GAD and N308PIF3 also were expressed in a TnT reaction separately and used to bind to the G-box DNA as homodimers. A total of 30,000 cpm of labeled probe was used in each lane. The binding conditions were as described by Huq and Quail (2002). pLUC control plasmid was translated in the TnT reaction and used as the TnT-only control. The samples were separated on a 5% gel, and the gels were dried and exposed to PhosphorImager (Molecular Dynamics, Sunnyvale, CA) or x-ray film for analysis. FP, free probe; mut, mutant; wt, wild type.
Because the analysis described above used only a single alignment method (Multalin), we also investigated the effect of another method on tree topology. Using CLUSTAL W for the alignment resulted in a neighbor-joining tree that was only minimally different from that shown in Figure 2 (data not shown). Of the 21 subfamilies, 19 remained unchanged in both trees. In the remaining two subfamilies, only seven genes clustered differently compared with the Multalin analysis, establishing that 95% of the AtbHLH proteins clustered in the same subfamilies by the two methods of alignment. Differences were observed in the deep nodes by the two methods, but the bootstrap values for these branches in the CLUSTAL W neighbor-joining analysis were low, like those for the Multalin neighbor-joining analysis, rendering these differences unreliable. We also investigated the effect of using an alternative method of phylogenetic tree construction. Using maximum parsimony analysis, we obtained a majority rule 50% consensus phylogenetic tree (of 1000 trees created) that was very similar to that obtained with the neighbor-joining method (see supplemental Figure 6 online). Of the 21 subfamilies identified by neighbor joining, 14 remained unchanged in the maximum parsimony analysis. In the remaining seven subfamilies, only 10 genes were clustered differently in the parsimony compared with the neighbor-joining analysis. Thus, 93% of the AtbHLH proteins were clustered into the same subfamilies by the two methods. We conclude that the neighbor-joining tree presented (Figure 2) provides a reliable indication of the likely phylogenetic relationships between the AtbHLH proteins within subfamilies.
Intron Distribution within the bHLH Domain
As part of our annotation verification process, we analyzed the intron distribution within the bHLH domain of all of the bHLH genes reported here. We observed nine different distribution patterns (designated A to I) ranging from three to zero introns within the domain (Figure 3). The results show that 80% of the identified members of the family have introns in their bHLH domains, and in most cases, the intron position is conserved, even though the number can vary. The most common pattern involves three introns in the bHLH region, as is the case with our reference protein, PIF3. Only 8% of the genes had introns in the bHLH domain at positions different from the rest of the members of the family (patterns F, G, and H), and 20% showed no introns in the bHLH region (pattern I). The supplemental data online includes a summary of the intron distribution pattern for each of the proteins identified in this study (see supplemental Figure 5 online).
We analyzed whether the intron/exon position and distribution patterns corresponded with the phylogenetic subfamilies defined in Figure 2. Indeed, a clustering of similar patterns within subfamilies was observable (Figure 2, intron pattern). For example, members of subfamily 8 have the same intron distribution pattern (pattern I), and this is different from the pattern shown by the members of subfamily 9 (pattern E). The same situation is observed for subfamilies 5 and 6, 10 and 11, 14 and 15, and 18 and 19. The three cases in which the intron position within the bHLH domain is different from the general patterns observed correspond to proteins in subfamilies 3 (pattern H), 6 (patterns G and H), and 11 (pattern F). For these proteins, even though the position of the intron is different, their fit to the bHLH consensus motif is good (with four to six mismatches). Therefore, the intron/exon distribution patterns give further support for the phylogenetic subfamilies defined here.
The AtbHLH Genes Are Distributed throughout the Arabidopsis Genome
Based on the chromosomal location information provided by the National Center for Biotechnology Information (NCBI; Arabidopsis genome update from August 2002), we localized the AtbHLH genes in the five Arabidopsis chromosomes and determined that the family is distributed across all of them (Figure 4). There are some areas with higher density of bHLH genes that include clusters of up to 11 genes, such as the bottom of chromosome II, the top of chromosome III, and the bottom of chromosome IV. Conversely, there are large regions that are apparently devoid of bHLH genes, including the top half of chromosome II and the central sections of chromosomes III and V. There are very few bHLHs (8%) in nonduplicated regions of the genome.
Analysis of the designated genome tandem arrays and intrachromosomally and interchromosomally duplicated areas (TIGR database) and their relationship to the localization of genes highly similar in their predicted bHLH domains (Figure 4) indicates that, overall, 38% of the AtbHLH proteins could have evolved from some type of genome duplication event. More specifically, there are 11 cases of duplicated genomic region tandem arrays that include homologous bHLH domains. These cases, encompassing 25 proteins, constitute 17% of the total number of AtbHLH proteins (for the specifics of each case, see supplemental Figure 7A online). Another 48 of the closely related AtbHLH genes (32% of the members of the family) can be grouped into putative intrachromosomal (4 cases involving 8 proteins) and interchromosomal (15 cases involving 40 proteins) duplication events (see supplemental Figures 7B and 7C online). Additional evidence that supports the common origin of closely related bHLHs from duplication events in the genome comes from the intron distribution patterns within the bHLH domain. As shown in Figure 4, there is a strong correlation between the examples of duplication discussed above and the conservation of the intron distribution pattern within the genes involved (indicated by the connecting lines).
For the remaining genes, even though they are localized mostly in putatively duplicated areas of the genome, there is no direct correlation between their localization and the degree of sequence relatedness in their bHLH domain amino acid sequences. However, exemplified by the bottom of chromosomes II and III, the intron pattern within the proteins localized in large duplicated areas tend to have some degree of conservation. Based on our analysis, we propose that some bHLHs might have a recent common evolutionary origin and that the large size of the bHLH protein family can be explained in part by the segmental and tandem duplications that occurred in the genome.
Predicted Functional Properties of the AtbHLH Proteins
There are two important functional activities determined by the amino acid sequence of the bHLH domain: DNA binding and dimerization.
DNA Binding Properties
The basic region in the bHLH domain determines the DNA binding activity of the protein (Massari and Murre, 2000). Therefore, the presence or absence of basic residues in the first 17 positions of the bHLH domain is the basis for defining the first two major categories of AtbHLH proteins in terms of DNA binding properties: (1) DNA binding bHLHs and (2) non-DNA binding bHLHs (Table 3). A total of 120 proteins are predicted to bind DNA, because they have an average of 6 basic residues in the first 17 positions, whereas 27 proteins are predicted not to bind DNA, because they have a “less basic region” (an average of 3.8 basic residues in the first 17 positions) (Table 3; see also supplemental Tables 1 and 2 online).
