Abstract
The recombination protein RAD51 is a component of the meiotic recombination pathway and has been proposed to play a role in the homology search, a process by which homologous chromosomes find each other before they pair in the prophase of meiosis. To study the relationship between recombination and chromosome pairing, we examined the distribution of RAD51 foci on meiotic chromosomes in maize mutants with defects in chromosome pairing. The patterns of RAD51 distribution showed dramatic variation among the meiotic mutants. The mutants generally exhibited significant decreases in the number of RAD51 foci at zygotene, corresponding to the degree of their pairing defects. These results provide evidence for a key role of RAD51 structures in the homology search.
INTRODUCTION
During early prophase of meiosis, homologous chromosomes find each other and pair (John, 1990). This process likely involves a step of DNA homology recognition (the homology search) and a subsequent step of intimate codirectional alignment of the homologous chromosomes (pairing). Because the genomes of most plants contain large fractions of repetitive sequences, the homology search mechanism has the difficult task of restricting pairing to truly homologous sequences and preventing ectopic pairing interactions. In most organisms, including maize and other plants, meiotic chromosome pairing is preceded by the formation of a polarized chromosome arrangement with clustered telomeres (“bouquet”), which is thought to facilitate homologous chromosome pairing (Bass et al., 2000; Niwa et al., 2000; Trelles-Sticken et al., 2000; Cowan et al., 2001; Scherthan, 2001). Despite great progress made in understanding many other processes that occur in early meiotic prophase (Roeder, 1997; Zickler and Kleckner, 1999; Anderson and Stack, 2003), the homology search process remains elusive.
In most organisms, including plants, successful pairing of homologous chromosomes depends on the meiotic recombination pathway initiated by the topoisomerase SPO11, which creates double strand breaks (DSBs) on meiotic chromosomes (Keeney et al., 1997; Baudat et al., 2000; Romanienko and Camerini-Otero, 2000; Grelon et al., 2001; Villeneuve and Hillers, 2001). In budding yeast, chromosome pairing also has been demonstrated to require passage through the early stages of the recombination pathway after the introduction of the DSBs (Peoples et al., 2002).
One of the components of the early meiotic recombination pathway that has been proposed, but not generally accepted, to play a major role in the homology search is RAD51, a eukaryotic homolog of the bacterial RecA protein (Ashley et al., 1995; Rockmill et al., 1995; Barlow et al., 1997; Franklin et al., 1999; Masson and West, 2001; Burgess, 2002; Moens et al., 2002). RAD51 is a recombination protein that binds to single-stranded DNA, forming a nucleoprotein filament, which then invades double-stranded DNA to form a heteroduplex joint (Masson and West, 2001; Shibata et al., 2001). To accomplish heteroduplex formation, the RAD51 nucleoprotein filament has the ability to recognize DNA sequence homology. In vitro, the RAD51 protein can, even in the absence of other proteins, promote the extensive and efficient pairing of DNA molecules several kilobases long (Eggler et al., 2002). Furthermore, when overexpressed in human cells, RAD51 significantly increases the efficiency of gene targeting, which highlights its ability to find homologous DNA sequences (Yanez and Porter, 1999).
RAD51 is required for meiotic recombination (Shinohara et al., 1992), but RAD51 mutants are also defective in the pairing of homologous chromosomes in addition to being defective in recombination (Weiner and Kleckner, 1994; Rockmill et al., 1995). RAD51, together with its meiosis-specific homolog DMC1, forms mixed protein complexes visible as foci on chromosomes during the meiotic prophase (Bishop, 1994; Terasawa et al., 1995; Tarsounas et al., 1999). These complexes are likely the early recombination nodules seen in immunoelectron microscopy, because RAD51 has been shown to be a component of these nodules (Anderson et al., 1997). In the mouse, DMC1, and presumably also RAD51, have been shown to accumulate at the sites of DSBs (Mahadevaiah et al., 2001). The appearance of RAD51 foci on chromosomes depends on the presence of DSBs; RAD51 foci are absent in yeast and mouse spo11 knockouts, which lack meiotic DSBs (Gasior et al., 1998; Baudat et al., 2000; Romanienko and Camerini-Otero, 2000).
The dynamics of RAD51 foci during meiosis have been studied in great detail using three-dimensional microscopy in wild-type maize (Franklin et al., 1999). RAD51 foci in maize are seen first at the beginning of zygotene, before the start of chromosome pairing, and reach their peak of ∼500 foci per nucleus at mid-zygotene. The foci are found mostly on unpaired chromosomes and disappear as chromosomes pair. Contiguous, dumbbell-shaped structures formed by two paired RAD51 foci frequently are found on recently paired chromosomes. These structures have been proposed to be sites where DNA sequence homology is compared (Franklin et al., 1999). During pachytene, the number of RAD51 foci decreases to ∼7 to 22 per nucleus, corresponding approximately to the number of chiasmata in maize meiosis ( Franklin et al., 1999).
The number of zygotene RAD51 foci differs from species to species and is considerably higher than the number of crossover exchanges during meiosis (Franklin et al., 1999; Moens et al., 2002). It has been suggested that the number of zygotene RAD51 foci is correlated with the length of the chromosomes, whereas the number of crossovers corresponds to the number of chromosome arms (Zickler and Kleckner, 1999; Anderson et al., 2001; Moens et al., 2002) . An intriguing question has been raised about the function of the excess RAD51 foci. It led to the suggestion that these foci may be involved in the homology search ( Rockmill et al., 1995; Franklin et al., 1999; Moens et al., 2002; Stack and Anderson, 2002) . Because the chromosome homology search has not been dissected at a molecular level, this hypothesis has not been verified experimentally.
Maize has an impressive collection of meiotic mutants, many of which are defective in the pairing of homologous chromosomes (Beadle, 1930; Golubovskaya and Mashenkov, 1975, 1976; Golubovskaya and Khristolyubova, 1985; Golubovskaya, 1989; Golubovskaya et al., 2003). To understand the relationship between homologous chromosome pairing and recombination, we studied the distribution patterns of the RAD51 recombination protein in seven maize meiotic mutants with defects in the pairing of homologous chromosomes. We found dramatic abnormalities in the distribution patterns of RAD51 foci in the meiotic mutants, especially with regard to the number of RAD51 foci, which correlated well with the degree of pairing defects in the mutants. By analyzing many mutants with pairing defects, we concluded that RAD51, and the protein complex with which it acts, is one of the key elements in the process of homologous chromosome pairing. These results provide novel evidence for a role of RAD51 in the meiotic homology search.
