Abstract
Replication of chloroplasts is essential for achieving and maintaining optimal plastid numbers in plant cells. The plastid division machinery contains components of both endosymbiotic and host cell origin, but little is known about the regulation and molecular mechanisms that govern the division process. The Arabidopsis mutant arc6 is defective in plastid division, and its leaf mesophyll cells contain only one or two grossly enlarged chloroplasts. We show here that arc6 chloroplasts also exhibit abnormal localization of the key plastid division proteins FtsZ1 and FtsZ2. Whereas in wild-type plants, the FtsZ proteins assemble into a ring at the plastid division site, chloroplasts in the arc6 mutant contain numerous short, disorganized FtsZ filament fragments. We identified the mutation in arc6 and show that the ARC6 gene encodes a chloroplast-targeted DnaJ-like protein localized to the plastid envelope membrane. An ARC6–green fluorescent protein fusion protein was localized to a ring at the center of the chloroplasts and rescued the chloroplast division defect in the arc6 mutant. The ARC6 gene product is related closely to Ftn2, a prokaryotic cell division protein unique to cyanobacteria. Based on the FtsZ filament morphology observed in the arc6 mutant and in plants that overexpress ARC6, we hypothesize that ARC6 functions in the assembly and/or stabilization of the plastid-dividing FtsZ ring. We also analyzed FtsZ localization patterns in transgenic plants in which plastid division was blocked by altered expression of the division site–determining factor AtMinD. Our results indicate that MinD and ARC6 act in opposite directions: ARC6 promotes and MinD inhibits FtsZ filament formation in the chloroplast.
INTRODUCTION
The division apparatus of plastids is derived from their ancestors, cyanobacterial endosymbionts, and some of its components are related to the cell division machinery in extant bacteria (Osteryoung and Vierling, 1995; Lutkenhaus and Addinall, 1997; Kuroiwa et al., 1998; Pyke, 1999; Rothfield et al., 1999; Osteryoung and McAndrew, 2001; Hashimoto, 2003). The first known step in the assembly of the bacterial cell division apparatus involves polymerization of the cytoskeletal tubulin-like protein FtsZ into a contractile ring at mid cell just beneath the cytoplasmic membrane (Bi and Lutkenhaus, 1991; Lutkenhaus and Addinall, 1997; Sun and Margolin, 1998). Other cell division proteins then are recruited to the division site and assembled in a hierarchical manner (for reviews, see Bramhill, 1997; Lutkenhaus, 1998; Rothfield et al., 1999; Addinall and Holland, 2002).
Bacterial cell division is governed in part through the timing and spatial control of FtsZ ring formation. Assembly of the FtsZ ring in Escherichia coli is restricted to the cell center by a dynamic system comprising the MinC, MinD, and MinE proteins (Margolin, 2001; Addinall and Holland, 2002; Lutkenhaus, 2002). MinC and MinD function together to prevent FtsZ ring formation, possibly by preventing FtsZ assembly or destabilizing nascent FtsZ polymers (Hu et al., 1999). At the cell center, MinC and MinD activity is inhibited by MinE, allowing FtsZ polymer assembly and ring formation to proceed at that position (Zhao et al., 1995; Raskin and de Boer, 1997; Fu et al., 2001; Shih et al., 2002). This initiates cell division. Overproduction of MinC or MinD abolishes FtsZ ring formation at all sites, thereby inhibiting cell division. Conversely, mutations in or deletion of MinC or MinD allow FtsZ rings to form and cell division to occur at improper positions (de Boer et al., 1989, 1992; Bi and Lutkenhaus, 1993; Yu and Margolin, 1999). The ZipA and FtsA proteins, which also are required for cell division in E. coli, are hypothesized to function in part by stabilizing the FtsZ ring at the division site (Hale and de Boer, 1997, 2002; RayChaudhuri, 1999; Pichoff and Lutkenhaus, 2002). In mutants that lack both ZipA and FtsA, FtsZ rings fail to assemble, and short, abnormal FtsZ filaments accumulate instead (Pichoff and Lutkenhaus, 2002). Another cell division protein that promotes FtsZ ring assembly is the recently discovered ZapA (Gueiros-Filho and Losick, 2002). Thus, both FtsZ-stabilizing and -destabilizing factors play central roles in FtsZ ring formation and the regulation of cell division in bacteria.
Compared with bacterial cell division, relatively little is known about the mechanisms that govern the division of chloroplasts, although some of the key players have been identified. The chloroplast division machinery is composed of elements of both eukaryotic (Gao et al., 2003; Miyagishima et al., 2003) and prokaryotic origin, among them nucleus-encoded, chloroplast-targeted homologs of FtsZ (Osteryoung and Vierling, 1995; Strepp et al., 1998). Plants and green algae have two distinct FtsZ protein families, FtsZ1 and FtsZ2 (Osteryoung and McAndrew, 2001; Wang et al., 2003; K. Stokes, unpublished data), both of which localize to a ring at mid plastid (Mori et al., 2001; Vitha et al., 2001) and are essential for plastid division (Osteryoung et al., 1998). However, factors that regulate FtsZ ring assembly in chloroplasts are largely unknown. Homologs of bacterial MinD and MinE, both of which mediate the positioning of the chloroplast division site (Wakasugi et al., 1997; Colletti et al., 2000; Kanamaru et al., 2000; Dinkins et al., 2001; Reddy et al., 2002), probably are involved, but plants seem to lack a homolog of the division inhibitor MinC. Other than FtsZ, MinD, and MinE, none of the known bacterial cell division genes from E. coli and Bacillus subtilis have easily recognizable counterparts in plants. Recently, however, a plastid and cell division protein called ARTEMIS was identified whose distribution appears to be restricted to plants and cyanobacteria (Fulgosi et al., 2002). This finding, together with the cyanobacterial ancestry of chloroplasts (Whatley, 1993; Douglas, 1998; Martin et al., 1998), suggests that other division proteins unique to plants and cyanobacteria probably exist.
The Arabidopsis arc mutants, which exhibit various defects in chloroplast size, shape, and number (Pyke and Leech, 1992; Pyke, 1997; Marrison et al., 1999), are a potentially rich resource of new plastid division genes in plants. Genetic analysis of the arc mutants indicates that there are at least 12 loci involved in plastid replication (Pyke, 1999). The most severe division defects are observed in the arc6 and arc12 mutants, whose leaf mesophyll cells contain only one or two grossly enlarged chloroplasts (Pyke et al., 1994; Robertson et al., 1995; Pyke, 1999). Here, we describe the identification of the lesion in arc6 and show that the wild-type gene product, ARC6, is a plastid-targeted DnaJ-like protein. ARC6 and its orthologs are found in plants and cyanobacteria but not in other prokaryotes. Analysis of the arc6 mutant phenotype provides evidence that ARC6 functions in the assembly of the FtsZ ring in chloroplasts.
RESULTS
arc6 Bears a Mutation in a Gene Related to the Cyanobacterial Cell Division Gene Ftn2
Three alleles of arc6, arc6-1, arc6-2, and arc6-3, all isolated from the same population of T-DNA insertion mutants (K. Pyke, unpublished data), were obtained from the ABRC. Previous studies of the mutant phenotype (Pyke and Leech, 1992; Pyke et al., 1994; Robertson et al., 1995; Pyke, 1997; Marrison et al., 1999) were performed using arc6-1, which was not tagged (Pyke et al., 1994; Rutherford, 1996). The mapping data for arc6-1 indicated that the mutation was located on chromosome 5, between markers m247 and DFR, close to marker g4028 (Rutherford, 1996; Marrison et al., 1999). One of the genes within this region, At5g42480, was significantly similar to the cyanobacterial cell division gene Ftn2 (Koksharova and Wolk, 2002), which was identified by transposon mutagenesis in Synechococcus PCC 7942. Like the E. coli fts (filamentation temperature sensitive) mutants (Bramhill, 1997; Rothfield et al., 1999), ftn2 (filamentation transposon) mutants display a filamentous morphology resulting from the cell division defect (Koksharova and Wolk, 2002). Functional studies of known chloroplast division genes (Osteryoung et al., 1998; Strepp et al., 1998; Colletti et al., 2000; Maple et al., 2002; Reddy et al., 2002) have shown that large chloroplasts, like those observed in the arc6 mutant, are the phenotypic equivalent of the filamentation observed in bacterial cell division mutants. Therefore, based on the proximity of the Arabidopsis Ftn2-like gene to the ARC6 locus, we sequenced this Arabidopsis gene in both the wild-type Wassilewskija (Ws) background and in the arc6-1, arc6-2, and arc6-3 mutant backgrounds.
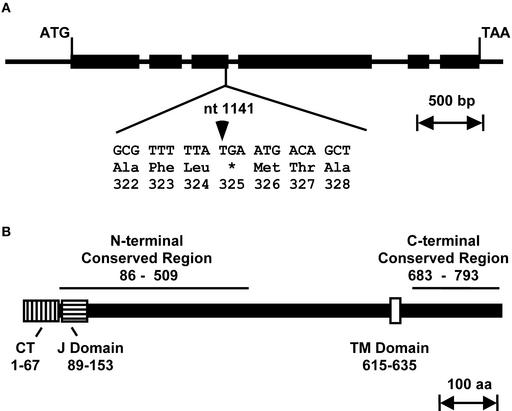
The sequence of the Ftn2-like gene in the arc6-1 mutant showed two differences compared with that in the wild type. First, an A-to-G transition occurred at position 1790 of the coding region, changing an Ala to a Thr at codon 513. This change probably represents a polymorphism between Ws-2, the genetic background of the arc6-1 mutant, and the wild-type background we used for sequencing (Ws, unknown subecotype), because the sequence from wild-type Columbia in this area is identical to that in the arc6-1 mutant. Second, a C-to-T transition occurred at position 1141 in the gene, close to the end of exon 3. This resulted in the introduction of a premature stop codon (TGA) that truncated the encoded protein from 801 to 324 amino acids (Figure 1A). The presence of this mutation suggested that the plastid defect in arc6-1 was caused by truncation of the Ftn2-like gene product and that the protein is required for plastid division. The wild-type allele corresponding to the Ftn2-like gene is referred to hereafter as ARC6.
