Abstract
ClpX and ClpA are molecular chaperones that interact with specific proteins and, together with ClpP, activate their ATP-dependent degradation. The chaperone activity is thought to convert proteins into an extended conformation that can access the sequestered active sites of ClpP. We now show that ClpX can catalyze unfolding of a green fluorescent protein fused to a ClpX recognition motif (GFP-SsrA). Unfolding of GFP-SsrA depends on ATP hydrolysis. GFP-SsrA unfolded either by ClpX or by treatment with denaturants binds to ClpX in the presence of adenosine 5′-O-(3-thiotriphosphate) and is released slowly (t1/2 ≈ 15 min). Unlike ClpA, ClpX cannot trap unfolded proteins in stable complexes unless they also have a high-affinity binding motif. Addition of ATP or ADP accelerates release (t1/2 ≈ 1 min), consistent with a model in which ATP hydrolysis induces a conformation of ClpX with low affinity for unfolded substrates. Proteolytically inactive complexes of ClpXP and ClpAP unfold GFP-SsrA and translocate the protein to ClpP, where it remains unfolded. Complexes of ClpXP with translocated substrate within the ClpP chamber retain the ability to unfold GFP-SsrA. Our results suggest a bipartite mode of interaction between ClpX and substrates. ClpX preferentially targets motifs exposed in specific proteins. As the protein is unfolded by ClpX, additional motifs are exposed that facilitate its retention and favor its translocation to ClpP for degradation.
ATP-dependent proteases combine a molecular chaperone and a self-compartmentalizing protease to create a complex that catalyzes regulated degradation of specific proteins (1–4). In Clp proteases and the proteasome, the proteolytic sites lie within internal aqueous chambers formed by the isologous interactions between two rings of seven subunits each (5–7). Because the proteolytic chambers are accessible only through narrow axial channels, only short unstructured polypeptides can be degraded by the proteolytic component acting alone. Degradation of proteins with significant secondary and tertiary structure requires the molecular chaperone activity of the regulatory ATPases that associate with the proteolytic core.
Escherichia coli ClpA and ClpX belong to the Clp/Hsp100 family of ATP-dependent molecular chaperones and can function as autonomous chaperones (1, 8). Our laboratories have been investigating the mechanism by which the chaperone activities of ClpA and ClpX contribute to the degradation of proteins by the holoenzyme complexes, ClpAP and ClpXP (9–11). ClpA and ClpX are related but quite distinct ATPases (12). ClpA is a fusion of two nonhomologous ATPase domains, whereas ClpX has a single ATPase domain. Also, the N-terminal regions of ClpA and ClpX do not share sequence homology and differ substantially in size and predicted structure. The predicted secondary structures of ClpA and ClpX in the ATPase and C-terminal domains are very similar and place them within the AAA superfamily (13).
ClpA and ClpX interact directly with proteins and function in substrate discrimination. Mutations in ClpX block degradation of such proteins as E. coli RpoS (14), phage P1 PhD (15), and λO protein (16, 17), whereas mutations in ClpA specifically stabilize E. coli MazE (18) and engineered N-end rule substrates, such as Leu-β-galactosidase (19). There is some overlap in substrate recognition between ClpA and ClpX; both promote degradation of proteins carrying a C-terminal extension of 11 amino acids encoded by the ssrA transfer mRNA (20, 21). SsrA tags are added cotranslationally to incomplete polypeptides bound to stalled ribosomes, and the resulting tagged protein is released from the ribosome and degraded by ClpXP or ClpAP (22, 23). The differences in sequence and structure of ClpA and ClpX not only should contribute to recognition of different substrates but also may reflect a difference in the mechanisms of interaction with proteins and delivery of the proteins to ClpP.
Because access to the active sites of ClpP is limited by narrow axial channels, it has been assumed that the function of the chaperone activity of ClpA or ClpX is to unfold protein substrates. The ability of ClpA to promote unfolding of a stable folded protein was recently shown by the Horwich group, using the green fluorescent protein carrying an SsrA C-terminal extension (GFP-SsrA) (24). Unfolded GFP-SsrA released from ClpA was trapped by using a mutant of GroEL that tightly binds unfolded proteins even in the presence of ATP. While unfolding and release of unfolded proteins supported a role for this activity in protein degradation, it was important to demonstrate that the ClpA complexed with ClpP could carry out protein unfolding. In this study, we have used GFP-SsrA to show that ClpX, as well as ClpA, catalytically unfolds GFP-SsrA and can translocate the unfolded protein to proteolytically inactive ClpP, where it remains in an unfolded state.
Experimental Procedures
Reagents.
ATP and ADP were obtained from Sigma. Adenosine-5′-O-(3-thiotriphosphate) (ATP[γS]) was obtained from Boehringer. Carbobenzoxy (Cbz)-Leu-Tyr-chloromethyl ketone (CMK) was purchased from Bachem. [3H]Succinimidylpropionic acid and [3H]formaldehyde were obtained from DuPont/NEN. Monoclonal anti-His6 antibody was from CLONTECH. Peroxidase-conjugated anti-rabbit and anti-mouse IgGs were obtained from Life Technologies.
Protein Preparations.
