Abstract
The assembly and maintenance of cilia require intraflagellar transport (IFT), a microtubule-dependent bidirectional motility of multisubunit protein complexes along ciliary axonemes. Defects in IFT and the functions of motile or sensory cilia are associated with numerous human ailments, including polycystic kidney disease and Bardet–Biedl syndrome. Here, we identify a novel Caenorhabditis elegans IFT gene, IFT-associated gene 1 (ifta-1), which encodes a WD repeat-containing protein with strong homology to a mammalian protein of unknown function. Both the C. elegans and human IFTA-1 proteins localize to the base of cilia, and in C. elegans, IFTA-1 can be observed to undergo IFT. IFTA-1 is required for the function and assembly of cilia, because a C. elegans ifta-1 mutant displays chemosensory abnormalities and shortened cilia with prominent ciliary accumulations of core IFT machinery components that are indicative of retrograde transport defects. Analyses of C. elegans IFTA-1 localization/motility along bbs mutant cilia, where anterograde IFT assemblies are destabilized, and in a che-11 IFT gene mutant, demonstrate that IFTA-1 is closely associated with the IFT particle A subcomplex, which is implicated in retrograde IFT. Together, our data indicate that IFTA-1 is a novel IFT protein that is required for retrograde transport along ciliary axonemes.
INTRODUCTION
Cilia and flagella are highly conserved cellular appendages found on many, if not most, mammalian cell types as well as on cells of countless multi- and unicellular lower eukaryotes, including Caenorhabditis elegans and Chlamydomonas reinhardtii. Cilia are grossly characterized as motile (e.g., Chlamydomonas flagella) or nonmotile (e.g., primary cilia of mammals and C. elegans). Motile cilia enable fluid/cell movement, whereas nonmotile cilia are implicated in sensory processes such as olfaction, chemosensation, mechanosensation, and photoreception (Davenport and Yoder, 2005). The physiological importance of cilia function is highlighted by the existence of numerous human ciliopathies, including polycystic kidney disease, nephronophthisis, retinal dystrophy, organ laterality defects, and phenotypically complex disorders such as Bardet–Biedl syndrome (BBS), Alström syndrome, and Meckel syndrome (Ansley et al., 2003; Otto et al., 2003; Afzelius, 2004; Pazour, 2004; Hearn et al., 2005; Kyttala et al., 2006; Smith et al., 2006).
The assembly and maintenance of the microtubule-dependent cilia and flagella structures depends on intraflagellar transport (IFT). Initially described in Chlamydomonas flagella, IFT is a bidirectional motility of multisubunit protein complexes, which deliver various cargo proteins that serve to build and maintain the structure and function of cilia and flagella (Kozminski et al., 1993; Rosenbaum and Witman, 2002; Scholey 2003). Anterograde IFT from the basal body, at the base of cilia, to the distal ciliary tip is driven by microtubule plus-ended kinesin-2 motors (Cole et al., 1993; Signor et al., 1999a; Snow et al., 2004), and an IFT-dynein motor directs retrograde IFT back to the basal body (Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999b; Wicks et al., 2000). Whereas anterograde IFT serves as a means to deliver proteins within cilia, retrograde IFT is used to recycle the IFT machinery (and presumably turnover products) back to the ciliary base (Rosenbaum and Witman, 2002; Scholey, 2003). IFT motor complexes are associated with the IFT particle, which consists of at least 17 proteins that can be biochemically separated into two distinct subcomplexes, A and B, of at least six and 11 different proteins, respectively (Piperno and Mead, 1997; Cole et al., 1998; Piperno et al., 1998).
The multisubunit assembly of motor complexes and IFT particles constitute the core IFT machinery, and functional loss of individual components leads to abrogation of IFT and consequently, defects in cilia structure and function. Findings in C. elegans and Chlamydomonas demonstrate that IFT particle B (IFT-B) subcomplex proteins play important roles in anterograde transport, because their loss results in ciliary structures that are severely truncated (C. elegans) or absent altogether (Chlamydomonas), with the core IFT machinery largely excluded from the stumpy ciliary axonemes of the nematode (Perkins et al., 1986; Collet et al., 1998; Fujiwara et al., 1999; Pazour et al., 2000; Brazelton et al., 2001; Haycraft et al., 2001, 2003). In contrast, IFT particle A (IFT-A) subcomplex proteins seem to function within the retrograde arm of the IFT pathway, because loss-of-function IFT-A subcomplex mutants possess only partially truncated cilia and flagella, which contain large accumulations of IFT particles that are brought in by anterograde transport but are unable to be recycled back via retrograde transport (Perkins et al., 1986; Piperno et al., 1998; Schafer et al., 2003).
