Abstract
The CYP2C subfamily of cytochrome P450 monoxygenases is responsible for the metabolism of approximately 20% of therapeutic drugs and many endogenous compounds in humans. These enzymes can be induced by prior treatment with drugs, resulting in changes in drug efficacy. Induction of human CYP2C enzymes by xenobiotics occurs at the transcriptional level and is reported to involve the constitutive androstane receptor (CAR) and the pregnane X receptor (PXR). In the present study, we report that murine CYP2C37 mRNA is induced by phenobarbital and phenytoin. In contrast, the mouse PXR (mPXR) agonist PCN did not induce CYP2C37 mRNA suggesting that PXR does not regulate this gene. The induction of CYP2C37 mRNA by phenobarbital and phenytoin is essentially abolished in CAR-null mice, thus induction of Cyp2c37 by these xenobiotics is CAR dependent. A functional CAR response element (CAR-RE) was identified at -2791 bp from the translation start site of the Cyp2c37 gene. Mutation of this CAR-RE abolished mouse CAR (mCAR) transactivation of a Cyp2c37 -2.9 kb luciferase reporter construct in HepG2 cells.
Abbreviations: RXR, retinoid X receptor; DR-n, direct repeat spaced by n nucleotides; PHREM, phenytoin responsive module; CAR-RE, CAR responsive element; PCN, 5-pregnen-3β-ol-20-one-16α-carbonitrile; 1, 4-bis-[2-(3, 5,-dichloropyridyloxy)] benzene, TCPOBOP; Dimethyl-sulfoxide, DMSO; C3H/HeNCrlBR, C3H
The human CYP2C subfamily of cytochrome P450 monooxygenases (P450) is responsible for the metabolism of approximately 20% of all clinically prescribed drugs, other xenobiotics and many endogenous compounds. Clinically prescribed substances known to be metabolized by the human CYP2C enzymes include phenytoin (anticonvulsant), warfarin (anticoagulant), tolbutamide (antidiabetic), torsemide (diuretics), and numerous nonsteroidal anti-inflammatories (Goldstein and de Morais, 1994; Miners and Birkett, 1998). Recent work has shown that prior exposure to certain clinical drugs and herbal medicines can induce the expression of CYP2C8, CYP2C9, and CYP2C19 (Gerbal-Chaloin et al., 2001; Raucy et al., 2002; Chen et al., 2004; Ferguson et al., 2005). Drug induced expression of P450 enzymes leads to increased metabolic activity potentially altering the effectiveness of the inducing agent or co-administered drugs, resulting in adverse drug reactions.
P450 expression is frequently regulated at the transcriptional level and is mediated by receptors such as aryl hydrocarbon receptor (Ahr), constitutive androstane receptor (CAR), pregnane X receptor (PXR), and peroxisome proliferator-activated receptor (PPAR). Once activated these receptors bind response elements located within the 5′-flanking regions of target genes, thus inducing gene transcription (Nebert and Jones, 1989; Aldridge et al., 1995; Goodwin et al., 1999; Gerbal-Chaloin et al., 2002; Goodwin, 2002; Chen et al., 2003; Chen et al., 2004; Wang et al., 2004a; Ferguson et al., 2005). Induction of the CYP2B and CYP3A enzymes by clinically prescribed drugs such as phenobarbital and rifampicin has recently been attributed to CAR and PXR, respectively (Honkakoski et al., 1998; Sueyoshi et al., 1999; Wang et al., 2003; Chen et al., 2004; Ferguson et al., 2005). These xenobiotic signaling pathways are extensively reviewed in Pascussi et al. (2004), Honkakoski and Negishi (2000), and Sueyoshi and Negishi (2001).
The mouse is used increasingly as a model for human disease and is an excellent system to investigate the drug induced transcriptional regulation of P450 genes due to the availability of receptor null-mice. Presently, fifteen murine Cyp2c genes have been identified (Nelson et al., 2004; Wang et al., 2004b); however, only the Cyp2c29 and Cyp2c44 genes have been examined for drug induction (DeLozier et al., 2004; Jackson et al., 2004). Previous studies in our laboratory identified a phenytoin-responsive module (PHREM) located within the 5′-flanking sequence of the murine Cyp2c29 gene and demonstrated that the PHREM was necessary for induction by phenobarbital and phenytoin (Jackson et al., 2004). In contrast, studies in our laboratory showed that Cyp2c44 is not inducible by either the CAR activator phenobarbital or the PXR agonist pregnenolone 16α-carbonitrile (PCN) (DeLozier et al., 2004). An alignment of the 5′-flanking sequence of all presently known murine Cyp2c genes indicated that the PHREM which is present in Cyp2c29 was unique to this gene. Nevertheless, spilot studies indicated that Cyp2c37 was inducible by phenobarbital. Although the PHREM of Cyp2c29 was not conserved within the 5′-flanking sequence of Cyp2c37, several putative CAR response elements (CAR-REs) were identified by computational analysis. Using mutant and deletion Cyp2c37 luciferase promoter constructs, we identified a functional CAR-RE located ~2.8 kb from the translation start site. In vivo studies demonstrate that both, phenobarbital and phenytoin induce CYP2C37 mRNA. In contrast, the mPXR agonist PCN did not induce CYP2C37 mRNA suggesting that PXR does not regulate Cyp2c37. Induction of CYP2C37 mRNA by phenobarbital and phenytoin was essentially abolished in CAR-null mice, thus phenobarbital and phenytoin induction of Cyp2c37 is CAR dependent, similar to Cyp2c29.
Methods
Materials and Reagents
Phenobarbital (sodium salt), phenytoin (5,5-diphenylhydantoin, sodium salt), 5α-androstenol, 5-pregnen-3β-ol-20-one-16α-carbonitrile (PCN), and 1, 4-bis-[2-(3, 5,-dichloropyridyloxy)] benzene (TCPOBOP) were purchased from Sigma-Aldrich (St. Louis, MO). DMSO and all other common reagents not listed were purchased from Sigma-Aldrich or other common vendors. Cell culture media, fetal bovine serum, 100X Penicillin-streptomycin-glutamine solution and trypsin/EDTA were purchased from Invitrogen (Carlsbad, CA). All oligonucleotides were purchased from Sigma-Genosys (The Woodlands, TX) at 50 nmol scale and desalted. HepG2 cells were purchased from the American Type Culture Collection (Manassas, VA).