ClpA, ClpP, and the mutant ClpP-S111C were purified by published methods (25) and stored at −80°C in buffer B [50 mM Tris⋅HCl, pH 7.5/0.1 M KCl/1 mM EDTA/10% (vol/vol) glycerol]. For purification of His6-ClpP, cells in buffer B without EDTA were broken at 20,000 psi (138 kPa) in a French pressure cell and centrifuged at 30,000 × g for 45 min at 4°C. The supernatant extract was passed over Talon resin (CLONTECH), which was then washed with buffer containing 10 mM imidazole; the bound protein was eluted with 0.2 M imidazole in the same buffer. The His6-ClpP was further purified on a Mono Q (10/10) column (Amersham Pharmacia Biotech) in buffer B, from which it was eluted in 0.3 M KCl. His6-GFP-SsrA was purified on a Talon resin as described for His6-ClpP, without the final Mono Q step. His6-GFP-SsrA is a derivative of GFP with MRGSHHHHHH fused to the N terminus and GSAANDENYALAA fused at the C terminus. The clone for His6-GFP-SsrA and the His6-GFP-SsrA/DD protein, in which DD replaces the terminal AA residues, were provided by C. Herman (University of California, Berkeley). λO protein was purified as described (26) and stored at −80°C in buffer H (25 mM Hepes/KOH, pH 7.5/0.1 M KCl/10% glycerol). GroEL-trap was prepared as described (27) and stored at 4°C. The clone for expression of GroEL-D87K (GroEL-trap) was provided by A. Horwich (Yale University, New Haven, CT).
Protein Modification.
Proteolytically inactive ClpP-CMK was prepared by treating ClpP (3–5 mg/ml) in buffer H on ice with two separate aliquots of 100 μM Cbz-Leu-Tyr-CMK for 30 min each. Excess reagent was removed on a Sephadex G-50 column in buffer B. Inactive DIP-ClpP was prepared with diisopropyl fluorophosphate as described (28). Tritiated GFP-SsrA and CI-SsrA were prepared by treating 1–3 mg/ml solutions in buffer H with 100 μCi (1 μCi = 37 kBq) of [3H]succinimidylpropionate for 1 h on ice and removing unincorporated reagent on Sephadex G-50. λO and α-casein were labeled by reductive methylation as described previously (29).
Analytical Methods.
Fluorescence was measured at constant temperature with an Aminco–Bowman spectrofluorimeter (series 2). GFP-SsrA fluorescence was measured with excitation at 395 nm (4-nm band width) and emission at 509 nm (4- to 8-nm band width). Immunochemical detection was performed after transfer of proteins to 0.22-μm pore nitrocellulose membranes (Millipore) and blocking with 5% nonfat dried milk. Detection of peroxidase-conjugated second antibodies was by chemiluminescence with the ECL reagents (Amersham Pharmacia Biotech).
Assays.
Protein degradation was assayed by conversion of radioactively labeled proteins to products soluble in 10% trichloroacetic acid, using slight modifications of previous condition (29). Degradation was also measured by densitometry of Coomassie blue-stained bands after SDS/PAGE. Continuous assays for unfolding and degradation of GFP-SsrA were conducted by mixing 1–5 μM GFP-SsrA, various concentrations of ClpX or ClpA, and 1 μM ClpP (where appropriate) in 50 mM Tris⋅HCl, pH 7.5/0.2 M KCl/20–30 mM MgCl2/10% glycerol, and nucleotide as desired. When ATP regeneration was required, 30 mM creatine phosphate and 250 μg/ml creatine kinase were included. Prewarmed solutions were transferred to a microcuvette (12- to 25-μl volume) and readings were initiated within 15–20 sec of addition of the initiating reagent. For trapping assays, GFP-SsrA was treated with either 10–50 mM HCl (pH < 1.5) or 8 M guanidine⋅hydrochloride (Gdn⋅HCl). The denatured protein was diluted into assay solutions to initiate refolding with or without different proteins to trap the unfolded protein. Gdn⋅HCl concentrations did not exceed 0.25 M and the final pH was 7.5 in all cases.
Results
Trapping of Unfolded Substrates by ClpX.
The ability of ClpX to bind unfolded proteins was tested with GFP-SsrA unfolded in acid. When added to neutral pH buffer, acid-denatured GFP-SsrA rapidly refolded (t1/2 ≈ 90 sec) and regained fluorescence (Fig. 1A). Refolding was usually greater than 80%. When GroEL-trap, which binds unfolded proteins without releasing them, was present in the neutralization buffer, >90% of the unfolded GFP-SsrA was trapped in a nonfluorescent form (Fig. 1A). If ClpX and ATP[γS] were present, between 60% and 80% of unfolded GFP-SsrA was trapped in the unfolded state (Fig. 1A). ClpX did not trap unfolded GFP-SsrA in the absence of nucleotide (data not shown). ClpX added at any time during refolding trapped a portion of the unfolded GFP-SsrA (data not shown), but trapping did not exceed 80%. Thus, some GFP-SsrA folds by a pathway that generates intermediates not recognized by ClpX.
