Abstract
Deamination of DNA bases can occur spontaneously, generating highly mutagenic lesions such as uracil and hypoxanthine. In Escherichia coli two enzymes initiate repair at hypoxanthine residues in DNA. The alkylbase DNA glycosylase, AlkA, initiates repair by removal of the damaged base, whereas endonuclease V, Endo V, hydrolyses the second phosphodiester bond 3′ to the lesion. We have identified and characterised a mouse cDNA with striking homology to the E.coli nfi gene, which also has significant similarities to motifs required for catalytic activity of the UvrC endonuclease. The 37-kDa mouse enzyme (mEndo V) incises the DNA strand at the second phosphodiester bond 3′ to hypoxanthine- and uracil-containing nucleotides. The activity of mEndo V is elevated on single-stranded DNA substrate in vitro. Expression of the mouse protein in a DNA repair-deficient E.coli alkA nfi strain suppresses its spontaneous mutator phenotype. We suggest that mEndo V initiates an alternative excision repair pathway for hypoxanthine removal. It thus appears that mEndo V has properties overlapping the function of alkylbase DNA glycosylase (Aag) in repair of deaminated adenine, which to some extent could explain the absence of phenotypic abnormalities associated with Aag knockout in mice.
INTRODUCTION
Cellular genomes are exposed to a wide range of toxic agents including ionising radiation, sunlight, chemical agents and intracellular reactive oxygen species (ROS). The molecular basis for the mutagenic and carcinogenic potency of these agents lies in their ability to alter the structure of individual DNA bases or the sugar–phosphate backbone (1). The most efficient way of correcting DNA damage is direct reversal of the specific lesion. This repair strategy has been identified for UV-induced pyrimidine dimers and alkylated bases in DNA, in both microorganisms and mammals (2–7). Excision repair is the other major pathway for correcting damaged or inappropriate DNA bases (8). Damaged base residues in DNA can be removed by one of two principally different mechanisms. Lesions generated endogenously by hydrolysis or exposure to ROS are corrected by base excision repair (BER), where release of the altered base in free form is mediated by a DNA glycosylase (9,10). Dipyrimidine adducts generated by exposure to UV light and other types of base damage that cause major helix distortions are generally processed by nucleotide excision repair (NER), in which the DNA chain is incised on each side of the lesion to release an oligonucleotide containing the damaged residue (8,11,12).
DNA is subjected to deamination at a physiologically significant rate. Hypoxanthine is the result of adenine deamination and can also be introduced by misincorporation of dIMP. Hypoxanthine pairs with cytosine during replication resulting in AT to GC transitions at deaminated adenines (13,14). Specific mechanisms for removal of deaminated bases have evolved. Several bacterial, archaeal and eukaryotic organisms contain an evolutionary conserved enzyme that recognises deaminated bases in DNA. This 3′deoxyinosine endonuclease (Endo V), the product of the nfi gene in Escherichia coli, was first described by Gates and Linn and later extensively characterised by Yao and co-workers (15–17). Endo V was originally described as a nuclease cleaving near lesions that alters the secondary structure of DNA, and has subsequently been shown to have a broad substrate spectrum acting at AP sites (17), urea residues (17), base mismatches (18), flap DNA and pseudo Y structures (19) and loops and hairpins (19). The ability of Endo V to recognise all three deamination products in DNA, deoxyinosine, deoxyuridine and deoxyxanthosine, is not shared by any other known repair enzymes (20). Endo V incises the DNA at the second phosphodiester bond 3′ to the lesion, leaving a 3′ OH and a 5′ P termini. In E.coli there are three other known repair pathways initiated by cleavage of a phosphodiester bond (21). The UvrABC complex excises pyrimidine dimers and nucleotides containing bulky adducts (11,12). The MutSLH system removes regions containing mismatched bases (22), while the very short patch repair system removes thymine resulting from deamination of 5-methylcytosine (23). In the Endo V-mediated repair pathway additional enzymes are required to excise the damaged base and complete the repair process. Kow recently suggested the action of a specific 3′–5′ exonuclease followed by polymerisation and ligation (24).
DNA repair enzymes acting on endogenous DNA lesions often show strong evolutionary conservation, both in sequence and function (25). Nfi homologues from Archaeoglobus fulgidus (20) and Thermotoga maritima (26) have recently been characterised. Here we describe the identification and characterisation of this gene from mouse and human cells. Endo V represents one of the most conserved DNA repair enzymes so far identified with 32% sequence identity between the mouse and E.coli orthologues. The mammalian Endo V homologue possesses DNA repair activities that are similar, but more limited, than those of E.coli Endo V. This antimutator enzyme initiates DNA repair by endonuclease cleavage at the second phosphodiester bond 3′ to the base lesion.
MATERIALS AND METHODS
Identification of mammalian expressed sequences with homology to E.coli Endo V (nfi)
A similarity search using the E.coli Endo V amino acid sequence as a query in a database of expressed sequence tags (ESTs) was performed. A multiple sequence alignment was created using Clustal W (27). Candidate ESTs were identified and the murine clone (GenBank accession no. AI509431) ordered for further characterisation. The complete insert was sequenced without identification of the start codon. RML-Race (RNA Ligase Mediated Rapid Amplification of cDNA Ends; Ambion) was used in combination with 10 µg total RNA from normal mouse lung (Ambion) in order to clone the 5′ end of the cDNA. A full-length sequence carrying the complete ORF was then amplified with the sequence information from RML-Race and clone AI509431 by Qiagen One Step RT–PCR kit (Qiagen) using mRNA as template (primers; 5′-CTCA AAGCCTCGAGGCGCTA-3′ and 5′-TGTGACTGTGGTG AGGTCTC-3′).
