Abstract
The synthesis of 3′ subgenomic RNA4 (sgRNA4) by initiation from an internal sg promoter in the RNA3 segment was first described for Brome mosaic bromovirus (BMV), a model tripartite positive-sense RNA virus (W. A. Miller, T. W. Dreher, and T. C. Hall, Nature 313:68-70, 1985). In this work, we describe a novel 5′ sgRNA of BMV (sgRNA3a) that we propose arises by premature internal termination and that encapsidates in BMV virions. Cloning and sequencing revealed that, unlike any other BMV RNA segment, sgRNA3a carries a 3′ oligo(A) tail, in which respect it resembles cellular mRNAs. Indeed, both the accumulation of sgRNA3a in polysomes and the synthesis of movement protein 3a in in vitro systems suggest active functions of sgRNA3a during protein synthesis. Moreover, when copied in the BMV replicase in vitro reaction, the minus-strand RNA3 template generated the sgRNA3a product, likely by premature termination at the minus-strand oligo(U) tract. Deletion of the oligo(A) tract in BMV RNA3 inhibited synthesis of sgRNA3a during infection. We propose a model in which the synthesis of RNA3 is terminated prematurely near the sg promoter. The discovery of 5′ sgRNA3a sheds new light on strategies viruses can use to separate replication from the translation functions of their genomic RNAs.
Single-stranded positive-sense RNA viruses utilize various strategies for expression of their RNA genomes (36), most notably via subgenomic RNAs (sgRNAs). The Coronaviridae and Arteriviridae families of the order Nidovirales express 6 to 7 proteins via sgRNAs, while the Closteroviridae generate between 6 and 11 sgRNAs. In some viruses, e.g., in Brome mosaic bromovirus (BMV), sgRNAs arise via internal initiation by the viral RNA polymerase (RdRp) on genomic minus-sense RNAs (20, 30, 33, 38). It is also likely that other viruses, e.g., red clover necrotic mosaic virus (54), tomato bushy stunt virus (10), and flock house virus (17), copy their sgRNAs from prematurely terminated minus strands (62). In Nidovirales (46, 61, 72) the noncontiguous RNA leaders are joined to variously located sequences during minus-strand synthesis, followed by sgRNA transcription, whereas toroviruses combine discontinuous and nondiscontinuous processes to produce their sgRNAs (63). The formation of sgRNAs can also result from premature termination of positive-strand synthesis. In closteroviruses, a highly structured sequence region produces 5′-terminal sgRNAs by pretermination (20) and additionally serves as a promoter for synthesis of another downstream sgRNA, with possible overlapping of termination and initiation signals (20).
The previously described 3′ sgRNA4 of BMV is transcribed from an intergenic 100-nucleotide (nt) promoter (sgp) that consists of the core domain, a transcription enhancer, and the poly(A) tract (18, 38, 57). The more-upstream 150-nt sequence functions in cis as an internal replication enhancer (IRE) for positive-strand RNA3 amplification and carries a conserved B-box motif (18). In addition, we recently reported that the poly(A) tract of the sgp can function as an efficient replicase detachment/reattachment site, thereby contributing to the high-frequency homologous RNA3-RNA3 crossovers mapping within this region (5, 68, 69).
In this work, we characterize a novel BMV sgRNA, designated sgRNA3a, which represents the 5′ portion of genomic BMV RNA3. This sgRNA3a accumulates in BMV-infected tissues and carries the 3′ oligo(A) tail. By using a BMV replicase in vitro assay, we show that sgRNA3a is generated by pretermination at the oligo(U) tract on minus-strand RNA3 templates. The in vitro translation reactions in both the rabbit reticulocyte and the wheat germ extracts showed an increased synthesis of protein 3a by sgRNA3a compared to that for the full-length RNA3. In the BMV-infected tissues, sgRNA3a localized to the cellular polysome fractions, suggesting its translational activity. Taken together, our data suggest active roles for sgRNA3a in the BMV life cycle.
MATERIALS AND METHODS
Materials.
Plasmids pB1TP3, pB2TP5, and pB3TP7 (29) were used as templates to synthesize in vitro the infectious, capped, full-length transcripts of wild-type (wt) BMV RNA1, 2, and 3, respectively, by using the MEGAscript T7 kit (Ambion, Austin, Tex.). Moloney murine leukemia virus (MMLV) reverse transcriptase and Taq DNA polymerase were purchased from MBI Fermentas. Restriction enzymes, Pfu DNA polymerase, and deoxynucleoside triphosphates (dNTPs) were purchased from Promega Corporation, and the QIAGEN PCR cloning kit was from QIAGEN GmbH. The rabbit reticulocyte lysate and wheat germ in vitro translation systems were both from Promega Corporation. The [35S]methionine was purchased from ICN.
Detection and cloning of sgRNA3a.
Barley plants were inoculated with equal amounts (5 μg) of transcribed BMV RNA1, 2, and 3 (29). The inoculated plants were maintained in a greenhouse for 10 days, and total RNA was extracted as previously described (16, 40). The presence of positive-strand sgRNA3a was detected by Northern blotting (35) with a radioactive RNA probe complementary to the positive-strand RNA3 sequence between nt 962 and 1111. Similarly, to detect the putative minus-strand sgRNA3a, the probe used was complementary to nt 401 to 510 of the minus-strand RNA3.
Total RNA was extracted from BMV-infected plants (16, 40) and separated by agarose gel electrophoresis. The band containing the sgRNA3a fraction was cut out of the gel, and the RNA was eluted. For cDNA cloning, an in vitro-transcribed 5′-phosphorylated oligoribonucleotide RNA linker (RNA-lig; 5′-GGUACGAGAUCUCGCGAGAACUGCAGAACCUAUGCAUUGG-3′ [PstI restriction site is underlined], sequence not related to BMV RNA3) was 3′ ligated to sgRNA3a by using T4-RNA ligase. The ligation mixture contained 500 ng RNA-lig, 250 ng sgRNA3a, 1× T4 RNA ligase buffer, 40 U RNasin RNase inhibitor, 10 U T4 RNA ligase, and 20% polyethylene glycol (PEG) in a total volume of 40 μl. The reaction was performed overnight at 16°C. The RNA products were dissolved in 20 μl H2O and amplified by reverse transcription (RT)-PCR with oligonucleotides RW-rev-lig3, which is complementary to the RNA-lig linker, and RW-M1400 (5′-GCGTAATACGACTCACTATAGGGAGAACTGCAGTTACTCATGCGTATTGG-3′), which is homologous to nt 658 to 680 of the positive-strand RNA3 sequence (the PstI sites are underlined), or with oligonucleotides RW-rev-lig3 and MP1, which are homologous to nt 1 to 20 of positive-strand RNA3. The cDNA products were digested with PstI, ligated into the PUC19 vector, and sequenced.
Construction of mutagenized RNA3 templates.
RW-IDrev1 RNA was constructed as previously described (69). Briefly, the region between nt 1 and 1910 of pB3TP7 was amplified by PCR with primers 11 (5′-CCCAAGCTTGGGTAAAATACCAACTAATTCTCG-3′) and 12 (5′-AAAACTGCAGAACCTTAGCCAAAGTGTCCTAC-3′), carrying HindIII and PstI restriction sites, respectively (both restriction sites are underlined). The PCR products were ligated into plasmid pB3TP7, and the resulting plasmid was linearized with PvuII restriction enzyme and in vitro transcribed with a MEGAscript kit from Ambion (69). The integrity of the RNA preparation was confirmed electrophoretically.
