Abstract
The UL28 protein of herpes simplex virus type 1 (HSV-1) is one of seven viral proteins required for the cleavage and packaging of viral DNA. Previous results indicated that UL28 interacts with UL15 and UL33 to form a protein complex (terminase) that is presumed to cleave concatemeric DNA into genome lengths. In order to define the functional domains of UL28 that are important for DNA cleavage/packaging, we constructed a series of HSV-1 mutants with linker insertion and nonsense mutations in UL28. Insertions that blocked DNA cleavage and packaging were found to be located in two regions of UL28: the first between amino acids 200 to 400 and the second between amino acids 600 to 740. Insertions located in the N terminus or in a region located between amino acids 400 and 600 did not affect virus replication. Insertions in the carboxyl terminus of the UL28 protein were found to interfere with the interaction of UL28 with UL33. In contrast, all of the UL28 insertion mutants were found to interact with UL15 but the interaction was reduced with mutants that failed to react with UL33. Together, these observations were consistent with previous conclusions that UL15 and UL33 interact directly with UL28 but interact only indirectly with each other. Revertant viruses that formed plaques on Vero cells were detected for one of the lethal UL28 insertion mutants. DNA sequence analysis, in combination with genetic complementation assays, demonstrated that a second-site mutation in the UL15 gene restored the ability of the revertant to cleave and package viral DNA. The isolation of an intergenic suppressor mutant provides direct genetic evidence of an association between the UL28 and UL15 proteins and demonstrates that this association is essential for DNA cleavage and packaging.
There are seven viral genes expressed late in infection that are essential for the cleavage and packaging of the herpes simplex virus type 1 (HSV-1) genome. These are UL6, UL15, UL17, UL25, UL28, UL32, and UL33 (3, 4, 20, 21, 23, 26, 32, 35). By analogy with the mechanism of packaging in double-stranded DNA bacteriophages, the function for several of these proteins has been proposed (9, 12, 15). The UL15 protein is the most highly conserved protein in the herpesvirus family, and since it shows amino sequence similarity to bacteriophage terminases, it is likely to function as the catalytic subunit of the DNA terminase complex that cleaves replicated concatemeric viral DNA into unit length molecules. In bacteriophages, the terminase consists of at least two subunits (7). In HSV-1, the terminase is likely to be composed of a complex of three proteins consisting of UL15, UL28, and UL33 (1, 3, 6, 17, 18, 27, 37, 39, 40). The DNA enters the capsid through the portal vertex, which consists of 12 copies of the UL6 protein (25). In bacteriophages, the terminase complex binds to the portal vertex (7). The HSV-1 UL6 protein has been shown to interact with UL15 and UL28 (37). The amount of UL6 in the capsid is found to be constant in all capsid forms, while UL15 and UL28 only transiently associate with capsids and are not present in DNA-containing capsids (40). The loss of UL15 and UL28 following DNA packaging is similar to what is found with the double-stranded DNA bacteriophages, where the terminase complex also disassociates from the capsids after the DNA is packaged.
The function of UL32 in the cleavage/packaging reaction is unclear, although UL32 has been shown to be a zinc binding cytoplasmic protein (11, 20). The UL17 and UL25 proteins are found associated with capsids but are not required for capsid assembly (23, 26, 33, 36). Both proteins appear to be important in stabilizing capsids once DNA is packaged (23, 34, 36). A similar phenomenon has been reported in mutants defective in the gene products gp4, gp10, and gp26 of phage P22 (7, 28). The role of these proteins is thought to be “head completion” stabilizing of the DNA inside the capsid.
The importance of the UL28 protein in HSV-1 DNA packaging was initially shown in studies with UL28 temperature-sensitive or -null mutants (2, 10, 24, 35). Mutants with lesions in the UL28 gene are defective in both cleavage of concatemeric DNA and DNA packaging. Studies with UL28 expressed in mammalian cells from plasmid vectors or in insect cells using recombinant baculoviruses demonstrated that UL28 interacts with UL15, UL33, and UL6 (1, 6, 17, 18, 37). The HSV-1 UL28 and cytomegalovirus (CMV) UL56 (homolog of HSV UL28) proteins have been found to bind DNA sequences that are required for the cleavage of concatemeric viral DNA, and an endonuclease activity was found to be associated with the UL56 protein that specifically targets CMV cleavage/packaging sequences (8). We have previously reported that, in infected cell lysates, UL28 coimmunoprecipitates with either UL33 or UL15, whereas the coimmunoprecipitation of UL15 and UL33 was observed only when UL28 was present (38). In addition, the UL28 protein was found to protect the UL33 protein from proteosomal degradation in HSV-1-infected cells (38). Taken together, these results suggest that UL28 interacts directly with UL15 and UL33 and support a role for UL28 as part of a three-protein complex (terminase complex) involved in the cleavage of concatemeric DNA into unit length molecules that are then packaged into preformed capsids.
In the present study, we constructed a series of HSV-1 mutants which contained linker insertion and nonsense mutations scattered throughout the UL28 open reading frame and assayed the effect of these mutations on the ability of the mutants to produce infectious virus and to cleave and package viral DNA. The UL28 mutants were utilized to map regions of UL28 that are important for interaction with UL15 and UL33. During these studies, it was observed that one of the lethal UL28 linker insertion mutants rapidly generated spontaneous revertants. A detailed analysis of the revertant revealed that a second-site compensatory mutation in the UL15 gene restored the ability of the virus to cleave and package viral DNA. The isolation of this intergenic suppressor mutant provides genetic evidence of an association between UL15 and UL28 and that this association is essential for forming a functional DNA cleavage/packaging complex.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney cells (Vero; American Type Culture Collection, Manassas, VA) and UL28-transformed C1 and CV28 cells were propagated as previously described (35, 38). HSV-1 wild-type KOS and F and UL28-null virus GCB have been previously described (35). CV33, which expresses the UL33 gene, and CV28 cells were made as previously described using the flp-In-CV-1 system (38). CV1 cells are derivatives of Vero cells and contain no HSV-1 genes (38).
Plasmids.
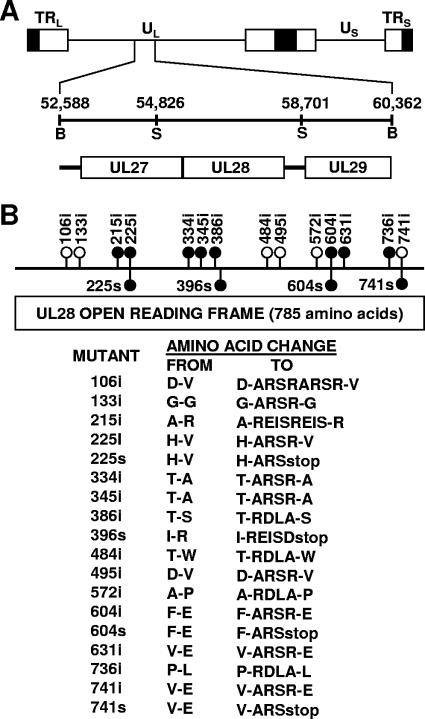
Plasmid pSG18B-SalI D was previously described (14) and contains a SalI insert that spans nucleotides 54826 to 58701 of the HSV-1 genome (22). The 785-amino-acid open reading frame for the UL28 gene extends from nucleotide 58159 to 55802. For the construction of linker insertion mutations in the UL28 open reading frame, pSG18B-SalI D was partially cleaved with one of several restriction endonucleases (AciI, BsaHI, MaeII, or TacI), all of which yield 5′ CG overhangs. The resulting linear molecules were purified by agarose gel electrophoresis and then ligated to a 12-bp linker (5′-CGCGAGATCTCG-3′) containing a novel BglII site. Annealing of the oligonucleotide results in a double-stranded molecule with 5′ CG overhangs. This construct was then digested with BglII to remove excess linkers, and linear molecules were again purified and then ligated to recircularize the plasmid and used to transform bacteria to ampicillin resistance. Clones were screened for the presence of the BglII restriction site within the UL28 open reading frame. Insertion mutations that mapped to within the UL28 coding region were sequenced to determine the exact position of the insertion (Fig. 1). Fourteen linker insertion mutations were obtained (Fig. 1). The net size of each insertion was either four or eight (due to the insertion of two linkers) amino acids. Four nonsense linker insertion mutants were made after the initial mutants were constructed and sequenced. Linker insertion mutations with BglII inserts after amino acid 225, 396, 604, or 741 were digested with BglII, and the ends were filled in using T4 DNA polymerase. The resulting linear molecules were purified by agarose gel electrophoresis and then blunt end ligated to a 14-bp double-stranded DNA molecule (5′-CTAGACTAGTCTAG-3′) containing a SpeI site and stop codons in all reading frames. The sample was then digested with SpeI, gel purified, and ligated to recircularize the plasmid, and the presence of the stop mutation was confirmed by DNA sequencing.