Northern blot hybridisation
Northern blots containing samples from multiple mouse tissues purchased from Clontech (Mouse MTN™ blot; 7762-1) were probed for mnfi expression. Northern blot hybridisation was carried out using ExpressHyb solution (Clontech) as recommended by the manufacturer. Probes were labelled with [α-32P]dCTP (3000 Ci/mmol, Amersham) using the Rediprime DNA labelling system (Amersham).
Protein over-expression and purification
The 1017 bp mEndo V sequence was amplified and inserted into the pET-28b vector (Navagen), resulting in the expression vector pET-28b/mnfi. This was obtained by introducing NcoI and HindIII restriction sites, at the start and stop, respectively, of the coding sequence using primers 5′-GGGCCCATGGC TCACACGGCCGCT-3′ and 5′-CGGCCGCAAGCTTGGGT GGTGACTGTCA-3′ in a PCR. The amplified fragment was ligated into the pET-28b vector resulting in the addition of a hexahistidine tag at the C-terminal end of the mEndo V protein. The plasmid carrying the mEndo V encoding gene was used to transform E.coli BL21 star (DE3) and positive colonies were inoculated in 500 ml of Luria–Bertani (LB) medium containing kanamycin (30 µg/ml). A negative control was generated by transforming the pET-28b vector into E.coli BL21 star (DE3). The cultures were incubated with shaking at 37°C until OD600 reached 0.4. IPTG was added to 1 mM and the cells were further grown for 4 h at 37°C. Cells were harvested by centrifugation at 4000 g for 6 min at 4°C, and resuspended in 10 ml of buffer A (50 mM Tris pH 8.0, 500 mM NaCl, 5 mM imidazole and 10 mM β-mercaptoethanol). Cells were disrupted by French press (800–1000 psig; Simoaminco) and the extract clarified by centrifugation at 17 000 g for 30 min at 4°C. A Ni2+ column, 6 mm in diameter, was packed with 1.0 ml of Ni2+-resin. The clarified extract containing histidine-tagged protein was loaded on the column and washed twice with 6× column volume, 4 and 15 mM imidazole, respectively, in buffer A. Bound protein was eluted with 500 mM imidazole dissolved in buffer A and collected in fractions of 0.5 ml. The fractions containing the protein of interest were desalted (50 mM Tris pH 8.0, 100 mM KCl and 20% glycerol) with a HiTrap column (Amersham). The protein was aliquoted into 10 µl fractions and stored at –20°C.
Enzyme assays
Single-stranded substrate preparation: a 24mer oligonucleotide containing a single hypoxanthine (5′-GGCGGCATG ACCC-hyp-GAGGCCCATC-3′) was 32P-labelled at the 5′ terminus with T4 polynucleotide kinase (MBI Fermentas) and [γ-32P]ATP (3000 Ci/mmol, Amersham). Double-stranded DNA substrate was produced by annealing a 49mer hypoxanthine containing oligonucleotide (5′-TAGACGGATGAAT ATTGAGG-hyp-CAGAAGTTGGATTTGGTAGT-3′), 32P-labelled at the 5′ terminus, to a complementary strand containing a thymine (T) opposite to the hypoxanthine base (5′-ACTACCAAATCCAACTTCTGTCCTCAATAT TCATCCGTCTA-3′). All oligos include phosphorothioate linkages at the ultimate and penultimate 5′ and 3′ ends to reduce exonucleolytic attack. Activity of mEndo V on DNA containing uracil residues was assayed on a single-stranded uracil containing oligonucleotide (5′-GCTCATGCG CAGXCAGCCGTACTCG-3′, X = uracil). A reaction mixture (10 µl) containing reaction buffer (2 mM MgCl2, 50 mM Tris pH 7.5, 50 mM KCl, 5% glycerol, 1 mM DTT and 100 ng/µl BSA) and ∼20 fmol 32P-labelled substrate was incubated at 37°C for 30 min with purified mouse Endo V. Reactions were terminated by addition of formamide loading mix. The sample was loaded onto a 6 or 20 cm 20% polyacrylamide gel containing 7 M urea and DNA molecules separated by electrophoresis. The gel was dried under vacuum and subjected to phosphorimaging for visualization.
Assay for mutation suppression in E.coli
The rate of spontaneous mutations was tested by growing cells on LB plates containing rifampicin (100 µg/ml). A wild type E.coli strain (TG:1) was compared with E.coli BK 2223 (alkA nfi in TG:1, this work). The open reading frame (ORF) of mEndo V gene was subcloned as described above and transformed into E.coli BK 2223 (BK2223/pET-28b/mnfi; alkA nfi mnfi+). For each experiment, an appropriate dilution of the bacterial culture was grown on LB plates to estimate the number of colony forming units. Mutated bacteria were identified by seeding on rifampicin plates. The plates were incubated overnight at 37°C and the number of colonies counted.
Site-specific mutagenesis
Site-specific mutations were introduced in the mEndo V wild-type sequence by a PCR based ‘Quick change’ (Stratagene) method. Primers containing the mutation of interest were as follows: mutation S93P; primer A: 5′-GTTGGCCTGAAGG CCCCCTATGTGCCAGGCTTCCTGGCCTTCCGAGAGG TCC-3′, primer B: 5′-GGACCTCTCGGAAGGCCAGGA AGCCTGGCACATAGGGGCCTTCAGGCCAAC-3′, and mutation Q133P; primer 1: 5′-GGATGGAAACGGGGT GCTTCACCCGCGAGGCTTCGGGGTGGCCTGCCACC- 3′, primer 2: 5′-GGTGGCAGGCCACCCCGAAGCCTCG CGGGTGAAGCACCCCGTTTCCATCC-3′. The PCR products were digested with DpnI and transformed into E.coli ER2566 by electroporation. Site-specific mutagenesis was verified by sequencing. The protocol described above was repeated for the construction of a double mutant with both amino acids changed. Prior to purification, the mutant sequences were transformed into E.coli BL21 star. Growth conditions and purification procedure were as described for the wild type protein.
