Abstract
Optimization of probe design for array-based experiments requires improved predictability of oligonucleotide hybridization behavior. Currently, designing oligonucleotides capable of interacting efficiently and specifically with the relevant target is not a routine procedure. Multiple examples demonstrate that oligonucleotides targeting different regions of the same RNA differ in their hybridization ability. The present work shows how thermodynamic evaluations of oligo-target duplex or oligo self-structure stabilities can facilitate probe design. Statistical analysis of large sets of hybridization data reveals that thermodynamic evaluation of oligonucleotide properties can be used to avoid poor RNA binders. Thermodynamic criteria for the selection of 20 and 21mers, which, with high probability, interact efficiently and specifically with their targets, are suggested. The design of longer oligonucleotides can also be facilitated by the same calculations of ΔG°T values for oligo-target duplex or oligo self-structure stabilities and similar selection schemes.
INTRODUCTION
Many techniques of molecular biology require interaction of oligonucleotides with DNA or RNA as a basic step. Oligonucleotide array gene expression monitoring or antisense-mediated gene down-regulation are examples. Poor interaction of an oligonucleotide with its target can significantly affect the efficiency of these processes.
Oligo intra- or inter-molecular structure can compete with oligo-target duplex formation and result in low hybridization intensity. Extensive secondary structure of the target can also limit this efficiency. The present work addresses whether thermodynamic consideration of the relative stability of oligo-target duplexes and both oligo intra- and inter-molecular self-structures, without consideration of target secondary structure, can be sufficient for selection of oligo-probes that are efficient target binders.
Oligonucleotide scanning arrays permit monitoring of the efficiency of hybridization simultaneously for many, or all, target regions of a particular RNA. RNA target affinity can also be measured for oligonucleotides of different length and self-structure in one hybridization experiment (1–7), so these arrays can be very useful for statistical study of oligonucleotide-related factors that influence an oligonucleotide’s ability to hybridize with target RNA or DNA.
Software for the calculation of the thermodynamic factors that are important for the prediction of oligonucleotide hybridization behavior was created some time ago (8). The program OligoWalk calculates thermodynamic factors related to stabilities of oligonucleotide-target duplex, oligonucleotide intra- or inter-molecular self-structures and target RNA or DNA secondary structure. However, a statistical study that demonstrates the usefulness of thermodynamic calculations for the prediction of array hybridization has not been reported for large sets of experimental data. This work fills this gap and suggests a new scheme for employment of thermodynamic calculations to predict the potential of an oligonucleotide to bind to its target.
MATERIALS AND METHODS
Oligonucleotide datasets of hybridization experiments
Three experimental datasets were used for statistical analysis. For obtaining dataset 1, Affymetrix GeneChip.TM.HIV PRT produced by Affymetrix Corporation, Santa Clara, CA was used. For obtaining datasets 2 and 3, a chip produced by Oxford Gene Technology, Oxford, UK was used. For all datasets, in vitro transcribed non-fragmented HIV-1 RNA was used for the hybridization experiments. The hybridization intensities of oligo probes targeting every overlapping 20 nucleotide fragments of the relevant RNA were collected for dataset 1. The hybridization intensities of oligo-probes targeting every overlapping 20 nucleotide fragments and every 21 nucleotide fragments of the relevant RNA were collected for dataset 2. The hybridization intensities of oligo-probes targeting every overlapping nucleotide fragment ranging in size from 3 to 21 nucleotides of the relevant RNA were collected for dataset 3. The experiments were performed with oligonucleotides immobilized on a solid support. The experimental conditions used to obtain the datasets are given in Table 1.