Animals
C3H/HeNCrlBR(C3H) mice were purchased from Charles Rivers Laboratory (Wilmington, MA). A CAR-null mouse (Ueda et al., 2002) was first crossbred with C3H to generate CAR heterozygous offspring. Subsequently, CAR heterozygous offspring were repeatedly backcrossed with C3H mice until the genetic background became over 95% C3H. The obtained heterozygous mice were bred to produce the wild-type and CAR-null C3H mice. PXR-null 129S1/Sv*129x1/SvJ*C57BL/6 and congenic wild-type mice were obtained from Jeff Staundinger (Staudinger et al., 2001b) and maintained at NIEHS. Mice were fed with a standard solid diet and tap water ad libitum for 5 days. Eight to fifteen week old male mice received corn oil (vehicle), phenobarbital (80 mg/kg), or phenytoin (80 mg/kg) once daily via gavage at a volume of 10 ml/kg for 4 consecutive days. Mice treated with PCN (80 mg/kg) were orally dosed once daily for 3 consecutive days using corn oil as vehicle. Animals were sacrificed 24 hrs after the last dose. The livers were removed for total RNA isolation and microsome isolation. The NIEHS Animal Care and Use Committee approved all animal procedures.
Total RNA Isolation & Quantitative RT-PCR
Total RNA was extracted using an ABI 6100 Nucleic Acid PrepStation. All chemicals for the ABI 6100 were purchased from Applied Biosystems (Foster City, CA). Total RNA from individual mice was isolated and stored at −80°C. Prior to reverse transcription, equal amounts of RNA from each individual RNA sample was pooled within each experimental group consisting of 3–5 mice according to treatment and genotype. Quantitative RT-PCR analysis was performed using a two step process. An initial reaction with MuLV Reverse Transcriptase (Applied Biosystems) followed by a subsequent quantitative PCR reaction using 2X SYBR Green Master Mix (Applied Biosystems). Reverse transcription was performed with 100 ng of total RNA combined with 1X PCR Buffer II, 0.4 μl (8 units) of Rnase inhibitor (Applied Biosystems), 5.5 mM MgCl2, 0.5 mM dATP, dCTP, dTTP, and dGTP (each), 2.5 μM random hexamers (Applied Biosystems), and 0.5 μl (25 units) of MuLV Reverse Transcriptase in a final volume of 20 μl. Reverse transcription reactions were incubated in a PCR System 9700 Thermocycler (Applied Biosystems) using the following cycling parameters: 25°C for 10 min, 42°C for 60 min, 95°C for 5 min (inactivation), and 4°C hold. The subsequent quantitative PCR reaction was performed on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). PCR reactions contained 0.5 μl of cDNA template, 1X SYBR Green Buffer Master Mix and 2.5 pmol of forward and reverse primer in a final volume of 10 μl. Gene specific primer set sequences are as follows: Cyp2b10 (Forward, 5′ACCCCACGTTCCTCTTCA3′; Reverse, 5′-CAGCAGGCGCAAGAACTGA-3′); Cyp2c29 (Forward, 5′-GTATTTGGGCTCAAAGCCTACTGTCA-3′; Reverse, 5′-CAGGGTCATGAGTGTAAATCGTCTCA-3′), Cyp2c37 (Forward, 5′-GATGGCAATCAACCATTGC-3′; Reverse, 5′-GCCGATCACATGCTCAATT-3′), Cyp3a11 (Forward, 5′-GTCAAACGCCTCTCCTTGCTG-3′; Reverse 5′-GGCTTGCCTTTCTTTGCCTTC-3′), and β-actin (Forward, 5′-CCTAGAAGCATTTGCGGTGCACGATG-3; Reverse, 5′-TCATGAAGTGTGACGTTGACATCCGT-3′). PCR cycling parameter were as follows: 50°C for 2 min hold, 95°C for 10 min hold, 94°C for 30 sec (denaturation), 60°C for 30 sec (annealing), and 72°C for 30 sec (extension). Denaturation, annealing, and extension temperatures and times were repeated for 42 cycles. PCR products were analyzed using gel electrophoresis, dissociation curve analysis, and by dye terminator DNA sequencing (Applied Biosystems) to determine single product formation and gene specificity. Standard curves (log of template dilution vs Ct value) for each gene specific primer set were used to determine relative mRNA content for each target gene. Each gene specific PCR reaction was performed in triplicate within each pooled experimental group. The triplicate values obtained from each gene specific PCR reaction was used to determine a relative starting template amount mean and standard error for each of the experimental groups.
Electrophoretic mobility Shift Assay (EMSA)
EMSA was performed on a 5% polyacrylamide gel using 0.5X TBE running buffer. Oligonucleotides were labeled with [α-32P]dCTP and probe was purified by Microspin G-25 columns (Amersham Biosciences) to remove unincorporated dNTP’s. Oligonucleotide sequences used in EMSA were as follows: CYP2C9 -1839 CAR-RE (Forward, 5′-CTAGACCAAACTCTTCTGACCTCT-3′; Reverse, 5′-CTAGAGAGGTCAGAAGAGTTTGGT-3′); Cyp2c37 -1890 CAR-RE (Forward, 5′-CTAGAGTTCTCTCCTGGATGAATTTGGGT-3′; Reverse, 5′-CTAGACCCAAATTCATCCAGGAGAGAACT-3′); Cyp2c37 -2065 CAR-RE (Forward, 5′-CTAGGTTACTGTGCTGGGTGAACTGTGTT-3′; Reverse, 5′-CTAGAACACAGTTCACCCAGCACAGTAAC-3′); Cyp2c37 -2791 CAR-RE (Forward, 5′-CTAGAAAAGCAAACTTTTCTGAACTCCATG; Reverse, 5′-CTAGCATGGAGTTCAGAAAAGTTTGCTTTT-3′); Cyp2c37 -2820 CAR-RE (Forward, 5′-CTAGAGCCCGTATCACAAAGTTCAACAAG-3′; Reverse, 5′-CTAGCTTGTTGAACTTTGTGATACGGGCT-3′); Cyp2c37 -3119 CAR-RE (Forward, 5′-CTAGATTAGTGAAATCAAAATGTGATGTATGAAATTCAAG-3′; Reverse, 5′-CTAGCTTGAATTTCATACATCACATTTTGATTTCACTAAT-3′); Cyp2c37 -3205 CAR-RE (Forward, 5′-CTAGCAAATAGAACAACATAAACTGAGAC; Reverse, 5′-CTAGGTCTCAGTTTATGTTGTTCTATTTG3′). Labeled probe (~100,000 cpm per rxn) was applied to each binding reaction in 2 μl of 5X binding buffer, 0.5 μg/ul poly(dI-dC), and 1 μl of each in vitro transcribed/translated protein (mCAR and hRXR) in a final volume of 10 μl. 5X binding buffer was composed of 20% glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 250 mM NaCl, and 50 mM Tris-HCl (pH 7.5). After addition of probe, the reactions were peformed at room temperature for 20 minutes before being loaded on a polyacrylamide gel and electrophoresed. Gels were then dried and exposed to film for 16 to 24 hours at −80°C.