Table 3.
Predicted DNA Binding Characteristics Based on the Amino Acid Sequence of the bHLH Domain of the AtbHLH Proteins
| Predicted Activity | Predicted Motif |
Number of Proteins |
|---|---|---|
| DNA binding | ||
| E-box binding | ||
| G-box binding | bHLH | 89 |
| Non-G-box binding | bHLH | 20 |
| Non-E-box binding | bHLH | 11 |
| Total | 120 | |
| Non-DNA binding | HLH | 27 |
The DNA binding bHLH category can be subdivided further into two subcategories based on the predicted DNA binding sequence: (1) the E-box binders and (2) the non-E-box binders (Table 3). This subdivision is based on the presence or absence of two specific residues in the basic region: Glu-13 and Arg-16 (position numbers are based on our alignment, corresponding to positions 9 and 12 in the motif described by Atchley et al. [1999]) (Table 1, Figure 1). These residues constitute the E-box recognition motif, because they are conserved in the proteins known to have E-box binding capacity (Fisher and Goding, 1992; Littlewood and Evan, 1998). The analysis of the crystal structures of USF, E47, Max, MyoD, and Pho4 (Ellenberg et al., 1994; Ferre-D'Amare et al., 1994; Ma et al., 1994; Shimizu et al., 1997; Fuji et al., 2000) has shown that Glu-13 is critical because it contacts the first CA in the E-box DNA binding motif (CANNTG). Site-directed mutagenesis experiments with Pho4, in which other residues (Gln, Asp, and Leu) were substituted for Glu-13, demonstrated that the substitution abolished DNA binding (Fisher and Goding, 1992). Meanwhile, the role of Arg-16 is to fix and stabilize the position of the critical Glu-13; therefore, it plays an indirect role in DNA binding (Ellenberg et al., 1994; Shimizu et al., 1997; Fuji et al., 2000).
In the AtbHLH protein family, 108 proteins have the conserved Glu-13/Arg-16 pair. In addition, one more (EN139/AtbHLH47) has Glu-13 but lacks Arg-16 and has a Lys in this position. Because this type of amino acid substitution is conservative, and animal proteins such as SREBP (Hua et al., 1993), although missing Arg-16, bind E-boxes, we considered EN139/AtbHLH47 part of this category. Experimental evidence is necessary to determine whether deviation from the consensus permits the retention of binding capacity. The predicted E-box binding bHLHs represent 74% of the total AtbHLHs reported in this study (Table 3). For a list of the proteins included in this category, see supplemental Table 1a online.
The E-box binding bHLHs can be categorized further into subgroups based on the type of E-box recognized. Crystal structures show that the type of E-box binding preferences are established by residues in the basic region, with the best understood case being that of the G-box binders (Ellenberg et al., 1994; Ferre-D'Amare et al., 1994; Shimizu et al., 1997). Therefore, we have further subdivided the Arabidopsis E-box binding bHLHs into (1) those predicted to bind G-boxes and (2) those predicted to recognize other types of E-boxes (non-G-box binders) (Table 3).
There are three residues in the basic region of the bHLH proteins: His/Lys, Glu, and Arg at positions 9, 13, and 17 (positions are relative to the alignment shown in Figure 1, which correspond to positions 5, 9, and 13 in the motif described by Atchley et al. [1999]), which constitute the classic G-box (CACGTG) recognition motif. Glu-13 is the key Glu involved in DNA binding, and analysis of the crystal structures of Max, Pho4, and USF indicates that Arg17 confers specificity for CACGTG versus CAGCTG E-boxes by directly contacting the central G of the G-box. His-9 has an asymmetrical contact and also interacts with the G residue complementary to the first C in the G-box (Ferre-D'Amare et al., 1994; Shimizu et al., 1997; Fuji et al., 2000). In Arabidopsis, 89 proteins (60% of the total number and 81% of the proteins predicted to bind DNA) have the conserved His/Lys-9, Glu-13, and Arg-17 residues and therefore would be predicted to be G-box binders (Table 3). The complete list and bHLH domain sequences of these proteins are provided in supplemental Table 3a online.
The rest of the AtbHLHs with E-box binding capacity but lacking the conserved residues to preferentially bind a G-box (20 proteins) (Table 3) were defined as non-G-box binders. For these proteins that lack the combination His/Lys-9 and Arg-17, the recognition mechanism of the central bases is not yet defined. The MyoD crystal structure showed no direct contact with the central bases, raising the possibility that the contacts could be water directed (Ma et al., 1994). The members of this category are listed in supplemental Table 3b online.
Apart from the described E-box binding proteins, the second subcategory of predicted DNA binding bHLHs is formed by 11 proteins (ENs 61, 67, 68, 111, 117, 123, 124, 131, 132, 142, and 143) that lack the E-box binding residues but that do have a considerable number of basic residues in their “basic region” (five to eight basic residues). These proteins with “unusual” basic regions might be able to bind DNA but lack the sequence specificity for E-boxes; therefore, they are defined as non-E-box binding proteins in this study (Table 3; see also supplemental Table 1b online). To date, DNA binding has not been tested experimentally for any of these proteins.
The non-DNA binding AtbHLHs (called simply HLH proteins) comprise 27 proteins with a “less basic region” that also lacks the Glu-13/Arg-17 necessary for binding to the E-box. The presence of Pro residues in the basic region of most of these proteins could indicate a differential positioning with respect to the DNA as a result of modified folding (Table 3). The non-DNA binding HLHs could have a function similar to that of the animal ID-HLH proteins, as negative regulators of E-box binding bHLHs through the formation of heterodimers that have lost the capacity to bind DNA (Fairman et al., 1993). The members of this category are summarized in supplemental Table 2 online.