Table 1. Summary of differences and similarities between hybridization experiments that were performed to obtain the datasets.
| Dataset 1 | Dataset 2 | Dataset 3 | |
|---|---|---|---|
| Target RNA length | 1041 nt | 290 nt | 290 nt |
| Temperature of hybridization | 37°C | 25°C | 25°C |
| Length of the oligo-probe | 20 nt | 20 and 21 nt | 3–21 nt |
| RNA target labeled with | fluorescein | P33 | P33 |
| Concentration of target RNA in experiment | 26.3 nM | 2.5 nM | 2.5 nM |
| Number of experimental data points in the dataset | 1021 | 541 | 6156 |
Thermodynamic calculations
Calculations of thermodynamic properties of oligonucleotides were done with the help of newly created and pre-existing software. For the oligonucleotides that were involved in the experiments performed at 37°C, the program OligoWalk from the package RNA structure 3.7 was used (8) (http://128.151.176.70/RNAstructure.html). For the oligonucleotides that were involved in the experiments performed at 25°C, Excel macro ‘OligoAnal’ was created (available for downloading at http://www.gesteland.genetics.utah.edu/members/olgaM/OligAnal.ZIP). Using thermodynamic parameters for the nearest neighbor model (9–16), this macro can produce relevant ΔG°T values (oligonucleotide inter-molecular and oligo-target pairing potentials) for each analyzed oligonucleotide. For calculation of oligonucleotide intra-molecular pairing potentials at 25°C, the program mfold version 3.0 (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form4.cgi) with thermodynamic parameters from the version 3.1 was used (15) (http://www.bioinfo.rpi.edu/~zukerm/dna/credit.html). Nucleic acid conformation was assumed to be linear and the ionic conditions were set at 1 M Na+. In the program output, the positive values of ΔG°25 were changed to 0.
Statistical analysis
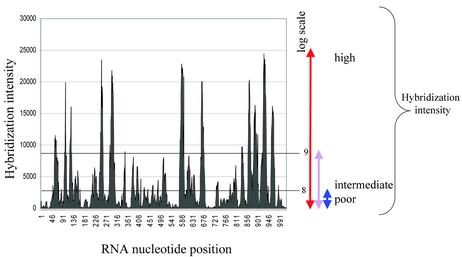
Statistical tools from Excel (Microsoft, Inc.) were used for correlation analysis (t-test) and scatter-plot data presentations. The oligonucleotides in both datasets were categorized into groups according to their hybridization intensity. Two thresholds for oligonucleotide categorizations were created: the upper threshold and the lower threshold. In both datasets the thresholds were set identically. The upper thresholds for logarithmic values of RNA hybridization intensity were set as 9, the lower thresholds for logarithmic values of RNA hybridization intensity were set as 8.
Thermodynamic filtration
The process of selection of oligo-probe sets using several thermodynamic criteria was called thermodynamic filtration.
RESULTS AND DISCUSSION
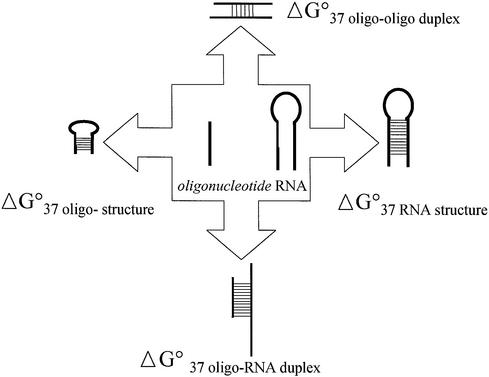
A schematic illustration of the competing molecular interactions relevant to oligo-RNA binding is shown in Figure 1. To estimate how thermodynamic evaluations of the stability of an RNA-DNA duplex and the stability of oligonucleotide self-structures can be related to oligonucleotide RNA binding properties, we analyzed two datasets of hybridization experiments performed with oligonucleotide scanning arrays.
Figure 1.
Scheme of oligonucleotide-target RNA interaction, which shows thermodynamic factors that can influence oligonucleotide RNA hybridization intensity.
Data for the first set were taken from the literature (17), while data for the second set were kindly provided by Dr Verhoef from Oxford Gene Technology. The differences and similarities between the two hybridization experiments that were performed to obtain the two datasets are summarized in Table 1 (see also Materials and Methods).