Expression Vectors and Cloning of the Cyp2c37 5′-Flanking Region
The expression construct pCR3-mCAR was previously described (Sueyoshi et al., 1999) and pGEMT-hRXR was a kind gift by Ronald Evans. PCR was performed to isolate a 1.97 kb fragment of the Cyp2c37 5′-flanking sequence using the following primers: Forward, 5′-TGCTGTTAGCTGGTCTTGTTCTCTC-3′; Reverse, 5′-CCATGGAGATTCTTCTTACTGACACA-3′. The reverse primer introduced an Nco I restriction site at the 3′ end of the PCR product. The PCR fragment was generated using mouse genomic DNA from Promega (Madison, WI). This fragment was then subcloned into pCR2.1 Topo TA vector (Invitrogen) following manufacturer’s protocol. The endonucleases Bgl II and Nco I were used to remove a 1.8 kb fragment of the Cyp2c37 5′-flanking region from the pCR2.1 Topo TA subclone. This 1.8 kb fragment was then ligated into a Bgl II and Nco I digested pGL3Basic luciferase reporter vector (Promega) producing the Cyp2c37 -1.8 kb luciferase reporter. The Cyp2c37 5′-flanking sequence from -1842 bp to -2887 bp was amplified from murine genomic DNA (Promega) by PCR using forward primer 5′-ACGCGTGGGAAATATAGGGCAATGTGCATC-3′ and reverse primer 5′-GAAGATCTATAGCACAGGGTCAGCTC-3′. The forward primer introduced a Mlu I site in the 5′ end of the PCR product for Cyp2c37 5′-flanking sequence. This ~1Kb fragment was then subcloned into pCR2.1 Topo TA vector (Invitrogen), then digested by Bgl II to excise an ~876 bp fragment using interior Bgl II sites, and finally ligated into a Bgl II digested Cyp2c37 -1.8 kb luciferase reporter to create the Cyp2c37 -2.7 kb luciferase reporter. The subclone for the -1842 bp to -2887 bp Cyp2c37 5′-flanking sequence was digested by Stu I and Mlu I to remove an ~273 bp fragment (-2887 bp to -2614 bp). Subsequent ligation of this fragment into a Stu I and Mlu I digested Cyp2c37 -2.7 kb luciferase reporter created the Cyp2c37 -2.9 kb luciferase reporter. To create the Cyp2c37 -3.6 kb luciferase reporter, murine genomic DNA was again used as a template to amplify the Cyp2c37 5′-flanking sequence from -2540 bp to -3598 bp using a forward primer 5′-CGCGTTTGTAGAGTTAGAAAACACAATTTGC-3′ which also introduced a Mlu I site and the reverse primer 5′-CCCAGTTTCTCATGCTCAAATGGAA-3′. The PCR product -2540 bp to -3598 bp of the Cyp2c37 5′-flanking sequence was subcloned into pCR2.1 Topo TA vector (Invitrogen) and digested with Mlu I and Stu I to excise an ~1050 bp fragment of Cyp2c37 5′-flanking sequence. This fragment was then ligated into a Stu I and Mlu I digested Cyp2c37 -2.9 kb luciferase reporter, producing the Cyp2c37 -3.6 kb luciferase reporter. Mutations of putative binding sites in Cyp2c37 luciferase reporters were produced by site-directed mutagenesis using QuikChange (Stratagene, La Jolla, CA) on the Cyp2c37 -2.9 kb luciferase reporter background. Mutant oligonucleotides used in the site-directed mutagenesis reactions are as follows: -2791Bmut forward, 5′-ATAAAAGCAAACTTTTCCCCCCTCCATGCAATAAAAACAGTG-3′; -2791Bmut reverse, 5′-CACTGTTTTTATTGCATGGAGCCCCGAAAAGTTTGCTTTTAT-3′; -2791Amut forward, 5′-CAAGTTATAAAAGCCCCCCTTTCCCCCCTCCATGC-3′; -2791Amut reverse, 5′-GCATGGAGGGGGGAAAGGGGGGCTTTTATAACTTG-3′. (mutations underlined) Dye terminator DNA sequencing (Applied Biosystems) was used to verify all sequences.
Transcriptional Activation Assays
HepG2 cells were maintained in Eagle’s minimum essential media (Invitrogen) supplemented with 10% fetal bovine serum and 1X penicillin-streptomycin-glutamine solution (Invitrogen). Cells were plated into 24 well plates at a density of ~100,000 cells/well. Transfections included 100 ng of nuclear receptor (pCR3-mCAR), 100 ng of each luciferase promoter construct, and 1 ng of pRL-tk transfection control. Twenty-four hours after transfection, cells were treated with drugs or vehicle and incubated for an additional 24 hours. Cells were subsequently lysed with 100 μl of 1X passive lysis buffer (Promega) for 30 min at room temperature with gentle rocking, and dual luciferase assays (Promega) were then performed on cell lysates per manufacturer’s procedures.
Statistics
Results from luciferase assays and quantitative RT-PCR are expressed as mean +/− standard error of triplicate determinations. Single statistical comparisons were made using Dunnett’s Method; however, Tukey-Kramer HSD Test was utilized for multiple statistical comparisons when appropriate. The statistical tests were performed using JMP version 5.1 software (SAS Institute, Inc., Cary, NC) and the criterion of significance was set at p = 0.05.
Results
Induction of hepatic CYP2C37 mRNA in wild-type mice
C3H wild-type mice were initially treated with corn oil (10 ml/kg), phenobarbital (80 mg/kg), or phenytoin (80mg/kg) via gavage for four consecutive days. Twenty-four hours after the last dose was administered RNA was isolated from the liver and quantitative RT-PCR was performed. CYP2C37 mRNA was induced ~10 fold by phenobarbital and ~20 fold by phenytoin (Fig. 1).
Figure 1.