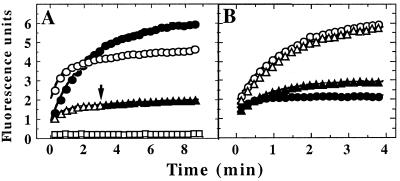
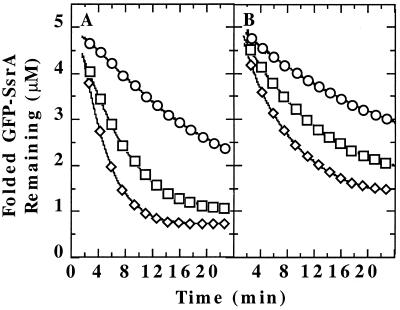
Figure 1.
Binding of specific unfolded proteins by ClpX. (A) GFP-SsrA was unfolded in acid and diluted to a final concentration of 0.5 μM in pH 7.5 buffer at 37°C with various additions; refolding was monitored by fluorescence expressed in arbitrary units. ●, No additions; ▵, 5 μM ClpX and 2 mM ATP[γS]; ○, 5 μM ClpX and 2 mM ATP[γS] with 8 μM λO added before GFP-SsrA; and □, 10 μM GroEL-trap. At the arrow, 8 μM λO was added to GFP-SsrA trapped with ATP[γS] and ClpX (▴. (B) GFP-UV (circles) or GFP-SsrA/DD (triangles) was denatured in acid and diluted into buffer with 2 mM ATP[γS] and either 5 μM ClpX (open symbols) or 5 μM ClpA (filled symbols). Both forms of GFP refolded with similar kinetics in the absence of ClpX and ClpA (data not shown).
Trapping of Unfolded Proteins by ClpX May Require the Same Recognition Motifs Exposed in Natural Substrates.
ClpX was not able to trap GFP that did not carry an SsrA tag (Fig. 1B). To test the specificity for the SsrA tag, we used GFP-SsrA/DD, in which the two C-terminal alanine residues are replaced with aspartate residues. Such derivatives have lower affinity for ClpX and ClpA and are not degraded by ClpAP or ClpXP (30). GFP-SsrA/DD could not be trapped by ClpX (Fig. 1B). Thus, ClpX has low affinity for unfolded regions of proteins, and the presence of a specific binding motif is needed to promote a binding mode characterized by slow release that allows trapping. In contrast, ClpA trapped unfolded GFP and GFP-SsrA/DD (Fig. 1B). The inability of ClpX to interact with unfolded proteins may be a general property, because ClpX was also inefficient in binding Gdn⋅HCl-denatured α-casein, and ClpXP degraded <5% of unfolded rhodanese (data not shown). In contrast, ClpAP degrades unfolded rhodanese (31), indicating that ClpA interacts more strongly than ClpX with unfolded proteins.
Release of Unfolded Substrates from ClpX Requires ATP Hydrolysis.
Trapping of unfolded GFP-SsrA was blocked by prior addition of excess competitive substrate to ClpX (Fig. 1A). However, when the competitor was added after trapping in the presence of ATP[γS], no change in fluorescence was observed (Fig. 1A), indicating that the rate of release of the unfolded protein from ClpX was very low. To investigate the effect of ATP hydrolysis on binding of unfolded proteins to ClpX, we first formed trapped complexes in ATP[γS] (Fig. 2A, ▴) and then added ATP; no change in fluorescence was observed (Fig. 2A, ▵). Addition of excess λO after the ATP led to a release of the trapped GFP-SsrA (Fig. 2A, ○). When ATP[γS] was omitted entirely and only ATP was present, unfolded GFP-SsrA appeared to be trapped by ClpX (Fig. 2A, ▴) and could be released upon addition of excess λO (Fig. 2A, □). We concluded that in the presence of ATP, equilibrium favors binding of the unfolded protein to ClpX, but the unfolded protein was continually released and rebound; excess competitive substrate prevented reassociation of the released protein.
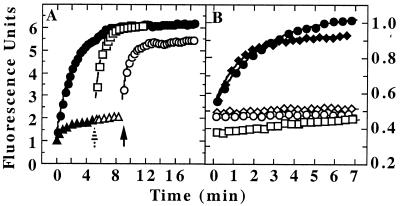
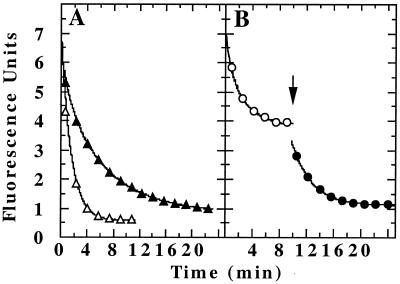
Figure 2.
ATP-dependent release of unfolded GFP-SsrA from ClpX. (A) Refolding of acid-denatured GFP-SsrA was monitored by fluorescence after dilution into neutral buffer (●). For trapping, the buffer contained 5 μM ClpX and either 2 mM ATP[γS] or 5 mM ATP plus an ATP-regenerating system. The initial fluorescence increase and plateau were essentially the same with ATP and ATP[γS] (▴). After 5 min (striped arrow), 7 μM λO was added to the sample trapped in ATP, and fluorescence was monitored (□). Also after 5 min (striped arrow), 8 mM ATP was added to the sample trapped in ATP[γS] (▵), and after an additional 4 min (solid arrow), 7 μM λO was added to the same sample and measurements were continued (○). Fluorescence was corrected for dilution caused by the additions. (B) Unfolded GFP-SsrA was trapped with ClpX plus ATP[γS] as in A. Aliquots were diluted into buffer containing 2 mM ATP[γS] (□); 5 mM ADP (♦); 5 mM ADP plus 10 μM GroEL-trap (⋄); 5 mM ATP plus 7 μM λO (●); or 5 mM ATP plus 7 μM λO plus 10 μM GroEL-trap (○).