RESULTS
Maize Meiotic Prophase Mutants Can Be Ordered According to the Degree of Their Chromosome Pairing Defects
To study the interaction between chromosome pairing and the RAD51-mediated recombination events during meiosis, we selected seven maize meiotic mutants (Table 1) with defects in homologous chromosome pairing. Six of the mutants, as1, dsy1-1, dsy9901, dsyCS, mtm99-25, and segII-513, appear to have primary defects in this process. The seventh mutant, afd1, also is defective in homologous pairing, but its primary defect is more likely related to the abnormal chromosome structure and the chromatin condensation pattern at the leptotene-zygotene transition as well as to the premature loss of sister chromatid cohesion during prophase I (Golubovskaya and Mashenkov, 1975; Golubovskaya and Khristolyubova, 1985) (Table 1).
Table 1.
Maize Meiotic Mutants with Defects in Homologous Chromosome Pairing and Synapsis
| Mutant | Phenotype | References |
|---|---|---|
| afd1 | Premature condensation of chromosomes during early prophase I. No pairing of homologous chromosomes, only univalents at metaphase I. Defects in sister chromatid cohesion; centromeres precociously divide at metaphase I, resulting in equational segregation of chromosomes in meiosis I. Only short, highly abnormal fragments of synaptonemal complex are formed; synapsed regions are only ∼12% of total synaptonemal complex length. |
Golubovskaya and Mashenkov, 1975; Golubovskaya and Khristolyubova, 1985 |
| as1 | Incomplete pairing of homologs at pachytene. Univalents and bivalents present at metaphase I. Meiotic defects variable and seen in ∼50% of meiocytes, whereas the other 50% of meiocytes undergo normal meiosis. Axial elements form normally, but synaptonemal complex found predominantly in terminal regions of chromosome pairs, whereas the interstitial regions have incomplete and abnormal synapsis. |
Beadle, 1930; Miller, 1963; Maguire, 1978a, 1978b; Maguire and Riess, 1991; I.N. Golubovskaya, unpublished data |
| dsy1-1 | High frequency of univalents but bivalents occasionally also present at metaphase I. Axial elements form normally, but synapsis is mostly abnormal: unsynapsed chromosomes, promiscuous synapsis (chromosomes switching pairing partners along their lengths), and intrachromosomal synapsis frequent at pachytene. |
Golubovskaya and Mashenkov, 1976; Timofeeva and Golubovskaya, 1991; Maguire et al., 1993 |
| dsy9901 | Mild defects in chromosome pairing: on average, eight bivalents present at metaphase I. | Golubovskaya et al., 2003 |
| dsyCS | Severe defects in chromosome pairing: almost exclusively univalents at metaphase I. | Staiger, 1990; Golubovskaya et al., 2003 |
| mtm99-25 | Severe defects in chromosome pairing: mostly univalents and only occasionally bivalents present at metaphase I. |
Golubovskaya et al., 2003 |
| segII-513 | Severe defects in chromosome pairing: almost exclusively univalents at metaphase I. | Golubovskaya et al., 2003 |
The defects in chromosome pairing lead to improper synapsis, i.e., the process of “zipping-up” the paired chromosomes by the synaptonemal complex proteins, because homologous chromosomes are not aligned correctly. As a result, defects in homologous chromosome pairing can be seen as incomplete synapsis and synapsis between nonhomologous chromosomes on synaptonemal complex spreads (Table 1). Pairing defects also are manifested by the presence of unpaired chromosomes during pachytene and the absence of chiasmata, because the chromosomes do not undergo crossing over. The lack of chiasmata leads to the presence of univalents instead of bivalents at metaphase I (Beadle, 1930; Miller, 1963; Golubovskaya and Mashenkov, 1975, 1976; Golubovskaya et al., 2003).
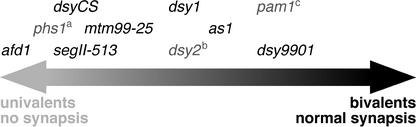
The seven maize meiotic mutants were selected so that they display a range of phenotypes with regard to the severity of their pairing defects. Using information available in the literature (Beadle, 1930; Miller, 1963; Golubovskaya and Mashenkov, 1976; Maguire, 1978a, 1978b; Golubovskaya and Khristolyubova, 1985; Golubovskaya, 1989; Staiger, 1990; Maguire and Riess, 1991; Timofeeva and Golubovskaya, 1991; Maguire et al., 1993; Golubovskaya et al., 2003), we ordered the mutants relative to each other according to the severity of their phenotypes (Figure 1). As the main criterion, we used the average number of bivalents at metaphase I (Miller, 1963; Golubovskaya and Mashenkov, 1976; Golubovskaya and Khristolyubova, 1985; Golubovskaya et al., 2003). We also considered data, where they were available, on the degree of abnormalities in synapsis observed in synaptonemal complex spreads (i.e., the presence and frequency of incomplete or nonhomologous synapsis) (Table 1).
Figure 1.
Ordering of Maize Meiotic Mutants with Regard to the Severity of Their Defects in Homologous Chromosome Pairing.
To order the mutants relative to each other, we used the average number of bivalents at metaphase I reported in the literature (Miller, 1963; Golubovskaya and Mashenkov, 1976; Golubovskaya and Khristolyubova, 1985; Golubovskaya et al., 2003) and the occurrence of incomplete or nonhomologous synapsis in synaptonemal complex spreads (see Table 1). Mutants whose RAD51 phenotypes are described elsewhere are shown in gray: a W.P. Pawlowski, I.N. Golubovskaya, and W.Z. Cande, unpublished data; b Franklin et al., 2003; c Golubovskaya et al., 2002.
Ordering of the seven meiotic mutants (Figure 1) according to these criteria indicated that afd1, dsyCS, and segII-513 exhibit the most severe pairing defects and have almost exclusively univalents at metaphase I (Golubovskaya and Mashenkov, 1975; Golubovskaya et al., 2003). dsy1-1 and mtm99-25 also display severe pairing defects, although less severe than the previous group (Timofeeva and Golubovskaya, 1991; Golubovskaya et al., 1997, 2003). In these two mutants, mostly univalents but also a few bivalents are found at the end of prophase I. as1 and dsy9901 show the least severe pairing defects, as manifested by the presence of a moderate to high number of bivalents at metaphase I (Beadle, 1930; Miller, 1963; Golubovskaya et al., 2003). We also include in Figure 1 three other maize meiotic prophase mutants, pam1, dsy2, and phs1, that were analyzed for RAD51 distribution but the results of which are being published elsewhere (Golubovskaya et al., 2002; Franklin et al., 2003; W.P. Pawlowski, I.N. Golubovskaya, and W.Z. Cande, unpublished data).