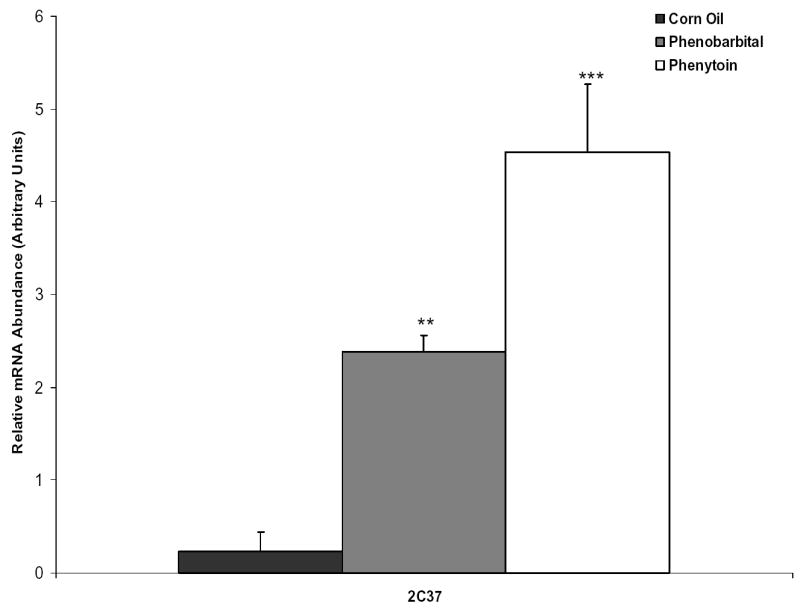
Drug response of hepatic CYP2C37 mRNA in C3H mice. Mice were treated via gavage with corn oil, phenobarbital (80 mg/kg), or phenytoin (80 mg/kg) for four consecutive days and sacrificed 24 hrs after last dose for isolation of hepatic total RNA. Quantitative RT-PCR was performed to determine hepatic CYP2C37 mRNA content in response to phenobarbital and phenytoin. Target gene was normalized to a reference gene, β-actin. Values expressed above represent the relative abundance of gene specific mRNA +/− SE. P-values were determined using the Dunnett’s Method comparing treated to corn oil control. ** p ≤ 0.01 *** p ≤ 0.001.
Element identification and binding analysis
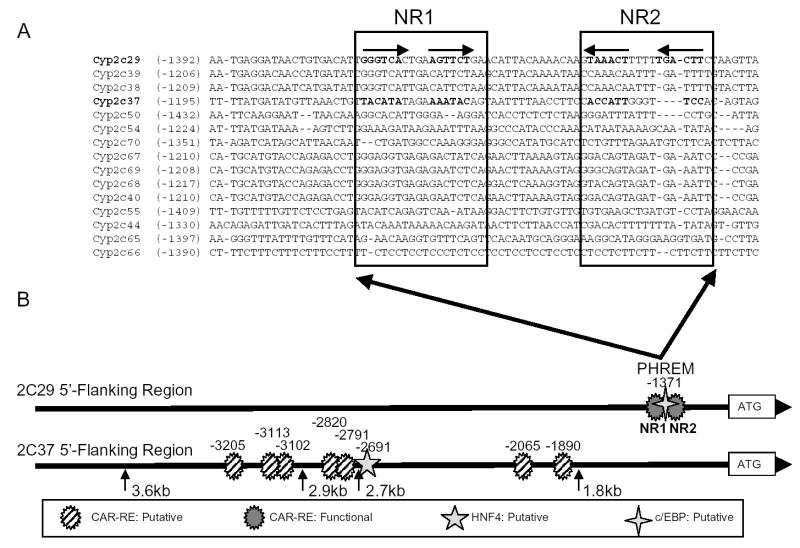
An alignment of ~5 kb of all presently known murine Cyp2c 5′-flanking regions was conducted using Vector NTI Advance 9.0 (Invitrogen) to investigate the conservation of the previously identified PHREM within this murine gene subfamily. The alignment indicated that the PHREM is not conserved within the corresponding region of the Cyp2c37 gene nor is it conserved in the other Cyp2c genes thus, it is unique to Cyp2c29 (Fig. 2A). These findings suggested that induction of Cyp2c37 by phenytoin and phenobarbital is mediated by an alternative responsive region.
Figure 2.
Alignment of CYP2C 5′-flanking regions and summary illustration of Cyp2c37 and Cyp2c29 5′-flanking regions A: Alignment of Cyp2c29 PHREM and surrounding sequence with corresponding 5′-flanking sequence of all presently known murine CYP2C genes B: A schematic representation of the Cyp2c37 and Cyp2c29 5′-flanking region providing a summary of putative and functional transcription factor binding sites.
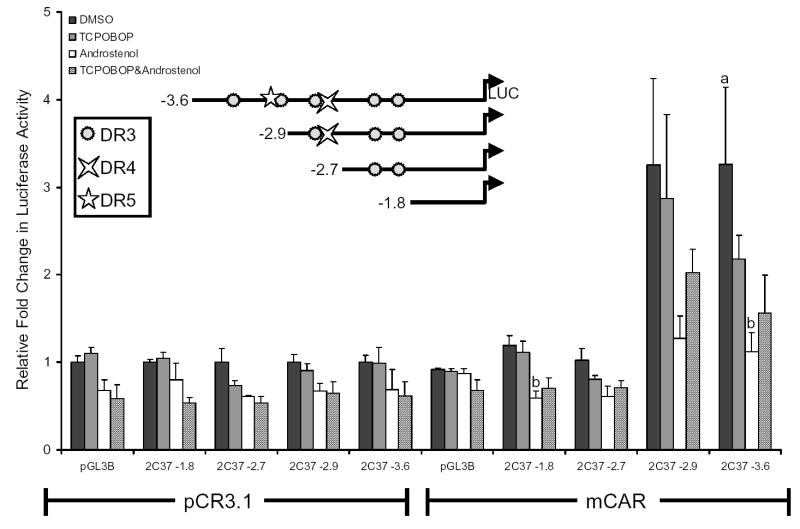
SeqLab GCG (Accelrys, San Diego, CA) was used to search 10 kb of the Cyp2c37 5′-flanking region for CAR/PXR response elements, defined as imperfect direct repeats of AGGTCA spaced by 3 to 5 nucleotides (DR-n). Several putative response elements were found upstream of the Cyp2c37 translation start site, including DR-3 motifs at -3205 bp, -3102 bp, -2820 bp, -2065 bp, and -1890 bp; one DR-5 motif at -3113 bp; and one DR-4 motif at -2791 bp (Fig. 2B). Luciferase reporters containing various lengths of the Cyp2c37 5′-flanking region were constructed and cotransfected with mCAR into HepG2 cells. The transfected cells were treated with mCAR activity modulators TCPOBOP and androstenol to facilitate the identification of CAR responsive regions. Cyp2c37 -1.8 kb and -2.7 kb luciferase reporter constructs were not activated by the presence of mCAR regardless of whether ligands were added (Fig. 3). However, the Cyp2c37 -2.9 kb and -3.6 kb luciferase reporters exhibited constitutive mCAR activation of ~3 fold by mCAR that was not significantly induced further by TCPOBOP. The constitutive transactivation of the -2.9 kb and the -3.6 kb Cyp2c37 luciferase reporters was repressed by the addition of 10 μM androstenol (Forman et al., 1998) (Fig. 3). This effect could be partially reversed by the addition of 250 nM TCPOBOP, a mCAR specific activator (Tzameli et al., 2000). These data indicated the presence of a CAR responsive region between -2.7 kb and -3.6 kb within Cyp2c37 5′-flanking sequence.
Figure 3.