RESULTS
Mouse and human proteins with extensive homology to E.coli Endo V
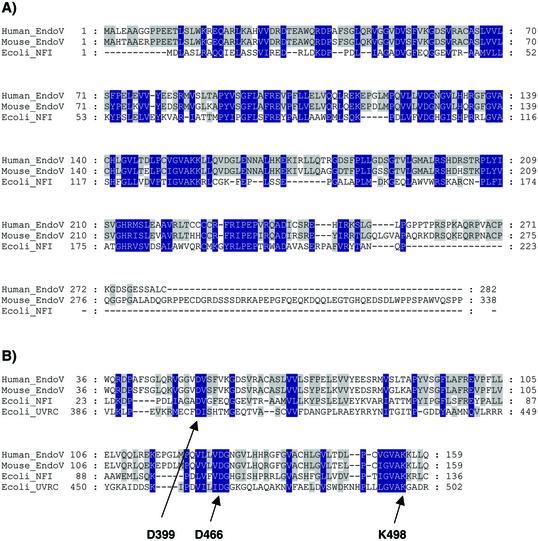
We have identified entries in the human and mouse EST database with striking sequence similarities to the E.coli nfi gene, termed hnfi and mnfi, respectively. The mouse clone was sequenced and the mRNA sequence was found to be completely identical to the GenBank sequence with accession number XM_203558. The corresponding protein has the interim name LOC277019 and accession number XP_203558. The ORF of mnfi encodes a putative polypeptide of 338 amino acids with a predicted molecular weight of 37.2 kDa. The amino acid sequence of mEndo V showed 32% sequence identity and 49% sequence similarity to E.coli Endo V (Fig. 1). The active site IGVAKK is conserved from E.coli to mouse and man. To our knowledge, a higher degree of conservation for a DNA repair function is seen only within the superfamily of uracil-DNA glycosylases (UDG/UNG) (28). The murine gene consists of nine exons spanning ∼7.5 kb of chromosome 11. The sequenced 5′ flanking region contains two putative TATA boxes, a CpG island and possible binding sites for several transcription factors and enhancer motifs [BioInformatics and Molecular Analysis Section (BIMAS), NIH]. Domains conserved between the mammalian and bacterial enzyme include homology identified previously between E.coli Endo V and UvrC (Fig. 1B) (25).
Figure 1.
Conserved sequence of mammalian Endo V species. (A) Amino acid sequences of human, murine and E.coli Endo V were aligned by the program Clustal W (Materials and Methods). (B) Conserved domains of human, murine and E.coli Endo V and uvrC. Identity/similarity between all four or three of the sequences are boxed in blue and grey, respectively (25).
Over-expression and purification of mEndo V
Several approaches were used to express and purify mEndo V. Expression and immediate purification of a C-terminal His-tagged fusion protein was found to give highest yield and reproducible activity. A hexahistidine (His6) tag was attached to the C-terminal end of mEndo V by subcloning in the pET-28b vector. The protein was expressed at high levels following IPTG induction of E.coli BL21 star transformed with the plasmid carrying the mEndo V sequence, pET-28b/mnfi (Fig. 2, lane 2). The C-terminal histidine tag enabled purification by Ni2+ affinity chromatography. Elution from the column with 500 mM imidazole revealed a single major protein band with a molecular mass of ∼37 kDa (Fig. 2, lane 3). The over-expressed and purified protein (lanes 2 and 3) also tested positively with a polyclonal antibody raised against a synthetic polypepdide of mEndo V (data not shown).
Figure 2.
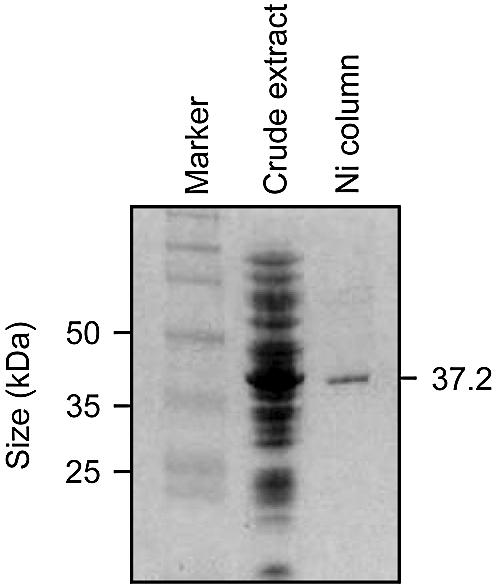
Purification of recombinant mEndo V protein. The protein was over-expressed in E.coli, and proteins were visualised on a 10% SDS– polyacrylamide gel by Coomassie blue staining. Lane 1 shows a protein marker (full-length rainbow, Bio-Rad); lane 2 shows crude extract from BK 2223/pET-28b/mnfi cells grown at 37°C (20 µg); lane 3 shows 1 µg of peak mEndo V protein fraction eluted with 0.5 M imidazole from a Ni2+ column loaded with the crude extract shown in lane 2. Positions of protein size markers in kDa are indicated on the left, and the predicted size of mEndo V is indicated on the right.