Recent studies in C. elegans have uncovered a number of novel IFT proteins, including DYF-1, DYF-3, and DYF-13, which because of their roles in the assembly of sensory cilia in nematodes and the fact that strong homologues exist in other organisms, are likely components of the core IFT machinery (Murayama et al., 2005; Blacque et al., 2005; Ou et al., 2005a; Ou et al., 2005b). The observation that DYF-1 and DYF-13 are specifically required for assembling ciliary distal segments (Blacque et al., 2005; Ou et al., 2005a) is of significant interest, because many cilia, including those present on vertebrate renal cells and olfactory neurons, those of C. elegans amphid channel neurons as well as those of Chlamydomonas engaged in mating, possess bipartite structures (Reese, 1965; Mesland et al., 1980; Perkins et al., 1986). In the nematode, the bipartite structure of certain sensory cilia consists of a “middle” segment, which forms the proximal section and is built of doublet microtubules, and a “distal” segment, which forms the distal end of the ciliary axoneme and is comprised of singlet microtubules (Perkins et al., 1986). Two kinesin-2 motors (heterotrimeric kinesin-II and homodimeric OSM-3-kinesin) cooperatively drive the assembly of C. elegans middle segments, whereas OSM-3-kinesin alone is involved in the biogenesis of distal segments (Snow et al., 2004; Ou et al., 2005a). Two C. elegans Bardet–Biedl syndrome proteins, BBS-7 and BBS-8, which also undergo IFT, have been shown to stabilize interactions between the two kinesin-2 motors as well as the association of the IFT-A and IFT-B subcomplexes (Blacque et al., 2004; Ou et al., 2005a).
Although significant strides have been made in dissecting the mechanisms of IFT, it remains a poorly understood process. For example, the full complement of its components is not yet known and the organization, regulation, and specific functions of the IFT machinery are incompletely understood. To identify novel ciliary genes, including new IFT components, numerous genomics- and proteomics-based approaches have been conducted with a view to identifying evolutionarily conserved ciliary transcriptomes and proteomes (Ostrowski et al., 2002; Avidor-Reiss et al., 2004; Li et al., 2004; Blacque et al., 2005; Efimenko et al., 2005; Pazour et al., 2005; Smith et al., 2005; Stolc et al., 2005). In addition, bioinformatic screening for C. elegans X-boxes, which are promoter elements that bind the ciliogenic RFX transcription factor DAF-19, have drawn up large data sets of candidate ciliogenic genes (Swoboda et al., 2000; Blacque et al., 2005; Efimenko et al., 2005). Together with time-lapse fluorescence microscopy assays in C. elegans, which track the motility of fluorescently tagged IFT components in live animals (Orozco et al., 1999; Signor et al., 1999a; Snow et al., 2004), the above-mentioned approaches have been successful in identifying a number of novel IFT proteins, including the aforementioned BBS, DYF-1, DYF-3, and DYF-13 proteins (Blacque et al., 2005; Efimenko et al., 2005; Murayama et al., 2005; Ou et al., 2005a).
In this present study, we sought to further our understanding of the IFT process by identifying and functionally characterizing a novel C. elegans IFT protein, IFT-associated protein 1 (IFTA-1). We find that IFTA-1 is an evolutionarily conserved protein that localizes to ciliary structures, undergoes IFT, and is required for building structurally intact and functional cilia. We also demonstrate that loss of IFTA-1 function abrogates IFT and causes the ciliary accumulation of core IFT machinery components. Together, our data indicate that IFTA-1 is a component of the core IFT machinery and suggest that this protein plays important roles in retrograde IFT, likely via a close association with the IFT-A subcomplex.