Two oligo(A) deletion RNA3 constructs (Δ17 and Δ26) were generated by replacing the wt sequence in pB3TP7 between the SalI and ClaI restriction sites with a deletion-carrying DNA insertion. The insertion was amplified from pB3TP7 with 3′ primers, either DelA18-2 (5′-GAAGTCGACATTATTAATACGCTGAATTAGGACATAGTTTTAATAATAACTCAGACACACAACATA GAATATCCACAAAC-3′) for Δ17 or DelA18-1 (5′-GAAGTCGACATTATTAATACGCTGAATTAGGACATAGATCCTCAGACACACAACATAGAAT ATCCACAAAC-3′) for Δ26, both carrying the SalI restriction site (underlined), and with the 5′ primer Cla-5′ (5′-GATAGCGGTAGGAATCGATGTTTTGGGATAGC-3′), carrying the ClaI site (underlined). The PCR products were religated into the SalI/ClaI-cut plasmid pB3TP7, and the resulting plasmid was linearized with EcoRI restriction enzyme and in vitro transcribed. The transcripts were analyzed electrophoretically as described above.
In vitro generation of sgRNA3a by copying of minus strands with BMV RdRp.
To generate sgRNA3a in vitro, a standard RNA copying reaction was set up, containing 1 μg of RW-IDrev1 RNA and a BMV replicase preparation (extracted from BMV-infected barley seedlings) plus other components (35). The RNA products were separated in 4% polyacrylamide-7 M urea sequencing gel, the radioactive band of the sgRNA3a (size, ca. 1,200 nt) was cut out of the gel, and the RNA was electroeluted, followed by RNA precipitation with ethanol.
In vitro translation and polyribosome isolation.
BMV RNAs were translated in vitro in a nuclease-treated, rabbit reticulocyte lysate system or in a wheat germ system, according to the manufacturer's protocols, in the presence of [35S]methionine. The translation products were solubilized in a sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis solution, incubated at 95°C for 2 min, and subjected to electrophoresis in a polyacrylamide gradient gel (8 to 15%). The gel was dried under a vacuum and exposed to X-ray film.
The polysomes were isolated from BMV-infected barley, using 500 mg of the infected leaf tissue (27), the extract was fractionated by ultracentrifugation in a 2.5-ml 0 to 40% linear sucrose gradient, and total RNA was isolated from 300-μl fractions by phenol-chloroform extraction and ethanol precipitation, followed by gel electrophoresis and Northern blotting.
RESULTS
A novel sgRNA3a accumulates in BMV-infected plants.
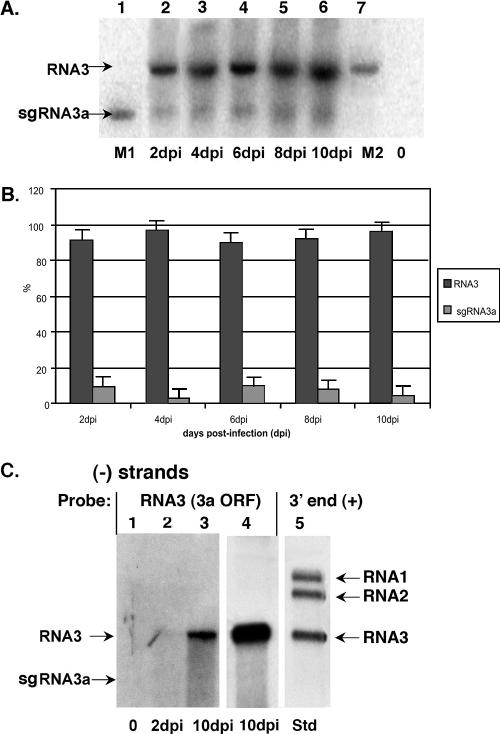
The multifunctional intercistronic region of BMV RNA3 can support the de novo synthesis of sgRNA4 (38), as well as high-frequency recombination events among BMV RNA3 molecules (68, 69). To test if this region supported the formation of 5′ sgRNA, total RNA was extracted from BMV-infected barley and the putative sgRNA was detected by Northern blotting with an RNA3-specific probe that was complementary to nucleotides 962 to 1111. As shown in Fig. 1A, two RNA3-related entities accumulated during infection of barley seedlings. The larger RNA represents the full-length BMV RNA3 (compare with the size marker), whereas the shorter RNA comigrated with a 1,221-nt in vitro-transcribed RNA standard. The shorter RNA was designated sgRNA3a because its size varied between those of RNA3 and sgRNA4. Time course analysis revealed that the fraction of sgRNA3a ranged from 2 to 10% between the two RNAs (Fig. 1B).
FIG. 1.
Northern blot analysis showing accumulation of RNA3-related products during BMV infection. (A and B) Accumulation of positive strands in barley plants inoculated in two separate experiments. Leaves were infected with BMV, and the RNA was extracted 2, 4, 6, 8 and 10 dpi. After being separated by electrophoresis in an agarose gel, the RNA material was blotted onto a nylon membrane and probed with a radioactive RNA probe complementary to an internal RNA3 sequence between nucleotides 962 and 1111. Lanes M1 and M2 carry the 1-to-1221-nt sequence and the full-length BMV RNA3 size standard transcripts, respectively. Line 0 represents RNA extracted from a healthy plant. (B) Accumulation of RNA3 and sgRNA3a was quantified by using Image Quant software in a Phosphoimager. (C) Accumulation of minus strands. RNA was extracted from plants infected with BMV at 2 (lane 2) and 10 (lanes 3 and 4; two separate extractions with lane 4 overexposed) days postinoculation. The probe represents the internal positive-strand RNA3 sequence between nucleotides 401 and 510. Lane 0, uninfected barley; lane 5, BMV RNA minus strands in total RNA from infected barley after 10 dpi, detected with BMV 3′-end positive-strand probe.
To test if sgRNA3a replicated through a plus- or minus-strand replication cycle, the RNA preparation obtained at 10 days postinoculation (dpi) was subjected to Northern blotting with a minus-strand RNA3-specific probe that represents the positive-strand RNA3 sequence between positions 401 and 510. This revealed only the band corresponding to the full-length RNA3 minus strand in the absence of a shorter putative minus-strand sgRNA3a product (Fig. 1C), suggesting that sgRNA3a was produced via pretermination rather than by RNA replication.
To characterize the 5′- and 3′-terminal sequences in the sgRNA3a molecule, a 5′-phosphorylated RNA linker was joined to its 3′ end, using RNA ligase, and the ligation product was amplified by RT-PCR, with the 5′ primer representing the wt RNA3 5′ end and the 3′ primer complementary to the linker. Sequencing of the cloned cDNA products revealed the presence of the heterogeneous 3′ oligo(A) tail (Table 1), ranging between 13 to 19 residues. Otherwise, all the clones carried the wt 5′ end and the wt sequence of the first 1,200 nt of BMV RNA3 (data not shown). Thus, sgRNA3a resembled a canonical mRNA molecule, carrying the 5′ noncoding region (presumably capped like the wt positive-strand RNA3), the 3a open reading frame, the 3′ noncoding region, and the 3′ oligo(A) tail.
TABLE 1.