Figure 1.
Structures of the Arabidopsis ARC6 Gene and the Encoded Protein.
(A) Genetic structure. Exons are depicted as black rectangles; ATG and TAA are the translation initiation and termination codons, respectively. The nucleotide sequence flanking the mutation in arc6 at position +1141 (arrowhead) and the C-to-T transition in codon 325, which introduced a premature stop codon (TGA), are shown. nt, nucleotide; bp, base pairs.
(B) ARC6 protein structure. Putative functional domains are depicted as rectangles, and their positions within the amino acid sequence are indicated: the chloroplast targeting signal (CT; vertically hatched box); the J domain (horizontally hatched box); and the transmembrane domain (TM; white box). Black lines above the diagram delineate the N- and C-terminal regions conserved among the ARC6-like proteins in plants and cyanobacteria. aa, amino acids.
Surprisingly, the other alleles of arc6 (arc6-2 and arc6-3) contained mutations identical to those found in arc6-1. This result was confirmed by sequencing the ARC6 locus from additional arc6-2 and arc6-3 plants grown from seeds obtained from the Nottingham Arabidopsis Stock Centre. These data indicate that the three arc6 alleles probably originated from the same initial mutation event.
The ARC6 coding region is 2406 nucleotides in length and contains six exons (Figure 1A). Both predicted and experimentally determined (see Methods) ARC6 cDNA sequences encode a protein of 801 amino acids with a molecular mass of 88.2 kD. The tissue sources from which an ARC6 EST, a full-length Arabidopsis cDNA already in the database, and an ARC6 cDNA isolated by us were derived indicated that ARC6 is expressed in leaves and/or stems of adult plants and in seedlings. Published reports further indicate that ARC6 is required for plastid division in leaf mesophyll and epidermal cells (Pyke et al., 1994), floral organs (Pyke and Page, 1998), shoot and root meristems (Robertson et al., 1995), and probably throughout the entire plant body (Yamamoto et al., 2002).
The arc6 Mutation Is Rescued by a Wild-Type Copy of ARC6
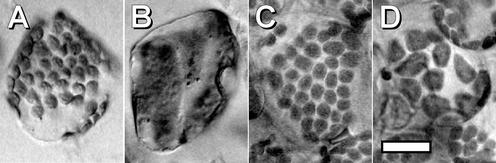
A genomic copy of ARC6 from wild-type Ws, flanked by 0.5 and 0.2 kb of the 5′ and 3′ regions, respectively, was introduced into the arc6-1, arc6-2, and wild-type plants, and the leaf mesophyll chloroplast numbers and sizes were assessed in T1 plants. The wild-type and arc6 mutant chloroplast phenotypes are shown in detail in the supplemental data online. Most (94%) of the transgenic arc6-1 and arc6-2 individuals showed plastid phenotypes that were wild-type-like or noticeably less severe than that in the mutant parent (Figures 2C and 2D, Table 1). Because chloroplast division is sensitive to altered levels of plastid division proteins (Osteryoung et al., 1998; Colletti et al., 2000; Stokes et al., 2000; Vitha et al., 2001; Reddy et al., 2002), the partial complementation in some of the transgenic lines was not surprising and probably resulted from ARC6 expression levels that were above or below the optimum. Complementation of the mutant phenotype by the wild-type gene confirmed that the lesion in ARC6 is responsible for the chloroplast division defect in the arc6 mutant and that ARC6 is essential for chloroplast division.
Figure 2.
Complementation of the arc6 Mutant Phenotype.
Chloroplasts in leaf mesophyll cells. Bar = 20 μm.
(A) Wild-type Ws.
(B) arc6-1 mutant.
(C) and (D) arc6-1 mutant transformed with the wild-type ARC6 transgene, showing full (C) or partial (D) complementation based on chloroplast size and number.
Table 1.
Leaf Mesophyll Chloroplast Phenotypes in T1 Plants Carrying the Wild-Type ARC6 Transgene
| Number of T1 Plants
|
||||
|---|---|---|---|---|
| Genetic Background | Total | Wild-Type-Like | Intermediate | arc6-Like |
| Wild-type Ws | 205 | 203 | 0 | 2 |
| arc6-1 | 120 | 97 | 18 | 5 |
| arc6-2 | 107 | 86 | 13 | 8 |
Phenotypes were scored as arc6-like (1 to 4 grossly enlarged chloroplasts per cell), intermediate (10 to 30 somewhat enlarged chloroplasts per cell), or wild-type-like (50 or more wild-type-sized chloroplasts per cell).
ARC6 and Its Orthologs Encode J-Domain Proteins Unique to Plants and Cyanobacteria
Genes related to Arabidopsis ARC6 were identified in all available fully sequenced cyanobacterial genomes and in the rice nonannotated genomic DNA sequence from chromosome 2 (Table 2). Each of these organisms contains a single ARC6-like gene. The rice sequence is located on the reverse strand of the contig and is predicted to have seven exons that encode a putative protein of 760 amino acids. Additionally, a number of ESTs representing ARC6 genes from several angiosperms, a fern (Ceratopteris richardii), and a moss (Physcomitrella patens) were identified (Table 2). An ARC6-like sequence also was found in the genome of a green alga, Chlamydomonas reinhardtii (Table 2), although significant similarity was apparent only near its predicted N terminus, in a region corresponding to an N-terminally conserved region of ∼420 amino acids present in the plant and cyanobacterial proteins (Figure 1B). No ARC6-like sequences were evident in noncyanobacterial prokaryotes.
Table 2.
ARC6-Like Proteins and Their Accession Numbers
| Abbreviation | Organism | Accession Number/Open Reading Frame Name |
|---|---|---|
| Anabaena | Anabaena sp PCC 7120 | BAB74406 |
| Nostoc | Nostoc punctiforme ATCC 29133 | Contig 493, gene 84 |
| Syn_PCC6803 | Synechocystis sp PCC 6803 | BAA10060 |
| Scc_PCC7002 | Synechococcus sp PCC 7002 | Contig 051302-306 |
| Scc_WH8102 | Synechococcus sp WH8102 | Gene 3082 |
| Scc_PCC7942 | Synechococcus sp PCC 7942 | AAL16071 |
| Ths_BP-1 | Thermosynechococcus elongatus BP-1 | tlr0758 |
| Tre_IMS101* | Trichodesmium erythraeum IMS101 | Scaffold 21, genes 2839 to 2840* |
| Pm_MED4 | Protochlorococcus marinus MED4 | Contig 1, gene 533 |
| Pm_MIT9313 | Protochlorococcus marinus MT9313 | Contig 1, gene 2677 |
| — | Chlamydomonas reinhardtii | Complement; scaffold 294, nucleotides 47288 to 51078 |
| Solanum* | Solanum tuberosum | EST; BE472035* |
| Medicago* | Medicago truncatula | EST; BI268376, AW696905, AL382914, AL382915* |
| Arabidopsis | Arabidopsis thaliana | BAB10489 (ARC6) |
| Oryza | Oryza sativa | BK000999 |
| Zea* | Zea mays | EST; BM498278, BM498757, AW331058* |
| — | Ceratopteris richardii | EST; BE641509* |
| — | Physcomitrella patens | EST; BI437111* |
| At3g19180 | Arabidopsis thaliana | NP_188549a |
| Eco DnaJ | Escherichia coli | NP_414556 |
| Eco Hsc56 | Escherichia coli | NP_415182 |
Partial sequences (indicated by asterisks) were deduced from EST records; for these, EST accession numbers are given. Where accession numbers are not yet assigned, the gene/open reading frame name or DNA contig designation is shown instead. Because of missing sequence data, draft analysis of Trichodesmium erythraeum IMS101 incorrectly identifies the sequence as two consecutive genes. The coding sequence predictions for C. reinhardtii vary depending on the software tool used (see Methods). Also listed are two DnaJ proteins from E. coli.
This ARC6-like protein has less similarity to ARC6 in the J domain than do the other plant and cyanobacterial proteins listed.
Alignment among ARC6-like proteins reveals conserved regions near the N and C termini separated by a highly divergent central area and a predicted transmembrane domain positioned immediately upstream of the conserved C-terminal region (Figure 1B; see also supplemental data online). The plant proteins also contain N-terminal extensions not present in the cyanobacterial sequences that represent putative chloroplast transit peptides (see supplemental data online). Overall, the cyanobacterial ARC6-like (Ftn2) proteins are ∼22% identical and 43% similar to Arabidopsis ARC6, whereas the rice sequence is 47% identical and 68% similar to the Arabidopsis sequence. These comparisons do not include the predicted N-terminal transit peptides in the plant proteins.
The conserved N-terminal region in the ARC6-like proteins contains a putative J domain (Figure 3; see also supplemental data online), a motif characteristic of DnaJ cochaperones (Cheetham and Caplan, 1998; Walter and Buchner, 2002). DnaJ proteins deliver polypeptide substrates to Hsp70 chaperones for processing and regulate chaperone activity via the interaction of their J domains with their Hsp70 partners (Bukau and Horwich, 1998; Walter and Buchner, 2002). The J domain in nearly all known DnaJ and DnaJ-like proteins contains a central His-Pro-Asp (HPD) motif, which is believed to be crucial for the interaction between the J domain and Hsp70 (Hennessy et al., 2000). However, this motif is not fully preserved in the J domains of ARC6 and its orthologs, because only the central Pro is conserved uniformly (Figure 3). Conversely, other residues hypothesized to be essential for J-domain structure (Hennessy et al., 2000) are conserved (Figure 3), and a database search based on 3D-PSSM protein structure predictions (http://www.sbg.bio.ic.ac.uk/servers/3dpssm/) (Kelley et al., 2000) showed a very good fit between the ARC6-like J domains and the experimentally determined crystal structures of the J domains from human HSP40 and E. coli DnaJ. Thus, the bioinformatic analyses strongly support inclusion of the ARC6 proteins in the DnaJ-like class of molecular cochaperones.