Dilution of ClpX/GFP-SsrA complexes into excess ADP caused rapid release of the bound unfolded protein (Fig. 2B) even without added competitor, indicating that ADP promotes a conformation of ClpX that has low affinity for unfolded proteins. We confirmed by light scattering that ADP did not lead to dissociation of ClpX (data not shown). To address the question of whether GFP-SsrA was folded before release from ClpX, trapped complexes were diluted into either ADP or ATP plus λO in the absence (Fig. 2B, closed symbols) or presence (Fig. 2B, open symbols) of GroEL-trap. GroEL-trap completely blocked the increase in fluorescence in a single round of ADP- or ATP-mediated release, indicating that the protein is unfolded at the time of release from ClpX.
Slow Dissociation of Unfolded Substrates from ClpX.
Native GFP-SsrA binds weakly to ClpX (Kd ≈ 3 μM; data not shown) and dissociates during gel filtration in the presence of ATP[γS] (Fig. 3A). When complexes of ClpX and unfolded GFP-SsrA were passed through a gel filtration column in the presence of ATP[γS], GFP-SsrA was recovered in the fractions with ClpX hexamers (Fig. 3B). Thus, binding of unfolded GFP-SsrA to ClpX in the presence of ATP[γS] is very tight and release is slow. Fluorescence and absorbance measurements indicated that GFP-SsrA remained unfolded while bound to ClpX (data not shown). To determine whether slow release of unfolded proteins was a general property of ClpX, an unrelated substrate, λO, was tested. λO was unfolded in Gdn⋅HCl and diluted into buffer with ClpX and ATP[γS]. For comparison, binding of native λO was also measured. In the presence of ATP[γS], both native and denatured λO bound ClpX and were retained by ultrafiltration; prior addition of excess competitor prevented binding of both forms (Fig. 3C). However, if the competitor was added after binding of λO to ClpX, most unfolded λO remained bound to ClpX after 5 min (gray bar), whereas folded λO was completely exchanged in the same time (white bar). Thus, unfolding a protein allows it to bind more tightly to ClpX and causes it to be released very slowly in the presence of ATP[γS]. When ATP was present, both unfolded and folded λO were rapidly released from ClpX (Fig. 3C).
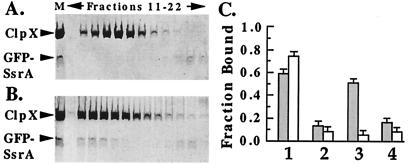
Figure 3.
Slow release of unfolded protein from ClpX in the presence of ATP[γS]. (A) ClpX (5 μM) was mixed with native GFP-SsrA (2.5 μM) in the presence of ATP[γS] and passed through a Superdex 200 gel filtration column (Amersham Pharmacia Biotech) in the presence of 1 mM ATP[γS]. Fractions (0.08 ml) were collected at 1-min intervals from 11 to 22 min, prepared for SDS/PAGE, separated on 10% polyacrylamide gels in SDS, and stained with Coomassie blue. Lane M, mixture before gel filtration. (B) ClpX (5 μM) was mixed with acid-denatured GFP-SsrA (2.5 μM) in the presence of ATP[γS], passed through Superdex 200, and analyzed as in A. (C) Ultrafiltration assays to compare ClpX binding of Gdn⋅HCl-denatured [3H]λO (gray columns) and folded native λO (white columns). ClpX (0.2 μM) was mixed with 0.1 μM λO in buffer with 2 mM ATP[γS] (column sets 1, 2, and 3) or 4 mM ATP (column set 4). The solutions were passed through a Microcon 100 ultrafiltration membrane (Amicon) by centrifugation for 5 min at 4°C, and the fraction of [3H]λO retained with ClpX was determined. Column set 1, binding of [3H]λO alone. Set 2, A 20-fold molar excess of unlabeled λO was added as a competitor at the same time as [3H]λO. Sets 3 and 4, after the [3H]λO had bound to ClpX for 5 min, the complex was incubated for 1 min with a 20-fold molar excess unlabeled λO before ultrafiltration.
Unfolding of GFP-SsrA by ClpX.
The protein unfolding activity of ClpX was tested by incubating GFP-SsrA with various amounts of ClpX in the presence of ATP. In each case, there was a time-dependent decrease in fluorescence which leveled off before all of the GFP-SsrA was unfolded (Fig. 4A). Unfolding was dependent on hydrolyzable forms of ATP (data not shown) and was blocked by prior addition of λO (Fig. 4B). Each reaction appeared to reach a steady state between ClpX-mediated unfolding and spontaneous refolding of GFP-SsrA. To confirm this possibility, GFP-SsrA unfolding was allowed to proceed until no further decrease was seen, and then excess competitor was added. Fluorescence increased rapidly (Fig. 4B), indicating that the competitor either blocked steady-state unfolding of GFP-SsrA or prevented rebinding of unfolded GFP-SsrA released from ClpX. The rate of refolding (t1/2 ≈ 45 sec) was slightly faster than the rate of spontaneous refolding of acid-denatured GFP-SsrA. Thus, in the steady state with ATP present, release of the unfolded protein from ClpX is not rate limiting and in fact must be very fast. Addition of ADP to steady-state reactions led to a rapid increase in fluorescence (data not shown). As seen with GFP-SsrA unfolded by ClpA, GroEL-trap could bind GFP-SsrA unfolded by ClpX. When GroEL-trap was added after the fluorescence had reached a plateau with ClpX, there was an immediate further decrease in fluorescence (Fig. 4B). The decrease in fluorescence without a lag confirms that, in the steady state, unfolded GFP-SsrA is rapidly released from ClpX, which is then free to interact with native GFP-SsrA for another cycle of unfolding. GroEL-trap also prevented any increase in fluorescence upon addition of λO (Fig. 4B), confirming that the GFP-SsrA was unfolded at the time of its release from ClpX.