MATERIALS AND METHODS
Strains and Genetic Crosses
All strains were maintained and cultured at 20°C. Standard mating procedures were used to cross fluorescence-tagged reporter constructs from wild-type worms to various mutant backgrounds, with single-worm polymerase chain reaction (PCR) used to follow the mutations in most cases. Deletions [ifta-1(nx61), bbs-8(nx77), and klp-11(tm324)] were detected using a single PCR reaction, where primers flanking the deletion distinguish wild-type and mutant copies of the gene. Two PCR reactions detected point mutations [osm-12(n1606), osm-3(p802)], where one reaction preferentially amplifies the wild-type gene and a second reaction preferentially amplifies the mutated gene. Primer sets used to detect mutations were as follows: ifta-1(nx61): AATATGGCTTCCACGCAAAC and ACAGGCGGAAGTAATGCAAC and CGCAATTTACGTGTCTATTTG; bbs-8(nx77): TTGAGCTGCCATGCAGCACAGCC and GAGCAGCACATCTGCAAGAAAC; klp-11(tm324): GTAAGTTCAGTGATATCTCA and AGTTGCCACAGCTTGTATA; osm-12(n1606): CCGACGTTGTATCAATTTCC and ATTGGGCAGTCATTAGATGG (wild-type primers) and CCGACGTTGTATCAATTTCT and ATTGGGCAGTCATTAGATGG (mutant primers); and osm-3(p802): GGATGCTCTTCTTCGAGAGTACC and CGAGACGATCCAGTTCGAATTC (wild-type primers) and GGATGCTCTTCTTCGAGAGTACT and CGAGACGATCCAGTTCGAATTC (mutant primers). che-11(e1810) mutations were followed using a dye-fill assay (see below), where heterozygous animals (che-11/+) are wild-type for dye-fill, and homozygous animals (che-11/che-11) animals possess a Dyf phenotype.
Construction of a Translational IFTA-1::GFP Reporter
A translational ifta-1::gfp fusion construct was constructed via fusion PCR as described previously (Blacque et al., 2004). Briefly, the entire exonic and intronic sequence of ifta-1, along with 511 base pairs of the upstream promoter sequence immediately 5′ to the start codon, was fused upstream of, and in frame with, the coding sequence for gfp. Transgenic animals expressing ifta-1::gfp as an extrachromosomal array in dpy-5(e907);Ex[dpy-5(+)] animals were generated as described previously (Ansley et al., 2003).
Localization of Homo sapiens IFTA-1 in Ciliated IMCD-3 Cells
The human orthologue of C. elegans IFTA-1, henceforth referred to as HsIFTA1, was PCR amplified from IMAGE clone 5266940 (ENSG00000118965) by using the aagcttgccaccatgttcttctacctg/gcggccgctcccactggactatggc primer pair. The resulting full-length fragment was cloned into HindIII and NotI sites of pCMV-H-myc after engineering a HindIII site into the pCMV-Myc vector (Clontech, Mountain View, CA) upstream of myc by using the gtaccaagcttgccaccatggcatcaatgcag/gtggcaagcttggtacaattccgcagcttttag primer pair. GFP was PCR amplified from the pEGFP-C2 vector (Clontech) with the cagcggccgccatggtgagcaag/cggccgtcacttgtacagctcgtc primer pair and inserted into the NotI site of pCMV-HsIFTA1 to generate pCMV-HsIFTA1::GFP. The HsIFTA1::GFP construct was transfected into IMCD-3 cells and grown on glass coverslips by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Twenty-four hours posttransfection, cells were fixed with 100% ice-cold methanol for 10 min at −20°C, rehydrated with Tris-buffered saline/Tween 20 (50 mM Tris base, pH 7.5, 150 mM NaCl, and 0.05% Tween 20), and immunostained for 1 h at room temperature with two primary antibodies, mouse anti-γ-tubulin (Sigma clone GTU-88) at 1:1000 dilution and mouse anti-acetylated-tubulin (Sigma clone 6-11B-1) at 1:10,000. A secondary antibody, goat-anti-mouse Alexa Fluor 488 (Invitrogen), was subsequently added at 1:2000 dilution and incubated for 1 h at room temperature. Imaging was performed with a Zeiss Axioskop2+ mot fluorescence microscope (Carl Zeiss, Thornwood, NY) mounted with a monochromatic charge-coupled device camera and Northern Eclipse software (Empix Imaging, Mississauga, Ontario, Canada).