Figure 3.
Sequence Alignment of J Domains.
J domains from E. coli DnaJ and Hsc56 (DnaJ homolog) and from plant and cyanobacterial ARC6-like proteins were aligned with the consensus sequence (Hennessy et al., 2000). Light-gray shading indicates 70% similarity, and black shading indicates 70% identity among all J domains in the Pfam database. Protein accession numbers, where available, and organism names are listed in Table 2. Symbols above the consensus indicate residues believed to be important, in E. coli DnaJ, for maintaining J-domain structure (x), for binding to Hsp70 chaperones (#), and for the specificity of this interaction (*) (Hennessy et al., 2000).
In addition to ARC6, the Arabidopsis genome contains a second ARC6-like gene of unknown function, At3g19180 (Table 2). The predicted protein is 970 amino acids in length and is 21% identical and 44% similar to ARC6. The similarity is more prominent at the C terminus than at the N terminus (Figure 3; see also supplemental data online), and the subcellular targeting predictions for this protein are inconclusive. A gene similar to At3g19180 and distinct from ARC6 also was identified in the rice nonannotated sequencing contig from chromosome 4 and in EST database records from maize, barley, sorghum, wheat, and tomato (data not shown). However, At3g19180-like sequences seem to be absent from C. reinhardtii and could not be identified in cyanobacteria or in any other prokaryotes.
arc6 Mutants Exhibit Abnormal FtsZ Filament Morphology and Reduced FtsZ Protein Levels
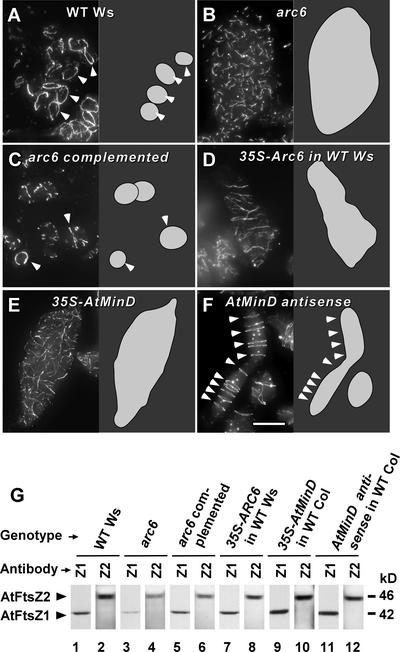
To further investigate ARC6 function, we examined FtsZ filament morphology and protein levels in the arc6 mutant background. Immunofluorescence microscopy of the leaf tissue revealed that arc6 chloroplasts contain numerous short, disorganized FtsZ filaments (Figure 4B) and lack the intact FtsZ rings typical in wild-type chloroplasts (Figure 4A). Previous double immunostaining experiments with arc6 indicated that these filaments contain both FtsZ1 and FtsZ2 (McAndrew et al., 2001), as do the FtsZ rings in wild-type plants (McAndrew et al., 2001; Vitha et al., 2001). In T1 mutant plants that express a wild-type copy of ARC6 (Figures 2C and 2D), FtsZ ring formation was restored partially, with most chloroplasts containing a partial or complete ring as well as several short FtsZ filaments (Figure 4C). These observations suggest a role for ARC6 in FtsZ ring assembly or maintenance.
Figure 4.
Immunofluorescence and Immunoblot Analyses of FtsZ.
(A) to (F) Localization of FtsZ2 in leaf mesophyll chloroplasts. Similar localization patterns also were obtained for FtsZ1 (data not shown). The immunofluorescence micrographs are shown at left, and the chloroplast shapes are drawn at right.
(A) Wild-type (WT) chloroplasts, each with a single FtsZ ring (arrowheads).
(B) A single, enlarged arc6 mutant chloroplast.
(C) Chloroplasts from an arc6 mutant plant complemented with a wild-type copy of the ARC6 gene. Partial and complete FtsZ rings are indicated by arrowheads.
(D) A single, enlarged chloroplast of an ARC6-overexpressing plant.
(E) Chloroplast from an AtMinD-overexpressing plant.
(F) Chloroplast from an AtMinD antisense plant. Arrowheads indicate multiple FtsZ rings in the enlarged chloroplast. Bar = 10 μm.
(G) Immunoblots of leaf extracts from wild-type Ws plants (lanes 1 and 2), arc6 mutant plants (lanes 3 and 4), arc6 mutant plants complemented with ARC6 (lanes 5 and 6), ARC6-overexpressing plants (lanes 7 and 8), AtMinD-overexpressing plants (lanes 9 and 10), and AtMinD antisense plants (lanes 11 and 12) probed with antibodies specific for AtFtsZ1 (Z1) or AtFtsZ2 (Z2). The identities of the immunoreactive bands are indicated at left, and their approximate molecular masses are indicated at right. Extracts from 1 mg of fresh leaf tissue were used in each lane. FtsZ levels in wild-type Columbia plants (data not shown) were identical to those in wild-type Ws (lanes 1 and 2).
In previous analyses of plants that express FtsZ1 and FtsZ2 antisense transgenes, we found that depletion of FtsZ proteins to nearly undetectable levels was accompanied by abnormalities in FtsZ localization and filament morphology (Vitha et al., 2001). To investigate FtsZ protein levels in arc6, we probed immunoblots of leaf extracts with antibodies specific for the recognition of FtsZ1 or FtsZ2 (Stokes et al., 2000; Vitha et al., 2001). Equal gel loading was confirmed by Coomassie blue staining, and samples normalized for fresh weight (Figure 4A), total protein, or chlorophyll content yielded similar results. FtsZ1 and FtsZ2 levels in the arc6 mutant (Figure 4G, lanes 3 and 4) were consistently lower than those in wild-type Ws (Figure 4G, lanes 1 and 2), although significant amounts of both proteins were detected. In mutant plants rescued by a wild-type copy of ARC6, FtsZ protein levels were closer to those in the wild type (Figure 4G, lanes 5 and 6).
The immunoblot analyses are consistent with the possibility that the fragmented FtsZ filament morphology in arc6 (Figure 4B) is a secondary effect of reduced FtsZ levels in the mutant. However, we do not favor this interpretation for several reasons: (1) partial depletion of FtsZ protein in FtsZ1 or FtsZ2 antisense plants did not fully abolish either plastid division or FtsZ ring formation (Osteryoung et al., 1998; S. Vitha and K.W. Osteryoung, unpublished results); (2) full depletion of either FtsZ1 or FtsZ2 in antisense plants blocked plastid division, but the FtsZ filament morphology differed from that in arc6; and (3) fragmented FtsZ filaments were observed in AtMinD overexpression lines (Figure 4E, described below) with wild-type levels of FtsZ protein (Figure 4G, lanes 9 and 10) (McAndrew et al., 2001). These data indicate that the FtsZ fragmentation phenotype is not correlated with reduced FtsZ levels. Based on these findings, we postulate that the partial reduction in FtsZ levels in arc6 is not the cause of the abnormal FtsZ filament morphology in the mutant. Additional experiments will be required to address this issue more thoroughly.
AtMinD RNA Levels Influence FtsZ Filament Morphology in Transgenic Plants but Are Unchanged in arc6
Kanamaru et al. (2000) reported that the arc6 mutants had increased levels of AtMinD RNA, and we and others have shown that overexpression of AtMinD confers a large chloroplast phenotype similar to that in arc6 (Colletti et al., 2000; Kanamaru et al., 2000; Dinkins et al., 2001). Because overproduction of MinD in E. coli inhibits FtsZ ring formation and causes abnormal FtsZ morphology (de Boer et al., 1989; Pichoff and Lutkenhaus, 2001), we wished to ascertain whether the fragmented FtsZ filaments in arc6 might be explained by increased AtMinD expression in the mutant. To this end, we asked whether FtsZ localization patterns in plants overexpressing AtMinD resemble those in arc6 and whether AtMinD RNA levels were increased in arc6, as reported by Kanamaru et al. (2000).
Immunofluorescence labeling in transgenic plants that express AtMinD under the control of the 35S promoter (Colletti et al., 2000) revealed that FtsZ1 and FtsZ2 colocalized to short filaments (Figure 4E) (McAndrew et al., 2001) similar to those observed in the arc6 mutant (Figure 4B). Immunoblot analysis indicated that FtsZ protein levels in the AtMinD overexpression lines (Figure 4G, lanes 9 and 10), unlike those in arc6 (Figure 4G, lanes 3 and 4), were not significantly different from those in the wild type (Figure 4G, lanes 1 and 2), supporting the idea that FtsZ fragmentation is not necessarily correlated with reduced FtsZ levels in plastid division mutants. In contrast to AtMinD overexpression, reduced expression of AtMinD in AtMinD antisense lines (Colletti et al., 2000) resulted in ectopic FtsZ ring assembly at multiple sites along the enlarged organelle (Figure 4F). FtsZ levels in these plants also were similar to those in the wild type (Figure 4G, lanes 11 and 12). RNA gel blot analysis confirmed that AtMinD RNA levels were increased or decreased in the overexpression and antisense lines, respectively (Colletti et al., 2000). Together, these findings indicate that MinD mediates the placement of the chloroplast division site by negatively regulating FtsZ ring assembly.