Figure 4.
Unfolding of GFP-SsrA by ClpX. (A) GFP-SsrA (5 μM) was incubated at 37°C with various amounts of ClpX plus 5 mM ATP and an ATP-regenerating system. Fluorescence traces with no ClpX (●) and 0.2 (⋄), 0.5 (□), 1.0 (○), and 3.5 μM ClpX (▵) are shown. (B) GFP-SsrA (1 μM) was incubated at 37°C with 1–2 μM ClpX and 5 mM ATP; a typical result of unfolding with 1 μM ClpX is shown (○). As a control, a competitive substrate, 7 μM λO, was mixed with 1 μM ClpX before adding the GFP-SsrA (□). At the arrow, one of the following was added to separate unfolding reaction mixtures: 8 μM λO (▴), 10 μM GroEL-trap (●), or 8 μM λO and 10 μM GroEL-trap (■).
Catalytic Unfolding of GFP-SsrA by ClpX and ClpA.
Catalytic unfolding of GFP-SsrA by ClpX was shown by using substoichiometric amounts of ClpX in the presence of excess GroEL-trap. ClpX was able to unfold ≥10-fold molar excess of GFP-SsrA within 15 min at 37°C. Under the conditions used in Fig. 5A, the rate of unfolding was proportional to the ClpX concentration. Since GFP-SsrA was about 60% saturating (data not shown), we estimated that the Vmax for unfolding and release is about one GFP-SsrA per min per ClpX hexamer. ClpA also unfolds GFP-SsrA catalytically (Fig. 5B). With 5 μM GFP-SsrA and 0.2 μM ClpA, about 10 molecules of GFP-SsrA were unfolded per ClpA hexamer within 20 min. The Km for GFP-SsrA was higher with ClpA (data not shown), and we estimated that the Vmax for unfolding by ClpA is comparable to that for ClpX.
Figure 5.
Catalytic unfolding by ClpX and ClpA. (A) GFP-SsrA (5 μM) was incubated in buffer containing ClpX, 5 mM ATP (plus regenerating system), and 10 μM GroEL-trap. ClpX concentrations were 0.2 (○), 0.5 (□), and 1 μM (⋄). (B) Catalytic unfolding of GFP-SsrA by ClpA. Conditions were the same as for A but 0.2 (○), 0.5 (□), and 1 μM (⋄) ClpA were used.
Degradation of GFP-SsrA by ClpXP and ClpAP.
When native GFP-SsrA was incubated with active ClpXP (Fig. 6A) or ClpAP complexes (data not shown), fluorescence was rapidly lost. Degradation of the protein was confirmed by monitoring loss of stained protein after SDS/PAGE (Fig. 6A Inset), and in separate experiments by acid solubilization of 3H-labeled GFP-SsrA by ClpXP (data not shown). The rate of degradation of GFP-SsrA by ClpXP was 3–4 times higher than the rate of unfolding and release to GroEL-trap by ClpX alone.
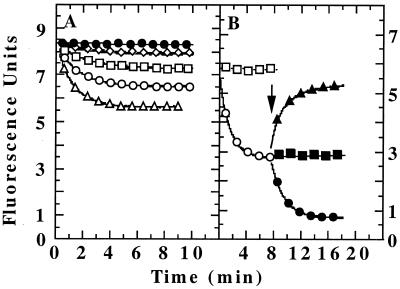
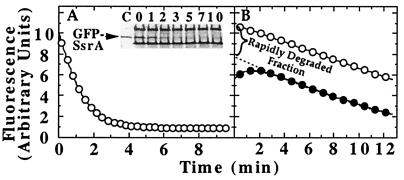
Figure 6.
Degradation of GFP-SsrA by ClpX. (A) Identical solutions with 1 μM each of GFP-SsrA, ClpX, and ClpP in degradation buffer with 10 mM ATP were incubated at 37°C. For one, the fluorescence was monitored, and, for the other, aliquots were withdrawn at various times and prepared for SDS/PAGE. The GFP-SsrA remaining was detected by silver staining (Inset). Numbers above lanes are the times of incubation with ClpXP. (B) GFP-SsrA was subjected to two different treatments, and the kinetics of degradation in the presence of 0.5 μM ClpP and 5 mM ATP was measured. In the first, GFP-SsrA was unfolded by 1 μM ClpX and diluted 10-fold into degradation buffer (●). In the second, GFP-SsrA was incubated without ClpX and then diluted into degradation buffer containing 0.1 μM ClpX in addition to the other components (○).