The distribution of these predicted DNA binding properties across the various phylogenetic subfamilies is shown color coded in the phylogenetic tree (Figure 2). Starting with the predicted DNA binding proteins in the E-box binding category of bHLHs, the predicted G-box binders form phylogenetic subfamilies 1, 2, 3, 5, 8, 10, 11, 15, 17, 18, 21, and part of 7 (Figure 2, pink). The non-G-box binders form phylogenetic subfamilies 4, 6, 9, and 16, with two forming part of subfamily 13 (Figure 2, blue). The non-E-box binders (Figure 2, green) form subfamily 20 and part of phylogenetic subfamilies 12, 13, 14, and 15. The other members of subfamilies 12 and 14 are HLH proteins, whereas the rest of the members of subfamily 13 are predicted to bind E-boxes (Figure 2, yellow and blue, respectively). The predicted non-DNA binding HLH proteins (Figure 2, yellow), form subfamily 19 and part of subfamilies 7, 12, 14, and 18 in the phylogenetic tree. Together, these data indicate that the different phylogenetic subfamilies may have evolved different functional activities based on their DNA binding capacities and sequence recognition specificities.
Dimerization
bHLH proteins are well known to dimerize, but the critical molecular determinants involved are not well defined (Shirakata et al., 1993; Littlewood and Evan, 1998; Ciarapica et al., 2003). On the other hand, the Leu residue at position 27 in our alignment has been shown to be structurally necessary for dimer formation in the mammalian Max protein (Brownlie et al., 1997). Therefore, it is notable that this is the only invariant residue in all 147 AtbHLH proteins (Figure 1, Table 1), consistent with a similar essential function in plant bHLH protein dimerization. Current information indicates that dimerization specificity is affected by multiple parameters, including hydrophobic interfaces, interactions between charged amino acids in the HLH region, and partner availability, but no complete explanation for partner recognition specificity has been documented (Ciarapica et al., 2003). Thus, although empirically it seems logical that bHLH proteins most closely related in sequence in the HLH region are the most likely to form heterodimers, there has been no systematic investigation of this possibility to date.
In plants, heterodimers between two members of the bHLH family, PIF3 and HFR1, have been reported (Fairchild et al., 2000). HFR1 is a bHLH protein with an atypical basic region that is associated in our phylogenetic analysis with the subfamily formed by the PIF3-like proteins. Based on the characteristics of HFR1, the dimer formed by HFR1 and PIF3 could act as a regulatory type of heterodimer either by preventing PIF3 from binding to an E-box or by targeting the dimer to a different type of DNA recognition motif. Moreover, in terms of interacting partners in a functional context, although HFR1 is unable to bind directly to phytochromes A and B, the heterodimers of PIF3 and HFR1 can form a ternary complex with phytochromes A and B (Fairchild et al., 2000).
To examine experimentally the question of whether related Arabidopsis bHLH proteins that are individually capable of DNA binding can form heterodimers that retain DNA binding activity, we investigated the interaction between PIF3 and PIF4 using a combination of different approaches. Data from a yeast two-hybrid β-galactosidase assay showed that GAL4 activation domain (GAD):PIF4 interacts strongly with GAL4 DNA binding domain (GBD):PIF3 (Figure 5A), consistent with heterodimerization, although this interaction is weaker than for GAD:PIF3-GBD:PIF3 homodimer formation in this assay. The interaction between PIF4 and PIF3 also was confirmed in vitro, where the two proteins were cotranslated in the TnT system (see Methods). PIF4:GAD and GAD:PIF3 reciprocally coimmunoprecipitated PIF3 and PIF4, respectively (Figure 5B). These results confirm that these two proteins can interact physically with each other.
We also investigated whether PIF4 and PIF3 can bind as dimers to the G-box DNA motif. We used PIF4:GAD and a truncated version of PIF3, which lacks the N-terminal 308 amino acids but contains the bHLH domain, including the C-terminal portion, for better separation of the heterodimer complex. The presence of a complex that migrates as an intermediately sized band between the presumptive N308PIF3-N308PIF3 homodimer complex and the PIF4:GAD-PIF4:GAD homodimer complex provides evidence that PIF3 and PIF4 can form heterodimers that are capable of recognizing the G-box motif in a sequence-specific manner (Figure 5C). Therefore, the data indicate that AtbHLH proteins have dimerization properties similar to those of their animal counterparts, because they have the potential to dimerize with more than one partner and to form heterodimeric molecules capable of sequence-specific DNA binding.
Comparison of AtbHLH Proteins with Those of Other Eukaryotes
Of the sequenced eukaryotic genomes, only the human genome is predicted to encode a greater total number of transcription factors than the Arabidopsis genome (Riechmann et al., 2000) (Table 4). For the bHLH proteins, the Arabidopsis genome encodes 2.6 times as many bHLHs as the Drosophila genome, 4.2 times more than the C. elegans genome, and 30 times more than the yeast genome (Table 4). A search by functional category of the human genome database indicates the presence of 174 bHLH proteins, and based on data from the Mouse Genome Sequencing Consortium (Waterston et al., 2002), we estimate the presence of ∼140 bHLHs. All of these numbers may represent an underestimation for all organisms, because many proteins have not been assigned yet to clear functional categories based on sequence similarities.
Table 4.
Comparison of bHLH Gene Occurrence in Arabidopsis and Other Eukaryotic Organisms Whose Genomes Have Been Sequenced
| Organism | Approximate Number of Genes |
Approximate Number of Transcription Factors |
Number of bHLH Proteins |
Percentage of Transcription Factors That Are bHLH Proteins |
|---|---|---|---|---|
| Saccharomyces cerevisiae* | 6,000 | 771 (12.9%) | 5 | 0.6 |
| Drosophila** | 14,000 | 635 (4.5%) | 56 | 8.8 |
| C. elegans** | 19,000 | 669 (3.5%) | 35 | 5.2 |
| Arabidopsis** | 26,000 | 1533 (5.9%) | 147 | 9.5 |
| Human*** | 35,000 | 1850 (5.4%) | 174a | 9.4 |
The asterisks indicate the source of the data for number of genes, number of transcription factors, and number of bHLH proteins (except for Arabidopsis, which was reported in this study):
Mewes et al. (2002) and MIPS Comprehensive Yeast Genome Database;
Number taken from a database search of the human genome database.