Expression of the RAD51 Protein during Meiosis Is Not Affected in the Meiotic Mutants
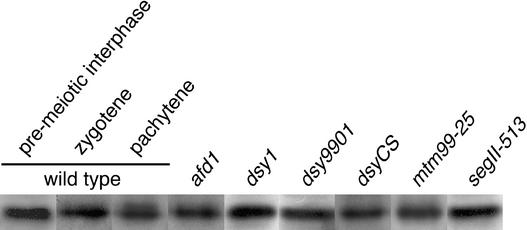
To determine whether the RAD51 protein was present during meiotic prophase in the mutants, we performed protein gel blot analysis with whole-protein extracts from meiotic anthers. Consistent with its status as an ancient tetraploid (Doebley et al., 1990), maize has two rad51 genes that encode proteins that are both ∼37 kD long and share 90% of their amino acid sequence (Franklin et al., 1999). Although antibodies generated against maize ZmRAD51 proteins are available, they do not recognize the native proteins in immunocytological preparations of maize meiocytes (Franklin et al., 1999). Therefore, we used a polyclonal antibody raised against the human HsRAD51 protein (Terasawa et al., 1995) that has been used successfully in maize immunocytological preparations (Franklin et al., 1999). The two ZmRAD51 proteins show nearly 100% identity in the region that is most similar to human RAD51 (Franklin et al., 1999) and most likely are both recognized by the anti-HsRAD51 antibody.
In wild-type maize anthers, Franklin et al. (1999) detected the RAD51 protein first during premeiotic interphase. Its level increased in leptotene and zygotene and remained high throughout the rest of meiosis. Our analyses of control wild-type maize anthers showed similar results (Figure 2, control lanes), except that we observed an increased level of the RAD51 protein even during premeiotic interphase.
Figure 2.
Immunoblot Detection of RAD51 Proteins in Normal and Mutant Maize Anthers.
In each lane, 50 μg of total protein was loaded.
We analyzed all meiotic mutants except as1 for the presence of RAD51 proteins in mid-zygotene anthers, in which RAD51 is present at the highest level in wild-type maize. RAD51 was detected in all mutants at approximately the same level as in the wild-type maize anthers (Figure 2).
Shape and Distribution of RAD51 Foci in Meiotic Mutants
We used three-dimensional immunofluorescence microscopy (Franklin et al., 1999) to analyze the nuclear localization of RAD51 proteins in meiotic stages ranging from early leptotene to meta- phase I. A traditional criterion used to distinguish between the zygotene and pachytene stages of meiotic prophase is based on the completion of homologous chromosome pairing by the end of zygotene and the presence of only paired chromosomes in pachytene. This criterion is not useful for staging meiotic mutants because most of them exhibit very reduced chromosome pairing at pachytene. However, the superb cytology of maize meiocytes allows the use of chromosome morphology—as defined by stage-specific changes in their thickness, compaction, and level of condensation—to stage mutant meiocytes (Golubovskaya et al., 2002).
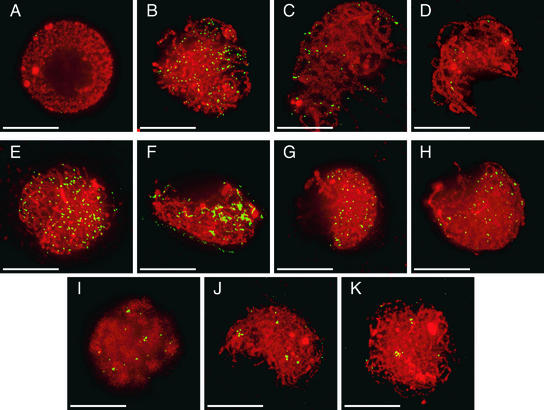
As a control, we analyzed the localization of RAD51 in normal meiocytes of the maize inbred line A344 (Figures 3A to 3D). We also used wild-type siblings from several of the mutants analyzed (data not shown). These two classes of normal meiocytes exhibited patterns of ZmRAD51 localization similar to those described previously (Franklin et al., 1999). RAD51 was diffuse within the nucleus of normal maize meiocytes during leptotene (Figure 3A). In zygotene, the protein formed distinct foci on chromosomes. The number of foci increased with the progression of zygotene, reaching its peak at mid-zygotene, with an average of ∼500 foci per nucleus (Figure 3B, Table 2). The number of foci decreased during late zygotene (Figure 3C). In pachytene, there were, on average, 12 RAD51 foci per nucleus (Figure 3D).
Figure 3.
Distribution of RAD51 Foci in Wild-Type and Mutant Maize Meiocytes.
(A) to (D) RAD51 immunostaining in wild-type maize meiocytes in leptotene (A), zygotene (B), late zygotene (C), and pachytene (D).
(E) to (K) RAD51 staining in zygotene nuclei of maize meiotic mutants dsy1-1 (E), as1 (F), dsy9901 (G), mtm99-25 (H), afd1 (I), segII-513 (J), and dsyCS (K).
4′,6-Diamidino-2-phenylindole–stained chromatin is shown in red, and RAD51 is shown in green. All images are flat projections from several consecutive optical sections through three-dimensional nuclei. The RAD51 staining on the outside of the nuclear envelope was detected in all stages of prophase I in most mutants as well as in the wild-type controls and was reported previously in wild-type maize meiocytes (Franklin et al., 1999). It likely represents RAD51 protein being imported into the nucleus or a contaminating nonspecific staining of the HsRAD51 antibody preparation (Franklin et al., 1999). Bars = 10 μm.
Table 2.