To test whether unfolded GFP-SsrA bound to ClpX was degraded more efficiently than free GFP-SsrA, we allowed the unfolding reaction with ClpX alone to proceed to a steady state and then diluted the mixture 20-fold into assay buffer containing ClpP (Fig. 6B). We compared the rate of degradation of this pretreated GFP-SsrA to that of native GFP added directly to ClpXP. A portion of GFP-SsrA unfolded by ClpX (≥60%) was degraded rapidly (i.e., in the time of mixing) determined by extrapolation of the linear degradation curve to time 0 (Fig. 6B). In contrast, there was no fast phase of degradation for native GFP-SsrA, and the initial degradation rate was similar to the slower second phase seen with ClpX-unfolded GFP-SsrA (Fig. 6B). Our data suggest that once proteins are unfolded translocation to ClpP for degradation is favored over dissociation from ClpX.
GFP-SsrA Remains Unfolded When Translocated to Inactive ClpP.
To measure the translocation of substrates to ClpP without degradation, we used ClpXP and ClpAP complexes made with proteolytically inactive ClpP. Fig. 7 shows the loss of fluorescence when GFP-SsrA was treated with ClpXP or ClpAP made with proteolytically inactive DIP-ClpP. Similar results were obtained with wild-type ClpP inactivated with a peptide chloromethyl ketone (ClpP-CMK) and with ClpP-S111C. Loss of fluorescence was faster and more extensive than seen with ClpX plus GroEL-trap (compare with Fig. 5A) but was slower than seen when active ClpP was used (compare with Fig. 6A).
Figure 7.
Translocation of unfolded GFP-SsrA to ClpP. (A) ClpXPin or ClpAPin was assembled by incubating ClpX (1 μM) or ClpA (1 μM) for 30 sec with 1 μM DIP-ClpP and 5 mM ATP (plus regenerating system). Fluorescence was monitored after addition of 1 μM GFP-SsrA to the ClpXPin complexes (▵) or the ClpAPin complexes (▴). (B) GFP-SsrA (5 μM) was unfolded with ClpXPin made with 1 μM ClpX and 1.2 μM inactive ClpP-CMK (○). After the fluorescence decrease reached a plateau, 10 μM GroEL-trap was added (●).
ClpXP Containing Translocated Substrate Can Continue to Unfold Proteins.
We wanted to know whether complexes of ClpXP with translocated substrate bound could continue to unfold proteins. We assembled ClpXP with 1 μM ClpX and 1.2 μM inactive ClpP-CMK. ClpP was in excess over ClpX to ensure that no free ClpX was available. GFP-SsrA (5 μM) was added and translocation was allowed to proceed until the fluorescence reached a plateau (Fig. 7B). The fluorescence decrease suggested that more than one equivalent of GFP-SsrA was unfolded by the complex. To confirm this possibility, GroEL-trap was then added. As seen in Fig. 7B, there was an immediate decrease in fluorescence, equivalent to 4.5 times the amount of ClpXP complex. Given that ClpXP complexes dissociate slowly under these conditions (S.K.S. and M.R.M., unpublished results), these data indicate that ClpXP complexes with substrate translocated to ClpP (see Fig. 8) can unfold GFP-SsrA and release the unfolded protein to GroEL-trap.
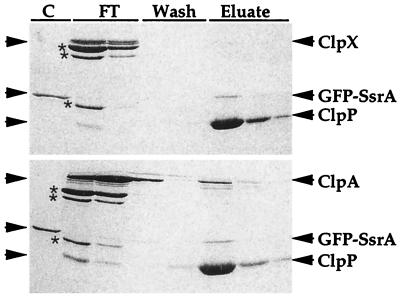
Figure 8.
Isolation of His6-ClpP-substrate complexes by metal-chelate chromatography. GFP-SsrA (1 μM) was incubated for 5 min at 37°C in 50 μl of buffer with 5 mM ATP and ClpXPin assembled with 1 μM ClpX and 1 μM His6-ClpP-CMK. The solution was diluted 20-fold in buffer without Mg and nucleotide and applied to a 200-μl bed of metal-chelate affinity gel. The column was washed once with 1 ml of buffer without Mg and nucleotide and twice with 1 ml each of the same buffer plus 10 mM imidazole. The bound protein was eluted with three washes with 200 μl of buffer with 200 mM imidazole. Proteins were precipitated with 10% trichloroacetic acid, prepared for SDS/PAGE, electrophoresed on a 10% polyacrylamide gel in SDS, and stained with Coomassie blue (Upper). The procedure was repeated but 1 μM ClpA was used instead of ClpX (Lower). Asterisks indicate proteins present in the creatine kinase added to reactions. Lane markings: C, GFP-SsrA; FT, flow-through of sample and buffer wash; Wash, 10 mM imidazole washes; Eluate, proteins eluted with 0.2 M imidazole.
Isolation of Unfolded GFP-SsrA Bound to ClpP in the Absence of ClpX.