Transcriptional activation analysis of the Cyp2c37 5′-flanking region by mCAR. HepG2 cells were cotransfected with pRL-Tk (internal transfection control), expression vectors mCAR (pCR3) or empty (pCR3.1), and pGL3 Basic luciferase vectors containing varying lengths of the Cyp2c37 5′-flanking region to evaluate mCAR effects on gene reporter activity. Androstenol (10 μM) and TCPOBOP (250 nM) were used to modulate mCAR activity. These data represent the results of three independent transfections. P-values were determined using the Tukey-Kramer HSD Test. a p≤ 0.05, mCAR significantly increased luciferase activity compared to no receptor DMSO control group. b p ≤ 0.05, androstenol significantly decreased luciferase activity compared to mCAR DMSO treatment group.
Electrophoretic mobility shift assays (EMSA) were performed to identify binding of mCAR to any of the previously identified putative response elements within this region. EMSA results of radiolabeled oligonucleotides indicated that only the putative DR-4 response element, located at -2791 bp from start of Cyp2c37 translation, bound mCAR (Fig. 4). This interaction was reduced with 25 fold molar excess of nonradiolabeled oligonucleotides. The -2791 element is similar to a CAR response element (CAR-RE) previously identified within the human CYP2C9 5′-flanking sequence at -1839 bp (Gerbal-Chaloin et al., 2002).
Figure 4.
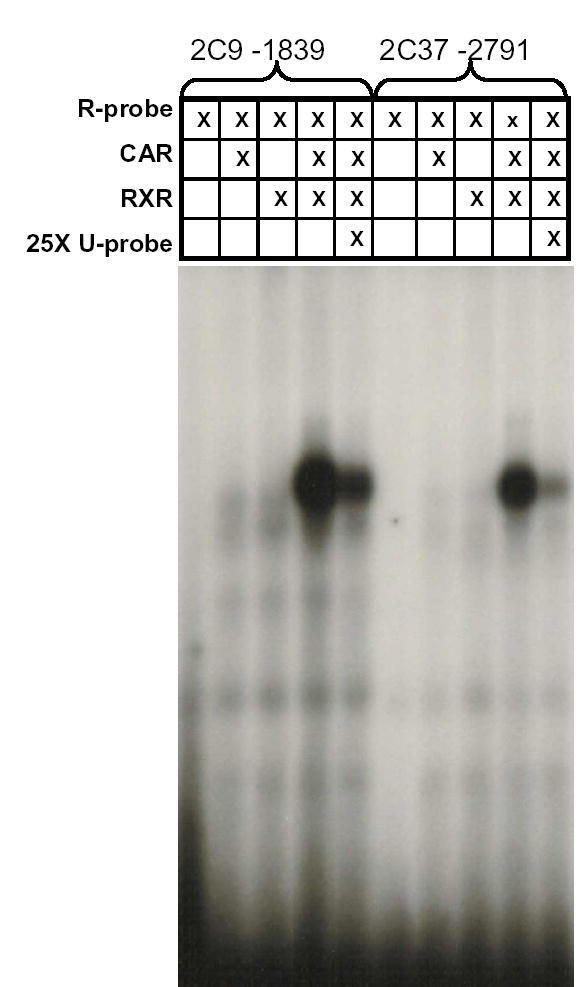
Analysis of mCAR binding to -2791 putative CAR-RE using EMSA. A: Radio-labeled -2791 putative CAR-RE (100,000 cpm 32P) oligonucleotides were incubated with in vitro transcribed and translated mCAR, hRXR or mCAR/hRXR proteins. In parallel experiments, incubation was performed in the presence of 25 fold molar excess of nonradiolabeled probe. EMSA data demonstrated that the Cyp2c37 putative -2791 CAR-RE binds mCAR/RXR as indicated by the presence of a band/shift. This interaction could be specifically competed out using nonradiolabeled Cyp2c37 -2791 CAR-RE oligonucleotides. The positive control, CYP2C9 -1839 CAR-RE, showed similar results. B: Illustration comparing CYP2C9 -1839 CAR-RE sequence to that of Cyp2c37 -2791 putative CAR-RE sequence.
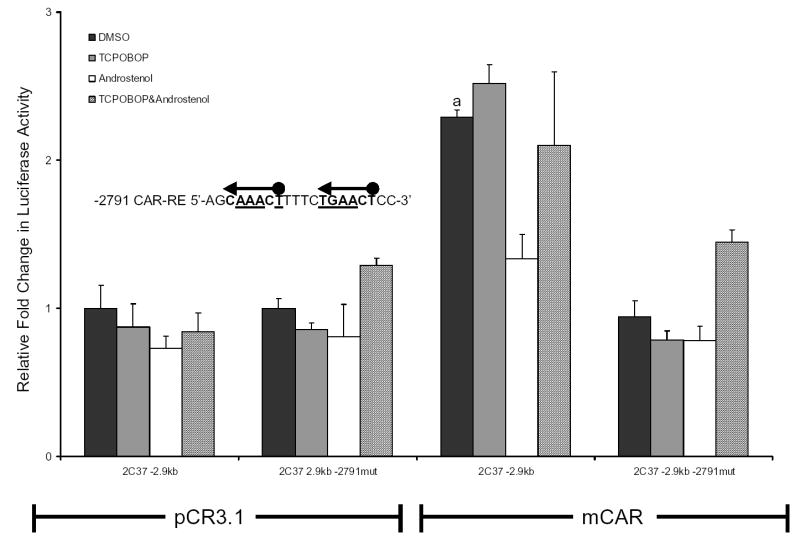
Cyp2c37 -2791 CAR-RE fuctional analysis
Mutagenesis was performed to determine if the Cyp2c37 -2791 CAR-RE site is necessary for mCAR constititutive transactivation of the Cyp2c37 -2.9 kb luciferase reporter. Mutation of the Cyp2c37 -2791 CAR-RE completely abolished mCAR transactivation of the Cyp2c37 -2.9 kb luciferase reporter (Fig. 5).
Figure 5.
Functional analysis of the Cyp2c37 -2791 CAR-RE mutant. HepG2 cells were cotransfected with pRL-Tk (internal transfection control), expression vectors mCAR (pCR3) or empty (pCR3.1), and Cyp2c37 wild-type or -2.9 kb Cyp2c37 mutant luciferase reporter to evaluate mCAR effects on gene reporter activity. Underlined nucleotides in Cyp2c37 -2791 putative CAR-RE sequence were mutated to deoxycytidines using site-directed mutagenesis (Stratagene). These data represent the results of three independent transfections. Mutation of -2791 imperfect DR-4 CAR-RE abolished mCAR transactivation. This reporter was also not responsive to androstenol (10 μM) repression. P-values were determined using the Tukey-Kramer HSD Test. a p≤ 0.05, mCAR significantly increased luciferase activity compared to no receptor DMSO control group. b p ≤ 0.05, androstenol significantly decreased luciferase activity compared to mCAR DMSO treatment group.