Heterogeneity of the 3′ oligo(A) tail in total RNA and in sgRNA3a virion moleculesa
| Source and no. of clone | 3′ Terminal sequence (no. of A residues) | % Total RNA | ||
|---|---|---|---|---|
| sgRNA3a from total RNA
|
||||
| 1 | poly(A) (13) | 10.0 | ||
| 6 | poly(A) (17) | 60.0 | ||
| 2 | poly(A) (18) | 20.0 | ||
| 1 | poly(A) (19) | 10.0 | ||
| Encapsidated sgRNA3a
|
||||
| 1 | poly(A) (14) | 5.5 | ||
| 6 | poly(A) (13) | 33.0 | ||
| 1 | poly(A) (9) | 5.5 | ||
| 2 | poly(A) (8) | 11.0 | ||
| 3 | poly(A) (7) | 17.0 | ||
| 1 | poly(A) (5) | 5.5 | ||
| 3 | poly(A) (3) | 17.0 | ||
| 1 | poly(A) (2) | 5.0 | ||
The number of poly(A) residues was determined by sequencing of RT-PCR-generated cDNA clones from either total RNA or virion BMV RNA preparations, as described in Materials and Methods.
sgRNA3a is packaged in BMV virions.
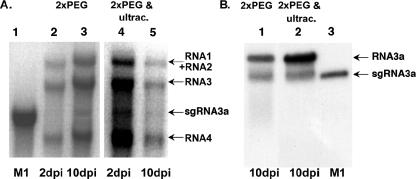
sgRNA4 coencapsidates with RNA3 in BMV virions (12, 51). To find out if sgRNA3a encapsidates, a BMV virus preparation was purified from infected barley leaves by two rounds of PEG precipitation or by two rounds of PEG precipitation followed by ultracentrifugation at 100,000 × g. The virion RNA was extracted from both virus preparations and analyzed by electrophoresis in denaturing agarose gel (Fig. 2A, lanes 1 to 5). This revealed the presence of two bands corresponding to RNA3 and sgRNA3a, with the former predominating over the latter. Northern blot analysis of BMV RNA samples (Fig. 2A, lanes 4 and 5) using the RNA3-specific probe confirmed the presence of both RNA entities (Fig. 2B). A lower concentration of sgRNA3a than sgRNA4 reflected their total relative abundances (Fig. 1 and Discussion).
FIG. 2.
Encapsidation of sgRNA3a. (A) BMV RNA was extracted from a purified virus preparation either by two cycles of PEG precipitation or by PEG precipitation and additional ultracentrifugation at 100,000 × g. The extracted RNA was separated in an agarose denaturing gel and stained with ethidium bromide. Lane 1, nt 1 to 1221 transcribed RNA3 size standard; lanes 2 and 3, virion RNA after two PEG precipitations at 2 and 10 days postinoculation; lanes 4 and 5, virion RNA after ultracentrifugation at 2 and 10 days postinfection. The encapsidated sgRNA3a and other BMV RNAs are indicated by arrows. (B) Northern blot analysis of BMV RNA extracted from a virus preparation 10 days postinoculation by using an RNA3-specific probe as for Fig. 1. Lanes 1 and 2, BMV RNA from a viral preparation after two PEG precipitations and after additional ultracentrifugation, respectively; lane 3, nt 1 to 1221 transcribed sgRNA3a size standard.
To characterize the nucleotide sequences of the encapsidated sgRNA3a, 18 cDNA clones were sequenced, demonstrating that they carried the full-length sgRNA3a sequence with the heterogeneous 3′ oligo(A) tail, varying between 2 to 14 A residues (Table 1), which was shorter than the tail in total RNA.
BMV replicase preterminates at the minus-strand oligo(U) tract and generates sgRNA3a in vitro.
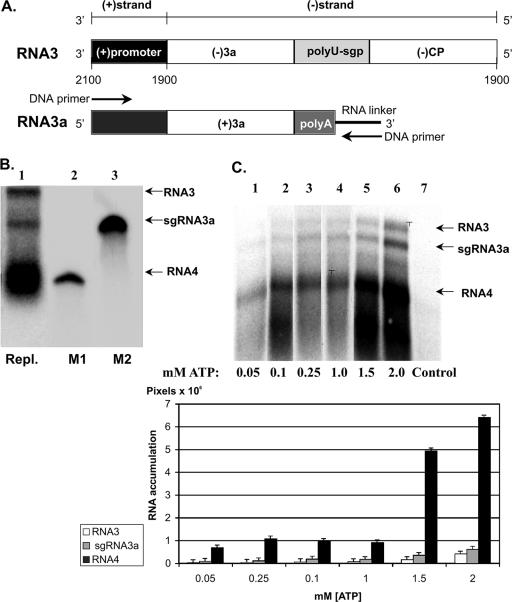
Because BMV replicase is not capable of 3′ initiation on wt minus-strand RNA3 sequences (67), a special RW-IDrev1 construct was designed. Its 3′ portion had the 3′ noncoding promoter region from BMV genomic RNA3 that was fused to the 5′ portion of the RNA3 minus-strand sequence in such a way that nucleotide 2100 of the minus-strand RNA3 joined nucleotide 1911 of the positive-strand 3′ sequence (Fig. 3A). In the in vitro replicase assay, the resulting minus-strand RNA3 molecule secured initiation of RNA synthesis from the positive-strand 3′ promoter, followed by copying of the minus-strand RNA3 template. As shown in lane 1 of Fig. 3B, among the three products of this template, the largest RNA was the full-length RNA3 (band RW-1Drev RNA3), and the shortest was sgRNA4 (band sgRNA4). The intermediate band represents sgRNA3a (band sgRNA3a), which comigrated with the corresponding synthetic RNA marker (Fig. 3B, lanes 4 and 5).
FIG. 3.
Copying in vitro of minus-strand [(-)strand] RNA3 templates with BMV replicase. (A) Diagram of RW-IDrev1 RNA template. The positive-sense RNA initiation sequence [(+)promoter] is represented by a solid bar, with flanking nucleotide positions corresponding to those of the wt RNA3 shown below. The sequences complementary to the 3a and CP ORFs are shown as open boxes, and the oligo(U)-containing subgenomic promoter as a shaded box. The sgRNA3a molecule (lower line) carries the 3′-oligo(A) tail that is represented by a gray box. The locations of the RNA linker and those of both DNA primers used for PCR amplification are shown as a black line and arrows, respectively. (B) Electrophoretic analysis of RNA products after in vitro copying of RW-IDrev1 RNA3. The RNA (1 μg) was copied in the RdRp assay reaction (see Materials and Methods) by using a BMV replicase preparation, and the radioactive products were separated by electrophoresis in a 4% polyacrylamide-12 M urea denaturing gel (lane 1). Besides a full-length copy of the input RW-IDrev1 RNA3 template, shorter products of the premature termination reaction at the oligo(U) tract (1,400 nt) and subgenomic sgRNA4 (800 nt) are visible. Lanes M1 and M2 show separate migrations of radioactive in vitro-transcribed RNAs used as size standards: the 800-nt RNA transcript the size of sgRNA4 and the 1,221-nt marker corresponding to the size of sgRNA3a. Repl., BMV replicase. (C) The effect of ATP on the copying of RW-IDrev1 RNA3. The copying reactions at increased ATP concentrations (indicated at the bottom) and analysis of the products were performed as described for panel B.