Expression patterns of mEndo V in mouse tissue
Expression patterns of mEndo V were investigated in several mouse tissues by northern blot hybridisation (Fig. 3). The labelled mEndo V cDNA probe consistently detected three different bands with the highest level in liver, and high levels in heart, kidney and testis. Several reports indicate high levels of DNA repair enzymes in these organs (29,30). The expected 1.8 kDa mRNA was detected in all tissues tested. Two larger transcripts, 6.4 and 3.6 kDa, were observed in all tissues examined and the nature of these are unknown, but are most probably unprocessed and partially processed mRNA. Northern analysis of brain mRNA indicates a low level of processed mRNA (Fig. 3). Positive entries for the mEndo V gene in the sequence database (NCBI, mouse EST database) includes lung, eyeball, brain, liver, testis, mammary and foetus, indicating a ubiquitous expression pattern. Similar results from the corresponding human EST database identified hEndo V expression in prostate, brain, skin, lung, kidney, eye, pancreas, spleen, placenta and uterus.
Figure 3.
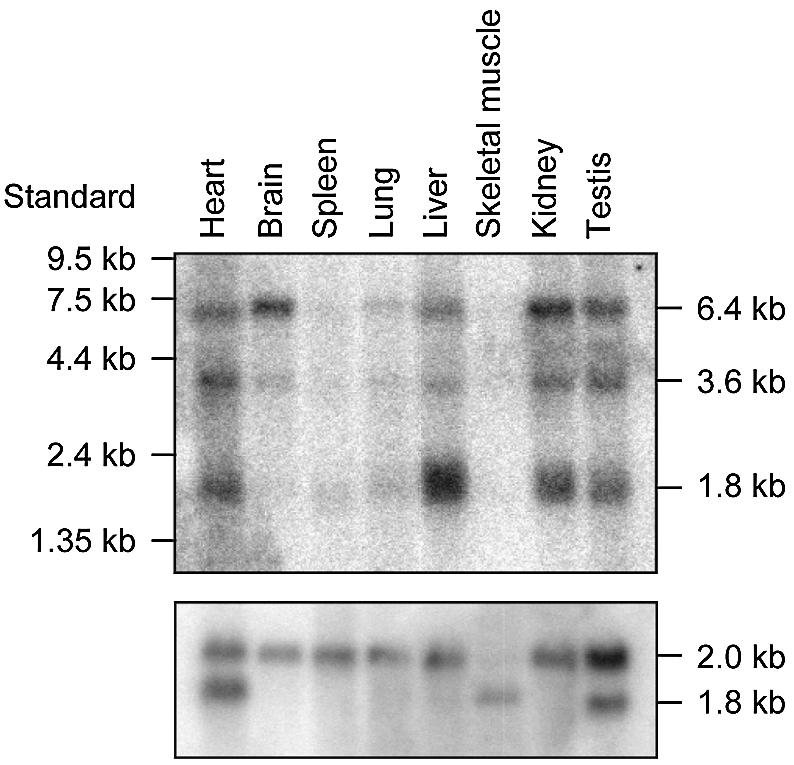
Tissue-specific expression of mEndo V by northern blot analysis. Northern blot containing 2 µg poly(A)+ mRNA purified from multiple normal mouse tissues (Clontech) were hybridised with 32P-labelled cDNA of mEndo V (upper panel) and human β-actin (lower panel).
Endonuclease activity on DNA containing hypoxanthine or uracil
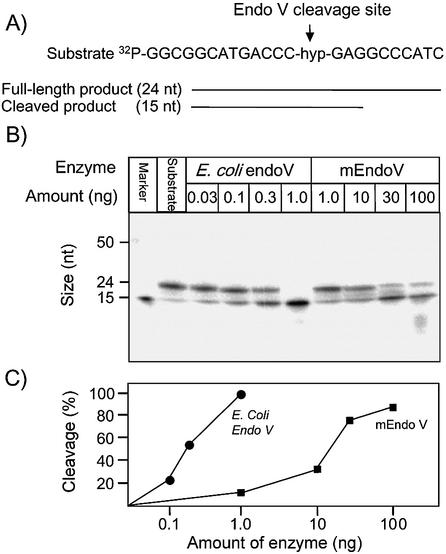
DNA containing hypoxanthine has been shown to be the main substrate for E.coli Endo V. We found significant endonuclease activity of such DNA also by the mEndo V enzyme (Fig. 4). The specific activity of the mouse homologue was approximately 50 times lower than for E.coli Endo V. From the data presented it is estimated that 2 fmol of E.coli Endo V and 100 fmol of mEndo V are required to cleave 1 fmol of a 32P-labelled single-stranded hypoxanthine-containing oligonucleotide in 30 min at 37°C. It should be noted that the background activity of non-specific nicking in DNA also seems to be conserved for the different Endo V orthologues (31). A possible role for this activity in vivo, e.g. in recombination, would explain its conservation. Higher activity was detected on single-stranded than on double-stranded substrate (Fig. 5) (16). Furthermore, the activity towards uracil is greatly reduced compared with hypoxanthine with the following substrate preference: ss DNA containing hypoxanthine > DNA ds containing hypoxanthine ∼ ss DNA containing uracil. The mouse homologue is dependent on Mg2+ and no significant activity was observed towards uracil residues in double-stranded DNA, nor against 8-oxoguanine, C:C mismatches, AP-sites or 5′ flap structures (data not shown).
Figure 4.
Activity of mEndo V on a single-stranded oligonucleotide containing hypoxanthine. (A) Oligonucleotide sequence and predicted reaction products. (B) The oligonucleotide was 32P-labelled at the 5′ end and incubated at 37°C for 30 min with purified mouse and E.coli Endo V at the concentrations indicated and as described in Materials and Methods. Reaction products were analysed by phosphorimaging after electrophoretic separation in a denaturing 20% polyacrylamide gel. Positions and sizes (nucleotides, nt) of reaction products are indicated on the left. Nucleotide residues are numbered from the 5′ end. (C) Cleaved and full-length material from (B) were measured and the fractions of cleaved DNA substrates are indicated.