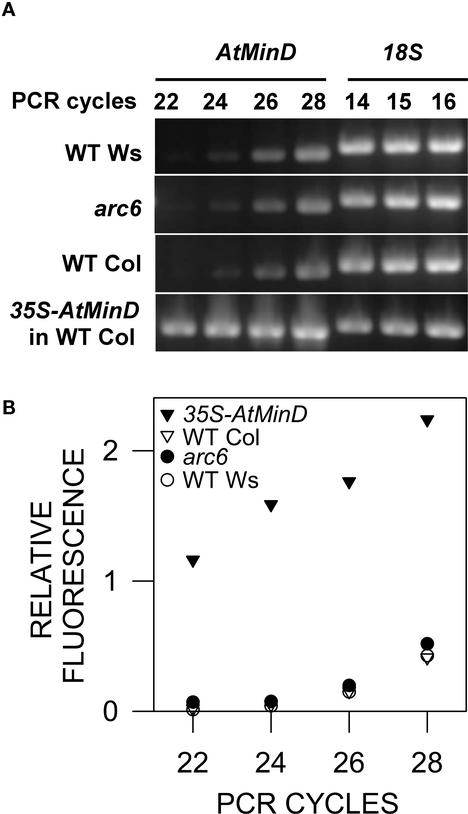
The FtsZ fragmentation phenotype in the AtMinD overexpression line (Figure 4E) is consistent with the possibility that the arc6 mutant phenotype is a consequence of AtMinD overexpression. Therefore, we analyzed AtMinD RNA levels in arc6 via RNA gel blot analysis. AtMinD transcript levels were very low and did not differ detectably among wild-type Ws, arc6, and wild-type Columbia but were increased significantly in AtMinD overexpression lines (data not shown). To more quantitatively assess AtMinD transcript levels in arc6, reverse transcription (RT)–PCR analysis was performed (Figures 5A and 5B). Total RNA was reverse transcribed using random primers, and an AtMinD cDNA fragment was amplified using gene-specific primers. The PCR product was normalized against an RT-PCR product amplified from 18S rRNA. Controls in which the reverse transcriptase was omitted from the RT reaction did not yield PCR products, indicating that the starting RNA was free of DNA contamination (data not shown). Repeated RT-PCR experiments supported the conclusion from the RNA gel blot analysis that AtMinD RNA levels in arc6 did not differ significantly from those in the wild type (Figures 5A and 5B). By contrast, RT-PCR assays detected increased levels of AtMinD RNA in plants carrying the 35S-AtMinD transgene (Figures 5A and 5B). Thus, in contrast to the results of Kanamaru et al. (2000), we detected no increased AtMinD RNA levels in arc6. Therefore, although fragmented FtsZ filaments are present in both the AtMinD overexpression lines and in arc6, we do not believe that this phenotype in arc6 is the result of increased AtMinD expression.
Figure 5.
AtMinD RNA Analysis.
(A) Ethidium bromide–stained gel with RT-PCR products amplified with primers specific for AtMinD or 18S rRNA. The number of amplification cycles and the identities of the amplified target are indicated at top, and the sources of RNA samples are shown at left. WT, wild type.
(B) Quantification of RT-PCR products based on fluorescence intensities of the bands shown in (A). The band intensity of the AtMinD PCR product is expressed relative to that of the 18S rRNA after 15 cycles of amplification.
Overexpression of ARC6 Blocks Chloroplast Division and Causes Excessive FtsZ Filament Formation
An ARC6 transgene driven by the 35S promoter of Cauliflower mosaic virus caused significant defects in chloroplast division in T1 plants when expressed in wild-type Ws and Columbia backgrounds (Table 3). Some of these plants contained as few as one large chloroplast in leaf mesophyll cells. The division defect probably was not caused by cosuppression, because the FtsZ localization pattern in the ARC6 overexpression lines (Figure 4D, described below) differed from that in the arc6 mutant (Figure 4B).
Table 3.
Leaf Mesophyll Chloroplast Phenotypes in T1 Plants Carrying the 35S-ARC6 Transgene
| Number of T1 Plants
|
||||
|---|---|---|---|---|
| Genetic Background | Total | Wild-Type-Like | Intermediate | arc6-Like |
| Wild-type Columbia | 15 | 0 | 6 | 9 |
| Wild-type Ws | 32 | 2 | 14 | 16 |
| arc6-1 | 12 | 1 | 4 | 7 |
See Table 1 for description of phenotype categories.
Four of the T1 plants carrying the ARC6 overexpression construct and exhibiting severe plastid division defects (Table 3) were analyzed for FtsZ protein levels and FtsZ localization patterns. All four plants had wild-type levels of FtsZ1 and FtsZ2 (Figure 4G, lanes 7 and 8), indicating that overexpression of ARC6 did not affect FtsZ protein levels. However, the FtsZ filaments in chloroplasts from these plants were long and numerous and occasionally formed a spiral or a ring around the enlarged chloroplast (Figure 4D). In the arc6 background, the 35S-ARC6 transgene partially or fully restored chloroplast division in 5 of the 12 T1 plants analyzed (Table 3). This construct was less effective in complementing the arc6 mutation than was the ARC6 gene controlled by its own promoter (Table 1), probably because transgene expression levels in the 35S-ARC6 plants were above the optimum. These findings are in agreement with previous results indicating that the levels of plastid division proteins are crucial for the proper functioning of the chloroplast division apparatus (Colletti et al., 2000; Kanamaru et al., 2000; Stokes et al., 2000; Dinkins et al., 2001; Vitha et al., 2001; Wang et al., 2002). The effect of the 35S-ARC6 transgene on FtsZ filament morphology and localization provides further evidence that ARC6 functions in wild-type plants by facilitating FtsZ polymer formation or stabilizing FtsZ filaments.
ARC6 Is an Inner Envelope Protein Localized at the Chloroplast Division Site
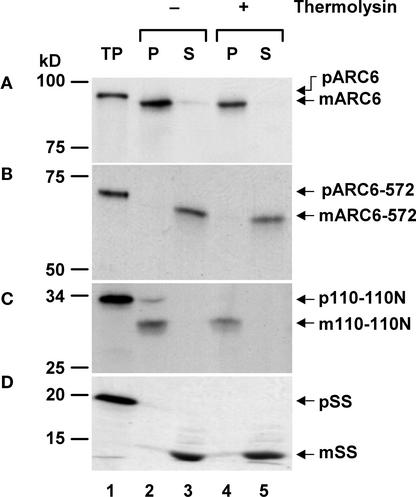
The full-length Arabidopsis and rice ARC6 sequences and a partial potato sequence contain putative N-terminal chloroplast-targeting signals, with TargetP prediction scores of 0.738, 0.961, and 0.583, respectively. To experimentally determine the localization of Arabidopsis ARC6, we performed an in vitro chloroplast import assay using radiolabeled ARC6 protein produced by coupled in vitro transcription/translation of the cDNA (Figure 6A, lane 1). Control assays were performed with the inner membrane–localized fragment of Tic110 that faces the stromal compartment (Jackson et al., 1998; McAndrew et al., 2001) (Figure 6C) and the soluble, stroma-localized small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Figure 6D). After incubation of the ARC6 translation product with isolated pea chloroplasts, the protein was protected from degradation by the protease thermolysin, which does not penetrate the outer plastid envelope membrane (Cline et al., 1984) (Figure 6A, lane 4), indicating that ARC6 was imported. The imported polypeptide migrated on SDS gels as a slightly smaller molecule than did the full-length translation product (Figure 6A, lanes 2 and 4), consistent with processing of the transit peptide. Chloroplast fractionation indicated that the import product was associated with the pellet fraction (Figure 6A, lanes 2 and 4), which is indicative of membrane localization. An additional set of import experiments using ARC6-572, a truncated form of ARC6 that lacks the putative transmembrane region (Figure 5B, lane 1), demonstrated that ARC6-572 also was imported into the chloroplast and processed, but it was localized in the soluble rather than the pellet fraction of the organelle after import (Figure 5B, lanes 3 and 5). This result confirmed the bioinformatic prediction that the full-length ARC6 is a membrane protein.
Figure 6.
Chloroplast Import Assay.
In vitro–synthesized, radiolabeled proteins (lane 1) were incubated with isolated pea chloroplasts. Chloroplasts then were incubated without (−, lanes 2 and 3) or with (+, lanes 4 and 5) the protease thermolysin for 30 min on ice and then quenched. After protease treatments, intact chloroplasts were recovered, lysed, and separated by centrifugation into total membrane (P) and soluble (S) fractions. Precursor protein (p) and mature protein (m) are indicated by arrows at right, and the positions of molecular mass markers are shown at left. TP, 10% of the radiolabeled translation product used in the import reaction (lane 1).
(A) Arabidopsis full-length ARC6.
(B) Truncated ARC6 (ARC6-572), representing the first 572 amino acids of the full-length protein.
(C) Truncated inner membrane–localized Tic110-110N, facing the stromal compartment (Jackson et al., 1998).
(D) Soluble, stroma-localized small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase.
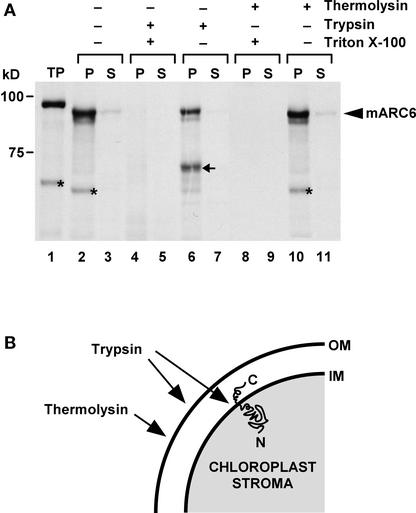
To investigate the membrane topology of ARC6, the import reaction was followed by treatment of the chloroplasts with the protease trypsin, which penetrates the outer but not the inner plastid envelope membrane and thus can digest proteins exposed in the intermembrane space but not in the stroma (Joyard et al., 1983; Cline et al., 1984). After trypsin treatment, a significant fraction of the ARC6 import product was truncated by ∼18 kD (Figure 7A, arrow in lane 6). This shift corresponds to the calculated molecular mass of the C-terminal region of ARC6 downstream of the predicted transmembrane domain (Figure 1B). Incomplete degradation of proteins exposed to the intermembrane space in trypsin-treated chloroplasts has been observed previously (McAndrew et al., 2001) and probably is caused by slow penetration of the protease across the outer envelope. Control reactions in which the chloroplasts were disrupted by detergent treatment before trypsin or thermolysin digestion confirmed that the protease was fully capable of digesting the ARC6 completely (Figure 7A, lanes 4, 5, 8, and 9). The low molecular mass bands indicated by asterisks in lanes 1, 2, and 10 probably represent degradation products unrelated to the protease treatments.
Figure 7.