Analysis of the predicted properties of these bHLHs shows that in Arabidopsis, the most abundant type are the putative G-box binders, as they are in animals, in which they are classified as phylogenetic group B (Atchley and Fitch, 1997). This observation correlates well with the proposal that group B is the ancestral bHLH type (Atchley and Fitch, 1997; Ledent and Vervoort, 2001). However, the AtbHLH G-box binders are distributed in different subfamilies and thus do not form a unique monophyletic clade, as do the animal proteins. We observed a similar situation for the non-E-box binding class of AtbHLHs. This type of bHLH does not have close homologs in the animal kingdom, but it might be equivalent to the animal proteins that bind preferentially to a noncanonical core sequence (such as the N-boxes). However, DNA binding properties have not been demonstrated for any of them yet. No AtbHLHs have the signature residues that define animal group A (Atchley and Fitch, 1997). Another type of bHLH represented in Arabidopsis and animals are the HLHs (non-DNA binders). However, we found no close sequence similarity between the AtHLHs and the animal ID-like proteins (group D) (Atchley and Fitch, 1997). Functional analysis will be necessary to determine if the AtHLHs function as negative regulators of bHLH proteins.
In other eukaryotes, apart from the bHLH domain, additional functional domains have been identified in the bHLH proteins. These additional domains play roles in protein–protein interactions (e.g., PAS, WRPW, and COE in groups C, E, and F, respectively) and in bHLH dimerization specificity (e.g., the zipper [ZIP] domain, part of group B). Even though we focused our analysis on the bHLH domain, we also surveyed the AtbHLHs for the presence of these other motifs. ZIP domains were detected using the program created by Bornberg-Bauer et al. (1998) in seven AtbHLH proteins, immediately C terminal to the bHLH domain, like the configuration in animal bHLH-Leu zipper (bHLHZIP) proteins (Dang et al., 1992). These AtbHLHZIP proteins (ENs 133 through 139) cluster tightly together in subfamily 6 in the phylogenetic tree (Figure 2), suggesting that they arose by relatively recent gene duplication events. The only other AtbHLH protein that has been predicted in the literature to have a ZIP domain is AtMYC2, previously called Rd22BP1 (EN38/AtbHLH6) (Abe et al., 1997, 2003). This protein shows three Leu residues in a row, but the spacing is not strictly six amino acids apart and no coiled-coil structure is predicted for it. Therefore, we do not classify this protein as a bHLHZIP.
The PAS-bHLHs are important regulatory components in the regulation of circadian clocks, hypoxia, and toxin metabolism in other organisms (Ledent and Vervoort, 2001). They are characterized by the presence of a pair of PAS domains on the C-terminal side of the protein, after the bHLH domain. The exact position of the PAS domains is variable. We made use of the NCBI Consensus Domain Search program to search for the presence of PAS domains in the predicted full-length AtbHLH proteins (NCBI protein database). This program recognizes canonical PAS domains represented in the database, such as those in Per, Arnt, and Sim proteins, but it did not detect any similarity in the AtbHLHs. A BLAST search with Per-Arnt-Sim PAS domains also reported no hits in Arabidopsis. However, considering the sequence variability within PAS domains, a more refined method might be necessary to detect them. Two AtbHLHs, PIF3 and PIF4, have been proposed to have a region with limited similarity to the PAS motif (Ni et al., 1998; Huq and Quail, 2002). However, recent more comprehensive computational analysis, benefiting from the accumulation of a large number of PAS-related sequences not available when PIF3 was identified initially (Ni et al., 1998), concludes that the PIF3 sequence is insufficiently related to the currently defined consensus motif to be classified as a bona fide PAS domain (Iyer et al., 2003). In addition, even if these PIF sequences were in fact PAS-like domains, there are two clear structural differences between these proteins and the animal PAS-bHLHs: first, PIF3 and PIF4 each have only one such domain; and second, it is located on the N-terminal side with respect to the bHLH domain. Together, these data suggest that plant bHLH proteins lack PAS domains.
None of the predicted AtbHLH proteins have a WRPW motif at the C-terminal end of the protein, nor do they display the atypical HLH motif with a duplicated helix 2, characteristic of the COE-bHLHs (Crozatier et al., 1996; Dubois and Vincent, 2001). Therefore, except for the few members with ZIP domains, none of the classic domains associated with some other eukaryotic bHLHs appear to be present in the AtbHLHs. Together, these data suggest that the plant bHLH family has not evolved the same degree of diversity within the bHLH domain or the same set of additional recognizable motifs as its counterparts in other organisms.
The PIF3-Like Proteins
As indicated, our initial interest in the AtbHLH proteins came from the proposed central role of PIF3 in phytochrome signaling and the possibility that other members of the family may have a similar function (Quail, 2000). The phylogenetic analysis presented here indicates that the members of subfamily 15 are the most closely related to PIF3 (Figure 2). The experimental data obtained to date indicate that of the 15 proteins in this subfamily, three members, PIF3, PIF4, and HFR1, are involved in phytochrome-regulated responses (Ni et al., 1998; Fairchild et al., 2000; Huq and Quail, 2002), whereas two others, ALC and SPT, are not. The latter two proteins instead are involved in gynoecium development (Heisler et al., 2001; Rajani and Sundaresan, 2001). On the one hand, although the data verify that proteins with closely related bHLH domains can have similar biological functions, they likewise demonstrate that such related proteins can have very divergent functions. Thus, although the present phylogenetic analysis clearly provides a certain degree of predictive value, continued systematic forward- and reverse-genetics analyses will be necessary to define the functional activities of these proteins and to refine our understanding of the relationship between the predicted bHLH sequence and these activities.
DISCUSSION
The identification of 147 bHLH-encoding genes in Arabidopsis establishes this as the second largest transcription factor family in the genome (constituting 9.5% of the total number of transcription factors present), behind only the MYB superfamily of 190 members (Riechmann et al., 2000), and as one of the larger gene families overall in this species. Similar systematic analyses of some of the other large Arabidopsis transcription factor families have been reported, including the R2R3-MYB family (125 members) (Stracke et al., 2001), the bZIP family (75 members) (Jakoby et al., 2002), and the WRKY superfamily (61 members) (Eulgem et al., 2000). However, many of the remaining plant transcription factor families that have been identified (Riechmann et al., 2000) have not been analyzed in depth. Although several other sequenced eukaryotes also have large bHLH families, when expressed as a percentage of the total genes present in the genome, Arabidopsis has the largest relative representation at 0.56% of the identified genes, compared with yeast (0.08%), C. elegans (0.20%), Drosophila (0.40%), Takifugu rubripes (0.40%), human (0.40%), and mouse (0.50%) (Riechmann et al., 2000; Ledent and Vervoort, 2001; Mewes et al., 2002; Waterston et al., 2002). This observation suggests that the bHLH factors have evolved to assume a major role in plant transcriptional regulation. On the other hand, plant bHLHs appear to have evolved a narrower spectrum of variant sequences within the bHLH domain than those of the mammalian systems and appear to lack some of the various ancillary signature motifs, such as the PAS and WRPW domains, found in certain bHLH protein subclasses in other organisms.