The sgRNA3a band was cloned, and the 3′ ends of 13 clones were sequenced. Ten clones carried the oligo(A) tail (77%) (Table 2), while three were terminated at upstream positions 1011 to 1014 (23%) in the IRE region (17, 32). These results suggested that the oligo(U) tract served as an efficient termination site, while upstream IRE was much less efficient. The length of the oligo(A) tail ranged between 1 and 13 A residues (Table 2), suggesting the termination range within the oligo(U) tract. The lack of sgRNA3a clones with the 3′ tail exceeding the length of the wt oligo(A) tract (18 A residues) suggested effective termination at the oligo(U) sequence.
TABLE 2.
Distribution of termination sites during in vitro copying of minus-strand RNA3 templates with the BMV replicase enzymea
| Termination site (wt RNA3)b | Site within the INT sequencec | Number of clones | Frequency of termination (%) |
|---|---|---|---|
| 1204 | Oligo(U) (3rd U) | 2 | 15.4 |
| 1216 | Oligo(U) (13th U) | 1 | 7.7 |
| 1208 | Oligo(U) (7th U) | 1 | 7.7 |
| 1203 | Oligo(U) (1st U) | 1 | 7.7 |
| 1210 | Oligo(U) (8th U) | 3 | 23.1 |
| 1211 | Oligo(U) (10th U) | 1 | 7.7 |
| 1207 | Oligo(U) (5th U) | 1 | 7.7 |
| Total | 77.0 | ||
| 1011 | IRE (minus-strand) | 2 | 15.4 |
| 1014 | IRE (minus-strand) | 1 | 7.7 |
The 3′ termini were determined by sequencing of RT-PCR-generated cDNA clones from copying reaction products of minus-strand RNA3 templates, as specified in Fig. 3 and described in Materials and Methods.
The numbers refer to nucleotide positions in the published sequence of BMV RNA3 (1).
INT, intergenic region.
The ATP concentration has different effects on sgRNA3a/RNA3 and sgRNA4/RNA3 ratios in vitro.
BMV replicase assays performed at ATP concentrations ranging between 0.05 mM and 2.5 mM revealed a similar RNA3/RNA3a ratio (Fig. 3C, compare lanes 1 through 6). This suggested that sgRNA3a was not preterminated due to the deficiency of ATP. However, sgRNA4 increased faster than RNA3, suggesting that sgRNA4 has different requirements for synthesis than RNA3 or sgRNA3a (see Discussion).
Deletions in the oligo(A) tract inhibit synthesis of sgRNA3a in vivo.
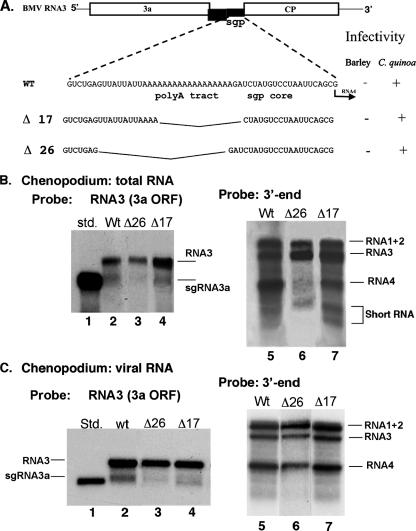
We tested whether the oligo(A) tract was relevant for the production of sgRNAs during infection. By deletion of the entire oligo(A) tract plus the upstream UUAUUAUU region (Fig. 4A), both sgRNAs were reduced in total RNA extracts compared to those for wt RNA3 (Fig. 4B, lanes 2 and 5 versus lanes 3 and 6). By leaving four A residues in the oligo(A) tract and the upstream UUAUUAUU region (in construct Δ17), fewer reductions in sgRNA3a and especially in sgRNA4 were observed (Fig. 4B, lanes 4 versus 7). Similar trends in RNA3/sgRNA3a ratios were observed in virion BMV RNA for both constructs (Fig. 4C, lanes 2 to 4). These results confirmed our in vitro data (Fig. 3) indicating that the oligo(A) tract is important for the formation of sgRNA3a as well as previous in vivo observations that oligo(A) is involved in sgRNA4 synthesis (55). In contrast, concentrations of encapsidated BMV RNA1 to RNA4 were similar among constructs Δ26, Δ17, and wt RNA (Fig. 4C, lanes 5 to 7), showing different preferences in packaging between sgRNA3a and sgRNA4 (see Discussion).
FIG. 4.
Effect of deletions in the oligo(A) tract on accumulation of sgRNA3a and sgRNA4 in vivo. (A) Schematic representation of the oligo(A) deletion constructs. The upper scheme shows the genomic RNA3 molecule and its sgp region sequence, while the lower part depicts deletion mutants. Δ26 lacks the entire oligo(A) sequence plus a short upstream region, and construct Δ17 carries only four A residues in the oligo(A) tract. (B) Northern blot analysis of accumulation of sgRNAs in total RNA extracts from Chenopodium quinoa. The RNA, extracted after 14 dpi as described in Materials and Methods, was separated in 1% denaturing agarose gel, transferred to a nylon membrane, and probed with an RNA3-specific probe (to detect sgRNA3a) or with a 3′-end probe (to detect sgRNA4). The inoculated RNA3 deletion constructs are indicated by the numbers on top, and the positions of individual RNAs are indicated on the right. (C) Northern blot analysis of accumulation of sgRNAs in the encapsidated BMV RNA from C. quinoa after 14 days psi, using two different probes, as specified. Std., in vitro transcribed 5′ 1,200 nt of RNA3.
sgRNA3a is more efficient in translation than RNA3.
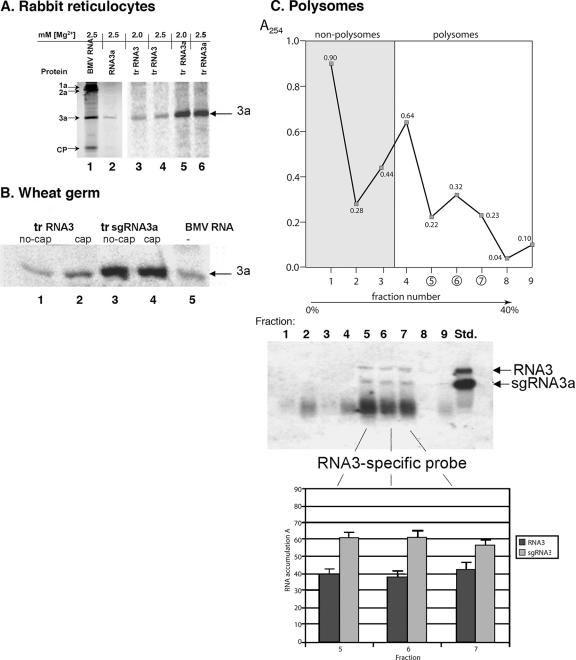
The translational competency of sgRNA3a was tested in vitro in rabbit reticulocyte and in wheat germ extracts. When 0.1 μg of gel-purified sgRNA3a was incubated with the reticulocyte lysate system, a radioactive protein that comigrated with protein 3a from the control translation with the wt BMV RNA was synthesized (Fig. 5A, lanes 1 and 2). To compare the translation efficiency of sgRNA3a with that of full-length RNA3, equimolar amounts (0.5 μg of RNA3 and 0.23 μg of sgRNA3a) of in vitro-transcribed and -purified RNA templates were translated at two different concentrations of MgCl2. sgRNA3a produced more protein 3a (by severalfold) than RNA3 at both 2.0 mM and 2.5 mM MgCl2 (Fig. 5A, compare lanes 3 and 4 with lanes 5 and 6), showing that sgRNA3a was more efficient in translation than RNA3. Similarly, higher translation activity was observed for sgRNA3a in the wheat germ system (Fig. 5B, lanes 1 to 4), but no visible differences existed between capped and uncapped RNAs. Overall, these results showed that sgRNA3a was a better template for protein 3a synthesis than full-length RNA3.