Number of RAD51 Foci at Mid-Zygotene in Nuclei of Normal and Mutant Maize Meiocytes
| Genotype | No. of RAD51 Foci per Nucleus (mean ± se) |
Percent of Wild Type | Shape of Foci | Paired Foci | No. of Nuclei Examined |
|---|---|---|---|---|---|
| Wild type | 500 ± 47 | 100 | Round | Many | 9 |
| dsy1-1 | 503 ± 28 | 100 | Round | Some | 28 |
| as1 | 327 ± 35 | 65 | Elongated | Very few | 12 |
| dsy9901 | 237 ± 18 | 47 | Round | Few | 17 |
| mtm99-25 | 140 ± 10 | 28 | Round | Few | 17 |
| afd1 | 53 ± 2 | 11 | Round | Few | 28 |
| segII-513 | 13 ± 1 | 3 | Round | Many double and multiple | 12 |
| dsyCS | 10 ± 1 | 2 | Round | Many double and multiple | 46 |
As predicted by the presence of ZmRAD51 proteins in anthers in protein gel blot analysis, we detected RAD51 foci in prophase I meiocytes in all seven meiotic mutants. At leptotene, RAD51 was diffuse in the nucleus in all mutants studied (data not shown), just as it was in control wild-type meiocytes. At zygotene and pachytene, we observed dramatic differences in the patterns of RAD51 foci among the meiotic mutants with regard to the number and shape of foci (Figures 3E to 3K, Table 2).
Below, we describe the patterns of RAD51 foci in the mutants with the order of description based on the average number of foci in zygotene nuclei.
dsy1-1
In dsy1-1 mutant meiocytes, the shape and number of RAD51 foci at zygotene were very similar to those of wild-type zygotene meiocytes, with ∼500 foci per nucleus (Figure 3E, Table 2). The number of foci decreased in late zygotene and pachytene, albeit more slowly than in wild-type maize, and an average of 76 RAD51 foci per nucleus were observed at pachytene. In pachytene meiocytes, some double (paired) foci located on apparently paired chromosomes were present. However, unlike in wild-type maize, the two foci in the doublets most often were distinctly separated rather than joined together into dumbbell-shaped structures. A normal number of zygotene RAD51 foci also was observed previously in pam1, a mutant defective in telomere bouquet formation (Golubovskaya et al., 2002).
as1
In the as1 meiocytes, the number of RAD51 foci in zygotene was decreased by ∼30% compared with that in the wild type (Figure 3F, Table 2). We encountered only a few meiocytes at the pachytene stage in this mutant. Interestingly, they contained large numbers of RAD51 foci, 260 on average. In contrast to wild-type meiocytes and to the other meiotic mutants examined here, in as1, we noticed many unusually extended, “worm-like” RAD51 foci (Figures 3F and 4B; cf. wild-type foci in Figure 4A). We measured the length of a large number of RAD51 foci in as1 and found that they were, on average, three times as long as the foci in wild-type meiocytes (Table 3). Most of the lengthening of the RAD51 foci in as1 occurred during early zygotene, when they increased in length from an average of 0.6 ± 0.1 μm in early zygotene to an average of 1.1 ± 0.1 μm in mid-zygotene. Very few double foci were observed in as1. Similar RAD51 behavior, with the presence of worm-like foci and a decrease in the number of foci, was observed previously in another maize mutant, dsy2 (Franklin et al., 2003).
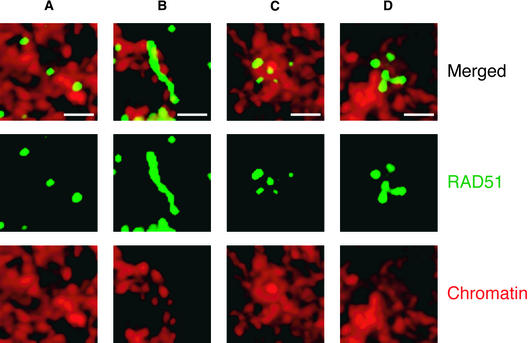
Figure 4.
Shape of RAD51 Foci in Wild-Type and Mutant Maize Meiocytes.
(A) Small, round RAD51 foci in normal maize.
(B) Extended foci in as1.
(C) and (D) Clusters of foci in dsyCS (C) and segII-513 (D). 4′,6-Diamidino-2-phenylindole–stained chromatin is shown in red, and RAD51 is shown in green. All images are flat projections from several consecutive optical sections through three-dimensional nuclei. Bars = 1 μm.
Table 3.
Length of RAD51 Foci in as1 and Wild-Type Meiocytes
| Variable | as1 | Wild Type |
|---|---|---|
| Average length of RAD51 foci (mean ± se) |
1.0 ± 0.1 μm | 0.3 ± 0.0 μm |
| Length of the longest RAD51 focus in the cell (mean ± se) |
3.1 ± 0.2 μm | 1.5 ± 0.2 μm |
| Number of foci measured per cell | 54 | 24 |
| Number of cells | 16 | 26 |
dsy9901
In the dsy9901 mutant, the number of foci at mid-zygotene was lower by ∼50% compared with the wild type (Figure 3G, Table 2). RAD51 foci tended to persist in greater numbers into pachytene, during which, on average, 61 foci per nucleus were present. Few double foci, either separate or joined together into the characteristic dumbbell-shaped structures, were found in zygotene and pachytene meiocytes in the dsy9901 mutant.
mtm99-25
The number of RAD51 foci in mtm99-25 mutant meiocytes at mid-zygotene was lower by ∼70% compared with that in the wild type (Figure 3H, Table 2). The foci also persisted in greater numbers into pachytene, although not as much as in dsy9901; the average number of RAD51 foci in pachytene nuclei was 41. Few double foci, either separate or joined together, were present in zygotene and pachytene mtm99-25 mutant meiocytes.
afd1, dsyCS, and segII-513
afd1, dsyCS, and segII-513 displayed dramatic reductions in the number of RAD51 foci, to ∼10% or less the number of foci seen in wild-type plants (Figures 3I to 3K, Table 2). In afd1, few double foci, separate or joined together, were found. In dsyCS and segII-513, on average, ∼30% of all foci were paired, similar to wild-type meiocytes, although the two foci in the doublets were almost never joined into contiguous RAD51 structures. Interestingly, dsyCS and segII-513 also exhibited large numbers of clusters of multiple (three to five) RAD51 foci in addition to the double foci. On average, 15% of RAD51 foci in dsyCS and 46% in segII-513 were found in such clusters (Figures 4C and 4D). The clusters were groups of foci that were close to each other in three-dimensional space. These foci were separate from each other and were never joined into contiguous structures; thus, they were clearly different from the worm-like elongated RAD51 foci observed in the as1 mutant.