Figure 5.
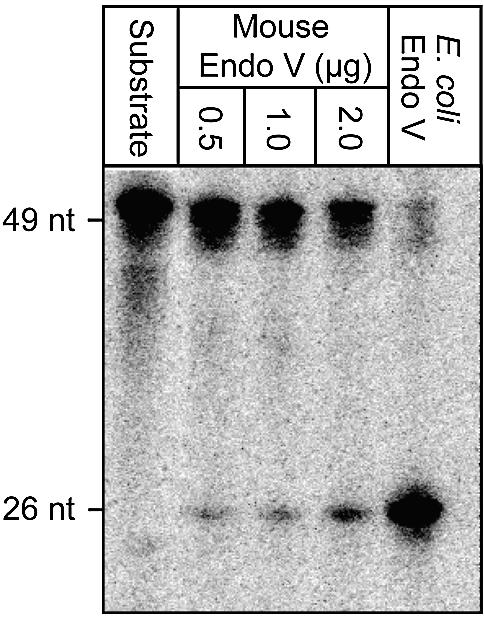
Activity of mEndoV on a double-stranded oligonucleotide containing hypoxanthine paired with thymine. The oligonucleotide was 32P-labelled at the 5′ end of the hypoxanthine-containing strand and incubated at 37°C for 30 min with purified mouse and E.coli endo V at the concentrations indicated and as described in Materials and Methods. Purified endo V from E.coli was used as a positive control for verification of correct endonuclease cut 1 base 3′ to hypoxanthine. The positive controls were cleaved with 10 ng E.coli Endo V. Reaction products were analysed by phosphorimaging after electrophoretic separation in a denaturing 20% polyacrylamide gel. Positions and sizes (nucleotides, nt) of reaction products are indicated on the left. Nucleotide residues are numbered from the 5′ end.
Two proline residues that are conserved from E.coli to Schizosaccharomyces pombe are located within domains required for enzymatic activity (32). These two prolines are replaced with serine (S93) and glutamine (Q133) in the wild type mouse protein. To test whether these changes could affect the enzymatic activity, we designed primers for site-specific mutations converting S93 and Q133 to prolines. Three mutant proteins were produced, the single mutants S93P and Q133P and the double mutant S93P/Q133P. Mutant proteins remained active. However, the activity was not increased relative to wild type (data not shown).
Suppression of the mutator phenotype of E.coli BK 2223 (alkA nfi) by mnfi
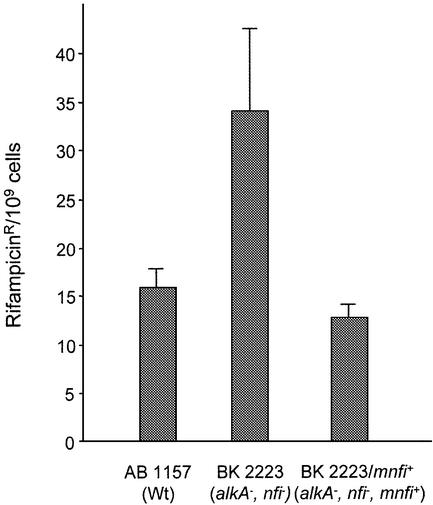
Nitrosative deamination by nitrous acid induces a moderate increase of mutations in an E.coli nfi mutant, whereas there is no increase in spontaneous mutations (33; data not shown). To block both pathways in the repair of hypoxanthine we constructed a double mutant of nfi and alkA, BK2223. AlkA also removes deamination products from DNA and previous experiments have suggested a role of AlkA in mutation avoidance following deamination (34,35). BK2223 (alkA nfi) showed a mild, but significant, mutator phenotype (Fig. 6). The mnfi cDNA, subcloned and introduced into BK2223, caused a complete reversion of the mutator phenotype. The antimutator effect was correlated with expression of mEndo V. However, it is unclear whether hypoxanthine or other unrecognized mEndo V substrates are the origin for the mutations.
Figure 6.
Antimutator effect of mEndo V cDNA in E.coli BK 3222 (alkA mnfi). Mutant E.coli BK 3222 (alkA nfi) cells transformed with pET-28b/mnfi (carrying the mEndo V cDNA) were plated both in the presence and absence of rifampicin. Results are from four independent experiments. Error bars are indicated.
DISCUSSION
Nature of mEndo V
Deamination of adenine to hypoxanthine induces AT to GC transitions in DNA (24). Here, we have described the cloning of a murine cDNA encoding an Endo V homologue that incises DNA at hypoxanthine residues. The enzyme is highly conserved from bacteria to mammals and, to our knowledge, uracil DNA glycosylase is the only other repair enzyme showing a similar degree of conservation. Aravind and co-workers recently identified significant similarity between Endo V and certain motifs of the UvrC protein (Fig. 1B) (25). They further hypothesised potential structural elements and a requirement of two aspartates (D52 and D126 of the murine sequence) and a lysine (K155) for catalysis, and these residues are also conserved in the mouse enzyme. Both UvrC and mEndo V represent damage-specific endonucleases cleaving near, but not next to, the DNA lesions. By analogy to UvrC, which only works efficiently together with UvrA and UvrB, mEndo V could also potentially be stimulated by other protein partners (36). Such stimulation is also observed for several other DNA repair enzymes (37).