FIG. 1.
(A) Physical map of the HSV-1 genome showing the location of the UL27, UL28, and UL29 genes. UL and US refer to the long and short unique region sequences. On the next line, the BamHI region located between nucleotides 52588 and 60362 of the HSV-1 genome are expanded. The locations of the UL27, UL28, and UL29 genes are indicated, with the boxed regions representing the open reading frames for these genes. (B) Insertion mutations within the HSV-1 UL28 gene. Numbers above the line indicate the location (amino acid number of UL28 open reading frame) where 12 or 24 bp was added to the UL28 gene, resulting in the addition of four or eight amino acids. Numbers below the line indicate the location where a nonsense linker was added to the UL28 gene. Filled circles indicate mutants which grow on only UL28-complementing cells; white circles indicate mutants which grow on noncomplementing Vero cells. At the bottom, the locations of the insertions and the predicted amino acid changes are shown. The number denotes the last unchanged amino acid before the insertions.
Construction of UL28 mutants by marker transfer.
Marker transfer experiments were carried out as previously described (16). C1 cells were transfected with one of the linker insertion or nonsense plasmid constructs and infectious GCB DNA, which contains a deletion that removes 1,881 bp (nucleotides 57923 to 56042) of the UL28 open reading frame (35). The resulting virus stocks were then screened for the ability to hybridize with a restriction fragment (537-bp XhoI fragment; nucleotides 56042 to 56579) specific for the region deleted in the GCB virus by an in situ hybridization procedure (16). Hybridizing plaques were purified three times in C1 cells.
Southern blot assays.
A 60-mm plate of Vero cells was infected with virus at a multiplicity of infection (MOI) of 5 PFU per cell. At 18 h postinfection, the medium was removed and the cells were washed in 1× phosphate-buffered saline (PBS), scraped off the plate, and pelleted. The cells were lysed, and viral DNA was prepared as previously described (16). The final DNA was digested with either SalI plus BglII or SalI plus SpeI to confirm the mutations or BamHI to look for the cleavage of viral DNA. The DNA was electrophoresed, transferred to Gene Screen Plus, and hybridized as previously described (16, 35).
Marker rescue.
Viral DNA was prepared from KOS, GCB, 334i, and 334iR1. KOS, 334i, and 334iR1 DNA was digested with EcoRI, and the EcoRI F fragment, which contains the UL28 gene, was cloned into pUC18 at the EcoRI site (13). The 3.88-kb SalI fragment that contains the UL28 gene (Fig. 1) was isolated from the EcoRI clones of KOS, 334i, and 334iR1 and cloned into the SalI site of pUC18. Marker rescue was performed by transfecting C1 cells with infectious GCB viral DNA along with SalI-digested plasmid DNA (UL28 SalI clones from KOS, 334i, or 334iR1). When the maximum cytopathic effect was observed (3 to 4 days posttransfection), the cultures were frozen, and then the titers were determined on Vero and C1 cells.
Immunoprecipitations.
Confluent CV-1 cells grown in a 10-cm dish (8.8 × 106 cells) were infected with wild-type HSV-1(F) or UL28 mutant viruses at an MOI of 5 for 2 h at 37°C. After virus adsorption, the overlying medium was replaced with fresh growth medium and the cells were allowed to incubate at 37°C for 16 additional hours. The cells were removed by scraping into 10 ml of cold PBS and pelleted at 3,200 × g in an Eppendorf tabletop centrifuge for 5 min at 4°C. The cell pellets were resuspended into 800 μl of lysis buffer [50 mM Tris, pH 7.4, 1 mM EDTA, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 2 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 1 mM benzamidine, 5 μg/ml leupeptin, 10 μg/ml pepstatin, 20 mM NaF, 0.1 mM Na3VO4] and incubated 30 min on ice. The whole lysates were centrifuged at 16,000 × g for 15 min in a microcentrifuge. The supernatants were precleared by adding preimmune serum and 30 μl of a 50% slurry of GammaBind G-Sepharose beads (Amersham Pharmacia Biotech) for 2 h at 4°C with rotation. The resulting supernatants were then incubated with the relevant antisera for 2 h at 4°C with rotation before adding 30 μl of 50% slurry of GammaBind G-Sepharose beads and incubating overnight at 4°C with rotation. The beads were then washed four times with lysis buffer, and immune complexes were eluted in sodium dodecyl sulfate (SDS)-containing buffer and boiled for 10 min.
Immunoblotting.
Protein extracts or immunoprecipitates were electrophoretically separated on a 12% SDS-polyacrylamide gel, and proteins were transferred electrically to nitrocellulose. The nitrocellulose was washed twice in PBS and blocked overnight in PBS supplemented with 10% nonfat dry milk (Carnation). Primary anti-UL15 or anti-UL28 rabbit polyclonal antisera were diluted at 1:1,000 in PBS supplemented with 2% bovine serum albumin (BSA), whereas anti-UL33 rabbit polyclonal antisera were diluted 1:400 as previously described (6, 30, 31, 35). The diluted antisera were reacted with the blocked nitrocellulose for 2 h at room temperature and washed, and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (Bio-Rad Laboratories) at a 1:5,000 dilution in PBS plus 2% BSA was added at room temperature. The bound immunoglobulins were revealed by enhanced chemiluminescence (Amersham Pharmacia Biotech).
UL33 stability assay.
CV33 cells (2 × 106 cells/100-mm dish) were transfected with 12 μg of plasmid DNA and 30 μl of Lipofectamine 2000 (Invitrogen) diluted in 1 ml of serum-free medium. The DNA-lipid complexes were allowed to form at room temperature for 20 min and then added to the cells and incubated at 37°C for 24 h. The next day the cells were collected and washed once with PBS, and the cell pellets were resuspended in 400 μl of loading buffer (62.5 mM Tris, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 12.5% glycerol), sonicated, and boiled for 10 min. The lysates were clarified at 14,000 rpm for 15 min at 4°C. The proteins were separated on a 12% SDS-polyacrylamide gel electrophoresis (PAGE) gel and then transferred electrically to nitrocellulose. Anti-UL33 rabbit polyclonal antiserum was diluted 1:400 as previously described, whereas anti-actin antibody (Santa Cruz Biotechnology) was diluted 1:200 according to the manufacturer's protocol. The diluted antisera were reacted with the blocked nitrocellulose for 2 h at room temperature and washed, and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G diluted 1:5,000 in PBS plus 2% BSA was added for 2 h at room temperature. The bound immunoglobulins were revealed by enhanced chemiluminescence.
Complementation assay.
Vero cells (1.0 × 105 cells/well in a 24-well plate) were transfected with 0.5 μg of plasmid DNA and 1 μl of Lipofectamine 2000 (Invitrogen) diluted in 100 μl of serum-free medium. The DNA-lipid complexes were allowed to form at room temperature for 20 min and incubated with the cells overnight. The next day the cells were infected with GCB at an MOI of 5 PFU/cell. After 90 min at 37°C, unabsorbed virus was inactivated by washing the cells three times with citrate buffer, pH 3. The cultures were then incubated for an additional 18 h and then frozen. The virus titers were then determined on CV28 cells and Vero cells.