The results of the oligonucleotide scanning array hybridization experiment that were used for creation of dataset 1 are presented graphically in Figure 2. A sharp contrast is evident between different oligonucleotides in their ability to hybridize with target RNA. By statistical analysis, we explored if this hybridization intensity contrast can be related to oligonucleotide thermodynamic properties.
Figure 2.
RNA hybridization intensity profile for the set of oligonucleotides (20mers) that was used for creation of the first dataset. The hybridization intensity is shown for each oligonucleotide in relation to its position in the target RNA. For statistical analysis, the oligonucleotides were categorized into groups according to hybridization intensity. Blue represents the group with low hybridization intensity; violet, intermediate; and red with high.
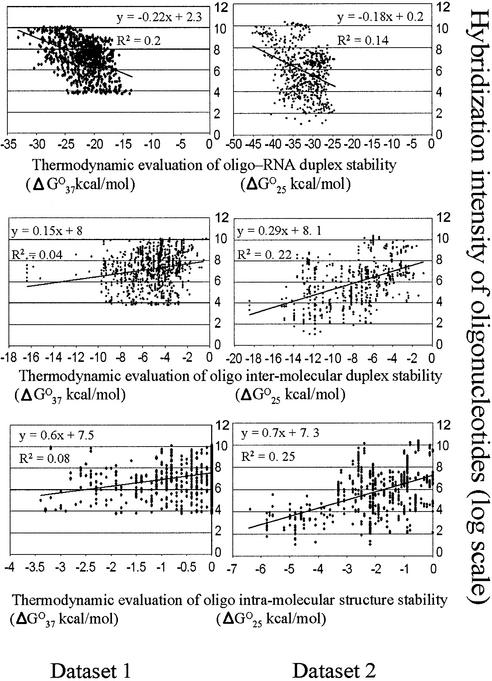
ΔG°T values for competing molecular interactions relevant to oligo-RNA binding were calculated for each oligonucleotide in the datasets based on thermodynamic parameters of the nearest neighbor model (see thermodynamic calculations in Materials and Methods). Correlation analyses (t-tests) of both datasets were performed (Table 2). For datasets 1 and 2, significant correlations (P < 0.01) were detected between the experimental hybridization intensity and the theoretical ΔG°T values associated with stability of oligonucleotide self- structures and oligonucleotide-RNA duplexes.
Table 2. Correlations between thermodynamic properties of oligonucleotides and their experimental RNA affinity.
| correlation coefficients for absolute values | Dataset 1 | Dataset 2 |
|---|---|---|
| ΔG°T oligo duplex with RNA versus ln(hybridization intensity) | 0.46 | 0.30 |
| ΔG°T oligo intra-molecular structure versus ln(hybridization intensity) | –0.28 | –0.52 |
| ΔG°T oligo inter-molecular structure versus ln(hybridization intensity) | –0.2 | –0.40 |
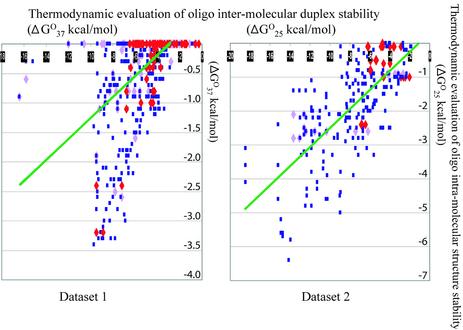
Scatter plots (Fig. 3) illustrate the relationship between the experimental intensity of hybridization signals and thermodynamic properties of oligonucleotides from the two datasets. Since the slope of the trend line in scatter plots indicates the existence of a correlation between two variables, a positive correlation is evident between the absolute value of the thermodynamic evaluation of oligonucleotide-RNA duplex stability and intensity of DNA-RNA hybridization (Fig. 3, top plots). In contrast, the slopes of the trend lines indicate that there is a negative correlation between the absolute ΔG°T values of oligonucleotide self-pairing and the intensity of DNA-RNA hybridization (Fig. 3, middle and bottom plots). An attempt to adjust mfold program input to improve evaluation of oligonucleotide intra-molecular self-structure by changing sodium or magnesium concentrations was not successful. Surprisingly, even though the experiments were performed at 100 mM Na+, the best correlations between theoretical and experimental values were achieved when the ionic conditions in the program input were set at 1 M Na+.