Unexpectedly from the high degree of sequence conservation, mEndo V did not show a broad substrate specificity as has been described for E.coli Endo V. Similar data have been described by Liu and Roy for Endo V from A.fulgidus (38). They suggested that the ability of E.coli Endo V to recognize other DNA lesions was acquired later during the course of evolution. Huang and co-workers cloned and expressed a thermostable Endo V homologue in T.maritima and detected some activity on a uracil substrate (26). They propose a less important role of Endo V on these types of lesions compared with the much more active uracil glycosylase that is specific for uracil lesion, and out-compete Endo V for repair.
Base excision repair versus alternative excision repair
Several proteins required for removal of BER intermediates are essential for viability as judged by the embryonic lethality of gene-targeted mice (39). In contrast, no dramatic phenotypes have been produced by targeting DNA glycosylases that initiate the BER pathway by removing the damaged base (40–44). However, all these studies have been followed by discoveries of alternative DNA repair pathways for specific DNA lesions. For example, alternative repair of uracil residues in DNA is accomplished by the SMUG enzyme (43,45), and a novel nuclear and mitochondrial glycosylase for thymine glycols (Tg) was revealed by disruption of the mouse Nth1 gene (44).
There are two major pathways for repair of deaminated bases in DNA: BER initiated by a damage-specific DNA glycosylase, and the AER initiated by Endo V (24). In the case of uracil, which is the most abundant deamination product, the two highly active and abundant glycosylases UDG and SMUG will account for almost all measurable activity for removal of uracil in DNA (43,45). Alkylpurine DNA glycosylases from mammalian cells (Aag/ANPG) efficiently excise hypoxanthine from DNA (34). As judged from in vitro assays of cell-free extracts from mice with a targeted deletion of Aag/ANPG, this is the major, or only, hypoxanthine DNA glycosylase (46,47). Nevertheless, Aag/ANPG null mice are viable with no apparent phenotypic abnormalities. For hypoxanthine removal, mEndo V could represent an important alternative repair pathway. Also, the Ogg1 mice lacking 8-oxoG DNA glycosylase activities appears to have alternative pathways for 8-oxoG repair (42,48).
Repair of deaminated purines is initiated by mEndo V which generates a nick in DNA, leaving the damaged hypoxanthine-containing nucleotide and an intact nucleotide penultimate and ultimate at the 3′ side of the nick, respectively. The subsequent removal of the deaminated purine requires the excision of a small patch of DNA of at least 2 nt. Recent studies have identified 3′–5′ mismatch-specific exonuclease activities associated with mammalian AP-endonuclease I (APE1), and a 3′ flap-specific endonuclease activity associated with Mus81 (49,50). As indicated in Figure 7 [slightly modified from Kow (24)], both APE1 and Mus81 should have the potential of removing a short 3′ mispaired/flapped DNA structure containing the hypoxanthine base. Finally, the gap is sealed by a DNA polymerase and a DNA ligase.
Figure 7.
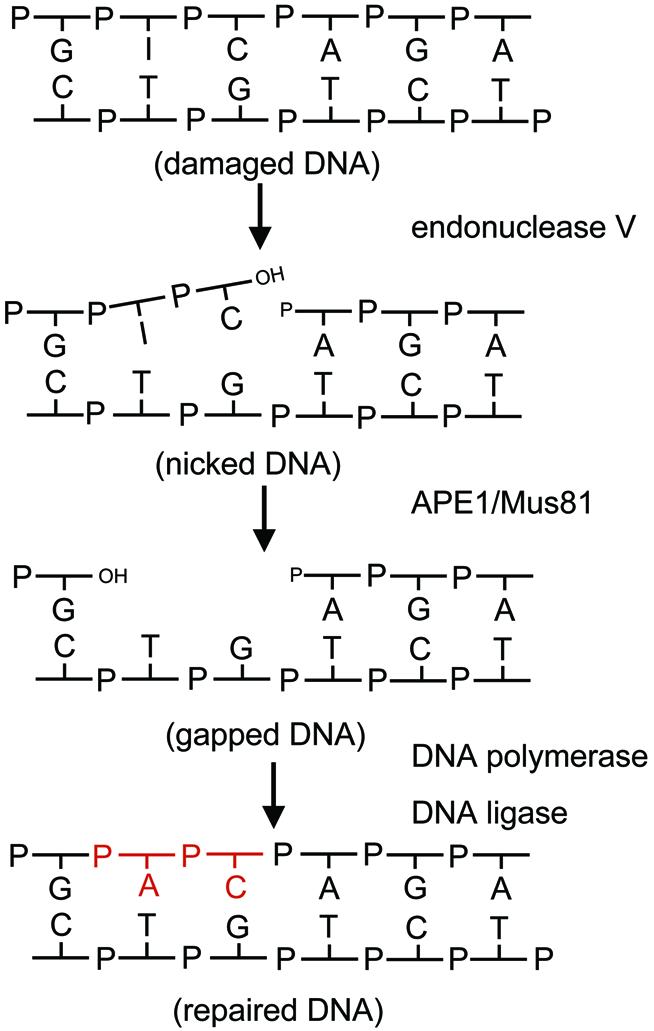
Proposed pathway for mEndo V-initiated DNA repair. The proteins believed to be responsible for the different steps are indicated. The repair process is initiated by cleavage at the second phosphodiester bond 3′ to the lesion by mEndo V. The damaged nucleotide is removed by the 3′ mismatch-specific exonucleolytic activity of the apurinic/apyrimidinic endonuclease APE1 (49), or alternatively by the 3′ flap-specific activity of Mus81 (50). Repair is completed by polymerisation and ligation. Repair synthesis is in red.