Previous studies had shown that substrates could be co-immunoprecipitated with DIP-ClpP after translocation by ClpAPin and removal of the ClpA (9). To isolate complexes of bound substrate with ClpP completely free of ClpX, we used ClpP carrying a C-terminal His6 extension and inactivated it with peptide CMK. After incubation of ClpX-(His6-ClpP-CMK) (Fig. 8 Upper) or ClpA-(His6-ClpP-CMK) (Fig. 8 Lower) complexes with GFP-SsrA in the presence of ATP, the mixtures were diluted to dissociate the ClpAP or ClpXP complexes and passed over a metal-chelate affinity column. After washing, the columns were treated with 0.2 M imidazole to elute bound protein. Both ClpX and ClpA were quantitatively separated from His6-ClpP, and GFP-SsrA was eluted in the fraction with His6-ClpP (Fig. 8). In control experiments, free GFP-SsrA, which has a His6-tag at the N terminus, was eluted in the 10 mM imidazole wash (data not shown). The major portion (60–90%) of the GFP-SsrA was recovered with His6-ClpP, and the bound GFP-SsrA was not fluorescent, indicating that it remained in an unfolded state.
Discussion
The ATPase components of ATP-dependent proteases have evolved to express both autonomous chaperone activity and an unfolding activity that is coupled to protein translocation and degradation by associated proteases. The present study demonstrates that ClpX, as has been shown for ClpA, functions independently to unfold proteins, which can be released and allowed to refold. The released protein can be captured by another chaperone, GroEL, and thus is unfolded at the time of release. This capturing mechanism was exploited for practical purposes in our study, but proteins unfolded and released from ClpX in vivo could, in principle, interact with any of a variety of chaperones or proteases within the cell. Our data suggest that, in the presence of ClpP, most substrates unfolded by ClpX will be degraded.
In this study, we have shown that ClpX binds specific unfolded substrates in the presence of a poorly hydrolyzable ATP analog, ATP[γS]. Binding properties were similar whether GFP-SsrA was unfolded by ClpX or ClpX was used to trap acid-denatured GFP-SsrA. The spectroscopic properties of the bound unfolded protein resemble those of acid-denatured GFP-SsrA, indicating that proteins unfolded by ClpX lose some or all of their tertiary structure. When released by ClpX the protein can be trapped by GroEL, indicating that it is still in a nonnative state, although we cannot rule out some degree of refolding while the protein is bound. Release of the unfolded protein occurs when ATP hydrolysis is allowed. ClpX may resemble other molecular chaperones, which cycle between an “open” state with low affinity for substrates and a “closed” high-affinity state in response to ATP binding and hydrolysis (32). As with ClpA, however (33), it is the ATP-liganded state of ClpX, mimicked in our experiments by ATP[γS], that appears to bind proteins. Another important difference is the release of bound unfolded protein from ClpX upon ADP addition. Since the ADP state is thought to favor retention of unfolded proteins on chaperones such as DnaK and GroEL (32), the effect of ADP on ClpX may reflect an influence on binding sites for specific recognition motifs that make a unique contribution to substrate interactions with ClpX. Alternatively, ADP could weaken hexamer interactions, thereby lowering affinity for bound protein.
ClpX interaction with both folded and unfolded proteins is facilitated by specific recognition motifs. The slower release from ClpX when proteins are unfolded implies that additional regions exposed in the unfolded state can interact with ClpX and contribute to the overall binding energy. Competition between specific substrates and unfolded proteins for binding to ClpX could reflect both proteins binding to the same site or one protein sterically hindering binding of another. Thus, at present we cannot distinguish whether the additional interactions upon substrate unfolding involve unique sites on ClpX subunits or the same site on different subunits of the ClpX oligomer. ClpX disassembles complexes of MuA tetramers bound to DNA (34, 35) and can dissolve λO aggregates (36). In each case, ClpX may recognize an exposed motif in either the N-terminal (37) or the C-terminal region (38) and begin to unfold the protein from the end. These data imply that ClpX may not function as a general chaperone for unfolded proteins but rather may be targeted to a subset of proteins that have recognition motifs of various degrees of integrity at their termini. In fact, ClpX makes a relatively minor contribution to degradation of abnormal protein in E. coli (17).
Earlier studies showed that the rate-limiting step in the degradation pathway precedes peptide bond cleavage (28). In our experiments, the rate of unfolding by ClpX or ClpA alone is slower than the rate of degradation of GFP-SsrA by ClpXP and ClpAP. Because unfolding is necessary for degradation, these data imply that ClpP stimulates the unfolding reaction. From our data it is not possible to distinguish between unfolding and translocation as the rate-limiting step. Translocation measured by degradation was 2–3 times faster than translocation measured by transfer to inactive ClpP. One possibility is that cleavage of the protein and dissociation of the products clear the active site chamber and the access channel, allowing more efficient translocation of the protein. An implication of this result is that in the presence of excess substrate in vivo, some unfolded protein could be released from ClpXP without degradation. In support of this idea, we have found that addition of excess competitive substrate after translocation of CI-SsrA results in release of a fraction (up to 50%) of the protein associated with ClpP (R.G. and M.R.M., unpublished results). A major unresolved issue concerning the function of Clp proteases is the balance between release and degradation of bound unfolded proteins. Our data suggest that for unfolded GFP-SsrA bound to ClpX, translocation to ClpP and degradation occurs much faster than release.