Figure 3.
Relationship between calculated thermodynamic parameters and hybridization intensity of the oligonucleotides with their target RNA.
The question of the reliability of mfold and how well it actually predicts the experimental melting curves of any self-complementary sequence is still open. The existence of a significant correlation between mfold calculated ΔG°T values of oligonucleotide self-pairing and the intensity of DNA-RNA hybridization indicates that mfold can be employed for the prediction of stability of oligo probe self-structures. Can this prediction be optimized? The current version of mfold complies nearest-neighbor as well as hairpin, bulge, internal and multi-branched loop parameters from different sources (http://www.bioinfo.rpi.edu/~zukerm/dna/credit.html). Perhaps thermodynamic parameters derived from one reliable modern source would be better. Obtaining optimized thermodynamic parameters can likely lead to a significant improvement of mfold prediction performance.
The next issue is how to employ the statistical findings described above and how to find thermodynamic thresholds for selection of oligonucleotide sets with a high proportion of efficient RNA binders. In this work, a trial and error approach was used. Variable, arbitrarily chosen cut-off points for all three thermodynamic criteria were applied, and the proportions of efficient RNA binders in the filtered oligo subset were determined for each combination. A combination that delivered the oligo subset with a high proportion of efficient RNA binders was found.
In future, when more experimental data will be available for statistical analysis, trial and error approaches for finding thermodynamic cut-off points, as used in this study, will likely be substituted by rational weighting of each thermodynamic parameter employing an equation suggested in an earlier publication (8).
In this study, the oligonucleotides in both datasets were categorized into groups according to the experimental intensity of DNA-RNA hybridization using certain arbitrarily chosen thresholds as described in Materials and Methods (Fig. 2). The group of efficient RNA binders includes oligonucleotides with DNA-RNA hybridization intensity higher than the upper threshold. The group of poor binders includes oligonucleotides with values worse than the lower threshold. Finally, the group of intermediate binders includes oligonucleotides with DNA-RNA hybridization intensity between the two thresholds.
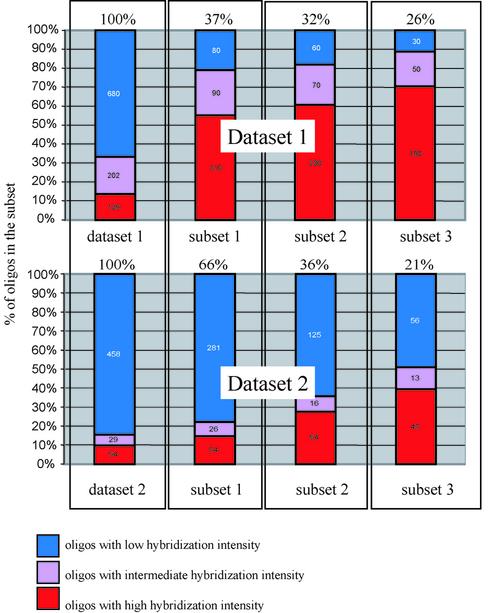
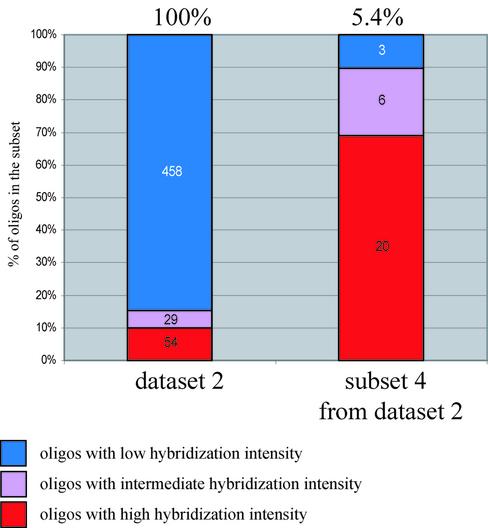
The proportions of efficient RNA binders among oligonucleotides were calculated in both datasets (Fig. 4). These proportions were also calculated for the probe subsets that were created using only oligonucleotides with certain thermodynamic properties. The proportions of efficient RNA binders were larger in the subsets that were predicted to form more stable oligonucleotide-RNA duplexes in comparison with the datasets of all probes (Fig. 4). These proportions become even larger if oligonucleotides that are able to form self-structures of specified stability are excluded (Fig. 4). The process of selection of oligo-probe sets using several thermodynamic criteria can deliver a high proportion of efficient RNA binders. This process can be called thermodynamic filtration.