Membrane Topology of Arabidopsis ARC6.
(A) After the in vitro import of radiolabeled ARC6 (lanes 2 and 3), pea chloroplasts were incubated with (+) or without (−) the protease trypsin (lanes 4 to 7) or thermolysin (lanes 8 to 11). As a control for protease activity, the treatments were preformed in the presence (+) of Triton X-100 (lanes 4, 5, 8, and 9). After protease treatments, intact chloroplasts were recovered, except from the Triton X-100–treated samples, and separated by centrifugation into total membrane (P) and soluble (S) fractions. Mature protein (mARC6) is indicated by an arrowhead at right, and the positions of molecular mass markers are shown at left. The arrow in lane 6 indicates the position of a truncated ARC6 protein band resulting from trypsin treatment of intact chloroplasts. The lower molecular mass bands, unrelated to protease treatment, are indicated with asterisks in lanes 1, 2, and 10. TP, 10% of the radiolabeled translation product used in the import reaction (lane 1).
(B) Proposed membrane topology of ARC6 in chloroplasts. The C and N termini of ARC6 are labeled as C and N, respectively. The accessibility of the outer, cytosolic chloroplast surface of and the envelope intermembrane space to proteases is indicated by arrows. OM and IM, outer and inner envelope membranes, respectively.
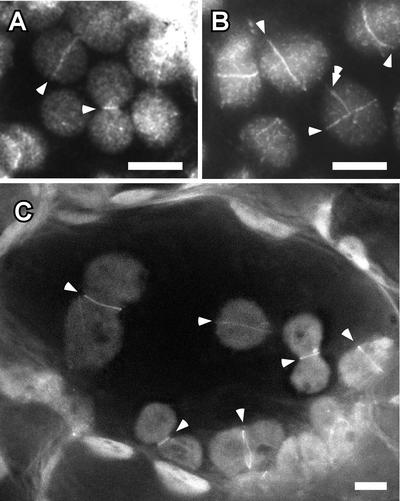
To investigate the suborganellar localization of ARC6, we expressed in both wild-type (Columbia and Ws) and arc6 mutant backgrounds a chimeric protein in which green fluorescent protein (GFP) was fused to the C terminus of ARC6. Because overexpression of ARC6 on the 35S promoter inhibited chloroplast division (Figure 4D), we used the native ARC6 promoter to create the ARC6-GFP transgene. Inspection by fluorescence microscopy of six independent transgenic lines from each genetic background indicated that the majority of the fusion protein was localized to a ring at the center of the chloroplasts (Figure 8, single arrowheads). Like the untagged ARC6 transgene (Figures 2C and 2D), the ARC6-GFP transgene restored plastid division in the arc6 background, as shown by the presence of multiple chloroplasts in all six transgenic lines tested (Figure 8C). These findings indicate that the ARC6-GFP fusion protein is functional. Based on the topological model shown in Figure 7B, the GFP portion of the fusion protein presumably was localized in the intermembrane space. The ARC6-GFP ring could be detected in both unconstricted and deeply constricted chloroplasts (Figure 8), suggesting that ARC6 functions throughout plastid division. This localization pattern is nearly identical to that observed for FtsZ1 and FtsZ2 (Vitha et al., 2001). In some chloroplasts, additional fusion protein strands also were detected (Figure 8B, double arrowhead). Because overexpression of FtsZ or FtsZ-GFP produces FtsZ filament networks not observed in wild-type plants (Kiessling et al., 2000; Vitha et al., 2001), it is possible that these strands reflect slight overexpression of ARC6-GFP in the transgenic plants. Immunofluorescence localization of the endogenous ARC6 protein, once antibodies become available, will clarify this issue.
Figure 8.
Localization of an ARC6-GFP Fusion Protein in Transgenic Arabidopsis Plants, and Rescue of the arc6 Mutant Phenotype by the Fusion Protein.
Arrows indicate concentrated areas of GFP fluorescence. The shapes of the chloroplasts are apparent from the dim background fluorescence. Single arrowheads indicate ARC6-GFP localized to the plastid midpoint. The double arrowhead in (B) indicates an additional ARC6-GFP strand not associated with the midpoint. Bars = 5 μm.
(A) and (B) GFP fluorescence in leaf mesophyll cell chloroplasts from two independent T1 plants expressing the ARC-GFP transgene in a wild-type (Columbia) background. A similar localization pattern was observed in the wild-type Ws background (data not shown).
(C) GFP fluorescence in a cotyledon mesophyll cell from a 6-day-old T1 plant expressing the ARC-GFP transgene in the arc6 mutant background. The presence of multiple chloroplasts in cotyledons and in true leaves (data not shown) indicates complementation of the arc6 chloroplast division defect by the fusion protein.
The results from these localization studies indicate that ARC6 is a chloroplast-targeted, integral membrane protein localized to the chloroplast division site. The proteolytic data strongly support a topological model in which ARC6 spans the inner envelope membrane, with the large N-terminal region upstream of the transmembrane domain, including the J domain, extending into the stroma and the smaller C-terminal region downstream of the transmembrane domain extending into the intermembrane space (Figure 7B). This topology and localization are consistent with a role for ARC6 in FtsZ function during chloroplast division.
DISCUSSION
ARC6 Orthologs in Plants and Cyanobacteria
The sequencing data and complementation of arc6 demonstrated that ARC6 is a nuclear gene that encodes a novel plastid division protein closely related to the cyanobacterial cell division protein Ftn2 (Koksharova and Wolk, 2002). These results indicate that ARC6, like most of the previously identified plastid division genes, probably is endosymbiotic in origin and evolved from a related cell division gene present in the cyanobacterial ancestor of chloroplasts. However, unlike FtsZ, MinD, and MinE, which are found in many prokaryotic organisms, ARC6 and Ftn2 appear to be unique to plants and cyanobacteria, as is the plastid division protein ARTEMIS (Fulgosi et al., 2002). This finding suggests that cyanobacteria, in which cell division has not been studied thoroughly, may have more utility than E. coli or B. subtilis as a model system for homology-based identification of new plastid division genes.
In addition to the ARC6-like sequences in plants, the recently released genome sequence of the green alga C. reinhardtii (http://genome.jgi-psf.org/chlre1/chlre1.home.html) also contains an ARC6-like gene. This finding suggests that orthologous genes probably are present in all plants and green algae. The role of the Arabidopsis protein homologous with ARC6, At3g19180, is unknown, but its divergence from ARC6 (Figure 3; see also supplemental data online) and the complete inhibition of plastid division in the arc6 mutant suggest that the two proteins are not functionally redundant.
FtsZ Levels in arc6 Chloroplasts
The arc6 mutant consistently showed reduced levels of FtsZ1 and FtsZ2 proteins (Figure 4G). The large chloroplasts in the mutant represent a phenotype similar to that seen in plants in which FtsZ1 or FtsZ2 levels are suppressed by antisense transgenes (Osteryoung et al., 1998). However, we do not believe that the severe plastid division defects and disruption of FtsZ localization are caused directly by the low FtsZ concentration in arc6, because, unlike in the FtsZ antisense plants (Vitha et al., 2001), arc6 chloroplasts contain significant amounts of FtsZ proteins (Figure 4G, lanes 3 and 4). Furthermore, the FtsZ localization pattern in arc6 chloroplasts (Figure 4B) is sharply different from that in AtFtsZ1 or AtFtsZ2 antisense plants: chloroplasts that lack FtsZ1 have multiple long FtsZ2 filaments, whereas chloroplasts that lack FtsZ2 have no or only a few short FtsZ1 filaments (Vitha et al., 2001). Less severe reduction in FtsZ2 levels does not affect FtsZ1 or FtsZ2 localization into rings at mid plastid, even though it may cause moderate inhibition of plastid division (our unpublished results).
The decrease in FtsZ levels in arc6 could be a consequence of the slightly lower chloroplast volumes in the mutant relative to those in the wild type (Pyke et al., 1994) or of slightly lower expression levels. Alternatively, the reduced FtsZ levels in the mutant may be a secondary effect of the defective FtsZ ring formation, which could lead to enhanced FtsZ degradation. Further studies of the arc6 mutant should illuminate this issue.
ARC6 Localization and Membrane Topology
The protein import and GFP fusion protein experiments demonstrated that ARC6 is imported into isolated chloroplasts and localized to the chloroplast division site. This is consistent with the role of ARC6 as an essential chloroplast division protein (Pyke et al., 1994; Robertson et al., 1995; Marrison et al., 1999). We identified a putative transmembrane region in all full-length ARC6-like sequences from plants and cyanobacteria. The protease protection assays suggested that ARC6 spans the inner chloroplast envelope membrane, with its N terminus, including the conserved J domain, exposed to the stroma (Figure 7B). This topology, along with the localization of ARC6 at the division site, supports a role for ARC6 in FtsZ ring formation or maintenance, because FtsZ1, FtsZ2, MinD, and MinE also are localized in the stroma (Osteryoung and Vierling, 1995; Colletti et al., 2000; Fujiwara and Yoshida, 2001; Itoh et al., 2001; McAndrew et al., 2001; Maple et al., 2002). Future experiments with chloroplasts from ARC6-GFP transgenic plants should further confirm the localization of GFP and the C terminus of ARC6 in the intermembrane space. The significance of the C-terminal intermembrane space–localized region is unknown at present.