We suggest that mEndo V plays an important role in the repair of hypoxanthine in eukaryotic cells and is currently addressing this by constructing gene-targeted knockout mice lacking the mnfi gene. By crossing mnfi negative mice with Aag/ANPG mull mice it should be possible to address the relative importance of mEndo V and/or Aag/ANPG for the repair of deaminated adenines in DNA.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Elisabeth Larsen, Luisa Luna, Knut Ivan Kristiansen and Karen O. Stenersen for their contributions to different aspects of this work, Pål Falnes for invaluable comments and Bernard Weiss (Emory University School of Medicine, Atlanta, GA) for the generous gift of E.coli strains. This work was supported by the Norwegian Cancer Society, the Research Council of Norway, EU (contract QLK1999-2002) and the Norwegian ‘Advanced Research Program’.
REFERENCES
- 1.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 2.Dulbecco R. (1949) Reactivation of ultraviolet inactivated bacteriophage by visible light. Nature, 163, 949–950. [DOI] [PubMed] [Google Scholar]
- 3.Sedgwick B. (1983) Molecular cloning of a gene which regulates the adaptive response to alkylating agents in Escherichia coli. Mol. Genet. Genetics, 191, 466–472. [DOI] [PubMed] [Google Scholar]
- 4.Trewick S.C., Henshaw,T.F., Hausinger,R.P., Lindahl,T. and Sedgwick,B. (2002) Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature, 419, 174–178. [DOI] [PubMed] [Google Scholar]
- 5.Falnes P.O., Johansen,R.F. and Seeberg,E. (2002) AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature, 419, 178–182. [DOI] [PubMed] [Google Scholar]
- 6.Duncan T., Trewick,S.C., Koivisto,P., Bates,P.A., Lindahl,T. and Sedgwick,B. (2002) Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA, 99, 16660–16665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aas P.A., Otterlei,M., Falnes,P.Ø., Vågbø,C.B., Skorpen,F., Akbari,M., Sundheim,O., Bjørås,M., Slupphaug,G., Seeberg,E. and Krokan,H.E. (2003) Human and bacterial oxidative demethylases repair alkylation damage in both DNA and RNA. Nature, 421, 859–863. [DOI] [PubMed] [Google Scholar]
- 8.Lindahl T., Karran,P. and Wood,RD. (1997) DNA excision repair pathways. Curr. Opin. Genet. Dev., 7, 158–169. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl T. (1974) An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl Acad. Sci. USA, 71, 3649–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeberg E., Eide,L. and Bjørås,M. (1995) The base excision repair pathway. Trends Biochem. Sci., 20, 391–397. [DOI] [PubMed] [Google Scholar]
- 11.Sancar A. and Rupp,W.D. (1983) A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell, 33, 249–260. [DOI] [PubMed] [Google Scholar]
- 12.Seeberg E., Nissen-Meyer,J. and Strike,P. (1976) Incision of ultraviolet-irradiated DNA by extracts of E. coli requires three different gene products. Nature, 263, 524–526. [DOI] [PubMed] [Google Scholar]
- 13.Schouten K. and Weiss,B. (1999) Endonuclease V protects Escherichia coli against specific mutations caused by nitrous acid. Mutat. Res., 435, 245–254. [DOI] [PubMed] [Google Scholar]
- 14.Hill-Perkins M., Jones,M.D. and Karran,P. (1986) Site-specific mutagenesis in vivo by single methylated or deaminated purine bases. Mutat. Res., 162, 153–163. [DOI] [PubMed] [Google Scholar]
- 15.Gates F.T. III and Linn,S. (1977) Endonuclease V of Escherichia coli. J. Biol. Chem., 252, 1647–1653. [PubMed] [Google Scholar]
- 16.Demple B. and Linn,S. (1982) On the recognition and cleavage mechanism of Escherichia coli endodeoxyribonuclease V, a possible DNA repair enzyme. J. Biol. Chem., 257, 2848–2855. [PubMed] [Google Scholar]
- 17.Yao M., Hatahet,Z., Melamede,R.J. and Kow,Y.W. (1994) Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′endonuclease, from Escherichia coli. J. Biol. Chem., 269, 16260–16268. [PubMed] [Google Scholar]
- 18.Yao M. and Kow,Y.W. (1994) Strand-specific cleavage of mismatch-containing DNA by deoxyinosine 3′endonuclease from Escherichia coli. J. Biol. Chem., 269, 31390–31396. [PubMed] [Google Scholar]
- 19.Yao M. and Kow,Y.W. (1996) Interaction of deoxyinosine 3′-endonuclease from Escherichia coli with DNA containing deoxyinosine. J. Biol. Chem., 271, 30672–30676. [DOI] [PubMed] [Google Scholar]
- 20.Liu J., He,B., Qing,H. and Kow,Y.W. (2000) A deoxyinosine specific endonuclease from hyperthermophile, Archaeoglobus fulgidus: a homolog of Escherichia coli endonuclease V. Mutat. Res., 461, 169–177. [DOI] [PubMed] [Google Scholar]
- 21.Rupp W.D. (1996) DNA repair mechanisms. In Neidhardt,F.C., Curtiss,R.,III, Ingraham,J.L., Lin,E.C.C, Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd Edn. ASM Press, Washington, D.C., pp. 2277–2294. [Google Scholar]
- 22.Glickman B.W. and Radman,M. (1980) Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc. Natl Acad. Sci. USA, 77, 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieb M. (1987) Bacterial genes mutL, mutS, and dcm participate in repair of mismatches at 5-methylcytosine sites. J. Bacteriol., 169, 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kow Y.W. (2002) Repair of deaminated bases in DNA. Free Radical Biol. Med., 33, 886–893. [DOI] [PubMed] [Google Scholar]