Figure 4.
Categorization of oligonucleotides into subsets according to their thermodynamic properties. The percentage of oligonucleotides with RNA hybridization intensity higher than the defined threshold in each subset is shown. The color code is the same as in Figure 2. Numbers of oligonucleotides in each subgroup are printed on color-highlighted parts of the columns. The proportion of oligonucleotides in each subset versus the total number of oligonucleotides in the relevant dataset is shown above each column. Subset 1 contains oligo-probes that can form stable duplexes with RNA ΔG°25 ≤ –29 kcal/mol; subset 2 contains the oligo-probes that can form stable duplexes with RNA ΔG°25 ≤ –29 kcal/mol with unstable intermolecular oligo self-structures ΔG°25 ≥ –8 kcal/mol; and subset 3 contains oligo-probes that can form stable duplexes with RNA ΔG°25 ≤ –29 kcal/mol but which form both unstable inter- and intra-molecular self-structures (ΔG°25 ≥ –8 kcal/mol for inter-molecular structures and ΔG°25 ≥ –1.1 kcal/mol for intra-molecular structures).
It is interesting that filtering out of the oligonucleotides that form intermolecular structures of specified stability increases the proportion of efficient RNA binders. It likely indicates that oligo-oligo intermolecular interaction can occur during hybridization experiments even though the oligonucleotides are covalently attached through their ends to a solid support.
Both thermodynamic evaluations of oligonucleotide intra- and inter-molecular self-interacting properties are strongly correlated to each other. The steep slopes of the trend lines of both scatter plots (Fig. 5), and highly significant correlation co-efficients (0.54 for the first dataset and 0.66 for the second dataset, p < 0.001) demonstrate this point. Sometimes, if two variables are highly correlated, only one is sufficient for predictive purposes. However, it was found that both thermodynamic criteria for self-structure forming potentials are simultaneously useful for efficient discrimination into subsets that mainly contain efficient or poor RNA binders (Fig. 5).
Figure 5.
Relationship between thermodynamic evaluations of oligonucleotide inter- and intra-molecular pairing potentials (x and y axes, respectively). Blue squares represent the group with low hybridization intensity; violet, intermediate; and red with high.
Here we describe the analysis of experimental datasets that combine hybridization data for two different RNAs. The temperature used for the hybridization experiments that yielded dataset 1 was 37°C, and for datasets 2 and 3, it was 25°C. For the subsets with the highest proportion of efficient RNA binders, the filtration (ΔG°T) cut-offs for DNA-RNA duplex stability are different; –35 kcal/mol for the experiments that were performed at 25°C and –29 kcal/mol for the experiments that were performed at 37°C (Figs 4 and 6). Temperature, concentration of target RNA, and ionic conditions of hybridization are the factors that can influence optimal filtration cut-off points. This work, however, demonstrates that, regardless of differences in the experimental conditions, thermodynamic filtration involving criteria of oligo-RNA duplex and oligo self-structure stabilities can be helpful for efficient elimination of poor RNA binders.