RESULTS
Isolation of UL28 mutants.
In order to define the functional domains of UL28, we isolated HSV-1 mutants with linker insertion and nonsense mutations throughout the UL28 open reading frame. The insertions were introduced by partially digesting plasmid pSG18B-SalI D with one of four restriction enzymes (AciI, BsaI, MaeII, or TacI). The resulting linear plasmids were then gel purified and ligated to a 12-base oligonucleotide containing a BglII restriction site (5′-CGCGAGATCTCG-3′). The samples were then digested with BglII, and the linear plasmid was again gel purified, ligated, and used to transform bacteria. Clones were screened for the presence of a single BglII restriction site, and the position of each insertion was determined by restriction mapping. The exact location of the linker within the UL28 open reading frame was determined by DNA sequence analysis. Fourteen linker insertion mutations were obtained (Fig. 1). Twelve of the mutants contained a single and two contained a double linker insertion, resulting in UL28 proteins with either four- or eight-amino-acid insertions. The insertions were found to be distributed throughout the UL28 open reading frame. Mutants expressing C-terminally truncated forms of the UL28 protein were made by digesting the plasmids containing the 225i, 396i, 604i, and 741i linker insertions with BglII. The linear plasmid was then blunt end ligated to a linker containing a SpeI restriction site and stop codons in all three reading frames (5′-CTAGACTAGTCTAG-3′). The presence of the linker was confirmed by restriction mapping followed by DNA sequence analysis (Fig. 1).
Mutations were transferred from plasmids into the viral genome through homologous recombination in vivo. Briefly, linearized plasmid DNA containing the mutation of interest was cotransfected with infectious GCB DNA into UL28-complementing C1 cells. GCB is an HSV-1 mutant that contains a deletion that removes nearly all of the UL28 coding sequences (35). The progeny from the transfection were then plated on C1 cells and screened by a previously described in situ viral plaque hybridization procedure for the ability to hybridize with a restriction fragment specific for the region deleted from the GCB virus (16). Positively hybridizing plaques were chosen, plaque purified three times, and propagated in C1 cells.
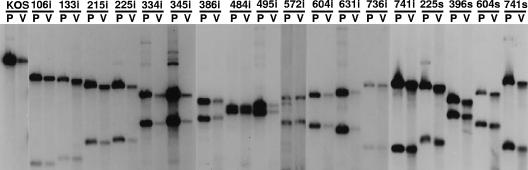
To confirm that the desired mutation had been transferred to each virus, we isolated viral DNA from Vero cells infected with each of the mutants. The DNA was digested either with SalI and BglII (linker insertion mutants) or with SalI and SpeI (nonsense mutants) and subjected to gel electrophoresis and Southern blot analysis (Fig. 2). Plasmids used to isolate each mutant were also digested with the appropriate enzymes and subjected to electrophoresis in parallel with the viral DNA. The filter was probed with the 3,800 bp UL28 SalI fragment (nucleotides 54826 to 58701) (Fig. 1). The digests for each of the mutants show two hybridizing bands in contrast to wild-type HSV DNA that has only one band. The blots demonstrate that the linker insertion mutants contain a BglII site within the UL28 gene that is not present in the KOS DNA. Similarly, the nonsense mutants contain a unique SpeI site within the UL28 gene. In addition, the size of the hybridizing fragments detected for each of the mutants is identical in size to the fragments generated from the plasmids used to construct each mutant. We concluded from these Southern blots that the desired UL28 mutants had been generated.
FIG. 2.
Southern blot analysis of DNAs from UL28 mutants confirming the locations of mutations. Total infected cell DNA was isolated from Vero cells infected with indicated viruses (V) or plasmid DNA (P). Total cell DNA and plasmid DNA were digested with either SalI plus BglII for insertion mutants or SalI plus SpeI for stop mutants. The blot was hybridized with a UL28-specific probe (SalI-D fragment, nucleotides 54826 to 58701).
Characterization of the UL28 mutants.
Virus stocks of the UL28 mutants grown on C1 cells were examined for their abilities to form plaques on UL28-complementing C1 cells and noncomplementing Vero cells (Table 1). KOS and six of the UL28 linker insertion mutants (106i, 133i, 484i, 495i, 572i, and 741i) showed comparable growth on Vero and C1 cells. A GCB rescued virus, GCB101, also formed plaques equally well on Vero and C1 cells. Most of the remaining eight linker insertion mutants (215i, 225i, 345i, 386i, 604i, 631i, and 736i) and all four of the stop mutants (225s, 396s, 604s, and 741s) were unable to form plaques on Vero cells, indicating that they harbored a functionally deficient UL28 protein. Interestingly, one of the linker insertion mutants (334i) showed low numbers of virus plaques on Vero cells, a point we will discuss later.
TABLE 1.
Results of virus plating assay
| Virus | Virus growth (PFU/ml) in:
|
DNA cleavage in Vero cellsa | |
|---|---|---|---|
| C1 cells | Vero cells | ||
| 106i | 4.0 × 106 | 4.0 × 106 | + |
| 133i | 2.2 × 107 | 1.8 × 107 | + |
| 215i | 1.3 × 107 | <1 × 102 | − |
| 225i | 3.6 × 106 | <1 × 102 | − |
| 334i | 2.4 × 106 | 1.8 × 104 | − |
| 345i | 1.6 × 106 | <1 × 102 | − |
| 386i | 1.5 × 106 | <1 × 102 | − |
| 484i | 1.3 × 107 | 1.8 × 107 | + |
| 495i | 2.5 × 107 | 2.8 × 107 | + |
| 572i | 1.2 × 107 | 1.4 × 106 | + |
| 604i | 1.2 × 106 | <1 × 102 | − |
| 631i | 1.7 × 106 | <1 × 102 | − |
| 736i | 2.4 × 106 | <1 × 102 | − |
| 741i | 2.0 × 107 | 1.8 × 107 | + |
| 225s | 3.0 × 106 | <1 × 102 | − |
| 396s | 1.8 × 106 | <1 × 102 | − |
| 604s | 1.1 × 106 | <1 × 102 | − |
| 741s | 3.5 × 106 | <1 × 102 | − |
| GCB | 5.0 × 106 | <1 × 102 | − |
| GCB101 | 2.3 × 106 | 3.0 × 106 | + |
| KOS | 2.5 × 108 | 2.5 × 108 | + |
The presence (+) or absence (−) of chromosomal termini was determined as described for Fig. 4.
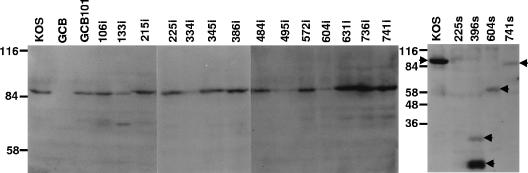
To determine that each mutant virus expressed a UL28 protein of the expected size, immunoblot analysis of Vero cells infected with each mutant virus as well as KOS, GCB, and GCB101 was performed as described in Materials and Methods. Figure 3 shows that the 14 UL28 BglII linker insertion viruses expressed a UL28 protein that was of a size similar to that of the UL28 protein expressed from wild-type KOS virus. A size difference would not be apparent since only four or eight amino acids were added to these mutant UL28 proteins. The apparent molecular mass is in agreement with the predicted molecular mass of 87 kDa.
FIG. 3.
Expression of UL28 protein from wild-type HSV-1 and UL28 mutant viruses. Vero cells were infected with the indicated virus at an MOI of 5 PFU per cell. At 18 h postinfection, cell lysates were prepared, separated by SDS-PAGE, and immunoblotted. The expression of the UL28 protein was detected using a polyclonal antisera raised against an Escherichia coli-expressed Cro-UL28 fusion protein (35). The left panel shows the insertion mutants. The right panel shows the stop mutants, and the locations of the UL28-specific polypeptides expressed from these mutants are indicated by arrowheads.