We analyzed only a few dsyCS and segII-513 meiocytes in stages equivalent to pachytene. These meiocytes had numbers of RAD51 foci similar to the numbers found in the zygotene meiocytes in the respective mutants. Another maize mutant severely deficient in homologous chromosome pairing, phs1, also exhibited a dramatic reduction in the number of RAD51 foci to an average of three per nucleus (W.P. Pawlowski, I.N. Golubovskaya, and W.Z. Cande, unpublished data).
DISCUSSION
Our analyses show dramatic abnormalities in the distribution patterns of RAD51 foci in maize meiotic mutants with defects in homologous chromosome pairing, suggesting that the RAD51 complex plays a central role in the process of homologous pairing. In large-genome organisms, such as maize, <5% of all RAD51 foci eventually become sites of crossovers (Franklin et al., 1999; Moens et al., 2002) that are essential for meiotic recombination and for holding homologs together until anaphase I. Most of the meiotic mutants in this study exhibited numbers of RAD51 foci far greater than that needed for crossing over and for the formation of chiasmata. Yet, all of these maize mutants have significant defects in homologous chromosome pairing. It is apparent from these data that a full complement of RAD51 foci is essential for proper chromosome pairing. This paradox provides support for a role of RAD51 in the homology search process in addition to its role in meiotic recombination.
Loss of RAD51 Foci Is Correlated with Pairing Defects in Maize Meiotic Mutants
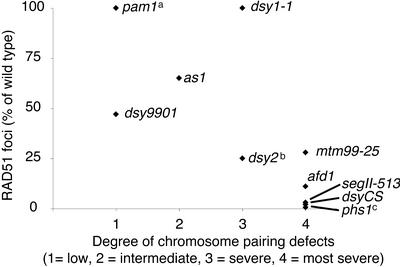
Because the maize meiotic mutants analyzed for RAD51 distribution exhibited a range of severity of defects in homologous pairing, we compared the patterns of RAD51 immunostaining in the mutants with the severity of their pairing defects (Figure 5). The severity of defects in the mutants generally corresponded to the reduction in the number of zygotene RAD51 foci. afd1, dsyCS, and segII-513 showed the most dramatic change in RAD51 distribution and were the most severely defective in chromosome pairing. Another maize mutant with a severe pairing defect, phs1, also belonged to this group and displayed an even more dramatic reduction in the number of RAD51 foci (W.P. Pawlowski, I.N. Golubovskaya, and W.Z. Cande, unpublished data). dsy9901, mtm99-25, and as1 as well as another recently studied maize mutant, dsy2 (Franklin et al., 2003), showed moderate pairing defects and exhibited an intermediate reduction in the numbers of RAD51 foci.
Figure 5.
Comparison of Defects in the Number and Shape of RAD51 Foci in Maize Meiotic Mutants with Defects in Homologous Chromosome Pairing.
To order the mutants relative to each other, we used the average number of bivalents at metaphase I as reported in the literature (Miller, 1963; Golubovskaya and Mashenkov, 1976; Golubovskaya and Khristolyubova, 1985; Golubovskaya et al., 2003) and the occurrence of incomplete or nonhomologous synapsis in synaptonemal complex spreads (see Table 1). Mutants in class 1 (low degree of pairing defects) show mostly bivalents in metaphase I and largely homologous synapsis. Class 2 (intermediate) mutants show, on average, 50% bivalents in metaphase I and some, albeit incomplete, synapsis. Mutants in class 3 (severe) show very few bivalents in metaphase I (on average, two per meiocyte) and mostly abnormal synapsis. Mutants in class 4 (most severe) have hardly any bivalents in metaphase I (<0.5 per meiocyte) and very little homologous synapsis, if any. Data for the indicated mutants are from the following sources: a Golubovskaya and Mashenkov, 1977; Golubovskaya et al., 2002; b Franklin et al., 2003; c W.P. Pawlowski, I.N. Golubovskaya, and W.Z. Cande, unpublished data.
dsy1-1 represents a clear exception to the correlation between the number of zygotene RAD51 foci and the severity of the pairing defect. This mutant shows strong defects in chromosome pairing but has a normal number of RAD51 foci. Its phenotype is reminiscent of that of another maize meiotic mutant, pam1, which is defective in the pairing of homologous chromosomes but also has a normal number of RAD51 foci (Golubovskaya et al., 2002). The pam1 mutant is known to be defective in telomere bouquet formation (Golubovskaya et al., 2002). It was proposed recently that dsy1-1 has a similar defect in telomere clustering (Bass et al., 2003). These results suggest that there are two general classes of meiotic prophase mutants in maize: those with defects in the early steps of the meiotic recombination pathway, and those with defects in telomere bouquet formation or other aspects of nuclear organization. The presence of the two classes of mutants underscores the importance of both the recombination pathway and prophase nuclear organization for the successful completion of chromosome pairing.
Completion of Chromosome Pairing Is Required for the Prompt Disappearance of RAD51 Foci in Pachytene
In pachytene, the number of RAD51 foci in wild-type maize decreased dramatically to ∼12 per nucleus. By contrast, many more RAD51 foci persisted into pachytene in the meiotic mutants. We observed this phenomenon previously in the bouquet-defective pam1 mutant (Golubovskaya et al., 2002). RAD51 foci also were shown to persist longer on asynapsed autosomes and segments of X and Y chromosomes in wild-type and triploid mouse cells (Barlow et al., 1997). Our data showing the persistence of RAD51 foci in pachytene in several meiotic mutants indicate that this is a general phenomenon and suggest that the completion of homologous pairing is required for the removal of RAD51 from chromosomes.
Abnormal Morphology of RAD51 Foci Is Associated with Pairing Defects
Paired, dumbbell-shaped RAD51 foci appear in significant numbers in wild-type maize meiocytes at mid-zygotene and represent, on average, 34% of all RAD51 foci in late zygotene (Franklin et al., 1999). This generally was not the case in the mutants. We found few or very few double foci in afd1, as1, dsy9901, and mtm99-25. In dsy1-1, the meiocytes exhibited some double foci; interestingly, however, the foci were never joined into contiguous, dumbbell-shaped structures. Because the double RAD51 foci have been proposed to be sites of the homology search (Franklin et al., 1999), their complete or nearly complete absence in meiotic mutants with defects in the homology search process are consistent with RAD51 playing an active role in homologous chromosome pairing.