Atypical J Domain of ARC6
ARC6 and its orthologs in plants and cyanobacteria contain a conserved J domain characteristic of DnaJ-like molecular cochaperones. DnaJ-like proteins are found in most organisms (Miernyk, 2001), and their J domains are responsible for their interaction with specific Hsp70 chaperones (Bukau and Horwich, 1998; Walter and Buchner, 2002). A conserved HPD motif located in the center of most J domains (Figure 3) is thought to be crucial for this interaction, because mutations in this motif abolish binding to Hsp70 (Bukau and Horwich, 1998; Hennessy et al., 2000). However, in all ARC6 orthologs, only the Pro of the HPD motif is conserved (Figure 3), suggesting that these proteins may interact with partners distinct from typical Hsp70 chaperones. On the other hand, the E. coli DnaJ-like protein Hsc56, which contains an HPE rather than an HPD motif, interacts with the Hsp70-like chaperone Hsc62 (Yoshimune et al., 2002). This finding indicates that noncanonical J domains can function in specific Hsp70 chaperone systems. Hsp70 proteins have been shown to interact with FtsZ in E. coli and may play a role in bacterial cell division and FtsZ ring formation (Bukau and Walker, 1989; McCarty and Walker, 1994; Uehara et al., 2001). If ARC6 acts as an Hsp70 cochaperone in the chloroplast, the chaperone system in which it functions probably is specific for plastid division, because plastid biogenesis and function are not impaired dramatically in the arc6 mutant (Pyke et al., 1994; Robertson et al., 1995; Rutherford, 1996; Yamamoto et al., 2002). Determining whether ARC6 interacts with a chloroplast Hsp70 will be critical for understanding its role in plastid division.
Opposite Effects of ARC6 and MinD on FtsZ Filament and Ring Formation
The FtsZ localization pattern in the arc6 mutant indicated that the arc6 defect is accompanied by the fragmentation of FtsZ polymers (Figure 4B) and that ARC6 is important for the stabilization or assembly of FtsZ rings. This finding was supported further by the analysis of ARC6-overexpressing plants, in which multiple, long FtsZ filaments were observed (Figure 4D), suggesting that excess ARC6 promotes FtsZ polymerization. In E. coli, two cell division proteins, FtsA and ZipA, are believed to function in stabilizing the assembled FtsZ, tethering it to the membrane, and recruiting downstream components to the midcell division site (Errington et al., 2003). Interestingly, E. coli mutants that lack both FtsA and ZipA are blocked in cell division and instead of FtsZ rings have numerous, short FtsZ arcs and dots (Pichoff and Lutkenhaus, 2002) similar to those seen in arc6 chloroplasts (Figure 4B). Homologs of FtsA and ZipA are not apparent in cyanobacteria or plants, and it is possible that ARC6 participates in a function related to those of FtsA and ZipA, anchoring and/or stabilizing the FtsZ ring at the chloroplast division site.
In contrast to ARC6, chloroplastic MinD has a negative effect on FtsZ ring formation. This conclusion is based on the results indicating that AtMinD overexpression lines lack FtsZ rings and have fragmented FtsZ filaments (Figure 4E), whereas AtMinD antisense lines have multiple, ectopically localized FtsZ rings (Figure 4F). Thus, MinD and ARC6 act in opposite directions: ARC6 promotes and MinD inhibits FtsZ ring formation. Experiments to further define the mechanisms by which these two proteins regulate FtsZ ring assembly and localization are in progress.
METHODS
Plant Material
All Arabidopsis thaliana plants, including wild-type ecotypes Wassilewskija (Ws; unknown subecotype) and Columbia, transgenic plants expressing antisense and sense constructs of AtMinD (Colletti et al., 2000) in ecotype Columbia background, the chloroplast division mutant lines arc6-1, arc6-2, and arc6-3 (Pyke et al., 1994; Robertson et al., 1995) in ecotype Ws-2 background, and the transgenic lines generated in this project, were grown for 5 weeks in a growth chamber as described previously (Osteryoung et al., 1998). Tissue for RNA isolation was obtained from seedlings grown for 10 days on Murashige and Skoog (1962) agar plates in a growth chamber (Osteryoung et al., 1998) or in a liquid culture (Zhang and Forde, 1998) for 12 days under continuous white light.
Seeds for the arc6 mutants were obtained from the ABRC (Ohio State University, Columbus). Additional seed stocks of arc6-1, arc6-2, and arc6-3 were kindly provided by the Nottingham Arabidopsis Stock Centre (University of Nottingham, Loughborough, UK).
Amplification and Sequencing of the ARC6 Gene
Genomic DNA was isolated from young leaf tissue using the Plant DNAzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The ARC6 gene flanked by 0.5-kb 5′ and 0.2-kb 3′ regions was amplified with the PfuTurbo DNA polymerase (Stratagene, La Jolla, CA) using the primers 5′-TGTCCAAATTTTATGTGACACTCC-3′ (forward) and 5′-TTGTGAAAGGCTTGAATGTAAGA-3′ (reverse). The ∼3.8-kb product was cloned into SmaI-digested pBluescript KS+ vector (Stratagene) and sequenced in both directions.
Plasmid Constructs, Plant Transformation, and Analysis of Chloroplast Phenotypes
For the ARC6 complementation construct, the PCR-amplified genomic fragment containing ARC6 flanked by 0.5-kb 5′ and 0.2-kb 3′ regions (see above) was cloned into the plant transformation vector pMLBART (obtained from Karl Gordon, Commonwealth Scientific and Industrial Research Organization, Canberra, Australia, via John Bowman, University of California, Davis), a derivative of pART27 (Gleave, 1992) that confers resistance to the herbicide glufosinate, as a selectable marker. To create an ARC6-overexpression construct, the PCR-amplified genomic fragment containing ARC6 (see above) was digested with NcoI at −1 nucleotide before the start codon, DNA overhangs were filled with Klenow fragment of DNA polymerase, and the entire ARC6 gene, including the 0.2-kb 3′ region, was cloned into the XbaI-digested and Klenow-treated pART7 vector (Gleave, 1992) behind the 35S promoter of Cauliflower mosaic virus. Orientation of the insert was confirmed by restriction analysis. The 35S-ARC6 construct then was excised with NotI and transferred into pMLBART.
To create a C-terminal GFP fusion of ARC6, the ARC6 genomic sequence was amplified as for sequencing (see above), except that a different reverse primer (5′-GAAGATCTGATGCAAGAACAGAGCCTTC-3′) was used to introduce a BglII site (underlined) in place of the ARC6 stop codon. The PCR product was digested with BglII and EcoRI, and a fragment representing the second half of ARC6 was isolated.
The plasmid used for sequencing (see above) was digested with KpnI and EcoRI to excise the fragment containing the ARC6 promoter (0.5 kb) and the first half of the ARC6 coding region, and both fragments were ligated into the binary vector pCAMBIA1302 (CAMBIA, Canberra, Australia), from which the 35S promoter had been excised with KpnI and BglII. The in-frame fusion of ARC6 with GFP and the lack of amplification errors were confirmed by sequencing. After Agrobacterium tumefaciens–mediated transformation of arc6-1, wild-type Columbia, and wild-type Ws plants, T1 seedlings were germinated on agar plates (Nakazawa and Matsui, 2003) supplemented with 20 mg/L hygromycin and 100 mg/L ampicillin. GFP fluorescence was visualized in leaves from 3-week-old T1 seedlings (in the wild-type Columbia background) or in cotyledons from 6-day-old T1 seedlings (in the arc6-1 and wild-type Ws backgrounds) as described below for immunofluorescence localization of FtsZ.
Agrobacterium-mediated transformation of wild-type, arc6-1, and arc6-2 plants with 35S-ARC6 and the complementation constructs, selection of the glufosinate-resistant T1 plants, and assessment of chloroplast size and number in tips from fully expanded leaves of 4-week-old plants were performed as described previously (Osteryoung et al., 1998; Vitha et al., 2001). Images of chloroplasts were recorded with a Coolpix 995 (Nikon, Tokyo, Japan) digital camera mounted on an Olympus BH2 microscope (Olympus America, Melville, NY) equipped with a ×40 objective. For detailed analysis of chloroplast shapes, images of chlorophyll autofluorescence were acquired as stacks of optical sections using a Zeiss LSM 5 PASCAL confocal microscope (Carl Zeiss, Oberkochen, Germany) and volume-rendered using ImageJ (http://rsb.info.nih.gov/ij/) and the VolumeJ plugin (http://www.isi.uu.nl/people/michael/vr.htm).
Isolation of cDNA and in Vitro Chloroplast Import Assay
Total RNA isolation from liquid culture–grown seedlings, reverse transcription reactions, and amplification of ARC6 cDNA were performed as described (McAndrew et al., 2001) using the following gene-specific primers: 5′-CATGGAAGCTCTGAGTCACGTC-3′ (forward) and 5′-GTAGCATGTCTGAGCTTGCG-3′ (reverse). The cDNA was amplified using Takara ExTaq DNA polymerase (PanVera, Madison, WI), cloned into pBluescript KS+ (Stratagene), and sequenced. The cDNA contains the entire coding sequence, 1 nucleotide of the 5′ region, and 31 nucleotides of the 3′ region. A truncated cDNA clone, pARC6-572, which corresponds to the first 572 amino acids of ARC6 and lacks the predicted transmembrane region (Figure 1B), was obtained in a similar manner using a different reverse primer, 5′-AAGACACCAGGCTCACCATC-3′.
The in vitro synthesis of the ARC6 protein, chloroplast import assays with isolated pea (Pisum sativum) chloroplasts, and protease treatment with trypsin and thermolysin were performed as described previously (McAndrew et al., 2001).
RNA Gel Blot Analysis and Reverse Transcription–PCR
Total RNA was isolated from plate-grown seedlings using a Plant RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA gel blot analysis was performed on two biological replicates as described by Fourney et al. (1988) using 20 μg of total RNA per lane (Figure 5B). The 32P-labeled AtMinD probe was prepared from 25 ng of the 1-kb fragment of AtMinD (Colletti et al., 2000), representing the entire coding sequence, using the Random Primers DNA Labeling System (Invitrogen). After overnight hybridization with the probe (Maniatis et al., 1982) and final washing with 1× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 60°C for 1 h, the membrane was exposed on a phosphor screen for 1 h and the signal was visualized with Personal Molecular Imager FX (Bio-Rad Laboratories, Hercules, CA). The membrane then was exposed on Kodak X-Omat AR film (Kodak, Rochester, NY) for 10 days.