The predicted sizes of the truncated UL28 proteins produced by the nonsense mutants 741s, 604s, 396s, and 225s are 81, 66, 44, and 25 kDa, respectively. The mutants 741s and 604s both expressed a UL28 product of the expected size. Two proteins, with apparent molecular masses of 33 and 17 kDa, were observed in the 396s lane, possibly the result of breakdown. No immunoreactive proteins were observed for 225s, indicating that this virus produced a peptide that was either unstable or not recognized by the UL28 polyclonal antisera (Fig. 3). The level of the UL28 expressed differs for several of the mutants. Since we did not compare the expression with that of another viral protein, we do not know if this variation is due to a difference in the expression of the mutant UL28 proteins or if it is due to how well these mutants infect.
Replication and cleavage of HSV-1 DNA.
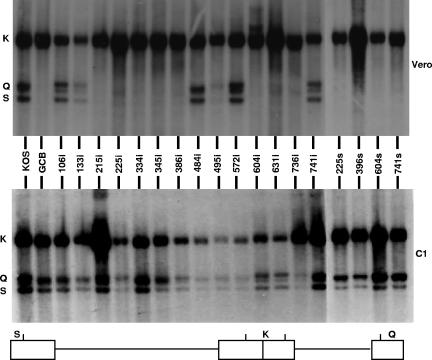
Previously, it has been shown that the UL28-null virus GCB was able to synthesize viral DNA but was not able to carry out cleavage and packaging (35). To determine whether viruses bearing linker insertions or nonsense mutation were also impaired in DNA cleavage, total DNA was isolated from Vero and C1 cells infected with wild-type and mutant viruses and subjected to Southern blot analysis. Viral DNA replication generates concatemers that are cleaved into unit length molecules and packaged into virions. The encapsidated viral DNA contains free chromosomal termini, whereas the nonpackaged DNA does not. The presence of chromosomal ends can easily be monitored by Southern blot analysis of total infected cell DNA digested with BamHI and probed with the HSV-1 BamHI K fragment. Only cleaved viral DNA with free chromosomal ends will give rise to the terminal BamHI Q and S fragments, while concatemeric DNA gives rise to only the joint-spanning BamHI K fragment (Fig. 4). All of the UL28 mutants were found to produce viral DNA when grown on either Vero or C1 cells, as assessed by the presence of the joint K fragments.
FIG. 4.
Processing of viral DNA. Vero cells or C1 cells were infected with the indicated virus at an MOI of 5 PFU per cell. At 18 h postinfection, total infected cell DNA was isolated, digested with BamHI, and subjected to Southern blot analysis using the BamHI K fragment (32P labeled) as a probe. Top panel, autoradiograph of DNA isolated from infected Vero cells. Bottom panel, autoradiograph of DNA isolated from infected C1 cells. The locations of the BamHI K joint-spanning fragment and the two end fragments, BamHI-Q and -S, in the HSV-1 genome are shown at the bottom.
On the other hand, the Q and S fragments were present in only DNA isolated from Vero cells infected with the wild type and UL28 mutants 106i, 133i, 484i, 495i, 572i, and 741i. In contrast, the terminal Q and S fragments were absent from DNA isolated from Vero cells infected with the UL28 mutants 215i, 225i, 334i, 345i, 386i, 604i, 631i, 736i, 225s, 396s, 604s, and 741s. When these viruses were grown in UL28-complementing C1 cells, the terminal Q and S fragments were detected, indicating that the UL28 proteins encoded by these viruses failed to support cleavage. Thus, the disruption of one of several regions throughout the UL28 protein is sufficient to prevent cleavage of the viral genome, which is necessary for the replication of the virus.
Interaction of UL28 with UL15 and UL33.
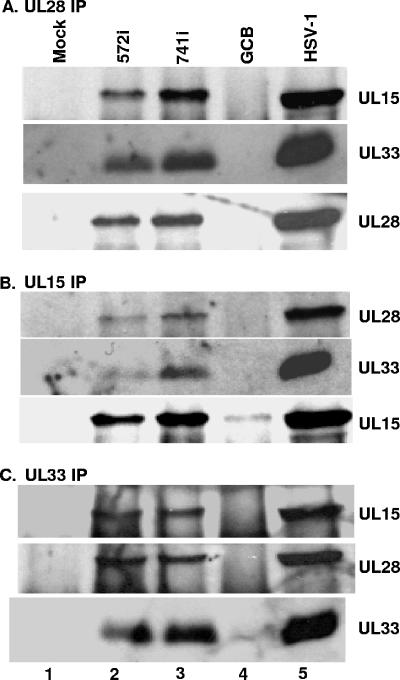
The UL15, UL28, and UL33 proteins have been shown to form a complex in HSV-1-infected cells (6). Results of a recent study (38) indicated that the UL15/UL28/UL33 complex is formed by the interaction of UL33 with UL28 and by the interaction of UL15 with UL28 and that there is no direct interaction between UL15 and UL33. Assuming that these interactions are essential, it would be expected that they could be detected in both wild-type-infected cells and cells infected with the UL28 insertion mutants that grow on Vero cells. To investigate whether this was the case, CV-1 cells were mock infected or infected with wild-type HSV-1 or with the UL28 mutant GCB, 572i, or 741i. At 18 h postinfection, the cells were lysed and the UL28, UL15, and UL33 proteins were separately immunoprecipitated from the lysates with the appropriate antisera. The immunoprecipitates were subjected to SDS-PAGE and transferred to a nitrocellulose membrane, and proteins were identified by immunoblotting with anti-UL15, anti-UL33, or anti-UL28 antisera. The UL28, UL15, and UL33 antisera caused the coimmunoprecipitation of all three proteins from lysates isolated from wild-type-infected cells and from lysates isolated from cells infected with the two UL28 insertion mutants, 572i and 741i (Fig. 5A to C, lanes 2, 3, and 5). As expected, UL28 was not immunoprecipitated from lysates of mock- or UL28-null (GCB) virus-infected cells (Fig. 5A, lanes 1 and 4). The UL15 and UL33 proteins were both immunoprecipitated from the UL28-null lysates (Fig. 5B and C, lanes 4). In contrast, the coimmunoprecipitation of UL15 and UL33 was not observed with either UL15 or UL33 antibodies from lysates of cells infected with the UL28-null virus (Fig. 5B and C, lanes 4). These results demonstrate that UL28 is necessary for UL33 and UL15 to coimmunoprecipitate and confirm previous observations that UL15 and UL33 interact with UL28 but not with each other (38).
FIG. 5.
Reciprocal immunoprecipitations of UL28 mutant viruses bearing nonlethal mutations. CV-1 cells were mock infected (lane 1) or infected with the indicated virus above each lane (lanes 2 to 5). Reciprocal immunoprecipitations were performed using anti-UL28 antisera (A), anti-UL15 antisera (B), or anti-UL33 antisera (C), followed by immunoblotting with anti-UL15, -UL28, or -UL33 antisera as indicated on the right. Signals on immunoblots were revealed by chemiluminescence.