Phylogenetic analysis of the bHLH domain allows division of the AtbHLH family into 21 subfamilies. The clustering of the members within these subfamilies is further supported by additional analysis with regard to other criteria, namely, predicted DNA binding capacity and sequence specificity, exon/intron organization and distribution pattern within the domain, and chromosomal location. These data support the general conclusion that members within subfamilies may have recent common evolutionary origins, resulting from various genomic duplication events, and may have related molecular functions. On the other hand, the strong sequence diversity outside of the bHLH domain across all members of the AtbHLH family suggests that the expansion of this family in Arabidopsis may have involved extensive domain shuffling after the duplication events, as in other organisms (Morgenstern and Atchley, 1999).
To date, the biological functions of only 14 members of the Arabidopsis family have been established, leaving >90% yet to be functionally characterized. Of the proteins characterized, three, PIF3, PIF4, and HFR1, are involved in phytochrome signaling (Ni et al., 1998; Fairchild et al., 2000; Huq and Quail, 2002), two, SPT and ALC, are involved in gynoecium development (Heisler et al., 2001; Rajani and Sundaresan, 2001), TT8 is involved in regulating flavonoid biosynthesis (Nesi et al., 2000), GL3 (the closest homolog of the maize R gene) is involved in trichrome differentiation (Payne et al., 2000), AMS is involved in microspore development (Sorensen et al., 2003), AtMYC2 is involved in abscisic acid–induced gene expression (Abe et al., 1997, 2003), ATR2 is involved in tryptophan biosynthesis (Smolen et al., 2002), BEE1, BEE2, and BEE3 are involved in brassinosteroid signaling (Friedrichsen et al., 2002), and ICE1 is involved in chilling and freezing tolerance responses (Chinnusamy et al., 2003) (summarized in supplemental Table 4 online). This analysis indicates that the bHLH family is likely to participate in regulating a broad range of growth and developmental processes at all phases of the plant life cycle.
The known molecular properties of bHLH proteins suggest a general mechanism by which such regulation may be accomplished. This mechanism involves the generation of a high degree of complexity and diversity in transcriptional regulatory activity through variation in the DNA sequence motif recognized by individual bHLH proteins, the capacity to combinatorially amplify the spectrum of possible specific protein–DNA interactions, through selective heterodimerization between bHLH proteins with different DNA sequence recognition specificity, and the capacity to interact with a network of transcriptional coactivators, corepressors, and signaling molecules through selective protein–protein interactions (Grandori et al., 2000; Baudino and Cleveland, 2001; Ciarapica et al., 2003; Levens, 2003).
The DNA sequence to which bHLH proteins bind appears to consist of a hierarchy of nucleotide sequence elements, progressing from those involved in recognition by many or most DNA binding members of the family to those potentially permitting highly specific discrimination between individual family members. The available data indicate that a hexanucleotide sequence is the core element recognized by all or most DNA binding members of the family. The nucleotide sequence within this core element provides both a common link between all DNA binding members of the family and the first level of binding selectivity. The core hexanucleotide sequence includes a range of motifs from the canonical E-box, CANNTG, and its variants, such as the G-box, CACGTG, to non-E-box motifs, such as the N-box variants CACGGC and CACGAC (Littlewood and Evan, 1998; Ledent and Vervoort, 2001). The presence of one or more of these motifs in different promoters potentially provides the first level of sequence-selective targeting of different subgroups of the bHLH family to different promoters. However, because of the limited number of sequence permutations within the hexanucleotide core and the large number of bHLH proteins known or predicted to bind to individual variants, such as a G-box (Table 3), two nonexclusive possibilities present themselves: (1) there may be high levels of redundancy, whereby large numbers of different bHLH proteins may bind to the same target site in a single promoter; and/or (2) other nucleotides outside of the hexanucleotide core may confer increased sequence recognition specificity up to the logical maximum of exclusive binding by a single bHLH family member.
There is some evidence that nucleotides outside of the core do in fact confer additional sequence specificity (Fisher and Goding, 1992; Littlewood and Evan, 1998), but experimental data are limited and essentially nonexistent for Arabidopsis. Currently, therefore, the potential exists for a spectrum of Arabidopsis gene promoters ranging from those targeted by multiple bHLH family members to those targeted uniquely by one member. The available genetic data are consistent with both scenarios. For example, some monogenic mutants, such as spt and alc, display visible phenotypes that indicate the absence of redundancy with any of the other 146 bHLH proteins (Heisler et al., 2001; Rajani and Sundaresan, 2001), whereas three bHLH proteins, BEE1, BEE2, and BEE3, appear to function redundantly in the brassinosteroid pathway (Friedrichsen et al., 2002).
The core DNA binding domain of the bHLH proteins encompasses the basic region of the bHLH domain and contains the residues that recognize and bind to the core hexanucleotide motif (Massari and Murre, 2000). The amino acid sequence in this region provides the first major subdivision of the bHLH family into those that are predicted to bind DNA and those that are not (Table 3). Key residues in this region also confer the capacity to discriminate between variants of the hexanucleotide motif, leading to the first level of subdivision of DNA binding bHLH proteins into those shown or predicted to bind to the canonical E-box motif, CANNTG, and those that are not (non-E-box binders) but that do bind to other variants of this sequence (Table 3). Additional residues within the basic region confer further DNA binding site sequence selectivity, permitting discrimination between, for example, G-box and non-G-box core motifs. Residues involved in potential higher order binding sequence specificity are poorly defined, but there is some evidence that residues in the loop region of the bHLH domain may contact nucleotides outside of the core motif, thereby conferring increased specificity of DNA sequence recognition (Nair and Burley, 2000). Thus, although much remains to be learned, the existing sequence information regarding the Arabidopsis bHLH proteins is consistent with a diversity of paired DNA binding site–bHLH protein combinations, ranging from exclusive to highly redundant.