These studies were initiated to define differences between ClpX and ClpA in the kinetics of unfolding, release, or translocation of substrates. ClpA has two nonhomologous functional ATPase domains, whereas ClpX has a single ATPase domain. The two ATPase domains of ClpA have been shown to make different contributions to chaperone activity and to activation of ClpP-dependent proteolysis (10, 39). A major difference we observed between ClpX and ClpA was in the ability to bind unfolded proteins. ClpX was inefficient at trapping unfolded proteins that did not also carry a specific recognition motif. ClpA was able to trap several different unfolded proteins (31). A more comprehensive survey of unfolded proteins will be needed before we can be certain that this difference in binding ability is general and whether it is related to the additional ATPase domain of ClpA. There was little difference in the proportions of bound substrate either released or degraded for the two enzymes. Thus, the single chaperone domain of ClpX appears to function as efficiently in translocating unfolded proteins to ClpP.
Note Added in Proof:
While this manuscript was under review another article appeared describing unfolding activity of ClpX (40).
Acknowledgments
We thank Yan Ning Zhou, National Cancer Institute, for providing the plasmid for expressing His6-ClpP and Christophe Herman for providing the GFP-SsrA/DD protein.
Abbreviations
- GFP
green fluorescent protein
- ATP[γS]
adenosine 5′-O-(3-thiotriphosphate)
- CMK
chloromethyl ketone
- Gdn⋅HCl
guanidine⋅hydrochloride
References
- 1.Schirmer E C, Glover J R, Singer M A, Lindquist S. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 2.Schmidt M, Lupas A N, Finley D. Curr Opinion Chem Biol. 1999;3:584–591. doi: 10.1016/s1367-5931(99)00013-7. [DOI] [PubMed] [Google Scholar]
- 3.Lupas A, Flanagan J M, Tamura T, Baumeister W. Trends Biochem Sci. 1997;22:399–404. doi: 10.1016/s0968-0004(97)01117-1. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman S, Wickner S, Maurizi M R. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 5.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 7.Beuron F, Maurizi M R, Belnap D M, Kocsis E, Booy F P, Kessel M, Steven A C. J Struct Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman S, Maurizi M R, Wickner S. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 9.Hoskins J R, Pak M, Maurizi M R, Wickner S. Proc Natl Acad Sci USA. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pak M, Hoskins J R, Singh S K, Maurizi M R, Wickner S. J Biol Chem. 1999;274:19316–19322. doi: 10.1074/jbc.274.27.19316. [DOI] [PubMed] [Google Scholar]
- 11.Singh S K, Guo F, Maurizi M R. Biochemistry. 1999;38:14906–14915. doi: 10.1021/bi991615f. [DOI] [PubMed] [Google Scholar]
- 12.Gottesman S, Wickner S, Jubete Y, Singh S K, Kessel M, Maurizi M R. Cold Spring Harbor Symp Quant Biol. 1995;60:533–548. doi: 10.1101/sqb.1995.060.01.057. [DOI] [PubMed] [Google Scholar]
- 13.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 14.Schweder T, Lee K H, Lomovskaya O, Matin A. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehnherr H, Yarmolinsky M B. Proc Natl Acad Sci USA. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojtkowiak D, Georgopoulos C, Zylicz M. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 17.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 18.Aizenman E, Engelberg-Kulka H, Glaser G. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tobias J W, Shrader T E, Rocap G, Varshavsky A. Science. 1991;254:1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 20.Gottesman S, Roche E, Zhou Y, Sauer R T. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman C, Thevenet D, Bouloc P, Walker G C, D'Ari R. Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roche E D, Sauer R T. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keiler K C, Waller P R, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 24.Weber-Ban E U, Reid B G, Miranker A D, Horwich A L. Nature (London) 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 25.Maurizi M R, Thompson M W, Singh S K, Kim S H. Methods Enzymol. 1994;244:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- 26.Grimaud R, Kessel M, Beuron F, Steven A C, Maurizi M R. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 27.Fenton W A, Kashi Y, Furtak K, Horwich A L. Nature (London) 1994;371:614–619. doi: 10.1038/371614a0. [DOI] [PubMed] [Google Scholar]
- 28.Thompson M W, Maurizi M R. J Biol Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- 29.Katayama-Fujimura Y, Gottesman S, Maurizi M R. J Biol Chem. 1987;262:4477–4485. [PubMed] [Google Scholar]
- 30.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 31.Hoskins J, Singh S K, Maurizi M R, Wickner S. Proc Natl Acad Sci USA. 2000;97:8892–8897. doi: 10.1073/pnas.97.16.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 33.Pak M, Wickner S. Proc Natl Acad Sci USA. 1997;94:4901–4906. doi: 10.1073/pnas.94.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levchenko I, Luo L, Baker T A. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 35.Jones J M, Welty D J, Nakai H. J Biol Chem. 1998;273:459–465. doi: 10.1074/jbc.273.1.459. [DOI] [PubMed] [Google Scholar]
- 36.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonciarz-Swiatek M, Wawrzynow A, Um S J, Learn B A, McMacken R, Kelley W L, Georgopoulos C, Sliekers O, Zylicz M. J Biol Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- 38.Levchenko I, Yamauchi M, Baker T A. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 39.Singh S K, Maurizi M R. J Biol Chem. 1994;269:29537–29545. [PubMed] [Google Scholar]
- 40.Kim Y I, Burton R M, Burton B M, Sauer R T, Baker T A. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]