FIG. 5.
Translation activity of sgRNA3a in vitro and in the presence of polysomes. (A) Translation in rabbit reticulocyte system. The RNAs were incubated in rabbit reticulocyte extract at two different concentrations of MgCl2, as indicated, and the products were separated in 12% SDS-polyacrylamide gel. The template RNAs are BMV RNA, total encapsidated viral RNA extracted from purified BMV preparation; sgRNA3a, the fraction of sgRNA3a purified from the encapsidated BMV RNA by cutting a band off the agarose formaldehyde/formamide gel; trRNA3 and trRNA3a, the RNA3 and sgRNA3a preparations synthesized by transcription in vitro and purified from unincorporated NTPs (on RNeasy columns; QIAGEN) prior to the translation reaction. (B) Translation in the wheat germ system. The RNAs were incubated in the wheat germ system with [35S]methionine, and the products were analyzed in polyacrylamide-SDS gel, as described for panel A. The synthetic RNA3 or sgRNA3a templates were either capped or uncapped (see Materials and Methods), as indicated (lanes 1 through 4), whereas standard BMV RNA (containing the same amount of the RNA3 component) was translated in lane 5. The migration of protein 3a is shown on the right. tr, transcribed. (C) Detection of BMV RNA3-related sequences in polysomes. The polysomal RNA was extracted from BMV-infected barley leaves as described in Materials and Methods and separated by ultracentrifugation in sucrose gradient. Individual fractions (numbered 1 to 8) were analyzed by Northern blotting in 1% denaturing agarose gel and with the RNA3-specific probe. The positions of RNA3 and sgRNA3a were confirmed by comigration with the corresponding size transcripts in the “Std.” lane on the right.
Both sgRNA3a and RNA3 are found on polysomes.
In order to determine the translation activity of sgRNA3a in vivo, a polyribosomal fraction was extracted from BMV-infected barley leaves and fractionated by sucrose gradient ultracentrifugation. Total RNA was isolated separately from each of the nine collected fractions, purified, separated by electrophoresis in denaturing agarose gel, blotted, and probed with the RNA3-specific probe. As shown in Fig. 5C, three fractions (5 to 7, within the polysomal portion of the gradient) contained both wt-size BMV RNA3 and sgRNA3a-size molecules (with a constant ratio of sgRNA3a/RNA3 of 60%/40%), confirming the presence of ribosome-bound sgRNA3a during infection. In addition, diffuse bands of short RNA3-related RNAs were detected in most of the fractions, reflecting the presence of ribosome-bound RNA-3-derived fragments. These data substantiated the notion that sgRNA3a was translationally active during infection.
DISCUSSION
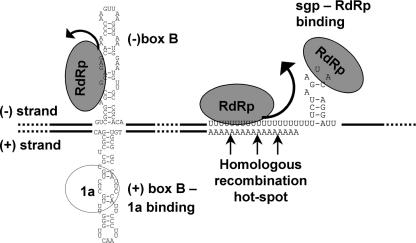
The phenomenon of 5′ sgRNA formation by premature termination has been described for several RNA viruses (for an example, see reference 72), but this work reports the first example of premature termination on the minus-strand oligo(U) tract. We present a model (Fig. 6) in which sgRNA3a is generated by the detachment at the minus-strand oligo(U) while sgRNA4 is made by de novo initiation at the sgp. With this discovery, there are now two known BMV sgRNAs.
FIG. 6.
Model illustrating the synthesis of sgRNA3a in view of multiple functions of the intergenic region in minus-strand RNA3. The BMV RdRp enzyme complex (represented by gray ovals) migrates alongside the minus-strand RNA template and pauses (represented by curved arrows) at the secondary structure or, most notably, at the oligo(U) tract, leading to the formation of subgenomic sgRNA3a. Yet another molecule of the RdRp enzyme binds to the sgp and initiates the de novo synthesis of sgRNA4. Also, the rehybridization of the sgRNA3a oligo(A) tail to the RNA3 minus template can resume full-length copying, which primes the observed RNA3-RNA3 recombination (5, 69). The positive and minus RNA strands are represented by thick lines and both the oligo(U) tract in the minus-strand template and the oligo(A) 3′-termini are exposed. The stem-and-loop structures adopted by the positive and minus strands upstream of their oligo(U) and oligo(A) tracts (3) are shown. The region that binds to protein 1a via the B box of the stem-loop structure in positive strands (28, 57) is shown.
Mechanism of sgRNA3a generation.
Previously, detachment of BMV replicase at the AU-rich regions was suggested (41, 42, 53), and in this work we have now mapped the BMV RNA3 crossovers to the oligo(A) tract (68). Data on bacterial systems reveal that poly(A) tracts (65) or AU-rich sequences (14, 25, 47) can destabilize the transcription complex. Similar factors could enhance premature termination at the BMV oligo(U) tract (Fig. 6). Both in vitro (Fig. 3) and in vivo (Fig. 1 and 4) experiments imply that sgRNA3a molecules arise by premature termination. Deletions of the oligo(A) tract significantly reduce the two sgRNAs (Fig. 4), verifying its role during RNA replication and transcription (22, 55).
We showed that the ATP concentration did not significantly affect RNA3/sgRNA3a ratios in vitro (Fig. 3C), implying that these two RNAs have common initiations. It is quite likely, then, that pretermination was not due to ATP deficiency, especially because of the fact that ATP usually reaches higher levels than other NTPs in the cell. Theoretically, higher levels of ATP might prevent termination, which was not observed here, suggesting a different mechanism of termination, e.g., the decreased template-nascent strand interactions at oligo(A) tracks. In contrast, the concentration of sgRNA4 increased faster with ATP than that of sgRNA3a. This demonstrated that sgRNA4 initiation was a different process than sgRNA3a termination. The latter starts with a G residue (24, 32), but apparently the ATP is also critical. Either the copying process of sgRNA4 utilizes the nucleotides more efficiently or the RNA4 transcription complex binds better to sgp (56, 57).
BMV replicase produced less sgRNA3a than sgRNA4 in vitro (Fig. 3). Like the previously reported interference between sgRNA4 transcription and RNA3 replication (22), pretermination of sgRNA3a may compete with the copying of full-length RNA3 for the replicase complex factors. However, interpretation of in vitro results must be done cautiously because the RNA3 minus-strand templates (Fig. 3) carry the tRNA-like promoter that normally occurs in positive strands. This was required to initiate RNA copying because the used BMV replicase could initiate only at tRNA-like structures. Thus, a factor could be lost which in turn could affect processivity and/or pretermination characteristics.