Figure 6.
Categorization of oligonucleotides into subsets according to their thermodynamic properties. Two sets of oligonucleotides in dataset 2 are shown. The first set represents all oligonucleotides in the dataset, while the second represents only the fraction with certain thermodynamic properties. The proportion of oligonucleotides in each subset versus the total number of oligonucleotides in dataset 2 is shown above each column. The percentage of oligonucleotides with RNA hybridization intensity higher than the defined threshold in each set is also shown. The color code is the same as in Figure 2. Numbers of oligonucleotides in each subgroup are printed on color-highlighted parts of the columns. Subset 4 contains oligo-probes that can form stable duplexes with RNA ΔG°25 ≤ –35 kcal/mol but which form both unstable inter- and intra-molecular self-structures (ΔG°37 ≥ –8 kcal/mol for inter-molecular structures and ΔG°37 ≥ –1.1 kcal/mol for intra-molecular structures).
Correlations between thermodynamic factors and experimental binding of oligonucleotides with RNA or DNA targets were found previously (8,18–21). This work demonstrates that selection of oligonucleotides using a thermodynamic filtration approach can increase, by several-fold, the proportion of DNA oligonucleotides that can bind RNA efficiently. For gene expression monitoring with the Affymetrix Chips, a similar approach can minimize the number of oligo-probes needed per gene, thereby increasing the number of different genes detectable on each chip. This should significantly raise the sensitivity and decrease the cost of such analyses.
This study suggests thermodynamic criteria for elimination of oligo-probes that are very likely poor RNA binders. The criteria are based on statistical analysis of hybridization of short 20 and 21mer probes. Longer oligo-probes in the range from 50 to 150mers can be also used for array experiments. Similar statistical analysis and thermodynamic filtration schemes can be applied to hybridization data produced with long oligo-probes. It can reveal optimal thermodynamic criteria for long oligo-probe design at different experimental conditions.
Target RNA secondary structure may play an important role in selection of the most potent RNA binders. Figure 4 demonstrates that many efficient RNA binders are lost during the steps of thermodynamic filtration performed in this study. It is likely that taking into consideration thermodynamic properties related to RNA secondary structure could diminish this loss. However, the analysis performed in this study reveals that oligo-probes with a high probability of being efficient RNA binders in array experiments can still be selected without consideration of the thermodynamic properties related to RNA secondary structure.
Thermodynamic filtration can dramatically increase the proportion of oligonucleotides with efficient RNA binding. As illustrated in Figures 4 and 6, the proportions of efficient binders among the oligonucleotides in both experimental datasets are small (∼14% for dataset 1 and 10% for dataset 2). However, these proportions can be increased up to 70%, or even more, if a set of oligonucleotides that form stable duplexes with RNA and little self-structure are selected.
Removing subsets of oligonucleotides with low probability of hybridizing efficiently with their RNA target is important but is not the only problem relevant to probe design algorithms. Another important issue is elimination of the oligonucleotides that can cross hybridize with other genes. Modern algorithms include a BLAST search for dealing with the problem. The limitations of BLAST or similar programs are due to the absence of well-defined criteria for the prediction of hybridization. For optimal solution of this problem, an efficient thermodynamic predictor of hybridization intensity is needed.