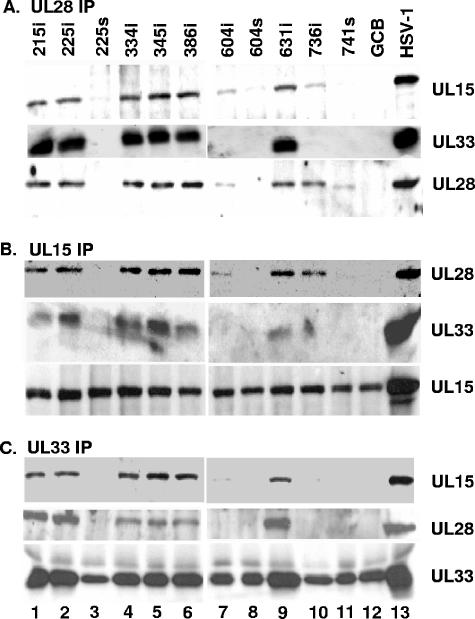
To test the possibility that some of the mutations in UL28 precluded viral replication because they interfered with UL28 interactions with UL15 or UL33, cells were infected with the viruses bearing lethal mutations in the UL28 gene and cell lysates were reacted with the anti-UL28 antibody. The immunoprecipitates were subjected to SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane, and proteins were identified by immunoblotting with anti-UL28, anti-UL15, or anti-UL33 antisera. Figure 6A shows that the UL28 protein was immunoprecipitated from the lysates of cells infected with UL28 mutant viruses 215i, 225i, 334i, 345i, 386i, 604i, 631i, 736i, and 741s and wild-type HSV-1. On the other hand, UL28 was not immunoprecipitated from lysates of mock- or 225s, 604s, and GCB virus-infected cells. Based on the Western blotting performed earlier (Fig. 3), we expected that a truncated 66-kDa UL28 protein would be immunoprecipitated from the 604s infection (Fig. 6, lane 8) but no UL28-specific bands were found following Western blot analysis. One possible explanation is that the truncated UL28 protein was not efficiently immunoprecipitated so there was little or no protein present on the immunoblot. The UL28 proteins produced by UL28 mutants 604i and 741s were immunoprecipitated less efficiently than the other UL28 mutants, suggesting that the truncation at amino acid 741 and insertion at amino acid 604 may have altered the antibody epitopes or rendered the protein less stable. As would be expected, the UL28 protein made by the 741s mutant is slightly smaller due to a C-terminal truncation. In nearly all of the immunoprecipitates that contained a UL28 protein, both UL33 and UL15 were coimmunoprecipitated with UL28. The exception were mutants 604i and 736i, in which some UL15 but no UL33 was coimmunoprecipitated by the UL28-specific antisera, and 741s (the only C-terminally truncated mutant that makes a detectable UL28 protein), where neither UL15 nor UL33 was coimmunoprecipitated (Fig. 6A, lanes 7, 10, and 11).
FIG. 6.
Reciprocal immunoprecipitation of UL28 mutant viruses bearing lethal mutations. CV-1 cells were infected with the virus indicated above each lane. Reciprocal immunoprecipitations were performed by anti-UL28 antisera (A), anti-UL15 antisera (B), or anti-UL33 antisera (C), followed by immunoblotting with anti-UL15, -UL28, or -UL33 antisera as indicated on the right. Signals on immunoblots were revealed by chemiluminescence.
The UL15 protein was immunoprecipitated with its cognate antibody in roughly equal amounts from all infected cell lysates tested (Fig. 6B). The UL15 antisera caused the coimmunoprecipitation of UL28 from lysates of cells infected with all of the lethal UL28 insertion mutants (215i, 225i, 334i, 345i, 386i, 604i, 631i, and 736i) but not from lysates of the three stop mutants (225s, 604s, and 741s) and GCB. UL33 was readily detectable in UL15 immunoprecipitates from lysates of cells infected with viruses 215i, 225i, 334i, 345i, 386i, and 631i and wild-type HSV-1 (Fig. 6B). In contrast, the UL33 protein was not coimmunoprecipitated with UL15 in lysates of cells infected with the linker insertion mutant 604i or 736i and the three stop mutants (225s, 604s, and 741s). These data suggest that the region of UL28 around amino acids 604 to 736 is essential for interaction with UL33 and that the extreme C terminus (amino acids 736 to 785) of UL28 is important for the interaction of UL28 with UL15.
The lysates of cells infected with wild-type and mutant viruses were also reacted with the UL33-specific antisera, and coimmunoprecipitated proteins were identified by immunoblotting with UL28-, UL15-, and UL33-specific antisera (Fig. 6C). The UL33 protein was immunoprecipitated with its cognate antibody from all of the mutants, but the amount of UL33 detected was smaller for mutants that either did not express a detectable UL28 protein (225s, 604s, and GCB) or expressed a UL28 protein (604i, 736i, and 741s) that did not react with UL33 (see above UL28 immunoprecipitations). This is consistent with previous studies which demonstrated that UL28 protects UL33 from proteosome-dependent degradation (38). Otherwise, the results with the UL33 antisera were similar to those obtained upon immunoprecipitation with the UL15 antibody. Specifically, the reaction with the UL33-specific antibody coimmunoprecipitated UL15 and UL28 from lysates of cells infected with 215i, 225i, 334i, 345i, 386i, and 631i and wild-type HSV-1, whereas these proteins were not coimmunoprecipitated from lysates of cells infected with 604i or 736i. Taken together, these data indicated that the C terminus of UL28 is required for the interaction with the UL15 and UL33 proteins. Moreover, different regions of UL28 are required for these interactions since insertions at amino acids 604 and 736 of the UL28 protein alter its interaction with UL33 but not UL15, while the removal of the C terminus of UL28 (amino acids 741 to 785) blocks its interaction with both proteins.
UL28/UL33 interaction protects UL33 from degradation.
As mentioned above, previous studies have demonstrated that the UL33 protein is rapidly degraded through a proteosomal pathway when expressed from a transformed cell line or a plasmid expression vector (38). These studies went on to demonstrate that the interaction of UL33 with the UL28 protein protected UL33 from proteosomal degradation. This hypothesis predicts that the UL28 insertion mutants that were shown to interact with UL33 in the immunoprecipitation assays would protect UL33 from degradation, while the insertion mutants that did not interact with UL33 would not. To test this possibility, a cell line (CV33) expressing the UL33 protein was transfected with expression plasmids containing either the wild-type UL28 protein or one of the UL28 mutants. The cells were harvested 24 h posttransfection, and the presence of UL33 and actin (gel loading control) in the lysates was determined by Western blot analysis. As shown in Fig. 7, the expression of either the wild-type UL28 protein or the UL28 insertion mutants 215i, 225i, 334i, 345i, 386i, and 631i all yielded higher levels of UL33 protein in CV33 cells compared to those of mock-transfected CV33 cells. In contrast, the expression of the UL28 mutants 225s, 604i, 604s, and 736i did not increase the amount of UL33 protein compared to that of mock-transfected CV33 cells. These data demonstrate that the UL28 mutants (215i, 225i, 334i, 345i, 386i, and 631i) that were found to interact with UL33 in the immunoprecipitation assays protected UL33 from degradation, while mutants (225s, 604i, 604s, and 736i) that failed to coimmunoprecipiate did not protect UL33 from degradation. The results confirm our earlier findings that UL28 is necessary for protecting UL33 from degradation and that the protection is a direct consequence of the interaction of these two proteins.
FIG. 7.
UL33 detected by immunoblot analysis. CV33 cells were mock transfected or transfected with plasmids expressing the wild-type UL28 protein (HSV-1) or the indicated UL28 mutant. Twenty-four hours later, lysates were prepared and separated by SDS-PAGE. After transfer to a nitrocellulose membrane, immunoblotting was performed using antibodies against UL33 or actin.
Characterization of a UL28 mutant that grows with reduced titers on Vero cells.