- 25.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J., Lu,J., Barany,F. and Cao,W. (2001) Multiple cleavage activities of endonuclease V from Thermotoga maritima: recognition and strand nicking mechanism. Biochemistry, 40, 8738–8748. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearl L.H. (2000) Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res., 460, 165–181. [DOI] [PubMed] [Google Scholar]
- 29.Olsen A.K., Bjørtuft,H., Wiger,R., Holme,J., Seeberg,E., Bjørås,M. and Brunborg,G. (2001) Highly efficient base excision repair (BER) in human and rat male germ cells. Nucleic Acids Res., 29, 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niederreither K., Harbers,M., Chambon,P. and Dolle,P. (1998) Expression of T:G mismatch-specific thymidine-DNA glycosylase and DNA methyl transferase genes during development and tumorigenesis. Oncogene, 17, 1577–1585. [DOI] [PubMed] [Google Scholar]
- 31.Huang J., Kirk,B., Favis,R., Soussi,T., Paty,P., Cao,W. and Barany,F. (2002) An endonuclease/ligase based mutation scanning method especially suited for analysis of neoplastic tissue. Oncogene, 21, 1909–1921. [DOI] [PubMed] [Google Scholar]
- 32.Huang J., Lu,J., Barany,F. and Cao,W. (2002) Mutational analysis of endonuclease V from Thermotoga maritima. Biochemistry, 41, 8342–8350. [DOI] [PubMed] [Google Scholar]
- 33.Weiss B. (2001) Endonuclease V of Escherichia coli prevents mutations from nitrosative deamination during nitrate/nitrite respiration. Mutat. Res., 461, 301–309. [DOI] [PubMed] [Google Scholar]
- 34.Saparbaev M. and Laval,J. (1994) Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc. Natl Acad. Sci. USA, 91, 5873–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sidorkina O., Saparbaev,M. and Laval,J. (1997) Effects of nitrous acid treatment on the survival and mutagenesis of Escherichia coli cells lacking base excision repair (hypoxanthine-DNA glycosylase-ALK A protein) and/or nucleotide excision repair. Mutagenesis, 12, 23–28. [DOI] [PubMed] [Google Scholar]
- 36.Kumura K., Sekiguchi,M., Steinum,A.L. and Seeberg,E. (1985) Stimulation of the UvrABC enzyme-catalyzed repair reactions by the UvrD protein (DNA helicase II). Nucleic Acids Res., 13, 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klungland A., Höss,M., Gunz,D., Constantiou,A., Clarkson,S.G., Doetsch,P.W., Bolton,P.H., Wood,R.D. and Lindahl,T. (1999) Base excision repair of oxidative DNA damage activated by XPG protein. Mol. Cell, 3, 33–42. [DOI] [PubMed] [Google Scholar]
- 38.Liu X. and Roy,R. (2002) Truncation of amino-terminal stimulates activity of human endonuclease III (hNTH1). J. Mol. Biol., 321, 265–276. [DOI] [PubMed] [Google Scholar]
- 39.Friedberg E.C. and Meira,L.B. (2000) Database of mouse strains carrying targeted mutations in genes affecting cellular responses to DNA damage. Version 4. Mutat. Res., 459, 243–274. [DOI] [PubMed] [Google Scholar]
- 40.Engelward B.P., Dreslin,A., Christensen,J., Huszar,D., Kurahara,C. and Samson,L. (1996) Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO J., 15, 945–952. [PMC free article] [PubMed] [Google Scholar]
- 41.Minowa O., Arai,T., Hirano,M., Monden,Y., Nakai,S., Fukuda,M., Itoh,M., Takano,H., Hippou,Y., Aburatani,H., Masumura,K., Nohmi,T., Nishimura,S. and Noda,T. (2000) Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl Acad. Sci. USA, 97, 4156–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klungland A., Rosewell,I., Hollenbach,S., Larsen,E., Daly,G., Epe,B., Seeberg,E., Lindahl,T. and Barnes,D.E. (1999) Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl Acad. Sci. USA, 96, 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsen H., Rosewell,I., Robins,P., Skjelbred,C.F., Andersen,S., Slupphaug,G., Daly,G., Krokan,H.E., Lindahl,T. and Barnes,D.E. (2000) Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell, 5, 1059–1065. [DOI] [PubMed] [Google Scholar]
- 44.Takao M., Kanno,S., Shiromoto,T., Hasegawa,R., Ide,H., Ikeda,S., Sarker,A.H., Seki,S., Xing,J.Z., Le,X.C. et al. (2002) Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J., 21, 3486–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haushalter K.A., Todd Stukenberg,M.W., Kirschner,M.W. and Verdine,G.L. (1999) Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol., 9, 174–185. [DOI] [PubMed] [Google Scholar]
- 46.Engelward B.P., Weeda,G., Wyatt,M.D., Broekhof,J.L., de Wit,J., Donker,I., Allan,J.M., Gold,B., Hoeijmakers,J.H. and Samson,L.D. (1997) Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl Acad. Sci. USA, 94, 13087–13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hang B., Singer.B., Margison,G.P. and Elder,R.H. (1997) Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc. Natl Acad. Sci. USA, 94, 12869–12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LePage F., Klungland,A., Barnes,D.E., Sarasin,A. and Boiteux,S. (2000) Transcription coupled repair of 8-oxoguanine in murine cells: the Ogg1 protein is required for repair in nontranscribed sequences but not in transcribed sequences. Proc. Natl Acad. Sci. USA, 97, 8397–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chou K.-M. and Cheng,Y.-C. (2002) An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature, 415, 655–659. [DOI] [PubMed] [Google Scholar]
- 50.Constantinou A., Chen,X.B., McGowan,C.H. and West,S.C. (2002) Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J., 21, 5577–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]