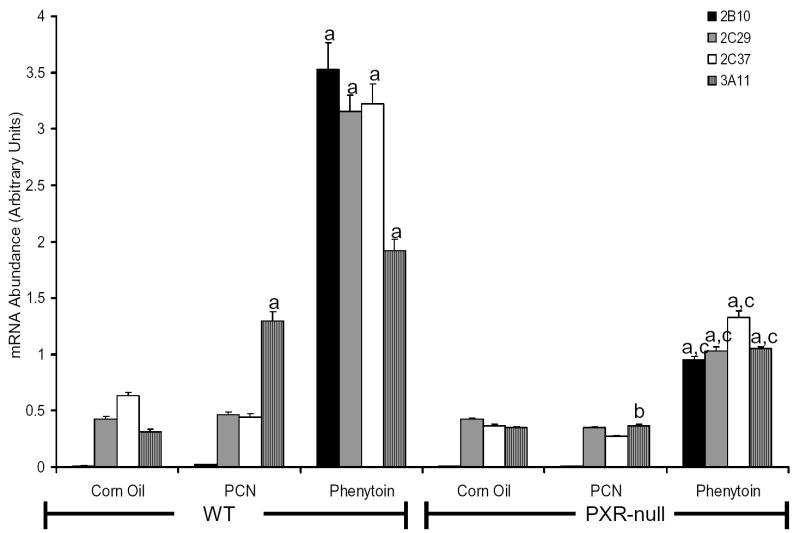
Evaluation of Cyp2c37 induction in CAR-null and PXR-null mice
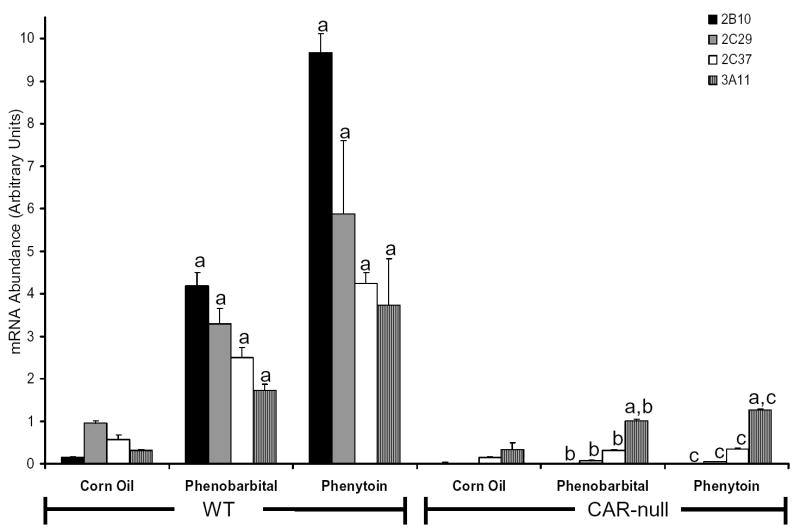
CAR-null (Fig 6) and PXR-null mice (Fig 7) were employed to examine whether induction of hepatic CYP2C37 mRNA by phenytoin is mediated by CAR or PXR in vivo. Hepatic CYP2B10 mRNA and CYP3A11 mRNA were evaluated as positive controls to evaluate xenobiotic activation by CAR and PXR, respectively. In CAR-null mice (Fig 6), induction of CYP2B10 mRNA by phenytoin and phenobarbital was completely abolished as expected. The induction of CYP2C29 and CYP2C37 mRNA by phenobarbital was dramatically reduced (~98% and ~87% respectively) in CAR-null mice compared with their congenic controls. Similarly induction of CYP2C29 and CYP2C37 mRNA by phenytoin in CAR-null mice was reduced by ~99% and ~92% respectively. However, the induction of CYP3A11 mRNA by phenytoin was only moderately decreased (~66%) in CAR-null mice.
Figure 6.
Evaluation of Cyp2c37 induction by phenytoin in CAR-null mice Mice were treated as described above. Quantitative RT-PCR was performed to evaluate target gene mRNA content in response to phenobarbital and phenytoin. Target genes were normalized to a reference gene, β-actin. CYP2B10, CYP2C29 and CYP2C37 mRNA was induced by phenobarbital and phenytoin in congenic wild-type mice, but induction was abolished in CAR-null mice. CYP3A11 mRNA was also induced by phenobarbital and phenytoin in wild-type mice; however, induction of CYP3A11 mRNA by phenytoin was not removed in CAR-null mice. Values expressed above represent the relative abundance of gene specific mRNA +/− SE. P-values were determined using the Tukey-Kramer HSD Test. a p< 0.05, significantly higher than corn oil controls. b p < 0.05, significantly lower than wild-type treated with phenobarbital. c p < 0.05, significantly lower than wild-type treated with phenytoin.
Figure 7.
Evaluation of Cyp2c37 induction by phenytoin and PCN in PXR-null mice. Mice were treated with phenytoin as described above. Mice treated with PCN (80 mg/kg) were treated orally for three consecutive days. Quantitative RT-PCR was performed to evaluate target gene mRNA content in response to PCN and phenytoin. Target genes were normalized to a reference gene, β-actin. CYP2B10, CYP2C29, and CYP2C37 mRNA induction by phenytoin was reduced, not abolished in PXR-null mice. CYP3A11 mRNA was induced by PCN in congenic wild-type mice, but was eliminated in PXR-null mice. In contrast, induction of CYP3A11 mRNA by phenytoin was reduced, not abolished in PXR-null mice. Values expressed above represent the relative abundance of gene specific mRNA +/− SE. P-values were determined using the Tukey-Kramer HSD Test. a p< 0.05, significantly higher than corn oil controls. b p < 0.05, significantly lower than wild-type treated with PCN. c p < 0.05, significantly lower than wild-type treated with phenytoin.
We next examined induction in PXR-null mice in parallel with the congenic wild-type mice (Fig 7). CYP3A11 mRNA was induced ~6-fold by PCN in wild-type mice, and this induction was completely abolished in PXR-null mice. In contrast, induction of CYP3A11 mRNA by phenytoin was reduced ~45% in PXR-null mice. Induction of CYP2B10, CYP2C29 and CYP2C37 mRNA by phenytoin was attenuated but not abolished in PXR-null mice, respectively (Fig. 7). Taken together, these data suggest that induction of CYP2C29, CYP2C37 and CYP2B10 mRNA by phenytoin and phenobarbital is mediated predominantly by CAR. This hypothesis is further supported by the observation that PCN, a known mPXR agonist, does not induce CYP2C29, CYP2C37, or CYP2B10 mRNA. In contrast, the induction of CYP3A11 mRNA by phenytoin appears to be mediated by CAR and PXR as suggested by the fact that induction is not abolished in either CAR-null or PXR-null mice.