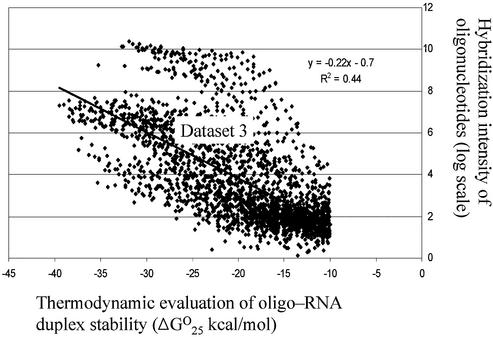
Statistical analysis was performed to find out what range of values of ΔG°T of DNA-RNA duplex stability of oligo-probes with little self-structure is optimal for this purpose. Two subsets from dataset 3 were created. Both subsets include only oligo-probes with little self-structure (ΔG°25 ≥ –8 kcal/mol for inter-molecular structures and ΔG°25 ≥ –1.1 kcal/mol for intra-molecular structures). The first subset includes oligo-probes with ΔG°25 values of DNA-RNA duplex stability ranging from 0 to –10 kcal/mol. The second subset includes oligo-probes with ΔG°25 values of DNA-RNA duplex stability ranging from –10 to –40 kcal/mol. The correlation between the values of hybridization intensities of the oligo-probes and the values of ΔG°25 of DNA-RNA duplex stability was absent in the first subset and was highly significant in the second with a correlation coefficient of 0.7. The scatter plot with correlation trend-line for subset 2 from dataset 3 is presented in Figure 7.
Figure 7.
Relationship between calculated values of ΔG°25 of DNA-RNA duplex stability and hybridization intensities of the oligonucleotides with their target RNA for the subset of oligo-probes with little self-structure from dataset 3.
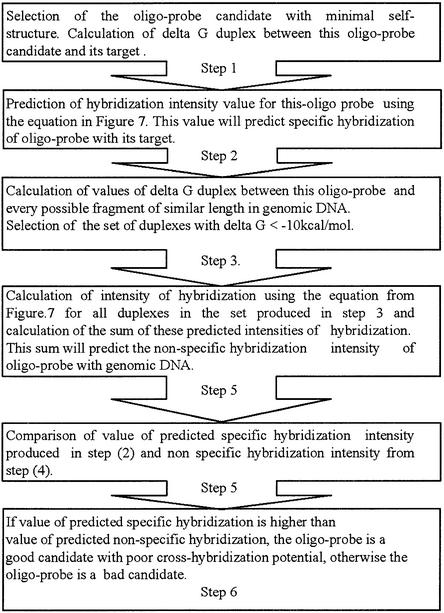
Statistical analysis reveals that the calculated value of ΔG°25 of DNA-RNA duplex stability in the range from –10 to –40 kcal/mol can be considered as a predictor of oligo-probe hybridization intensity for the molecules with minimum self-structure. So the intensity of cross hybridization between these oligo-probes and partially complementary target sequences can be predicted after calculation of thermodynamic values. The scheme for this prediction is shown in Figure 8. This scheme should be helpful for the discrimination of oligo-probes into candidates with strong or weak cross-hybridization potentials. The application of this scheme is limited to the conditions in which dataset 3 was obtained.
Figure 8.
Scheme for evaluation of cross-hybridization potentials of oligo-probe candidates.
In conclusion, statistical analysis of large sets of hybridization data suggests that thermodynamic evaluation of oligonucleotide properties can be used to avoid poor RNA binders. This analysis also indicates that thermodynamic evaluation of oligonucleotide properties can be directly linked to the solution of the cross-hybridization problem. So thermodynamic calculations can be helpful for optimization of hybridization sensitivity and specificity of the oligo-probes. However, much more experimental data and software optimization are needed before cross-hybridization potentials of the oligo-probes can be reliably calculated for the range of hybridization conditions.
Acknowledgments
ACKNOWLEDGEMENTS
We greatly appreciate the critical provision of unpublished data by Dr K. Verhoef and Professor E. Southern without which we would not have been able to perform this work. We are grateful to Dr Baranov for constructive comments and Mr B. Moore for invaluable computer help. R.F.G. was supported by NIH grant R01-GM61200 and J.F.A. by NIH grant GM48152.