Reverse transcription reactions with random primers and Superscript II reverse transcriptase (Invitrogen) were performed according to the manufacturer's protocol. In control reactions, the reverse transcriptase was omitted. PCR amplification then was performed in a 25-μL final volume using an amount of reverse transcription reaction that corresponds to 50 ng of total RNA and primers specific for AtMinD: 5′-TTGGTCTCCGTAACCTCGAT-3′ (forward) and 5′-CAAACCCGTAACCCTATCAG-3′ (reverse). As an internal standard, amplification also was preformed with primers specific for 18S ribosomal RNA: 5′-GTGCATGGCCGTTCTTAGTT-3′ (forward) and 5′-ACCGGATCATTCAATCGGTA-3′ (reverse). The numbers of amplification cycles were 22, 24, 26, and 28 for AtMinD and 14, 15, and 16 for 18S RNA. The ∼400-bp PCR products were quantified on ethidium bromide–stained agarose gels using Quantity One software (Bio-Rad). The intensity from each AtMinD band then was expressed relative to the band intensity of 18S RNA resulting from 15 amplification cycles.
Immunoblot and Immunofluorescence Analyses of AtFtsZ
Immunoblot analysis with leaf tissue extracts and immunofluorescence microscopy of leaf mesophyll chloroplasts with rabbit antipeptide antibodies specifically recognizing AtFtsZ1 or AtFtsZ2 were performed as described previously (Stokes et al., 2000; Vitha et al., 2001). For immunoblot analysis, extracts from 1 mg (fresh weight) of tissue were loaded per lane. Gel loading was assessed by Coomassie Brilliant Blue R250 staining. A control set of samples also was loaded to achieve a uniform amount (54 ng) of total protein per lane. Protein quantification in extracts was performed using the RC DC Protein Assay kit (Bio-Rad) according to the manufacturer's instructions.
Specimens were viewed with a Leica DMR A2 microscope (Leica Microsystems, Wetzlar, Germany) equipped with epifluorescence illumination, a ×100 oil-immersion objective, a fluorescein isothiocyanate fluorescence filter set (excitation, 460 to 500 nm; emission, 512 to 545 nm), and a cooled charge-coupled device camera, Retiga 1350 EX (Qimaging, Burnaby, Canada). Image stacks were acquired with 0.3-μm spaces between sections and combined to achieve extended depth of focus using ImageJ version 1.27 software (http://rsb.info.nih.gov/ij/) and Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA).
Databases and Software Tools
DNA and protein sequence databases were accessed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). Preliminary sequence data for most cyanobacterial genomes were obtained from the Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/JGI_microbial/html/index.html) and from the Kazusa DNA Research Institute of Japan (http://www.kazusa.or.jp/cyano/). The Chlamydomonas reinhardtii genomic sequence was accessed at http://www.biology.duke.edu/chlamy_genome/blast/blast_form.html. Protein and DNA similarity searches were performed using Basic Local Alignment Search Tool (TBLASTN and BLASTN; Altschul et al., 1990). For predictions of subcellular protein targeting, TargetP version 1.01 (Emanuelsson et al., 2000) and Predotar version 0.5 (http://www.inra.fr/Internet/Produits/Predotar/) were used. Prediction of transmembrane domains in protein sequences was performed with HMMTOP version 2.0 (Tusnady and Simon, 2001), TMHMM version 2.0 (Krogh et al., 2001), DAS (Cserzo et al., 1997), SOSUI (Hirokawa et al., 1998), Split (Juretic et al., 2002), TMPRED (http://www.ch.embnet.org/software/TMPRED_form.html), and TopPred2 (Claros and von Heijne, 1994).
Identification of conserved domains was facilitated by searches in the Pfam database (http://pfam.wustl.edu/index.html). The exon/intron analysis for the rice and C. reinhardtii ARC6 homologs used TBLASTN comparison of the genomic DNA sequence with the Arabidopsis ARC6 protein combined with the gene prediction results from GeneScan (Burge and Karlin, 1997), GrailEXP version 3.3 (Xu and Uberbacher, 1997), FGENESH 1.1 (http://genomic.sanger.ac.uk/gf/gf.shtml), and Genie (Kulp et al., 1996). Sequence manipulation, multiple alignments, and shading of aligned sequences were performed using BioEdit 5.09 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). For multiple protein alignment, the Gonnet series protein weight matrix was used, and the gap-opening and extension penalties were set to 10.0 and 0.20, respectively. DNA sequencing reads were processed using the Phred basecaller (Ewing et al., 1998), assembled with Phrap, and viewed with Consed (http://www.phrap.org/).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Katherine W. Osteryoung, osteryou@msu.edu.
Accession Numbers
The accession numbers for Arabidopsis ARC6 sequences were assigned as follows: wild-type (ecotype Ws) cDNA, AY221469; wild-type (Ws) genomic, AY221468; and arc6 mutant, genomic, AY221467. The predicted cDNA sequence data for rice ARC6 was deposited in the Third Party Annotation Section of the DDBJ/EMBL/GenBank databases (see Table 2). Other accession numbers are as follows: the ARC6 genomic sequence from wild-type Columbia, NM_123613; ARC6 EST, AI998415; a full-length Arabidopsis cDNA already in the database, AY091075; an ARC6 cDNA isolated by us, AY221469; a rice nonannotated sequencing contig from chromosome 4 containing a homolog of the Arabidopsis At3g19180, AL732351; and the binary vector pCAMBIA1302, AF234298.
Supplementary Material
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre (University of Nottingham, Loughborough UK; http://arabidopsis.org.uk) for providing seeds of arc6-1, arc6-2, and arc6-3 mutants and the Michigan State University Genomics Technology Support Facility for DNA sequencing. We thank Tanya Wagner and David Yoder for careful reading of the manuscript. This project was supported by grants from the National Science Foundation and the U.S. Department of Energy.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013292.
Footnotes
Online version contains Web-only data.
References
- Addinall, S.G., and Holland, B. (2002). The tubulin ancestor, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J. Mol. Biol. 318, 219–236. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bi, E., and Lutkenhaus, J. (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164. [DOI] [PubMed] [Google Scholar]
- Bi, E., and Lutkenhaus, J. (1993). Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill, D. (1997). Bacterial cell division. Annu. Rev. Cell Dev. Biol. 13, 395–424. [DOI] [PubMed] [Google Scholar]
- Bukau, B., and Horwich, A.L. (1998). The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366. [DOI] [PubMed] [Google Scholar]
- Bukau, B., and Walker, G.C. (1989). Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J. Bacteriol. 171, 2337–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Cheetham, M.E., and Caplan, A.J. (1998). Structure, function and evolution of DnaJ: Conservation and adaptation of chaperone function. Cell Stress Chaperon. 3, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros, M.G., and von Heijne, G. (1994). TopPred II: An improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10, 685–686. [DOI] [PubMed] [Google Scholar]
- Cline, K., Werner-Washburne, M., Andrews, J., and Keegstra, K. (1984). Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 75, 675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti, K.S., Tattersall, E.A., Pyke, K.A., Froelich, J.E., Stokes, K.D., and Osteryoung, K.W. (2000). A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10, 507–516. [DOI] [PubMed] [Google Scholar]
- Cserzo, M., Wallin, E., Simon, I., von Heijne, G., and Elofsson, A. (1997). Prediction of transmembrane α-helices in prokaryotic membrane proteins: The dense alignment surface method. Protein Eng. 10, 673–676. [DOI] [PubMed] [Google Scholar]
- de Boer, P.A.J., Crossley, R.E., and Rothfield, L.I. (1989). A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56, 641–649. [DOI] [PubMed] [Google Scholar]
- de Boer, P.A.J., Crossley, R.E., and Rothfield, L.I. (1992). Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkins, R., Reddy, M.S.S., Leng, M., and Collins, G.B. (2001). Overexpression of the Arabidopsis thaliana MinD1 gene alters chloroplast size and number in transgenic tobacco plants. Planta 214, 180–188. [DOI] [PubMed] [Google Scholar]
- Douglas, S.E. (1998). Plastid evolution: Origins, diversity, trends. Curr. Opin. Genet. Dev. 8, 655–661. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Errington, J., Daniel, R.A., and Scheffers, D.-J. (2003). Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- Fourney, R., Miyakoshi, J., Day, R., and Paterson, M. (1988). Northern blotting: Efficient RNA staining and transfer. Focus 10, 5–7. [Google Scholar]
- Fu, X., Shih, Y.L., Zhang, Y., and Rothfield, L.I. (2001). The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98, 980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, M., and Yoshida, S. (2001). Chloroplast targeting of chloroplast division FtsZ2 proteins in Arabidopsis. Biochem. Biophys. Res. Commun. 287, 462–467. [DOI] [PubMed] [Google Scholar]
- Fulgosi, H., Gerdes, L., Westphal, S., Glockmann, C., and Soll, J. (2002). Cell and chloroplast division requires ARTEMIS. Proc. Natl. Acad. Sci. USA 99, 11501–11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H., Kadirjan-Kalbach, D., Froehlich, J.E., and Osteryoung, K.W. (2003). ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 100, 4328–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, P. (1992). A versatile binary vector system with T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Gueiros-Filho, F.J., and Losick, R. (2002). A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, C.A., and de Boer, P.A.J. (1997). Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185. [DOI] [PubMed] [Google Scholar]
- Hale, C.A., and de Boer, P.A.J. (2002). ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 184, 2552–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, H. (2003). Plastid division: Its origins and evolution. Int. Rev. Cytol. 222, 63–98. [DOI] [PubMed] [Google Scholar]
- Hennessy, F., Cheetham, M.E., Dirr, H.W., and Blatch, G.L. (2000). Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chaperon. 5, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998). SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379. [DOI] [PubMed] [Google Scholar]
- Hu, Z., Mukherjee, A., Pichoff, S., and Lutkenhaus, J. (1999). The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, R., Fujiwara, M., Nagata, N., and Yoshida, S. (2001). A chloroplast protein homologous to the eubacterial topological specificity factor MinE plays a role in chloroplast division. Plant Physiol. 127, 1644–1655. [PMC free article] [PubMed] [Google Scholar]