Discussion
Several studies have examined the drug induced transcriptional regulation of the human CYP2C subfamily of P450 enzymes by CAR and PXR; however, relatively little is known of the regulation of their murine counterparts. Presently, a total of fifteen murine Cyp2c genes have been identified within a cluster on chromosome 19 (Nelson et al., 2004; Wang et al., 2004b). Within this large P450 gene subfamily, only the Cyp2c29 gene has been shown to be drug inducible thus far (Jackson et al., 2004). Herein, we show that hepatic CYP2C37 mRNA is induced by phenobarbital and phenytoin, becoming the second known drug inducible murine Cyp2c gene. We demonstrate using CAR-null mice that the induction of CYP2C37 mRNA is primarily CAR-dependent. Additionally, we identify a functional DR-4 CAR-RE at -2791 bp from the translation start site of the Cyp2c37 gene.
Alignments of the 5′-flanking regions of the murine Cyp2c genes indicate that particular subsets of Cyp2c members including Cyp2c29, Cyp2c39, and Cyp2c38 share high sequence homology; however, as a group the 5′-flanking regions are not highly conserved. The alignments show that the PHREM is exclusive to Cyp2c29 and is not conserved in any of the other corresponding 5′-flanking regions. These findings suggested that induction of hepatic CYP2C37 mRNA is mediated by an alternative drug responsive region. A number of putative CAR binding sites were identified; however, only the putative DR-4 CAR-RE (-2791) bound mCAR and mutagenesis of this site indicated that it is necessary for mCAR transactivation. These results are consistent with our previous studies examining transcriptional regulation of the Cyp2c29 gene by CAR (Jackson et al., 2004). Additionally, the alignment of the 5′-flanking regions of the murine Cyp2c genes indicated that the newly identified -2791 CAR-RE was unique to Cyp2c37 and was also not conserved among the other Cyp2c members (Fig. 8A). Although neither responsive region was conserved, the lack of conservation does not prove that the other subfamily members are non-inducible following exposure to phenobarbital or phenytoin.
Figure 8.

Alignment of CYP2C 5′-flanking regions and summary illustration of common mCAR and mPXR activators. A: Alignment of Cyp2c37 -2791 CAR-RE and surrounding sequence with corresponding 5′-flanking sequence of all presently known murine CYP2C genes. B: Ilustration showing the shared activators of the xenobiotic sensing nuclear receptors PXR and CAR and their respective genes that are induced.
Using Quantitative RT-PCR, we determined that CYP2C37 mRNA is induced by phenytoin and phenobarbital in wild-type mice, but in CAR-null mice the induction of CYP2C37 mRNA was essentially eliminated. These data indicate that phenobarbital and phenytoin induction is primarily CAR-dependent. Phenytoin induction of CYP2C37 and CYP2C29 mRNA was reduced moderately in PXR-null mice; however, a significant amount of induction remained. Although it is possible that that PXR is directly involved in the induction of Cyp2c37 and Cyp2c29 by phenytoin, its effects could be indirect. First, induction of Cyp2c37 and Cyp2c29 by phenytoin was essentially abolished in CAR-null mice suggesting minimal contribution of PXR. Secondly, the mPXR specific agonist PCN did not induce these genes in wild-type mice. A study by Maglich et al. (2002) demonstrated that expression of mCAR is induced by PCN in wild-type mice, but not in PXR-null mice. These results suggest that CAR expression is regulated by PXR in mice; therefore, CAR expression may be lower in PXR-null mice. A reduction in the expression of CAR or other nuclear receptors could indirectly attenuate the induction of CYP2C37 and CYP2C29 mRNA by phenytoin in PXR-null mice. Thus, PXR could be involved indirectly in regulating murine CYP2C gene regulation. In humans, both CAR and PXR appear to regulate the drug induced expression of CYP2C8 and CYP2C9 (Chen et al., 2004; Ferguson et al., 2005; Al-Dosari et al., 2006) by compounds like the CAR agonist CITCO and the PXR agonist rifampicin. In contrast, the present studies suggest that CAR is the primary regulator of murine Cyp2c37 and Cyp2c29 drug induction, suggesting a species difference in CYP2C regulation.
Although initially monitored to determine PXR activation by PCN, CYP3A11 mRNA was observed to be induced by phenytoin in wild-type mice. Induction of CYP3A11 mRNA by phenytoin was reduced, but not abolished in either CAR-null or PXR-null mice. In contrast, PCN induction of CYP3A11 mRNA was abolished in PXR-null mice consistent with PXR mediated induction (Xie et al., 2000; Staudinger et al., 2001a; Staudinger et al., 2001b; Goodwin et al., 2002). Thus, phenytoin appears to act as a mPXR and mCAR activator similar to dieldrin and clotrimazole (Fig. 8B) (Zhang et al., 2004).
In conclusion, we have demonstrated that CYP2C37 mRNA is induced by phenobarbital and phenytoin. We have shown that the induction of hepatic CYP2C37 mRNA by phenobarbital and phenytoin is CAR-dependent. We identified a functional DR-4 CAR-RE located at -2791 bp from the translation start site of Cyp2c37, which we propose mediates CAR-dependent drug induction of the Cyp2c37 gene. The mPXR agonist PCN does not induce murine Cyp2c37 or Cyp2c29. Thus, our studies also suggest that CAR is the predominate regulator of these murine Cyp2c genes (Fig 8B). Furthermore, our studies indicate that phenytoin probably activates both mCAR and mPXR in the induction of the Cyp3a11 gene.
Acknowledgments
We would like to especially thank Dr. Jeff Staudinger (Kansas University, Department of Pharmacology and Toxicology) for his gift of the PXR-null mice to Dr. Masahiko Negishi (NIEHS, Laboratory of Reproductive and Developmental Toxicology ). We would like to acknowledge Rick Moore (NIEHS, Laboratory of Reproductive and Developmental Toxicology ) for his excellence in maintaining and managing our CAR-null and PXR-null breeding programs. In addition, we would like to recognize Dr. Grace Kissling (NIEHS, Biostatistics Branch) for her expertise in statistical analysis. We would also like to thank Louise Harris (NIEHS, Animal Facility) for her expertise in animal dosing.