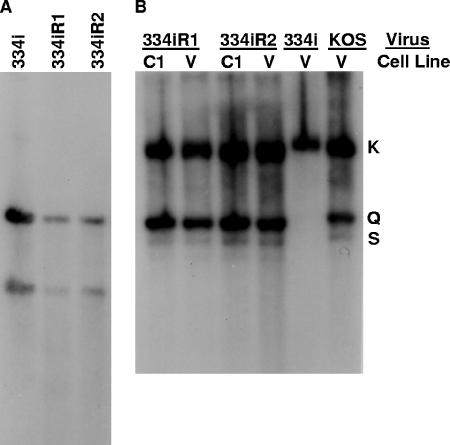
As shown in Table 1, the stock for one of the UL28 mutant viruses, 334i, contained virus that grew on the nonpermissive Vero cells but the titer on these cells was 2 log units lower than what was observed on C1 cells. The simplest explanation was that the 334i virus stock contained wild-type virus that was generated by recombination with the UL28 sequences present in the UL28-complementing C1 cell line. However, the fact that none of the other lethal UL28 linker insertion mutants grew on Vero cells suggested that the revertants found in the 334i virus stock may be the result of a second-site suppressor mutation. Two plaques from the 334i revertant viruses were isolated from Vero cells and designated 334iR1 and 334iR2. Each of the viruses was plaque purified twice, virus stocks were prepared on Vero cells, and both viruses were found to have equivalent titers on Vero and C1 cells (data not shown). Viral DNAs were examined by Southern blotting for the presence of the BglII restriction site associated with the 334i insertion mutation (Fig. 8A). Digests of viral DNA isolated from 334i, 334iR1, and 334iR2 with SalI and BglII resulted in two UL28-hybridizing fragments of 2.3 and 1.5 kbp, which were the predicted fragments if the BglII linker was retained in the UL28 gene. Therefore, the two revertants were due to mutations at a second site that overcame the lethal 334i insertion.
FIG. 8.
Southern blot analysis of DNA isolated from Vero and C1 cells infected with UL28-334i revertants. (A) Total infected-cell DNA isolated from Vero cells infected with indicated viruses was digested with SalI plus BglII and subjected to Southern blot analysis using the SalI fragment described in the legend for Fig. 2. (B) Total infected-cell DNA isolated from Vero (V) or C1 cells was digested with BamHI, Southern blotted, and probed with the BamHI K fragment.
The 334iR1 and 334iR2 viruses were examined for their abilities to cleave viral DNA. Figure 8B shows that the DNA of these two viruses is cleaved equally when grown on Vero cells and when grown on C1 cells. Theses results demonstrate that the suppressor mutation releases the block on cleavage of viral DNA that is associated with the four-amino-acid insertion in the UL28 gene of the 334i virus and allows the production of infectious virus.
The second-site mutation is extragenic.
Marker rescue experiments were performed in order to determine whether the second-site mutation mapped to the UL28 gene of 334iR1. The UL28 genes from KOS, 334i, and 334iR1 were cloned (SalI fragments) and then separately transfected into C1 cells along with infectious GCB viral DNA. The titers of the progeny from the transfections were then determined on Vero and C1 cells to test for the rescue of GCB. The plasmids containing the 334i and 334iR1 UL28 genes failed to rescue GCB, while the plasmid containing the wild-type UL28 gene efficiently rescued GCB (data not shown). These data are consistent with the conclusion that a mutation outside of the UL28 gene in 334iR1 is responsible for the ability of this virus to grow on Vero cells. In support of this finding, we have sequenced the entire coding region of the UL28 gene for both the 334i and 334iR1 clones and found the sequences of these two genes to be identical.
The suppressor mutation resides in the UL15 gene.
Because the UL28 protein has been shown to interact with UL6, UL15, and UL33, it is likely that the second-site mutation resides in the open reading frame for one of these proteins. To determine whether this was the case, we first tried to map the second-site mutation by marker transfer. Genomic DNA prepared from 334i was cotransfected into C1 cells along with plasmid DNA containing cloned restriction fragments (13 EcoRI fragments covering the entire HSV genome) from the 334iR1 revertant virus. Homologous recombination between a cloned restriction fragment containing the second-site mutation and the 334i genome would give rise to progeny viruses that should grow on Vero cells. Unfortunately, these studies were difficult to interpret due to the high spontaneous reversion frequency of the 334i virus which approached the marker transfer background. We then sequenced the entire open reading frames of the UL6, UL15, and UL33 genes isolated from 334i, 334iR1, and 334iR2 viruses. The DNA sequences of the genes from the two revertants were compared with the sequences of these genes from the original 334i mutant. There were no DNA sequence alterations in either the UL6 or the UL33 gene. In contrast, the UL15 genes for both 334iR1 and 334iR2 were found to contain a single base (G to A) change at nucleotide 634 (base 1 was A of the ATG start codon) compared to the UL15 gene from 334i. The base substitution altered the codon for amino acid 212 from GCC to ACC, resulting in an alanine-to-threonine amino acid change (A212T).
To determine whether the single amino acid change in the UL15 gene was responsible for the suppressor phenotype, transient complementation assays were performed. Vero cells were transfected with plasmids expressing the UL28-334i gene, along with a plasmid expressing either the wild-type UL15 gene or the UL15 gene containing the A212T amino acid change. The UL15 and UL28 genes were cloned into a pcDNA vector, in which the CMV promoter drove the expression of the mutant and wild-type genes. The next day the cells were infected with the UL28-null virus GCB. Following another day of incubation, the virus progeny yield was determined by plating the lysates on complementing CV28 cells. As shown in Table 2, the plasmid expressing the wild-type UL28 gene was able to efficiently complement the UL28-null mutant. The plasmid expressing the UL28-334i gene failed to complement when transfected alone or when it was cotransfected with a plasmid expressing the wild-type UL15 gene. In contrast, the UL28-334i plasmid was found to support the replication of the UL28-null virus when cotransfected with a plasmid expressing the UL15-A212T gene. The expression of the UL28-334i protein along with the UL15-A212T protein resulted in a virus yield that was approximately 20% of that found with the wild-type UL28 gene. The expression of either wild-type or mutant UL15 proteins alone failed to complement the UL28-null virus (Table 2). The A212T amino acid change had no apparent effect on UL15 function since the UL15-A212T gene efficiently complemented a UL15-null virus in a transient complementation assay (data not shown). These results demonstrate that the ability of the 334i revertants to grow on Vero cells was due to an extragenic suppressor mutation in the UL15 gene.
TABLE 2.
Results from the transient complementation assaya
| UL28 plasmid | UL15 plasmid | Titer (PFU/ml) on CV28 cells | No. of expts | CI |
|---|---|---|---|---|
| NP | NP | 3.4 ± 1.7 × 102 | 4 | 1.0 |
| pcDNA-UL28 | NP | 1.1 ± 0.4 × 105 | 4 | 323 |
| pcDNA-UL28/334i | NP | 4.4 ± 2.7 × 102 | 4 | 1.3 |
| NP | pcDNA-UL15 | 2.8 ± 1.7 × 102 | 4 | 0.8 |
| NP | pcDNA-UL15/A212T | 3.8 ± 3.3 × 102 | 4 | 1.1 |
| pcDNA-UL28/334i | pcDNA-UL15 | 5.8 ± 3.3 × 102 | 10 | 1.7 |
| pcDNA-UL28/334i | pcDNA-UL15/A212T | 2.3 ± 0.9 × 104 | 10 | 68 |
| pcDNA-UL28/215i | pcDNA-UL15/A212T | 4.1 ± 0.9 × 102 | 4 | 1.2 |
| pcDNA-UL28/225i | pcDNA-UL15/A212T | 4.5 ± 1.0 × 102 | 4 | 1.3 |
| pcDNA-UL28/345i | pcDNA-UL15/A212T | 3.9 ± 1.4 × 102 | 4 | 1.1 |
| pcDNA-UL28/386i | pcDNA-UL15/A212T | 5.6 ± 1.3 × 102 | 4 | 1.6 |
| pcDNA-UL28/604i | pcDNA-UL15/A212T | 2.2 × 102 | 2 | 0.6 |
| pcDNA-UL28/631i | pcDNA-UL15/A212T | 3.1 × 102 | 2 | 0.9 |
| pcDNA-UL28/736i | pcDNA-UL15/A212T | 4.4 ± 1.9 × 102 | 4 | 1.3 |
The transient complementation assay was performed as described in Materials and Methods. No virus was detected for any of the samples when titers were determined on Vero cells. The complementation index (CI) was determined by dividing the viral titer resulting from the transfection of each plasmid by the viral titer resulting when no plasmid was used. NP, no plasmid.