The high degree of divergence in amino acid sequence outside of the bHLH domain of the Arabidopsis bHLH proteins suggests significant diversity in the molecular functions of these domains. Two such potential formal functions are the detection of incoming regulatory signals from various cellular pathways and the direction of the transcriptional activity of the target gene. Evidence for the existence of at least one interacting signaling molecule in Arabidopsis has been provided in the case of PIF3, in which photoactivated phytochrome has been shown to bind specifically to DNA-bound PIF3 (Martinez-Garcia et al., 2000). That this binding involves a domain outside of the bHLH domain has been established by binding studies with deletion derivatives of PIF3 (Martinez-Garcia et al., 2000; Zhu et al., 2000). Evidence has been presented for the interaction of bHLH proteins with core transcriptional initiation complex proteins (Roy et al., 1991; Pscherer et al., 1996) and with certain transcriptional coactivators and/or repressors in other systems (Paroush et al., 1994; Puri et al., 1997; Dhordain et al., 1998; Grandori et al., 2000; Massari and Murre, 2000; Beischlag et al., 2002; Levens, 2003). However, to date, no data are available for any Arabidopsis bHLH proteins.
The HLH region of the bHLH domain is responsible for the dimerization of bHLH proteins, providing the potential for either homodimerization and/or heterodimerization (Grandori et al., 2000; Massari and Murre, 2000). The amino acid sequence in this region presumably dictates the specificity of the interaction, but relatively little is known about how the specificity is defined (Ciarapica et al., 2003). There is evidence, however, for a variety of interaction patterns among different family members in nonplant systems, including apparent obligate homodimerization or heterodimerization, or the dual capacity for both homodimerization and heterodimerization (Grandori et al., 2000; Baudino and Cleveland, 2001; Ciarapica et al., 2003; Levens, 2003). Clearly, the capacity to heterodimerize with other family members immediately expands the diversity of possible intermolecular interactions, potentially creating new functional activities, such as recognition of new hybrid DNA binding sites, and directing the convergence of separate signaling pathways to the same promoter via the pairing of domains that recognize distinct signaling molecules (Grandori et al., 2000; Massari and Murre, 2000).
The magnitude of the increase in the number of possible new combinations generated by heterodimerization will depend on the promiscuity of partner recognition between family members. This has been found to vary considerably in other systems (Grandori et al., 2000; Baudino and Cleveland, 2001; Levens, 2003). The large size of the Arabidopsis bHLH family presents the theoretical opportunity for an enormous number of combinatorial interactions. To date, there is limited evidence that closely related Arabidopsis bHLH family members can heterodimerize (Fairchild et al., 2000) (Figure 5), verifying, in principle, the notion that this mechanism may function in plants. More extensive analysis will be needed to define the extent to which this capacity extends across the family.
METHODS
Database Search and Annotation Verification
Multiple database searches were performed to identify members of the Arabidopsis thaliana basic/helix-loop-helix (AtbHLH) protein family. We used the Basic Local Alignment Search Tool (BLAST) search capabilities (TblastN and BlastP) available on the National Center of Biotechnology Information (http://www.ncbi.nlm.nih.gov) and TAIR (http://www.arabidopsis.org) databases to search the published sequence of the entire Arabidopsis genome. As a query sequence, we used the amino acid sequence of the PIF3-bHLH domain (58 amino acids). To increase the accuracy and extent of the database search results and to minimize as much as possible the exclusion of real hits caused by incorrect annotation (missing exons or introns annotated as exons), we retrieved the nucleotide sequence for each of the unique hits obtained and used the software NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/) (Brunak et al., 1991; Hebsgaard et al., 1996) to predict intron/exon boundaries in the putative bHLH domain for each protein. Sequences that appeared to be annotated incorrectly were corrected for subsequent analysis.
Sequence Pileups
We constructed a database consisting of the amino acid sequences of the bHLH domains of the 147 proteins reported in this study. The sequences were aligned using the program Multalin with the default parameters (http://prodes.toulouse.inra.fr/multalin/multalin.html) (Corpet, 1988) and were further adjusted visually. The MacBoxShade program (http://www.ch.embnet.org/software/Box_doc.html) was used to highlight conserved and similar amino acids. The alignment, shown in Figure 1, represents 50% conservation shading. We used CLUSTAL W (http://www.ebi.ac.uk/clustalw) (Thompson et al., 1994) as a second method to align sequences and to double check our phylogenetic analysis results (data not shown).
Segmental Duplication in the Arabidopsis Genome
For the detection of large segmental duplications, we used the duplicated blocks map provided by TIGR (http://www.tigr.org/tdb/e2k1/ath1/arabGenomeDups.html). The map was modified to use a color code in which the same color indicates duplicated regions. On this map, each of the bHLH genes was localized on the corresponding chromosome using the coordinates from the genome sequence data (August 2002 version).
Tree Building
For the creation of the phylogenetic tree reported in Figure 2, we used PAUP 4.0 (http://www.paup.csit.fsu.edu) and the neighbor-joining algorithm. The same program was used to perform the bootstrap analysis with 1000 replicates to test the significance of the nodes. The starting point for our tree construction was the bHLH domain amino acid multiple sequence alignment created with Multalin and verified by eye (Figure 1). The trees generated are unrooted. The distance parameters selected are total character difference, among-site rate variation, and random seed initiation. The tree shown in the figures has branch lengths not proportional to the distance between sequences (for the length values, a tree is provided in supplemental Figure 3 online). The tree with complete bootstrap values for each branch also is included in supplemental Figure 4 online. The bootstrap tree was constructed with the same distance parameters as the neighbor-joining tree and includes groups compatible with a 50% majority rule consensus tree. As a second method to validate the conclusions derived from the neighbor-joining tree, we also created, with a heuristic search of 1000 trees, a maximum parsimony majority rule 50% consensus tree, which is presented in supplemental Figure 5 online. The other parameters used were random seed initiation and stepwise addition, and the number of parsimony informative characters was 60. Gaps were treated as missing data. The tree is unrooted, and topological constraints were not enforced.