In the as1 mutant, we observed abnormally elongated, or worm-like, RAD51 foci. The substantial increase in the length of these RAD51 foci occurs in early zygotene, when chromosome pairing is just initiated and few double foci are present (Franklin et al., 1999). Therefore, we propose that the abnormally extended RAD51 structures are formed by the uncontrolled growth of RAD51 complexes, presumably when they are searching for homology, and not by stretching of the dumbbell-shaped paired foci. Similar worm-like RAD51 foci were described recently in another maize mutant, dsy2 (Franklin et al., 2003).
In two mutants, dsyCS and segII-513, we observed distinct clusters of RAD51 foci. These two mutants showed the most severe pairing defects and the most dramatically decreased numbers of RAD51 foci. Because very few foci were present in dsyCS and segII-513 nuclei, it is unlikely that several foci would be located so close to each other by chance. It also is unlikely that these clusters of RAD51 foci are equivalents of the dumbbell-shaped RAD51 foci that connect presumably homologous DNA sites in wild-type meiocytes, because there is very little homologous pairing in the two mutants (Golubovskaya et al., 2003). Moreover, the multiple foci clusters obviously cannot all represent homologous sites.
We propose that the presence of these clusters of RAD51 foci may be the result of a mechanism that brings RAD51 foci together. RAD51 foci may associate with each other when chromosomes move around in the nucleus during bouquet formation. Because unpaired chromosomes carry large numbers of RAD51 foci and the already paired chromosomes have very few foci, bringing RAD51 foci together also would bring together chromosomes searching for their pairing partners. If there was a bias in the distribution of RAD51 foci on chromosomes—for example, if they were more frequent near the clustered telomeres—transient RAD51 interactions would be very useful in coaligning chromosomes, even if these interactions were not dependent on homology. This homology-independent step would generate preliminary pairing associations of chromosomes that would facilitate the subsequent homology identification steps. However, it also is possible that the clusters of RAD51 foci are present only in these two mutants and result from the presence of very few DSBs per nucleus, all of which are located near each other. Because DSBs are prerequisites for the formation of RAD51 foci on chromosomes, the nonrandom distribution of DSBs could lead to apparent clusters of RAD51 foci.
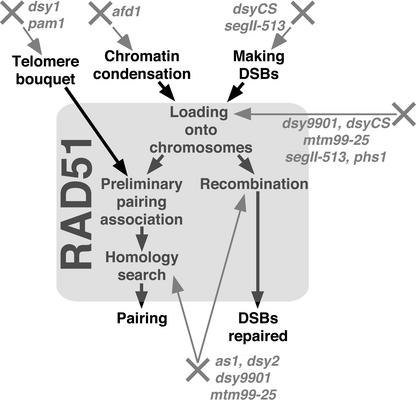
Our Model for RAD51 Function
Based on our analyses of maize mutants defective in homologous chromosome pairing, we propose that (1) pairing-defective maize mutants that are not defective in bouquet formation show abnormal distribution of RAD51 foci on zygotene and pachytene chromosomes; (2) the degree of chromosome pairing defects in these mutants generally correlates with the decrease in the number of RAD51 foci; and (3) there is a mechanism that controls the growth of the RAD51 foci as they search for homology. These findings, in conjunction with known aspects of the behavior of RAD51 and RAD51 foci, suggest that RAD51, and/or the recombination complex in which it acts, plays a key role in the process of the homology search in addition to its well-documented role in meiotic recombination.
We propose a model (Figure 6) in which RAD51 foci are installed on the sites of DSBs on zygotene chromosomes and the RAD51-carrying chromosomes interact with each other in a preliminary recognition-pairing association step. These interactions are stabilized when homology is found through the action of the RAD51 complexes as they repair the DSBs. The abnormal RAD51 distribution patterns and other specific defects in our maize meiotic mutants suggest that the mutants likely are defective in various aspects of this homology search/recombination pathway. Further characterization of these mutants, especially finding mutants defective in recombination but not in the homology search, will help dissect the role of RAD51 in this process. Maize is an ideal organism in which to conduct this type of study because it exhibits a large number of RAD51 foci that are amenable to quantitative analyses and at the same time is a genetically tractable organism.
Figure 6.
Proposed Roles of RAD51 in the Meiotic Pathway of Recombination and the Homology Search and Possible Defects in the Maize Meiotic Mutants Analyzed in This and Previous Studies.
Some mutants are listed as affecting more than one process because their primary defects are unknown.
METHODS
Plant Material
Maize (Zea mays) plants were grown in the greenhouse, and immature tassels were harvested before emergence. Mutant plants were identified and anthers were staged on squash slides of a light microscope according to criteria described in detail previously (Golubovskaya et al., 2002).
Protein Gel Blot Analyses
Anthers were dissected from preliminarily staged tassels. From each flower, two of the three synchronously developing anthers were collected, whereas the third anther was used for precise staging. Protein extracts were prepared from 6 to 10 anthers by grinding them in liquid nitrogen followed by extraction in buffer containing 8 M urea, 2% SDS, 50 mM Tris, pH 6.8, 2 mM EDTA, pH 8.0, and 2% 2-mercaptoethanol. Protein concentration in extracts was measured using the Bio-Rad Protein Assay (Bio-Rad, Richmond, CA). Approximately 50 μg of total protein was loaded per lane. After transfer of protein to a polyvinylidene difluoride membrane (Immobilon P; Millipore, Bedford, MA), the membrane was stained briefly with Ponceau S to reassess protein loading, incubated with an anti-HsRAD51 antibody (Terasawa et al., 1995) diluted 1:500, and incubated with an anti-rabbit antibody conjugated with horseradish peroxidase (Pierce Chemical, Rockford, IL). Protein bands were visualized using the Lumi-Light protein gel blot kit (Pierce Chemical).
Immunofluorescence Microscopy
Staged anthers were fixed for 45 min in buffer containing 4% formaldehyde to preserve the native structure of chromosomes in the meiocytes (Dawe et al., 1994). Meiocytes were extruded from cut anthers and embedded in 5% polyacrylamide to maintain the three-dimensional structure of the nuclei (Bass et al., 1997). Immunofluorescent detection of RAD51 in maize meiocytes was performed essentially as described previously (Franklin et al., 1999) with slight modifications. Polyacrylamide pads containing meiocytes were washed twice in 1× PBS for 10 min. The meiocytes then were permeabilized for 1 h in 1× PBS, 1% Triton X-100, and 1 mM EDTA. The samples were blocked for 2 h in 1× PBS, 3% BSA, 5% normal donkey serum, 1 mM EDTA, and 0.1% Tween 20 and incubated overnight in a humid chamber with 50 μL of a 1:500 dilution of the anti-HsRAD51 antibody (Terasawa et al., 1995) precleared for 30 min at 4°C with Escherichia coli XL1-Blue acetone powder.