- Jackson, D.T., Froehlich, J.E., and Keegstra, K. (1998). The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J. Biol. Chem. 273, 16583–16588. [DOI] [PubMed] [Google Scholar]
- Joyard, J., Billecocq, A., Bartlett, S.G., Block, M.A., Chua, N.H., and Douce, R. (1983). Localization of polypeptides to the cytosolic side of the outer envelope membrane of spinach chloroplasts. J. Biol. Chem. 258, 10000–10006. [PubMed] [Google Scholar]
- Juretic, D., Zoranic, L., and Zucic, D. (2002). Basic charge clusters and predictions of membrane protein topology. J. Chem. Inf. Comp. Sci. 42, 620–632. [DOI] [PubMed] [Google Scholar]
- Kanamaru, K., Fujiwara, M., Kim, M., Nagashima, A., Nakazato, E., Tanaka, K., and Takahashi, H. (2000). Chloroplast targeting, distribution and transcriptional fluctuation of AtMinD1, a eubacteria-type factor critical for chloroplast division. Plant Cell Physiol. 41, 1119–1128. [DOI] [PubMed] [Google Scholar]
- Kelley, L.A., MacCallum, R.M., and Sternberg, M.J. (2000). Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299, 499–520. [DOI] [PubMed] [Google Scholar]
- Kiessling, J., Kruse, S., Rensing, S.A., Harter, K., Decker, E.L., and Reski, R. (2000). Visualization of a cytoskeleton-like FtsZ network in chloroplasts. J. Cell Biol. 151, 945–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koksharova, O.A., and Wolk, P.C. (2002). A novel gene that bears a DnaJ motif influences cyanobacterial cell division. J. Bacteriol. 184, 5524–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kulp, D., Haussler, D., Reese, M.G., and Eeckman, F.H. (1996). A generalized hidden Markov model for the recognition of human genes in DNA. Proc. Int. Conf. Intell. Syst. Mol. Biol. 4, 134–142. [PubMed] [Google Scholar]
- Kuroiwa, T., Kuroiwa, H., Sakai, A., Takahashi, H., Toda, K., and Itoh, R. (1998). The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 181, 1–41. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus, J. (1998). The regulation of bacterial cell division: A time and place for it. Curr. Opin. Microbiol. 1, 210–215. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus, J. (2002). Dynamic proteins in bacteria. Curr. Opin. Microbiol. 5, 548–552. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus, J., and Addinall, S.G. (1997). Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66, 93–116. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J. (1982). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Maple, J., Chua, N.H., and Moller, S.G. (2002). The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J. 31, 269–277. [DOI] [PubMed] [Google Scholar]
- Margolin, W. (2001). Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4, 647–652. [DOI] [PubMed] [Google Scholar]
- Marrison, J.L., Rutherford, S.M., Robertson, E.J., Lister, C., Dean, C., and Leech, R.M. (1999). The distinctive roles of five different ARC genes in the chloroplast division process in Arabidopsis. Plant J. 18, 651–662. [DOI] [PubMed] [Google Scholar]
- Martin, W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M., and Kowallik, K.V. (1998). Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165. [DOI] [PubMed] [Google Scholar]
- McAndrew, R.S., Froehlich, J.E., Vitha, S., Stokes, K.D., and Osteryoung, K.W. (2001). Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127, 1656–1666. [PMC free article] [PubMed] [Google Scholar]
- McCarty, J.S., and Walker, G.C. (1994). DnaK mutants defective in ATPase activity are defective in negative regulation of the heat shock response: Expression of mutant DnaK proteins results in filamentation. J. Bacteriol. 176, 764–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk, J.A. (2001). The J-domain proteins of Arabidopsis thaliana: An unexpectedly large and diverse family of chaperones. Cell Stress Chaperon. 6, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima, S., Nishida, K., Mori, T., Matsuzaki, M., Higashiyama, T., Kuroiwa, H., and Kuroiwa, T. (2003). A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, T., Kuroiwa, H., Takahara, M., Miyagishima, S., and Kuroiwa, T. (2001). Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol. 42, 555–559. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nakazawa, M., and Matsui, M. (2003). Selection of hygromycin-resistant Arabidopsis seedlings. Biotechniques 34, 28–30. [DOI] [PubMed] [Google Scholar]
- Osteryoung, K.W., and McAndrew, R.S. (2001). The plastid division machine. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 315–333. [DOI] [PubMed] [Google Scholar]
- Osteryoung, K.W., Stokes, K.D., Rutherford, S.M., Percival, A.L., and Lee, W.Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10, 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung, K.W., and Vierling, E. (1995). Conserved cell and organelle division. Nature 376, 473–474. [DOI] [PubMed] [Google Scholar]
- Pichoff, S., and Lutkenhaus, J. (2001). Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183, 6630–6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff, S., and Lutkenhaus, J. (2002). Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke, K.A. (1997). The genetic control of plastid division in higher plants. Am. J. Bot. 84, 1017–1027. [PubMed] [Google Scholar]
- Pyke, K.A. (1999). Plastid division and development. Plant Cell 11, 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke, K.A., and Leech, R.M. (1992). Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol. 99, 1005–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke, K.A., and Page, A.M. (1998). Plastid ontogeny during petal development in Arabidopsis. Plant Physiol. 116, 797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke, K.A., Rutherford, S.M., Robertson, E.J., and Leech, R.M. (1994). arc6, a fertile Arabidopsis mutant with only two mesophyll cell chloroplasts. Plant Physiol. 106, 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin, D.M., and de Boer, P.A.J. (1997). The MinE ring: An FtsZ-independent cell structure required for selection of the correct division site in Escherichia coli. Cell 91, 685–694. [DOI] [PubMed] [Google Scholar]
- RayChaudhuri, D. (1999). ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18, 2372–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, S.M.S., Dinkins, R., and Collins, G.B. (2002). Overexpression of the Arabidopsis thaliana MinE1 bacterial division inhibitor homologue gene alters chloroplast size and morphology in transgenic Arabidopsis and tobacco plants. Planta 215, 167–176. [DOI] [PubMed] [Google Scholar]
- Robertson, E.J., Pyke, K.A., and Leech, R.M. (1995). arc6, an extreme chloroplast division mutant of Arabidopsis also alters proplastid proliferation and morphology in shoot and root apices. J. Cell Sci. 108, 2937–2944. [DOI] [PubMed] [Google Scholar]
- Rothfield, L., Justice, S., and Garca-Lara, J. (1999). Bacterial cell division. Annu. Rev. Genet. 33, 423–428. [DOI] [PubMed] [Google Scholar]
- Rutherford, S.M. (1996). The Genetic and Physical Analysis of Mutants of Chloroplast Number and Size in Arabidopsis thaliana. PhD dissertation (York, UK: University of York).
- Shih, Y.-L., Fu, X., King, G.F., Le, T., and Rothfield, L. (2002). Division site placement in E. coli: Mutations that prevent formation of the MinE ring lead to loss of the normal midcell arrest of growth of polar MinD membrane domains. EMBO J. 21, 3347–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, K.D., McAndrew, R.S., Figueroa, R., Vitha, S., and Osteryoung, K.W. (2000). Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 124, 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp, R., Scholz, S., Kruse, S., Speth, V., and Reski, R. (1998). Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc. Natl. Acad. Sci. USA 95, 4368–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q., and Margolin, W. (1998). FtsZ dynamics during the division cycle of live Escherichia coli cells. J. Bacteriol. 180, 2050–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady, G.E., and Simon, I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850. [DOI] [PubMed] [Google Scholar]
- Uehara, T., Matsuzawa, H., and Nishimura, A. (2001). HscA is involved in the dynamics of FtsZ-ring formation in Escherichia coli K12. Genes Cells 6, 803–814. [DOI] [PubMed] [Google Scholar]
- Vitha, S., McAndrew, R.S., and Osteryoung, K.W. (2001). FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi, T., et al. (1997). Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: The existence of genes possibly involved in chloroplast division. Proc. Natl. Acad. Sci. USA 94, 5967–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, S., and Buchner, J. (2002). Molecular chaperones: Cellular machines for protein folding. Angew. Chem. Int. Ed. Engl. 41, 1098–1113. [DOI] [PubMed] [Google Scholar]
- Wang, D., Kong, D., Wang, Y., Hu, Y., He, Y., and Sun, J. (2003). Isolation of two plastid division ftsZ genes from Chlamydomonas reinhardtii and its evolutionary implication for the role of FtsZ in plastid division. J. Exp. Bot. 54, 1115–1116. [DOI] [PubMed] [Google Scholar]
- Wang, D., Kong, D.D., Ju, C.L., Hu, Y., He, Y.K., and Sun, J.S. (2002). Effects of tobacco plastid division genes NtFtsZ1 and NtFtsZ2 on the division and morphology of chloroplasts. Acta Bot. Sin. 44, 838–844. [Google Scholar]
- Whatley, J.M. (1993). The endosymbiotic origin of chloroplasts. Int. Rev. Cytol. 144, 259–299. [Google Scholar]
- Xu, Y., and Uberbacher, E.C. (1997). Automated gene identification in large-scale genomic sequences. J. Comput. Biol. 4, 325–338. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K., Pyke, K.A., and Kiss, J.Z. (2002). Reduced gravitropism in inflorescence stems and hypocotyls, but not roots, of Arabidopsis mutants with large plastids. Physiol. Plant. 114, 627–636. [DOI] [PubMed] [Google Scholar]
- Yoshimune, K., Yoshimura, T., Nakayama, T., Nishino, T., and Esaki, N. (2002). Hsc62, Hsc56, and GrpE, the third Hsp70 chaperone system of Escherichia coli. Biochem. Biophys. Res. Commun. 293, 1389–1395. [DOI] [PubMed] [Google Scholar]
- Yu, X.C., and Margolin, W. (1999). FtsZ ring clusters in min and partition mutants: Role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32, 315–326. [DOI] [PubMed] [Google Scholar]
- Zhang, H., and Forde, B.G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. [DOI] [PubMed] [Google Scholar]
- Zhao, C.R., de Boer, P.A.J., and Rothfield, L.I. (1995). Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl. Acad. Sci. USA 92, 4313–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.