Generation of ifta-1 Mutant Alleles
A PCR screening strategy was used to screen mut-7(cxP5003::Tc1) worms, which contain a Tc1 insertion in ifta-1 (C54G7.4), for imprecise Tc1 excision events. Single mut-7(cxP5003::Tc1) animals (P0) were placed on 100 plates and allowed to lay 20–40 eggs (F1) at 23°C. After the removal of P0 animals, plates were placed at 15°C, and worms were grown to the F2 generation. Next, ∼50% of F2 worms from each plate were recovered, and crude DNA lysates (100 μl) were prepared as described at www.protocol-online.org To identify lysates harboring C54G7.4 deletions (caused by imprecise Tc1 excision), we performed PCR by using a primer set that flanks the cxP5003::Tc1 insertion site—AAGTGTCGGACGCTGAGAAG and ACAGGCGGAAGTAATGCAAC—and looked for rare small amplicons. Using this scheme, we isolated two ifta-1 alleles, nx34 and nx61, possessing 600- and 2009-base pair deletions in ifta-1, respectively. Both alleles were outcrossed five times to N2 worms.
Analysis of Cilia Structure and Function
Chemotaxis Assay.
Well-fed adult worms were washed four times with M9 solution and once with water. Approximately 50 worms were pipetted onto the center of a standard 9-cm Petri plate containing a standard assay surface (1.6% agar, 0.005 M K2PO4, 0.001 M CaCl2, and 0.001 M MgSO4); 1.5 cm from the center of the plate there are two circular zones, both of 1-cm radius. One zone contained the attractant (1μl of 1:100 isoamyl alcohol/ethanol solution), whereas the other zone contained the negative control (1 μl of ethanol). Both zones also contained 1 μl of 1 M sodium azide to anesthetize worms. After 1 h, worms were counted, and the chemotaxis index was calculated as follows: (no. worms in attractant zone − no. of worms in control zone)/(total worms counted).
Dye Uptake.
Fluorescent carbocyanine dye (DiI) uptake assays were performed as described previously (Blacque et al., 2004).
Visualization of Ciliary Structures and Cilia Length Measurements.
PHA/B and ASER ciliary structures were visualized using cell-specific transcriptional srb-6p::gfp, and gcy-5p::gfp reporters, respectively. GFP expressed from these reporters diffuses throughout the cell, illuminating the cell body, axon, dendrite, and cilium structures. Cilium length was measured as the distance from the distal end of the transition zone (denoted as a bulge of fluorescence at the base of ciliary axonemes) to the tip of the ciliary axoneme.
IFT Motility Assay
Worms expressing fluorescent IFT proteins were mounted on agarose pads and immobilized with 15 mM levamisole. Amphid cilia were examined on an Olympus microscope (Olympus America, Melville, NY) equipped with a 100×, 1.35 numerical aperture objective and an UltraVIEW spinning-disk confocal head at 0.3 s/frame for 2–3 min. Kymographs and movies were created from the resulting stacked tiff images by using MetaMorph software (Molecular Devices, Sunnyvale, CA), and the rates of fluorescent IFT particle motility along the middle and distal segments of the eight amphid channel neuron cilia (10 cilia in total) were measured as described previously (Snow et al., 2004).