Yeast Two-Hybrid Interaction, in Vitro Coimmunoprecipitation, and Electrophoresis Mobility Shift Assays
For the yeast two-hybrid interaction assay, GBD:PIF3 (amino acids 121 to 524 of PIF3 fused to the GAL4 DNA binding domain) and GAD:PIF4 (amino acids 59 to 430 of PIF4 fused to the GAL4 activation domain) were used. Yeast transformation and liquid β-galactosidase assays were performed according to the Yeast Protocol Handbook from Clontech (Palo Alto, CA). In vitro coimmunoprecipitation experiments were performed as described by Huq and Quail (2002). All proteins were expressed from T7 promoters in the TnT in vitro transcription/translation system (Promega) in the presence of 35S-Met. The constructs and procedure for expressing PIF3 are described by Fairchild et al. (2000), those for expressing GAD:PIF3 are described by Ni et al. (1999), and those for expressing PIF4:GAD are described by Huq and Quail (2002). The PIF4 open reading frame was cloned into pET17b to produce naked PIF4 protein and confirmed by sequencing. Both PIF3 and PIF4 proteins were coexpressed in vitro, and paramagnetic protein A beads (Dynabeads Protein A; Dynal, Oslo, Norway) and GAD antibody (Santa Cruz Biotechnology, Santa Cruz, CA) were used.
The binding buffer used contained 1× PBS, pH 7.2, 0.1% (v/v) Tergitol Nonidet P-40 (Sigma), 0.1% BSA, and 1× complete protease inhibitor (Roche, Indianapolis, IN). The same buffer was used for the first wash of the pellet, and the final wash was performed with the same buffer without BSA. Sample preparation and quantification were performed according to Huq and Quail (2002). Electrophoresis mobility shift assays were performed according to Martinez-Garcia et al. (2000). All of the proteins were synthesized using the TnT system (Promega). The truncated N308PIF3 construct was described by Zhu et al. (2000), and PIF4:GAD is described above. pLUC control plasmid was translated in TnT and used as a TnT control. A total of 30,000 cpm of labeled probe was used in each lane.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact P.H. Quail, quail@nature.berkeley.edu.
Supplementary Material
Acknowledgments
We thank Matthew Hudson for his help with bioinformatics; Antonio Izzo, John Taylor, Rachel Whitaker, and Lisa Grubisha for their help with the phylogenetic analysis; Giovanni Mele for his help with reformatting the figures; Elena Monte for her comments on the manuscript; and other laboratory members for discussion and support. G.T.-O. was the recipient of a fellowship from the Consejo Nacional de Ciencia y Tecnológia–Fulbright and the University of California–Mexus. This research was supported by National Institutes of Health Grant GM47475, Department of Energy Grant DE-FG03-87ER13742, U.S. Department of Agriculture–Agricultural Research Service Current Research Information System 5335-21000-017-00D, and the Torrey Mesa Research Institute (San Diego).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013839.
Footnotes
Online version contains Web-only data.
References
- Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid–regulated gene expression. Plant Cell 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley, W.R., and Fitch, W.M. (1997). A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. USA 94, 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley, W.R., Therhalle, W., and Dress, A. (1999). Positional dependence, cliques and predictive motifs in the bHLH protein domain. J. Mol. Evol. 48, 501–516. [DOI] [PubMed] [Google Scholar]
- Baudino, T.A., and Cleveland, J.L. (2001). The Max network gone mad. Mol. Cell. Biol. 21, 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beischlag, T.V., Wang, S., Rose, D.W., Torchia, J., Reisz-Porszasz, S., Muhammad, K., Nelson, W.E., Probst, M.R., Rosenfeld, M.G., and Hankinson, O. (2002). Recruitment of the NcoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol. Cell. Biol. 22, 4319–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornberg-Bauer, E., Rivals, E., and Vingron, M. (1998). Computational approaches to identify leucine zippers. Nucleic Acids Res. 26, 2740–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie, P., Ceska, T., Lamers, M., Romier, C., Stier, G., Teo, H., and Suck, D. (1997). The crystal structure of an intact human Max-DNA complex: New insights into mechanisms of transcriptional control. Structure 5, 509–520. [DOI] [PubMed] [Google Scholar]
- Brunak, S., Engelbrecht, J., and Knudsen, S. (1991). Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 220, 49–65. [DOI] [PubMed] [Google Scholar]
- Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B.-H., Hong, X., Agarwal, M., and Zhu, J.-K. (2003). ICE: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarapica, R., Rosati, J., Cesareni, G., and Nasi, S. (2003). Molecular recognition in helix-loop-helix and helix-loop-helix leucine zipper domains. J. Biol. Chem. 278, 12182–12190. [DOI] [PubMed] [Google Scholar]
- Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozatier, M., Valle, D., Dubois, L., Ibnsouda, S., and Vincent, A. (1996). collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr. Biol. 6, 707–718. [DOI] [PubMed] [Google Scholar]
- Dang, C.V., Dolde, C., Gillison, M.L., and Kato, G.J. (1992). Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc. Natl. Acad. Sci. USA 89, 599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhordain, P., Lin, R.J., Quief, S., Lantoine, D., Kerckaert, J.P., Evans, R.M., and Albagli, O. (1998). The LAZ3 (BCL-6) oncoprotein recruits SMRT/mSIN3A/histone deacetylase containing complex to mediate transcription. Nucleic Acids Res. 26, 4645–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois, L., and Vincent, A. (2001). The COE-Collier /Olf1/EBF transcription factors: Structural conservation and diversity of developmental functions. Mech. Dev. 108, 3–12. [DOI] [PubMed] [Google Scholar]
- Ellenberg, T., Fass, D., Arnaud, M., and Harrison, S. (1994). Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8, 970–980. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Fairchild, C.D., Schumaker, M.A., and Quail, P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14, 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fairman, R., Beran-Steed, R.K., Anthony-Cahill, S.J., Lear, J.D., Strafford, W.F., 3rd, DeGrado, W.F., Benfield, P.A., and Brenner, S.L. (1993). Multiple oligomeric states regulate the DNA binding of helix-loop-helix peptides. Proc. Natl. Acad. Sci. USA 90, 10429–10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D'Amare, A.R., Pognonec, P., Roeder, R.G., and Burley, S.K. (1994). Structure and function of the b/HLH/Z domain of USF. EMBO J. 13, 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, A., and Caudy, M. (1998). The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays 20, 298–306. [DOI] [PubMed] [Google Scholar]
- Fisher, F., and Goding, C.R. (1992). Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 11, 4103–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen, D.M., Nemhauser, J., Muramitsu, T., Maloof, J.N., Alonso, J., Ecker, J.R., Furuya, M., and Chory, J. (2002). Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuji, Y., Shimizu, T., Toda, T., Yaganida, M., and Hakoshima, T. (2000). Structural basis for the diversity of DNA recognition by bZip transcription factors. Nat. Struct. Biol. 7, 889–893. [DOI] [PubMed] [Google Scholar]
- Grandori, C., Cowley, S.M., James, L.P., and Eisenman, R.N. (2000). The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16, 653–699. [DOI] [PubMed] [Google Scholar]
- Hebsgaard, S.M., Korning, P.G., Tolstrup, N., Engelbrecht, J., Rouze, P., and Brunak, S. (1996). Splice site prediction in Arabidopsis thaliana DNA by combining local and global sequence information. Nucleic Acids Res. 24, 3439–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim, M.A., Jacoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Heisler, M.G.B., Atkinson, A., Bylstra, Y.H., Walsh, R., and Smith, D.R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128, 1089–1098. [DOI] [PubMed] [Google Scholar]
- Hua, X., Yokoyama, C., Wu, J., Briggs, M.R., Brown, M.S., Goldstein, J.L., and Wang, X. (1993). SREBP-2, a second basic-helix-loop-helix leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. USA 90, 11603–11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq, E., and Quail, P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21, 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, L.M., Aravind, L., Bork, P., Hofmann, K., Mushegian, A.R., Zhulin, I.B., and Koonin, E.V. (2003). Quoderat demostrandum? The mystery of experimental validation of apparent erroneous computational analyses of protein sequences. Genome Biol. 2, RESEARCH0051.1–RESEARCH0051.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedmann, J., Kroi, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Ledent, V., and Vervoort, M. (2001). The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 11, 754–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levens, D.L. (2003). Reconstructing MYC. Genes Dev. 17, 1071–1077. [DOI] [PubMed] [Google Scholar]
- Littlewood, T., and Evan, G.I. (1998). Helix-Loop-Helix Transcription Factors, 3rd ed. (New York: Oxford University Press).