The UL15 A212T mutation does not suppress any of the other lethal UL28 linker insertion mutants.
The ability of the UL15-A212T gene to act as a second-site suppressor of the other six lethal UL28 insertion mutants was tested. The UL28 genes from the 215i, 225i, 345i, 386i, 604i, 631, and 736i mutants were cloned into the pcDNA expression vector and then transfected into Vero cells along with the pcDNA-UL15-A212T vector, and the next day the cells were infected with GCB. As shown in Table 2, none of these UL28 insertion mutants were found to complement GCB, indicating that the suppressor phenotype of the UL15-A212T mutant was specific to the UL28-334i insertion mutant.
DISCUSSION
We have characterized the phenotypes of a panel of linker insertion mutations throughout the HSV-1 UL28 coding region. Six of the 14 mutants were able to produce infectious virus. Our results show that the C terminus of the UL28 protein is critical since all four nonsense mutants failed to produce infectious virus. Western blot analysis showed that full-size UL28 protein is produced in all of the linker insertion mutant viruses, demonstrating that the lack of UL28 function is due to the inserted mutation and not lack of protein expression. Western analysis of the nonsense mutants showed that the UL28 polypeptide truncated after amino acid 225 or 396 was not stable. The truncation of the polypeptide after amino acid 604 or 741 resulted in stable proteins of the expected size, but which were not capable of carrying out the UL28 function(s), indicating that the final 44 amino acids of UL28 are essential for function.
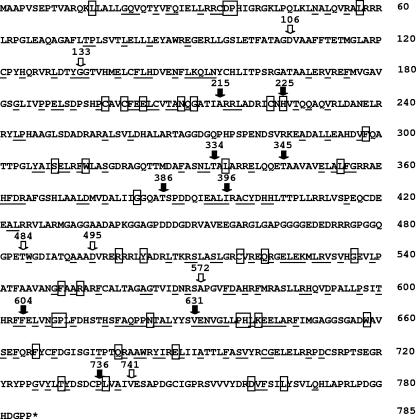
The insertions (106i, 133i, 484i, 495i, 572i, and 741i) that did not affect virus production appeared to cluster near the ends and middle of the UL28 coding region. The mutations in 106i, 484i, 495i, and 572i occur within regions where the predicted amino acid sequence is not strongly conserved within homologs of the UL28 gene of other herpesviruses (Fig. 9). Therefore, it is not surprising that insertions at these sites do not affect UL28 function. The insertion in 133i occurs in a region that is strongly conserved among alphaherpesviruses, but not among beta- and gammaherpesviruses. Interestingly, the disruption of this region does not appear to affect the ability of the virus to cleave and package its genome or to propagate in Vero cells. The apparent conservation among alphaherpesviruses suggests that there is some selective pressure to maintain the amino acid sequence found in this region. The 741i insertion occurs just prior to a strong conserved region but does not appear to affect UL28 function.
FIG. 9.
Amino acid sequence of HSV-1 UL28. The arrows show the sites of the linker insertions. Closed arrows indicate that the insertion mutation blocked the cleavage and packaging of viral DNA. Amino acids that are conserved in the alphaherpesviruses are underlined, and the amino acids that are boxed are conserved in all three herpesvirus families.
The mutations in 215i and 225i occur in the region of a putative zinc binding motif (19). A number of amino acids within this motif are conserved in most of the UL28 homologs from human and animal herpesviruses (Fig. 9). These include three cysteines and one histidine that are conserved in all UL28 homologs. The 215i mutation occurs within the 22-amino-acid zinc binding motif, and a four-amino-acid insertion might be expected to alter the conformation of this motif, rendering it nonfunctional. The 225i mutation occurs immediately after the final histidine residue of the motif, in a region where the amino acid sequence is also strongly conserved. Studies by Bogner et al. (8) and Adelman et al. (3) have shown that the CMV UL56 (CMV homolog of the HSV UL28 gene) and HSV-1 UL28 proteins are both sequence-specific DNA binding proteins. This metal binding motif could therefore be an important region of the UL28 protein for protein/DNA interaction. This region is also a target for a novel class of human CMV (HCMV) inhibitors (19). The benzimidazole ribonucleosides block the cleavage and packaging of HCMV DNA, and mutations that confer resistance to these drugs have been mapped to both the HCMV UL56 and UL89 genes. The amino acid change in the UL56 protein that conferred resistance was Q204R (Q210 in the HSV UL28 protein), which is located near the central region of the metal binding domain. It is not surprising, therefore, that the 215i and 225i insertions inhibit virus replication. Other viruses with mutations in regions of well-conserved amino acids and that fail to cleave and package their DNA are 334i, 345i, 604i, 631i, and 736i (Fig. 9).
It is of interest to note a couple of points regarding the DNA cleavage data in Fig. 4. First, in the case of the 495i mutant, there appears to be a significant decrease in “cleavage efficiency” (i.e., ratio of K to Q/S fragments) in Vero cells yet with no significant effect on virus titers. Although we have not done a quantitative analysis of the cleavage efficiency of this mutant, the implication is that cleavage can be substantially reduced without affecting the release of infectious virus. Second, with some of the truncation mutants, there appeared to be an altered ratio of Q to S fragments in C1 cells, with more Q formed than S. This may be the result of expressing the truncated forms of UL28 in the context of the full-length protein present in C1 cells. Also, additional Q/S fragments are present in the digests of DNA derived from infected Vero cells for some of the nonlethal insertion mutants (106i, 133i, 572i, and 741i). These fragments are most likely derived from termini with multiple “a” sequences. In HSV-1, multiple “a” sequences can occur at long-arm termini (S fragment) but not at short-arm termini (Q fragment). The differences between Vero cells and C1 cells might suggest that certain UL28 mutations could impact the cleavage reaction, resulting in L termini with either a single “a” sequence or termini with multiple “a” sequences.
We have not examined whether any of the lethal UL28 insertion mutants alter the ability of the UL28 protein to localize to the nucleus. The 724-amino-acid pseudorabies virus UL28 homolog requires the HSV-1 UL15 protein as a chaperone in order to reach the nucleus (18). A pseudorabies virus mutant lacking amino acids 570 to 724 of the UL28 protein failed to localize to the nucleus in the presence of HSV-1 UL15, suggesting that this region may be important for the interaction with the UL15 protein (18). C-terminal truncations of the HSV-1 UL28 protein were found to abolish UL28 function, and in addition, these mutants failed to coimmunoprecipitate with UL15, suggesting that the C terminus is important for UL28/UL15 interaction. The UL33 protein is also found in the nucleus and associates with capsids (5, 30). It therefore seems likely, given the capsid association and nuclear localization of all three proteins, that the complex of UL15/UL28/UL33 is also formed in the nucleus.
The immunoprecipitation studies confirm previous results showing that a UL15/UL28/UL33 complex can form in infected cells and that the complex appears to consist of UL33 and UL15 binding directly with UL28 and that UL15 and UL33 do not bind each other (38). The studies with the UL28 mutants demonstrate that the C terminus appears to be the region of the protein that UL33 and UL15 interact with and that the binding of UL33 is important for forming a stable complex. More importantly, UL15 was found to coimmunoprecipitate with all of the UL28 linker insertion mutants but not with any of the C-terminally truncated mutants. The UL28 741s mutant was missing amino acids 741 to 785, and this would suggest that this region of the UL28 protein either interacts directly with UL15 or is important for the proper folding of UL28. The 604i and 736i insertions were both found to interfere with the interaction of UL28 with UL33 but not with UL15, suggesting that these two regions are critical for UL33 binding. Since none of the insertions in the N terminus of UL28 were found to interfere with the interaction of UL28 with either UL33 or UL15 it appears that the binding sites for both proteins resides in the C-terminal end of UL28.