The 3′ oligo(A) tail could arise in sgRNA3a by detachment at the oligo(U) tract in the minus-strand template (Fig. 3) rather than by polyadenylation because of four factors: (i) RNA3 lacks the polyadenylation signal; (ii) canonical polyadenylation occurs in the nucleus; (iii) no sgRNA3a molecules with a longer oligo(A) tail were identified; and (iv) polyadenylation generates long poly(A) tails (up to 200 A's). However, cytoplasmic (48, 50) and viral (e.g., poxviruses; [26]) polyadenylation activities have been reported, and thus de novo polyadenylation cannot be ruled out.
Our data add one new function to the list of already known activities at the internal region of BMV RNA3. These previously known activities were the recruitment of RNA3 to the replicative spherules, 1a/2a interactions, regulation of the positive-strand/minus-strand ratio, initiation of sgRNA4, and RNA recombination (8, 28, 57). Figure 6 illustrates that premature termination occurs 5′ of the sgRNA4 de novo initiation. Similar observations have been made for Sindbis virus (66), citrus leaf blotch virus (64), grapevine virus A (19), and for a controller element of citrus tristeza closterovirus (CTV). CTV preterminates its 5′ sgRNA at a site located 30 to 50 nt upstream of another de novo-initiated 3′ sgRNA (2, 20). The CTV controller sequence carries two stem-loops (20), different from the BMV controller, which uses a combination of the oligo(U) and a stem-loop. All Bromoviridae and Togaviridae carry the internal poly(A) tract, implying that they could produce analogous 5′ preterminated sgRNAs.
No sgRNA3a clones that carry their 3′ ends beyond the length of the oligo(A) tract (no more than 18-residues long) were found. The oligo(U) tract must then act as an efficient termination signal, either by itself or via interaction with such elements as the sgp stem-loops, the upstream IRE hairpin (3, 8, 18, 28, 38, 57), or the packaging signal (12). 1a protein binding (57), replicase binding (11), or even some poly(U)-binding host proteins might further enhance pretermination. In potato virus X, the synthesis of sgRNAs is regulated by sgp signals that interact with the 5′ complementary elements (34). It is not known whether long-distance interactions (23, 39) can regulate sgRNA3a.
Similar to sgRNA3a (Fig. 1C) or sgRNA4, the above CTV sgRNAs did not have corresponding minus strands (20). However, a different set of CTV sgRNAs had both plus and minus strands, suggesting the orientation of their replication (20). In the cases of dianthoviruses (54), luteoviruses (39), and nodaviruses (17), it has been postulated that the prematurely terminated minus-strand sgRNAs were copied into the positive strands. It is not yet known if BMV could produce some other replicating sgRNAs in different hosts (15).
Our data cannot eliminate the possibility that sgRNA3a arose via cleavage of RNA3. Typically, the cleavage fragments have different levels of stability and thus accumulate to different levels in the cell. However, we did not detect any 3′-half RNA3 sequences among the cloned cDNAs by using 5′ rapid amplification of cDNA end experiments (data not shown). A prolonged incubation in water demonstrated that RNA3 was a stable molecule. One can speculate that sgRNA3a was uncommonly stable, but BMV RNAs treated with wheat germ extracts generated only fragments that were much shorter than the sgRNA3a (data not shown). Overall, we conclude that RNA cleavage is not a likely mechanism.
Putative function(s) of sgRNA3a.
One function of sgRNA3a could be during RNA3-RNA3 recombination (Fig. 6). We have recently mapped an RNA3 recombination hot spot to the oligo(A) tract (60, 68). This suggests that the predetached sgRNA3a intermediate functions as a primer (5, 68). Indeed, the previously described heterogeneity of the oligo(A) tract (1, 13) could be, at least in part, due to repriming at various positions alongside the minus-strand RNA3 oligo(U) region. The polymerase slippage could be another mechanism contributing to this heterogeneity (55).
The RdRps of such viruses as poliovirus and bacteriophage Qβ can support crossovers via primer extension (9, 49), while a role for sgp's in RNA recombination has been suggested for cucumoviruses (58) and carmoviruses (6, 7, 43, 44, 71). The RNA recombination of retroviruses usually occurs during reverse transcription (31). The BMV replication complex operates inside the membranous “spherules” (52), where the sgRNA3a primer could reach higher concentrations.
Another likely function of sgRNA3a is in translation. The sgRNA3a molecule served as a better template for 3a synthesis in both rabbit reticulocyte and wheat germ in vitro translation extracts (Fig. 5). This might be due to its less-stable structure that lacked the 3′ portion of wt RNA3. Neither in vitro extract is responsive to poly(A) tails on mRNAs, but in vivo the 3′-oligo(A) tail could further increase translation activity compared to that for the remaining non-3′-polyadenylated BMV RNA components. The lack of response to Mg+2 shown in Fig. 5A might be due to sufficient concentrations of endogenous Mg+2 (all commercially supplied kits contain about 2 mM Mg+2). Another possibility might be that stable folding of sgRNA3a or RNA3 did not change with Mg+2 (21, 23, 59). Additional experiments with protoplasts are needed to determine the effect of the poly(A) tail on sgRNA3a translation.
The presence of abundant amounts of sgRNA3a in polysomal fractions (Fig. 5C) provides strong evidence for its biological function. The ability to make 3a from two different RNAs (RNA3 or sgRNA3) could aid the virus in regulation of its spreading. It allows for separation of replication from translation functions, as postulated for CTV (20). That is, the sgRNAs could serve as templates during translation of RNA3-encoded proteins while the full-length RNA3 acts as a replicating (and recombining) genetic reservoir. Finally, translation from encapsidated sgRNA3a might boost up the synthesis of 3a to facilitate spreading of the virus within the infection foci.
Lower amounts of sgRNA3a compared to those of RNA3 in BMV virions followed their relative concentrations in the infected tissues (Fig. 1). The efficient encapsidation of RNA3 requires two signals (12), and only the upstream signal (between nt 624 and 811) is present in sgRNA3a. Similar to the RNA3-sgRNA4 pair (12), sgRNA3a could be packaged with RNA3 via RNA3-sgRNA3a interactions or with the aid of protein chaperones. Interestingly, the oligo(A) deletions reduced the amount of both sgRNAs in total RNA fractions (Fig. 4B) but only that of sgRNA3a among virion RNAs (Fig. 4C). This indicates that sgRNA4 is preferred in encapsidation over sgRNA3a.
Theoretically, the presence of a short oligo(A) 3′ tail (e.g., 20 A residues) may predestine sgRNA3a for the mRNA decay pathway (4, 37), and RNA decay may control the level of protein 3a. However, sgRNA3a is abundant in polysomes, suggesting some type of stabilization and compartmentalization. In general, there are examples of stable mRNAs with or without poly(A) tails (70). Along these lines, recent observations revealed that the deadenylation-dependent mRNA decapping complex requires elongation of translation of only replicating BMV RNAs (thus not of nonreplicating sgRNA4) (45). The pathway is apparently used to distinguish viral replication from nonreplication templates at the stage of translation before recruitment to replication. It would be interesting to find out if translation of sgRNA3a requires the mRNA decay pathway. Lack of such dependence might improve the economy of translation versus replication functions in BMV RNA3.
In summary, our data support the likely functions of sgRNA3a during RNA recombination and translation. More work is required to pinpoint the biochemical role(s) of sgRNA3a in the BMV life cycle.
Acknowledgments
Rafal Wierzchoslawski is on leave from the Institute of Plant Breeding and Acclimatization, Bydgoszcz, Poland. We thank Margaret Bujarska and Kathy Bujarska for excellent lab assistance.