The samples were washed in 1× PBS, 0.1% Tween 20, and 1 mM EDTA eight times, 1 h for each wash, with continued washing overnight. For detection with a secondary antibody, the samples were incubated overnight with a fluorescein isothiocyanate–labeled donkey anti-rabbit (Fab′)2 fragment (Jackson ImmunoResearch, West Grove, PA) at 1 μg/mL in blocking buffer. This step was followed by the same washing protocol used for the primary antibody incubation. The samples then were washed in 1× PBS twice for 10 min and stained with 10 μg/mL 4′,6-diamidino-2-phenylindole in 1× PBS for 30 min followed by three 10-min washes in 1× PBS. Prepared slides were mounted in 1,4-diazabicyclo-[2,2,2]octane, sealed with fingernail polish, and stored at −20°C.
Three-dimensional stacks of images were collected on a DeltaVision imaging station (Applied Precision, Seattle, WA) with optical sections 200 nm apart, subjected to constrained iterative deconvolution (Chen et al., 1995), and analyzed with DeltaVision/softWoRx software (Applied Precision). To count RAD51 foci, the foci were located automatically in the data sets by identifying local peak intensities in three dimensions. In nuclei with few foci (less than ∼50), the foci were counted manually. Only foci that were clearly above background and within the nucleus were counted.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact W.Z. Cande, zcande@uclink.berkeley.edu.
Acknowledgments
We thank Tomoko Ogawa (National Institute of Genetics, Mishima, Japan) for the gift of HsRAD51 antibodies. We are grateful to Lisa Harper, Ye Jin, and Scott Dawson for comments on the manuscript. This research was supported by a grant from the National Institutes of Health (RO1 48547-05) and by the Torrey Mesa Research Institute, Syngenta Research and Technology, San Diego, CA.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012898.
References
- Anderson, L.K., Hooker, K.D., and Stack, S.M. (2001). The distribution of early recombination nodules on zygotene bivalents from plants. Genetics 159, 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L.K., Offenberg, H.H., Verkuijlen, W.M.H.C., and Heyting, C. (1997). RecA-like proteins are components of early meiotic nodules in lily. Proc. Natl. Acad. Sci. USA 94, 6868–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, L.K., and Stack, S.M. (2003). Meiotic recombination in plants. Curr. Genomics 3, 507–525. [Google Scholar]
- Ashley, T., Plug, A.W., Xu, J., Solari, A.J., Reddy, G., Golub, E.I., and Ward, D.C. (1995). Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma 104, 19–28. [DOI] [PubMed] [Google Scholar]
- Barlow, A.L., Benson, F.E., West, S.C., and Hulten, M.A. (1997). Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J. 16, 5207–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, H.W., Bordoli, S.J., and Foss, E.M. (2003). The desynaptic (dy) and desynaptic1 (dsy1) mutations in maize (Zea mays L.) cause distinct telomere-misplacement phenotypes during meiotic prophase. J. Exp. Bot. 54, 39–46. [DOI] [PubMed] [Google Scholar]
- Bass, H.W., Marshall, W.F., Sedat, J.W., Agard, D.A., and Cande, W.Z. (1997). Telomeres cluster de novo before the initiation of synapsis: A three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J. Cell Biol. 137, 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, H.W., Riera-Lizarazu, O., Ananiev, E.V., Bordoli, S.J., Rines, H.W., Phillips, R.L., Sedat, J.W., Agard, D.A., and Cande, W.Z. (2000). Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J. Cell Sci. 113, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Baudat, F., Manova, K., Yuen, J.P., Jasin, M., and Keeney, S. (2000). Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998. [DOI] [PubMed] [Google Scholar]
- Beadle, G.W. (1930). Genetic and cytological studies of a Mendelian asynaptic in Zea mays. Cornell Agric. Exp. Stn. Mem. 129, 175–189. [Google Scholar]
- Bishop, D.K. (1994). RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79, 1081–1092. [DOI] [PubMed] [Google Scholar]
- Burgess, S.M. (2002). Homologous chromosome associations and nuclear order in meiotic and mitotically dividing cells of budding yeast. Adv. Genet. 46, 49–90. [DOI] [PubMed] [Google Scholar]
- Chen, H., Swedlow, J.R., Grote, M., Sedat, J.W., and Agard, D.A. (1995). The collection, processing, and display of digital three-dimensional images of biological specimens. In Handbook of Biological Confocal Microscopy, J. Pawley, ed (New York: Plenum Press), pp. 197–210.
- Cowan, C.R., Carlton, P.M., and Cande, W.Z. (2001). The polar arrangement of telomeres in interphase and meiosis: Rabl organization and the bouquet. Plant Physiol. 125, 532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R.K., Sedat, J.W., Agard, D.A., and Cande, W.Z. (1994). Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell 76, 901–912. [DOI] [PubMed] [Google Scholar]
- Doebley, J., Stec, A., Wendel, J., and Edwards, M. (1990). Genetic and morphological analysis of a maize-teosinte F2 population: Implications for the origin of maize. Proc. Natl. Acad. Sci. USA 87, 9888–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggler, A.L., Inman, R.B., and Cox, M.M. (2002). The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J. Biol. Chem. 277, 39280–39288. [DOI] [PubMed] [Google Scholar]
- Franklin, A.E., Golubovskaya, I.N., Bass, H.W., and Cande, W.Z. (2003). Improper chromosome synapsis is associated with elongated RAD51 structures in maize desynaptic2 mutant. Chromosoma 10.1007/S00412-003-0242-8. [DOI] [PubMed]
- Franklin, A.E., McElver, J., Sunjevaric, I., Rothstein, R., Bowen, B., and Cande, W.Z. (1999). Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 11, 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior, S.L., Wong, A.K., Kora, Y., Shinohara, A., and Bishop, D.K. (1998). Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes. Genes Dev. 12, 2208–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya, I.N. (1989). Meiosis in maize: mei genes and conception of genetic control of meiosis. Adv. Genet. 26, 149–192. [Google Scholar]
- Golubovskaya, I.N., Grebennikova, Z.K., Auger, D.L., and Sheridan, W.F. (1997). The maize desynaptic1 mutation disrupts meiotic chromosome synapsis. Dev. Genet. 21, 146–159. [Google Scholar]
- Golubovskaya, I.N., Harper, L.C., Pawlowski, W.P., Schichnes, D., and Cande, W.Z. (2002). The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics 162, 1979–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya, I.N., and Khristolyubova, N.B. (1985). The cytogenetic evidence of the gene control of meiosis: Maize meiosis and mei-genes. In Plant Genetics, M. Freeling, ed (New York: Alan R. Liss), pp. 723–738.