Studies of the revertant 334iR1 and its parent 334i confirm that both viruses contained the constructed linker insertion mutation. Marker rescue studies demonstrated that the revertant contained a second-site mutation that mapped outside of the UL28 gene. DNA sequence analysis, in combination with genetic complementation studies, demonstrated that a single amino acid change of A212T in the UL15 protein was responsible for suppressing the lethal UL28 linker insertion mutation. The 334i mutant was the only one of the lethal UL28 linker insertion mutants that formed plaques on noncomplementing cells. We attempted to isolate spontaneous revertants of the other seven lethal UL28 linker insertion mutants by passaging each virus multiple times in cell culture and were unsuccessful in isolating a virus that would grow on Vero cells. Based on the average titer of several 334i virus stocks (titers were determined on Vero- versus UL28-complementing cells), the reversion frequency for this mutant corresponded to approximately 1 revertant/1,000 total PFU. The selective isolation of a revertant that contained a suppressor mutation in the UL15 protein would suggest that the site of the insertion in the 334i mutant is within a region of the UL28 protein that is important for forming a functional terminase complex. The fact that the 334i insertion did not interfere with the interaction of UL28 with UL15 or UL33 suggests that the mutation at amino acid 212 may be located in an important region of the UL15 protein that is either directly or indirectly involved in the DNA cleavage reaction. The A212T suppressor mutation is located within a conserved region of the UL15 protein (29). This region of the UL15 protein is distinct from where the two proposed ATP binding domains (Walker A and B boxes) are located and where the benzimidazole resistance mutation maps (19, 29). The critical nature of this region of the UL15 protein was demonstrated by the fact that a recombinant virus containing a Q205E amino acid change can be propagated only on a UL15-complementing cell line (29). In an attempt to determine whether revertants with mutations other than the A212T would compensate for the UL28 334i insertion, we repassaged the 334i mutant and isolated additional revertants. Before passaging, the 334i virus was plaque purified three times to ensure that the stock was free of the original revertant. Following serial passage, revertants (three independent isolates) were isolated and the presence of the UL28 334i insertion was confirmed. DNA sequencing of the UL15 gene revealed that the three revertants all contained the same A212T change as that of the original 334i revertant. Thus, there appears to be strong selective pressure for this specific amino acid change.
In summary, the mutational analysis described here has resulted in the identification of specific regions of the UL28 protein that are critical for its cleavage/packaging function. The C-terminal end of UL28 appears to be critical for interaction with UL15 and UL33 to form a functional terminase complex. Insertions located between amino acids 200 and 400 did not interfere with the interaction of UL28 with either UL15 or UL33, but these mutants failed to cleave and package viral DNA. This region of UL28 contains a highly conserved putative metal binding domain which may be important for the interaction of UL28 with the HSV DNA cleavage and packaging sites on the viral genome. Further mutational studies targeting the conserved cysteines and histidine residues within this conserved domain should aid in determining the importance of this region in the cleavage/packaging reaction. Finally, the isolation of a spontaneous second-site revertant provides clear genetic evidence that UL28 and UL15 interact. It would be interesting to generate a similar set of UL15 linker insertion mutants and determine whether it is possible to isolate UL28 second-site suppressor revertants.
Acknowledgments
We thank Jamie Huffman for technical assistance and Jay Brown and Shelley Cockrell for critically reading the manuscript.
This work was supported by Public Health Services grants GM50740 (J.D.B.) and AI060836 (F.L.H.) from the National Institutes of Health.
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Abbotts, A. P., V. G. Preston, M. Hughes, A. H. Patel, and N. D. Stow. 2000. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J. Gen. Virol. 81:2999-3009. [DOI] [PubMed] [Google Scholar]
- 2.Addison, C., F. J. Rixon, and V. G. Preston. 1990. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J. Gen. Virol. 71:2377-2384. [DOI] [PubMed] [Google Scholar]
- 3.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180:380-388. [DOI] [PubMed] [Google Scholar]
- 5.Beard, P. M., and J. D. Baines. 2004. The DNA cleavage and packaging protein encoded by the U(L)33 gene of herpes simplex virus 1 associates with capsids. Virology 324:475-482. [DOI] [PubMed] [Google Scholar]
- 6.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76:4785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black, L. 1988. DNA packaging in dsDNA bacteriophages, p. 321-373. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Press, New York, N.Y. [Google Scholar]
- 8.Bogner, E., K. Radsak, and M. F. Stinski. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, J., M. A. McVoy, and F. L. Homa. 2002. Packaging DNA into herpesvirus capsids, p. 111-155. In A. H. E. Bogner (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 10.Cavalcoli, J. D., A. Baghian, F. L. Homa, and K. G. Kousoulas. 1993. Resolution of genotypic and phenotypic properties of herpes simplex virus type 1 temperature-sensitive mutant (KOS) tsZ47: evidence for allelic complementation in the UL28 gene. Virology 197:23-34. [DOI] [PubMed] [Google Scholar]
- 11.Chang, Y. E., A. P. Poon, and B. Roizman. 1996. Properties of the protein encoded by the UL32 open reading frame of herpes simplex virus 1. J. Virol. 70:3938-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 13.Goldin, A. L., R. M. Sandri-Goldin, M. Levine, and J. C. Glorioso. 1981. Cloning of herpes simplex virus type 1 sequences representing the whole genome. J. Virol. 38:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland, L. E., R. M. Sandri-Goldin, A. L. Goldin, J. C. Glorioso, and M. Levine. 1984. Transcriptional and genetic analyses of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J. Virol. 49:947-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 16.Homa, F. L., T. M. Otal, J. C. Glorioso, and M. Levine. 1986. Transcriptional control signals of a herpes simplex virus type 1 late (γ2) gene lie within bases −34 to +124 relative to the 5′ terminus of the mRNA. Mol. Cell. Biol. 6:3652-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koslowski, K. M., P. R. Shaver, J. T. Casey II, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenney, and N. E. Pederson. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koslowski, K. M., P. R. Shaver, X. Y. Wang, D. J. Tenney, and N. E. Pederson. 1997. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J. Virol. 71:9118-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W. Feng, R. K. Jain, R. G. Ptak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drach. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 22.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 23.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mettenleiter, T. C., A. Saalmuller, and F. Weiland. 1993. Pseudorabies virus protein homologous to herpes simplex virus type 1 ICP18.5 is necessary for capsid maturation. J. Virol. 67:1236-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel, A. H., and J. B. MacLean. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206:465-478. [DOI] [PubMed] [Google Scholar]
- 27.Pederson, N. E., J. T. Casey II, K. M. Koslowski, and P. R. Shaver. 1998. The UL6 locus of pseudorabies virus and its homology to oncogenic herpesviruses. Oncol. Rep. 5:115-119. [DOI] [PubMed] [Google Scholar]
- 28.Prevelige, P. E., Jr., and J. King. 1993. Assembly of bacteriophage P22: a model for ds-DNA virus assembly. Prog. Med. Virol. 40:206-221. [PubMed] [Google Scholar]
- 29.Przech, A. J., D. Yu, and S. K. Weller. 2003. Point mutations in exon I of the herpes simplex virus putative terminase subunit, UL15, indicate that the most conserved residues are essential for cleavage and packaging. J. Virol. 77:9613-9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds, A. E., Y. Fan, and J. D. Baines. 2000. Characterization of the U(L)33 gene product of herpes simplex virus 1. Virology 266:310-318. [DOI] [PubMed] [Google Scholar]
- 31.Salmon, B., and J. D. Baines. 1998. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of UL15-encoded proteins with B capsids requires at least the UL6, UL17, and UL28 genes. J. Virol. 72:3045-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tengelsen, L. A., N. E. Pederson, P. R. Shaver, M. W. Wathen, and F. L. Homa. 1993. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 67:3470-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, K., and J. D. Baines. 2006. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J. Virol. 80:5733-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, D., and S. K. Weller. 1998. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 243:32-44. [DOI] [PubMed] [Google Scholar]
- 40.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]