This work was supported by grants from the National Science Foundation (MCB-0317039) and the National Institutes of Health (G1A62203), by the Plant Molecular Biology Center of Northern Illinois University, and by the Polish government through grants (6 P04C 046 19 and 3 P04 A 039 25) from the State Committee for Scientific Studies, awarded to J.J.B.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Ahlquist, P., R. Dasgupta, and P. Kaesberg. 1984. Nucleotide sequence of the Brome mosaic virus genome and its implications for viral replication. J. Mol. Biol. 172:369-383. [DOI] [PubMed] [Google Scholar]
- 2.Ayllon, M. A., S. Gowda, T. Satyanarayana, and W. O. Dawson. 2004. cis-acting elements at opposite ends of the Citrus tristeza virus genome differ in initiation and termination of subgenomic RNAs. Virology 322:41-50. [DOI] [PubMed] [Google Scholar]
- 3.Baumstark, T., and P. Ahlquist. 2001. The Brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TpsiC-stem loop and is modified in vivo. RNA 7:1652-1670. [PMC free article] [PubMed] [Google Scholar]
- 4.Beelman, C., and R. Parker. 1995. Degradation of mRNA in eukaryotes. Cell 81:179-183. [DOI] [PubMed] [Google Scholar]
- 5.Bruyere, A., M. Wantroba, S. Flasinski, A. Dzianott, and J. J. Bujarski. 2000. Frequent homologous recombination events between molecules of one RNA component in a multipartite RNA virus. J. Virol. 74:4214-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascone, P. J., C. D. Carpenter, X. H. Li, and A. E. Simon. 1990. Recombination between satellite RNAs of turnip crinkle virus. EMBO J. 9:1709-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cascone, P. J., T. F. Haydar, and A. E. Simon. 1993. Sequences and structures required for recombination between virus-associated RNAs. Science 260:801-805. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chetverin, A. B., D. S. Kopein, H. V. Chetverina, A. A. Demidenko, and V. I. Ugerov. 2005. Viral RNA-directed RNA polymerases use diverse mechanisms to promote recombination between RNA molecules. J. Biol. Chem. 280:8748-8755. [DOI] [PubMed] [Google Scholar]
- 10.Choi, I. R., M. Ostrovsky, G. Zhang, and K. A. White. 2001. Regulatory activity of distal and core RNA elements in tombusvirus subgenomic mRNA2 transcription. J. Biol. Chem. 276:41761-41768. [DOI] [PubMed] [Google Scholar]
- 11.Choi, S.-K., M. Hema, K. Gopinath, J. Santos, and C. Kao. 2004. Replicase-binding sites on plus- and minus-strand Brome mosaic virus RNAs and their roles in RNA replication in plant cells. J. Virol. 78:13420-13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, Y. G., and A. L. Rao. 2003. Packaging of Brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 77:9750-9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dasgupta, R., and P. Kaesberg. 1982. Complete nucleotide sequences of the coat protein messenger RNAs of Brome mosaic virus and cowpea chlorotic mottle virus. Nucleic Acids Res. 10:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng, L., and S. Shuman. 1997. Elongation properties of vaccinia virus RNA polymerase: pausing, slippage, 3′ end addition, and termination site choice. Biochemistry 36:15892-15899. [DOI] [PubMed] [Google Scholar]
- 15.Dzianott, A., and J. J. Bujarski. 2004. Infection and RNA recombination of Brome mosaic virus in Arabidopsis thaliana. Virology 318:482-492. [DOI] [PubMed] [Google Scholar]
- 16.Dzianott, A., N. Rauffer-Bruyere, and J. J. Bujarski. 2001. Studies on functional interaction between Brome mosaic virus replicase proteins during RNA recombination, using combined mutants in vivo and in vitro. Virology 289:137-149. [DOI] [PubMed] [Google Scholar]
- 17.Eckerle, L. D., and L. A. Ball. 2002. Replication of the RNA segments of a bipartite viral genome is coordinated by a transactivating subgenomic RNA. Virology 296:165-176. [DOI] [PubMed] [Google Scholar]
- 18.French, R., and P. Ahlquist. 1988. Characterization and engineering of sequences controlling in vivo synthesis of Brome mosaic virus subgenomic RNA. J. Virol. 62:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galiakparov, N., D. E. Goszczynski, X. Che, O. Batuman, M. Bar-Joseph, and M. Mawassi. 2003. Two classes of subgenomic RNA of grapevine virus A produced by internal controller elements. Virology 312:434-448. [DOI] [PubMed] [Google Scholar]
- 20.Gowda, S., M. A. Ayllon, T. Satyanarayana, M. Bar-Joseph, and W. O. Dawson. 2003. Transcription strategy in a Closterovirus: a novel 5′-proximal controller element of Citrus tristeza virus produces 5′- and 3′-terminal subgenomic RNAs and differs from 3′ open reading frame controller elements. J. Virol. 77:340-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray, N. K., and M. W. Hentze. 1994. Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep. 195:195-200. [DOI] [PubMed] [Google Scholar]
- 22.Grdzelishvili, V. Z., H. Garcia-Ruiz, T. Watanabe, and P. Ahlquist. 2005. Mutual interference between genomic RNA replication and subgenomic mRNA transcription in Brome mosaic virus. J. Virol. 79:1438-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo, L., E. M. Ellen, and W. A. Miller. 2001. Base-pairing between untranslated regions facilitates translation of uncapped, nonpoly(A)denylated viral RNA. Mol. Cell 7:1103-1109. [DOI] [PubMed] [Google Scholar]
- 24.Hasnoot, P. C., P. C. Olsthoorn, and J. F. Bol. 2002. The Brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA 8:110-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hausmann, S., J. P. Jacques, and D. Kolakofsky. 1996. Paramyxovirus RNA editing and the requirement for hexamer genome length. RNA 2:1033-1045. [PMC free article] [PubMed] [Google Scholar]
- 26.Howard, S. T., C. A. Ray, D. D. Patel, J. B. Antczak, and D. J Pickup. 1999. A 43-nucleotide RNA cis-acting element governs the site-specific formation of the 3′ end of a poxvirus late mRNA. Virology 255:190-204. [DOI] [PubMed] [Google Scholar]
- 27.Jackson, A. O., and B. A. Larkins. 1976. Influence of ionic strength, pH, and chelation of divalent metals on isolation of polyribosomes from tobacco leaves. Plant Physiol. 57:5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned Brome mosaic virus cDNA and effects of 5′ extensions on the transcripts infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 30.Jaspars, E. M. 1998. A core promoter hairpin is essential for subgenomic RNA synthesis in alfalfa mosaic alfamovirus and is conserved in other Bromoviridae. Virus Genes 17:233-422. [DOI] [PubMed] [Google Scholar]
- 31.Joshi, P. J., T. W. North, and V. R. Prasad. 2005. Aptamers directed to HIV-1 reverse transcriptase display greater efficacy over small hairpin RNAs targeted to viral RNA in blocking HIV-1 replication. Mol. Ther. 11:677-686. [DOI] [PubMed] [Google Scholar]
- 32.Kao, C. C. 2002. Lessons learned from the core RNA promoters of Brome mosaic virus and cucumber mosaic virus. Mol. Plant Pathol. 3:53-59. [DOI] [PubMed] [Google Scholar]
- 33.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 34.Kim, K. H., and C. L. Hemenway. 1999. Long-distance RNA-RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA 5:636-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroner, P., D. Richards, P. Traynor, and P. Ahlquist. 1989. Defined mutations in a small region of the Brome mosaic virus 2 gene cause diverse temperature-sensitive RNA replication phenotypes. J. Virol. 63:5302-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maia, I. G., K. Seron, A. L. Haenni, and F. Bernardi. 1996. Gene expression from viral RNA genomes. Plant Mol. Biol. 32:367-391. [DOI] [PubMed] [Google Scholar]