- Golubovskaya, I.N., and Mashenkov, A.S. (1975). A mutation causing the absence of the first division in meiosis. Maize Genet. Coop. Newsl. 49, 97. [Google Scholar]
- Golubovskaya, I.N., and Mashenkov, A.S. (1976). Genetic control of meiosis. II. A desynaptic mutant in maize induced by N-nitroso-n-methylurea. Genetika 12, 7–14. [Google Scholar]
- Golubovskaya, I.N., and Mashenkov, A.S. (1977). Multiple disturbances of meiosis in corn caused by a single recessive mutation pamA-A344. Genetika 13, 1910–1921. [Google Scholar]
- Golubovskaya, I.N., Sheridan, W.F., Harper, L., and Cande, W.Z. (2003). New meiotic mutants of maize identified from Mu transposon and EMS mutant screens. Maize Genet. Coop. Newsl. 77, in press.
- Grelon, M., Vezon, D., Gendrot, G., and Pelletier, G. (2001). AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, B. (1990). Meiosis. (Cambridge, UK: Cambridge University Press).
- Keeney, S., Giroux, C.N., and Kleckner, N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384. [DOI] [PubMed] [Google Scholar]
- Maguire, M.P. (1978. a). Evidence for separate genetic control of crossing-over and chiasma maintenance. Chromosoma 64, 371–392. [Google Scholar]
- Maguire, M.P. (1978. b). A possible role for the synaptonemal complex in chiasma maintenance. Exp. Cell Res. 112, 297–308. [DOI] [PubMed] [Google Scholar]
- Maguire, M.P., and Riess, R.W. (1991). Synaptonemal complex behavior in asynaptic maize. Genome 34, 163–168. [DOI] [PubMed] [Google Scholar]
- Maguire, M.P., Riess, R.W., and Paredes, A.M. (1993). Evidence from a maize desynaptic mutant points to a probable role of synaptonemal complex central region components in provision for subsequent chiasma maintenance. Genome 36, 797–807. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah, S.K., Turner, J.M.A., Baudat, F., Rogakou, E.P., de Boer, P., Blanco-Rodriguez, J., Jasin, M., Keeney, S., Bonner, W.M., and Burgoyne, P.S. (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27, 271–276. [DOI] [PubMed] [Google Scholar]
- Masson, J.-Y., and West, S.C. (2001). The Rad51 and Dmc1 recombinases: A non-identical twin relationship. Trends Biochem. Sci. 26, 131–136. [DOI] [PubMed] [Google Scholar]
- Miller, O.L. (1963). Cytological studies in asynaptic maize. Genetics 48, 1445–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, P.B., Kolas, N.K., Tarsounas, M., Marcon, E., Cohen, P.E., and Spyropoulos, B. (2002). The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115, 1611–1622. [DOI] [PubMed] [Google Scholar]
- Niwa, O., Shimanuki, M., and Miki, F. (2000). Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 19, 3831–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples, T.L., Dean, E., Gonzalez, O., Lambourne, L., and Burgess, S.M. (2002). Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev. 16, 1682–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill, B., Sym, M., Scherthan, H., and Roeder, G.S. (1995). Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 9, 2684–2695. [DOI] [PubMed] [Google Scholar]
- Roeder, G.S. (1997). Meiotic chromosomes: It takes two to tango. Genes Dev. 11, 2600–2621. [DOI] [PubMed] [Google Scholar]
- Romanienko, P.J., and Camerini-Otero, R.D. (2000). The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975–987. [DOI] [PubMed] [Google Scholar]
- Scherthan, H. (2001). A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2, 621–627. [DOI] [PubMed] [Google Scholar]
- Shibata, T., Nishinaka, T., Mikawa, T., Aihara, H., Kurumizaka, H., Yokoyama, S., and Ito, Y. (2001). Homologous genetic recombination as an intrinsic dynamic property of a DNA structure induced by RecA/Rad51-family proteins: A possible advantage of DNA over RNA as genomic material. Proc. Natl. Acad. Sci. USA 98, 8425–8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, A., Ogawa, H., and Ogawa, T. (1992). Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69, 457–470. [DOI] [PubMed] [Google Scholar]
- Stack, S.M., and Anderson, L.K. (2002). Crossing over as assessed by late recombination nodules is related to the pattern of synapsis and the distribution of early recombination nodules in maize. Chromosome Res. 10, 329–345. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J. (1990). The Use of Maize Meiotic Mutants to Study Cell Division and the Cytoskeleton in Flowering Plants. PhD dissertation (Berkeley: University of California).
- Tarsounas, M., Morita, T., Pearlman, R.E., and Moens, P.B. (1999). RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J. Cell Biol. 147, 207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasawa, M., Shinohara, A., Hotta, Y., Ogawa, H., and Ogawa, T. (1995). Localization of RecA-like protein in chromosomes of the lily at various meiotic stages. Genes Dev. 9, 925–934. [DOI] [PubMed] [Google Scholar]
- Timofeeva, L.P., and Golubovskaya, I.N. (1991). A new type of desynaptic gene revealed by the microspreading method of synaptonemal complexes. [In Russian.] Cytologia 33, 3–8. [Google Scholar]
- Trelles-Sticken, E., Dresser, M.E., and Scherthan, H. (2000). Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 151, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve, A.M., and Hillers, K.J. (2001). Whence meiosis? Cell 106, 647–650. [DOI] [PubMed] [Google Scholar]
- Weiner, B.M., and Kleckner, N. (1994). Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell 77, 977–991. [DOI] [PubMed] [Google Scholar]
- Yanez, R.J., and Porter, A.C.G. (1999). Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther. 6, 1282–1290. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and Kleckner, N. (1999). Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33, 603–754. [DOI] [PubMed] [Google Scholar]