- Ludwig, S.R., Habera, L.F., Dellaporta, S.L., and Wessler, S.R. (1989). Lc, a member of the maize R gene family responsible for tissue specific anthocyanin production, encodes a protein similar to transcriptional activators and contains a myc-homology region. Proc. Natl. Acad. Sci. USA 86, 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, P.C.M., Rould, M.A., Weintraub, H., and Pabo, C.O. (1994). Crystal structure of MyoD bHLH domain-DNA recognition and implications for transcriptional activation. Cell 77, 451–459. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia, J., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Massari, M.E., and Murre, C. (2000). Helix-loop-helix proteins: Regulators of transcription in eukaryotic organisms. Mol. Cell. Biol. 20, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewes, H.W., Frishman, D., Guldener, U., Mannhaupt, G., Mayer, K., Mokrejs, M., Morgenstern, B., Munsterkotter, M., Rudd, S., and Weil, B. (2002). MIPS: A database for genomes and protein sequences. Nucleic Acids Res. 30, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, B., and Atchley, W.R. (1999). Evolution of bHLH transcription factors: Modular evolution by domain shuffling? Mol. Biol. Evol. 16, 1654–1663. [DOI] [PubMed] [Google Scholar]
- Murre, C., McCaw, P.S., and Baltimore, D. (1989). A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD and myc proteins. Cell 56, 777–783. [DOI] [PubMed] [Google Scholar]
- Nair, S.K., and Burley, S.K. (2000). Recognizing DNA in the library. Nature 404, 715–717. [DOI] [PubMed] [Google Scholar]
- Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400, 781–784. [DOI] [PubMed] [Google Scholar]
- Paroush, Z., Finley, R.L., Jr., Kidd, T., Wainwright, S.M., Ingham, P.W., Brent, R., and Ish-Horowicz, D. (1994). Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79, 5805–5815. [DOI] [PubMed] [Google Scholar]
- Payne, C.T., Zhang, F., and Lloyd, A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through the interaction with GL1 and TTG1. Genetics 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pscherer, A., Dorflinger, U., Kifel, J., Gawas, K., Ruschoff, J., Buettner, R., and Schule, R. (1996). The helix-loop-helix transcription factor SEF-2 regulates the activity of a novel initiator element in the promoter of the human somatostatin receptor II gene. EMBO J. 15, 6680–6690. [PMC free article] [PubMed] [Google Scholar]
- Puri, P.L., Avantaggiati, M.L., Balsano, C., Sang, N., Graessmann, A., Giordano, A., and Levrero, M. (1997). p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 16, 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2000). Phytochrome interacting factors. Semin. Cell Dev. Biol. 11, 457–466. [DOI] [PubMed] [Google Scholar]
- Rajani, S., and Sundaresan, V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11, 1914–1922. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2109. [DOI] [PubMed] [Google Scholar]
- Robinson, K.A., Koepke, J.I., Kharodawala, M., and Lopes, J.M. (2000). A network of yeast basic helix-loop-helix interactions. Nucleic Acids Res. 28, 4460–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, A.L., Meisterernst, M., Pognonec, P., and Roeder, R.G. (1991). Cooperative interaction of an initiator-binding transcription factor and the basic-helix-loop-helix activator USF. Nature 354, 245–248. [DOI] [PubMed] [Google Scholar]
- Shimizu, T., Toumoto, A., Ihara, K., Shimizu, M., Kyogoku, Y., Ogawa, N., Oshima, Y., and Hakoshima, T. (1997). Crystal structure of PHO4 bHLH domain-DNA complex: Flanking base recognition. EMBO J. 16, 4689–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakata, M., Friedman, F.K., Wei, Q., and Patterson, B. (1993). Dimerization specificity of myogenic helix-loop-helix DNA binding factors directed by nonconserved hydrophilic residues. Genes Dev. 7, 2456–2470. [DOI] [PubMed] [Google Scholar]
- Smolen, G.A., Pawlowski, L., Wilensky, S.E., and Bender, J. (2002). Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics 161, 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, A.M., Krober, S., Unte, U.S., Hujser, P., Dekker, K., and Saedler, H. (2003). The Arabidopsis Aborted Microspores (AMS) gene encodes a MYC class transcription factor. Plant J. 33, 413–423. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., et al. (2001). The sequence of the human genome. The Celera Genomics Human Genome Sequencing Consortium. Science 291, 1304–1350.11181995 [Google Scholar]
- Waterston, R.H., et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Mouse Genome Sequencing Consortium. Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- Winston, R.L., and Gottesfeld, J.M. (2000). Rapid identification of key amino-acid-DNA contacts through combinatorial peptide synthesis. Chem. Biol. 7, 245–251. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Tepperman, J.M., Fairchild, C.D., and Quail, P.H. (2000). Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 21, 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.