- 37.Meyer, S., C. Temme, and E. Wahle. 2004. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 39:197-216. [DOI] [PubMed] [Google Scholar]
- 38.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of Brome mosaic virus subgenomic RNA in vitro by internal initiation on (-)-sense genomic RNA. Nature 313:68-70. [DOI] [PubMed] [Google Scholar]
- 39.Miller, W. A., S. Liu, and R. Becket. 2002. Barley yellow dwarf virus: Luteoviridae or Tombusviridae? Mol. Plant Path. 3:177-183. [DOI] [PubMed] [Google Scholar]
- 40.Nagy, P. D., and J. J. Bujarski. 1992. Genetic recombination in Brome mosaic virus: effect of sequence and replication of RNA on accumulation of recombinants. J. Virol. 66:6824-6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagy, P. D., and J. J. Bujarski. 1996. Homologous RNA recombination in Brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J. Virol. 70:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy, P. D., and J. J. Bujarski. 1997. Engineering of homologous recombination hotspots with AU-rich sequences in Brome mosaic virus. J. Virol. 71:3799-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagy, P. D., and A. E. Simon. 1998. In vitro characterization of late steps of RNA recombination in turnip crinkle virus. II. The role of the priming stem and flanking sequences. Virology 249:393-405. [DOI] [PubMed] [Google Scholar]
- 44.Nagy, P. D., and A. E. Simon. 1998. In vitro characterization of late steps of RNA recombination in turnip crinkle virus. I. Role of motif1-hairpin structure. Virology 249:379-392. [DOI] [PubMed] [Google Scholar]
- 45.Noueiry, A. O., J. Diez, S. P. Falk, J. Chen, and P. Ahlquist. 2003. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for Brome mosaic virus genomic RNA translation. Mol. Cell. Biol. 23:4094-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pathak, V. K., and W. S. Hu. 1997. “Might as well jump!” template switching bt retroviral reverse transcriptase, defective genome formation, and recombination. Semin. Virol. 8:141-159. [Google Scholar]
- 48.Pellisier, T., C. Bosquet-Antonelli, L. Lavie, and J. M. Deragon. 2004. Synthesis and processing of tRNA-related SINE transcripts in Arabidopsis thaliana. Nucleic Acids Res. 32:3957-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pierangeli, A., M. Bucci, M. Forzan, P. Pagnotti, M. Equestre, and R. Perez Bercoff. 1999. ‘Primer alignment-and-extension’: a novel mechanism of viral RNA recombination responsible for the rescue of inactivated poliovirus cDNA clones. J. Gen. Virol. 80:1889-1897. [DOI] [PubMed] [Google Scholar]
- 50.Read, R. L., and C. J. Norbury. 2002. Roles for cytoplasmic polyadenylationin cell cycle regulation. J. Cell. Biochem. 87:258-265. [DOI] [PubMed] [Google Scholar]
- 51.Schmitz, I., and A. L. Rao. 1998. Deletions in the conserved amino-terminal basic arm of cucumber mosaic virus coat protein disrupt virion assembly but do not abolish infectivity and cell-to-cell movement. Virology 248:323-331. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 53.Shapka, N., and P. D. Nagy. 2004. The AU-rich RNA recombination hot spot sequence of Brome mosaic virus is functional in tombusviruses: implications for the mechanism of RNA recombination. J. Virol. 78:2288-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 55.Smirnygina, E., Y. H. Hsu, N. Chua, and P. Ahlquist. 1994. Second-site mutations in the Brome mosaic virus RNA3 intercistronic region partially suppress a defect in coat protein mRNA transcription. Virology 198:427-436. [DOI] [PubMed] [Google Scholar]
- 56.Stawicki, S. S., and C. C. Kao. 1999. Spatial perturbations within an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase (RdRP) reveal that RdRp can adjust its promoter binding sites. J. Virol. 73:198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan, M. L., and P. Ahlquist. 1999. A Brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki, M., T. Hibi, and C. Masuta. 2003. RNA recombination between cucumoviruses: possible role of predicted stem-loop structures and an internal subgenomic promoter-like motif. Virology 306:77-86. [DOI] [PubMed] [Google Scholar]
- 59.Tanguay, R. L., and D. R. Gallie. 1996. The effect of the length of the 3′-untranslated region on expression in plants. FEBS Lett. 394:285-288. [DOI] [PubMed] [Google Scholar]
- 60.Urbanowicz, A., M. Alejska, P. Formanowicz, J. Blazewicz, M. Figlerowicz, and J. J. Bujarski. 2005. Homologous crossovers among molecules of Brome mosaic bromovirus RNA1 or RNA2 segments in vivo. J. Virol. 79:5732-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van den Born, E., C. C. Posthuma, A. P. Gultyaev, and E. J. Nijder. 2005. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 79:6312-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Marle, G., L. C. van Dinten, W. J. Spaan, W. Luytjes, and E. J. Snijder. 1999. Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J. Virol. 73:5274-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Vliet, A. L. W., S. L. Smits, P. J. M. Rottier, and R. J. de Groot. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 21:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vives, M. C., L. Galipienso, L. Navarro, P. Moreno, and J. Guerri. 2002. Characterization of two kinds of subgenomic RNAs produced by citrus leaf blotch virus. Virology 295:328-336. [DOI] [PubMed] [Google Scholar]
- 65.von Hippel, P. H. 1998. An integrated model of the transcription complex in elongation, termination, and editing. Science 281:660-665. [DOI] [PubMed] [Google Scholar]
- 66.Wielgosz, M. M., and H. V. Huang. 1997. A novel viral RNA species in Sindbis virus-infected cells. J. Virol. 71:9108-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wierzchoslawski, R., and J. J. Bujarski. 2006. An efficient in vitro system of homologous recombination in Brome mosaic bromovirus. J. Virol. 80:6182-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wierzchoslawski, R., A. Dzianott, and J. J. Bujarski. 2004. Dissecting the requirement for subgenomic promoter sequences by RNA recombination of Brome mosaic virus in vivo: evidence for functional separation of transcription and recombination. J. Virol. 78:8552-8564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wierzchoslawski, R., A. Dzianott, S. Kunimalayan, and J. J. Bujarski. 2003. A transcriptionally active subgenomic promoter supports homologous crossovers in a plus-strand RNA virus. J. Virol. 77:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zanier, K., I. Luyten, C. Crombie, B. Müller, D. Schümperli, J. P. Linge, M. Nilges, and M. Sattlercell. 2002. Cycle regulation of human histone H1 mRNA. RNA 8:29-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, C. X., P. J. Cascone, and A. E. Simon. 1991. Recombination between satellite and genomic RNAs of turnip crinkle virus. Virology 184:791-794. [DOI] [PubMed] [Google Scholar]
- 72.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 287:57-94. [DOI] [PMC free article] [PubMed] [Google Scholar]