REFERENCES
- 1.Williams J.C., Case-Green,S.C., Mir,K.U. and Southern,E.M. (1994) Studies of oligonucleotide interactions by hybridization to arrays: the influence of dangling ends on duplex yield. Nucleic Acids Res., 22, 1365–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Southern E.M., Case-Green,S.C., Elder,J.K., Johnson,M., Mir,K.U., Wang,L. and Williams,J.C. (1994) Arrays of complementary oligonucleotides for analyzing the hybridization behavior of nucleic acids. Nucleic Acids Res., 22, 1368–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Southern E.M. (2001) History and overview. Methods Mol. Biol., 170, 1–15. [DOI] [PubMed] [Google Scholar]
- 4.Sohail M., Akhtar,S. and Southern,E.M. (1999) The folding of large RNAs studied by hybridization to arrays of complementary oligonucleotides. RNA, 5, 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sohail M. and Southern,E.M. (2001) Using oligonucleotide scanning arrays to find effective antisense reagents. Methods Mol. Biol., 170, 181–199. [DOI] [PubMed] [Google Scholar]
- 6.Sohail M., Hochegger,H., Klotzbucher,A., Guellec,R.L., Hunt,T. and Southern,E.M. (2001) Antisense oligonucleotides selected by hybridization to scanning arrays are effective reagents in vivo. Nucleic Acids Res., 29, 2041–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Southern E., Mir,K. and Shchepinov,M. (1999) Molecular interactions on microarrays. Nature Genet., 21, 5–9. [DOI] [PubMed] [Google Scholar]
- 8.Mathews D.H., Burkard,M.E., Freier,S.M., Wyatt,J.R. and Turner,D.H. (1999) Predicting oligonucleotide affinity to nucleic acid targets. RNA, 5, 1458–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.SantaLucia J. Jr, Allawi,H.T. and Seneviratne,P.A. (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry, 35, 3555–3562. [DOI] [PubMed] [Google Scholar]
- 10.Allawi H.T. and SantaLucia,J.,Jr (1997) Thermodynamics and NMR of internal G.T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 11.Allawi H.T. and SantaLucia,J.,Jr (1998) Thermodynamics of internal C.T mismatches in DNA. Nucleic Acids Res., 26, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest neighbor thermodynamic parameters for internal G.A mismatches in DNA. Biochemistry, 37, 2170–2179. [DOI] [PubMed] [Google Scholar]
- 13.Allawi H.T. and SantaLucia,J.,Jr (1998) Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: sequence dependence and pH effects. Biochemistry, 37, 9435–9444. [DOI] [PubMed] [Google Scholar]
- 14.Peyret N., Seneviratne,P.A., Allawi,H.T. and SantaLucia,J.,Jr (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 15.SantaLucia J. Jr, (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugimoto N., Nakano,S., Katoh,M., Matsumura,A., Nakamuta,H., Ohmichi,T., Yoneyama,M. and Sasaki,M. (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry, 34, 11211–11216. [DOI] [PubMed] [Google Scholar]
- 17.Shannon K. and Wolber,P. (2001) Method for evaluating oligonucleotide probe sequences. US patent 6,251,588.
- 18.Walton S.P., Stephanopoulos,G.N., Yarmush,M.L. and Roth,C.M. (1999) Prediction of antisense oligonucleotide binding affinity to a structured RNA target. Biotechnol. Bioeng., 65, 1–9. [PubMed] [Google Scholar]
- 19.Jayaraman A., Walton,S.P., Yarmush,M.L. and Roth,C.M. (2001) Rational selection and quantitative evaluation of antisense oligonucleotides. Biochim. Biophys. Acta, 1520, 105–114. [DOI] [PubMed] [Google Scholar]
- 20.Walton S.P., Stephanopoulos,G.N., Yarmush,M.L. and Roth,C.M. (2002) Thermodynamic and kinetic characterization of antisense oligodeoxynucleotide binding to a structured mRNA. Biophys. J., 82, 366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luebke K.J., Balog,R.P. and Garner,H.R. (2003) Prioritized selection of oligodeoxyribonucleotide probes for efficient hybridization to RNA transcripts. Nucleic Acids Res., 31, 750–758. [DOI] [PMC free article] [PubMed] [Google Scholar]