Footnotes
Send reprint requests to: Joyce Goldstein, National Institute of Environmental Health Sciences, MD A3-02, P.O. Box 12233, Research Triangle Park, North Carolina 27709
This research in entirety is a portion of the dissertation research in progress by Jonathan P. Jackson and was supported by the Intramural Research Program of the NIH and NIEHS. This work was previously presented at Experimental Biology 2006 meeting submitted to the ASPET
References
- Al-Dosari MS, Knapp JE, Liu D. Activation of human CYP2C9 promoter and regulation by CAR and PXR in mouse liver. Mol Pharm. 2006;3:322–328. doi: 10.1021/mp0500824. [DOI] [PubMed] [Google Scholar]
- Aldridge TC, Tugwood JD, Green S. Identification and characterization of DNA elements implicated in the regulation of CYP4A1 transcription. Biochem J. 1995;306 ( Pt 2):473–479. doi: 10.1042/bj3060473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ferguson SS, Negishi M, Goldstein JA. Identification of constitutive androstane receptor and glucocorticoid receptor binding sites in the CYP2C19 promoter. Mol Pharmacol. 2003;64:316–324. doi: 10.1124/mol.64.2.316. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ferguson SS, Negishi M, Goldstein JA. Induction of human CYP2C9 by rifampicin, hyperforin, and phenobarbital is mediated by the pregnane X receptor. J Pharmacol Exp Ther. 2004;308:495–501. doi: 10.1124/jpet.103.058818. [DOI] [PubMed] [Google Scholar]
- DeLozier TC, Tsao CC, Coulter SJ, Foley J, Bradbury JA, Zeldin DC, Goldstein JA. CYP2C44, a new murine CYP2C that metabolizes arachidonic acid to unique stereospecific products. J Pharmacol Exp Ther. 2004;310:845–854. doi: 10.1124/jpet.104.067819. [DOI] [PubMed] [Google Scholar]
- Ferguson SS, Chen Y, LeCluyse EL, Negishi M, Goldstein JA. Human CYP2C8 is transcriptionally regulated by the nuclear receptors constitutive androstane receptor, pregnane X receptor, glucocorticoid receptor, and hepatic nuclear factor 4alpha. Mol Pharmacol. 2005;68:747–757. doi: 10.1124/mol.105.013169. [DOI] [PubMed] [Google Scholar]
- Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, Moore DD. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- Gerbal-Chaloin S, Daujat M, Pascussi JM, Pichard-Garcia L, Vilarem MJ, Maurel P. Transcriptional regulation of CYP2C9 gene. Role of glucocorticoid receptor and constitutive androstane receptor. J Biol Chem. 2002;277:209–217. doi: 10.1074/jbc.M107228200. [DOI] [PubMed] [Google Scholar]
- Gerbal-Chaloin S, Pascussi JM, Pichard-Garcia L, Daujat M, Waechter F, Fabre JM, Carrere N, Maurel P. Induction of CYP2C genes in human hepatocytes in primary culture. Drug Metab Dispos. 2001;29:242–251. [PubMed] [Google Scholar]
- Goldstein JA, de Morais SM. Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics. 1994;4:285–299. doi: 10.1097/00008571-199412000-00001. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol. 1999;56:1329–1339. doi: 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, D"Costa DJ, Robertson GR, Liddle C. Transcriptional Regulation of the Human CYP3A4 Gene by the Constitutive Androstane Receptor. Molecular Pharmacology. 2002;62:359–365. doi: 10.1124/mol.62.2.359. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Redinbo MR, Kliewer SA. Regulation of cyp3a gene transcription by the pregnane x receptor. Annu Rev Pharmacol Toxicol. 2002;42:1–23. doi: 10.1146/annurev.pharmtox.42.111901.111051. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. Biochem J. 2000;347:321–337. doi: 10.1042/0264-6021:3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Ferguson SS, Moore R, Negishi M, Goldstein JA. The constitutive active/androstane receptor regulates phenytoin induction of Cyp2c29. Mol Pharmacol. 2004;65:1397–1404. doi: 10.1124/mol.65.6.1397. [DOI] [PubMed] [Google Scholar]
- Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol. 2002;62:638–646. doi: 10.1124/mol.62.3.638. [DOI] [PubMed] [Google Scholar]
- Meyer RP, Hagemeyer CE, Knoth R, Kurz G, Volk B. Oxidative hydrolysis of scoparone by cytochrome p450 CYP2C29 reveals a novel metabolite. Biochem Biophys Res Commun. 2001;285:32–39. doi: 10.1006/bbrc.2001.5111. [DOI] [PubMed] [Google Scholar]
- Miners JO, Birkett DJ. Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol. 1998;45:525–538. doi: 10.1046/j.1365-2125.1998.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Jones JE. Regulation of the mammalian cytochrome P1-450 (CYP1A1) gene. Int J Biochem. 1989;21:243–252. doi: 10.1016/0020-711x(89)90182-1. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Pascussi JM, Gerbal-Chaloin S, Drocourt L, Assenat E, Larrey D, Pichard-Garcia L, Vilarem MJ, Maurel P. Cross-talk between xenobiotic detoxication and other signalling pathways: clinical and toxicological consequences. Xenobiotica. 2004;34:633–664. doi: 10.1080/00498250412331285454. [DOI] [PubMed] [Google Scholar]
- Raucy JL, Mueller L, Duan K, Allen SW, Strom S, Lasker JM. Expression and induction of CYP2C P450 enzymes in primary cultures of human hepatocytes. J Pharmacol Exp Ther. 2002;302:475–482. doi: 10.1124/jpet.102.033837. [DOI] [PubMed] [Google Scholar]
- Staudinger J, Liu Y, Madan A, Habeebu S, Klaassen CD. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane X receptor. Drug Metab Dispos. 2001a;29:1467–1472. [PubMed] [Google Scholar]
- Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001b;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Negishi M. Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu Rev Pharmacol Toxicol. 2001;41:123–143. doi: 10.1146/annurev.pharmtox.41.1.123. [DOI] [PubMed] [Google Scholar]
- Tzameli I, Pissios P, Schuetz EG, Moore DD. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor CAR. Mol Cell Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Moore R, Sueyoshi T, Negishi M, LeCluyse E. Human constitutive androstane receptor mediates induction of CYP2B6 gene expression by phenytoin. J Biol Chem. 2004a;279:29295–29301. doi: 10.1074/jbc.M400580200. [DOI] [PubMed] [Google Scholar]
- Wang H, Faucette S, Sueyoshi T, Moore R, Ferguson S, Negishi M, LeCluyse EL. A novel distal enhancer module regulated by pregnane X receptor/constitutive androstane receptor is essential for the maximal induction of CYP2B6 gene expression. J Biol Chem. 2003;278:14146–14152. doi: 10.1074/jbc.M212482200. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhao Y, Bradbury JA, Graves JP, Foley J, Blaisdell JA, Goldstein JA, Zeldin DC. Cloning, expression, and characterization of three new mouse cytochrome p450 enzymes and partial characterization of their fatty acid oxidation activities. Mol Pharmacol. 2004b;65:1148–1158. doi: 10.1124/mol.65.5.1148. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-Tetri BA, Brunt EM, Guzelian PS, Evans RM. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang W, Qatanani M, Evans RM, Moore DD. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem. 2004;279:49517–49522. doi: 10.1074/jbc.M409041200. [DOI] [PubMed] [Google Scholar]