RESULTS
Identification of ifta-1 as a Strong Candidate for a Core IFT Machinery Gene
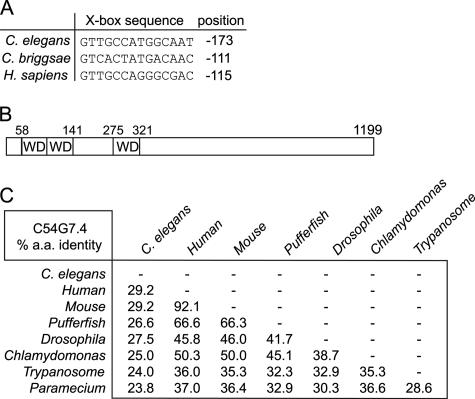
An excellent candidate IFT machinery component to arise from previous studies is the protein product of the C. elegans gene, C54G7.4, which we term IFTA-1. Findings in both C. elegans and Drosophila have shown that ifta-1 is an X-box–containing gene that is expressed specifically in ciliated cells (Avidor-Reiss et al., 2004; Blacque et al., 2005; Efimenko et al., 2005). The ifta-1 gene X-box is nearly canonical in sequence and position (near −100 base pairs from the start codon) in C. elegans and the related nematode Caenorhabditis briggsae, and it is notably also present within the promoter region of the human ifta-1 gene orthologue, ENSG00000118965/WDR35 (Figure 1A). In addition, the inclusion of ifta-1 in numerous data sets enriched for ciliary genes (Avidor-Reiss et al., 2004; Li et al., 2004; Pazour et al., 2005), and the finding that the Drosophila IFTA-1 protein (termed OSEG4) localizes at ciliary structures (Avidor-Reiss et al., 2004) further suggest that IFTA-1 may be an integral part of the core IFT machinery. The C. elegans ifta-1 gene encodes a protein of 1199 amino acids (∼140 kDa) that possesses several N-terminal WD repeats (Figure 1B). It is found in ciliated unicellular and multicellular organisms, including mammals, pufferfish, and Drosophila, but it is absent from nonciliated organisms such as S. cerevisiae and Arabidopsis thaliana (Figure 1C). Supplemental Figure 1 shows an amino acid alignment between C. elegans and H. sapiens IFTA-1, which displays ∼29% sequence identity throughout the entire protein.
Figure 1.
ifta-1 (C54G7.4) is an X-box–containing gene that is conserved in ciliated organisms. (A) RFX transcription factor-binding X-boxes are found upstream of ifta-1 in C. elegans, C. briggsae, and H. sapiens. (B) Schematic of the IFTA-1 protein, which consists of 1199 amino acids (in C. elegans) and is predicted to possess three WD protein motifs. (C) Cross-species comparison of the percentage of amino acid identity between IFTA-1 homologues in C. elegans, H. sapiens, Mus musculus, pufferfish, Drosophila melanogaster, C. reinhardtii, Trypanosoma brucei, and Paramecium. Note that IFTA-1 homologues are not present in nonciliated organisms such as A. thaliana and S. cerevisiae.
IFTA-1 Localizes to Basal Bodies and Undergoes IFT in C. elegans
To assess the cellular localization of IFTA-1, transgenic worms harboring an ifta-1::gfp transgene were constructed. Similar to C. elegans IFT and BBS proteins, we found that GFP-tagged IFTA-1 localizes strongly at the base of cilia (i.e., transition zones, which are akin to basal bodies) and along ciliary axonemes, as shown here for the phasmid (Figure 2A) and amphid (Figure 2B) sensory cilia. The ciliary localization of IFTA-1::GFP is highly specific, because GFP signals in the cell bodies, axons, and dendrites were found to be weak or absent. Consistent with this localization pattern, and similar to Drosophila OSEG4, a GFP-tagged version of the human IFTA-1 homologue (ENSP00000314444) localizes at the centrosomal/basal body region of a mouse IMCD3 ciliated cell line (Figure 2C). The localization of human IFTA-1 is essentially the same as many other known basal body/ciliary proteins, including BBS proteins (Ansley et al., 2003; Li et al., 2004).
Figure 2.
IFTA-1 localizes to ciliary structures in C. elegans and to the basal body/centrosomal region of mammalian kidney cells. (A and B) GFP-tagged IFTA-1 localizes specifically at the base of cilia (transition zones or basal bodies) and along the ciliary axonemes of C. elegans ciliated neurons. Shown are fluorescence images of one set of phasmid neurons in the nematode tail (A), and one set of amphid cilia in nematode head (B), with the cell bodies (cb), dendrites (den), transition zones (tz), and ciliary axonemes (cil) indicated. (C) GFP-tagged H. sapiens IFTA-1 localizes at the basal bodies and centrosomes of mammalian kidney cells. Shown are coimmunofluorescence images of ICMD3 cells expressing an H. sapiens ifta1::gfp transgene. Top, antibodies to endogenous γ-tubulin and acetylated α-tubulin detect centrosomes/basal bodies (red, bracket) and ciliary axonemes (red, arrowhead), respectively. Middle, GFP-tagged IFTA1 displays punctate fluorescence (brackets), consistent with its localization at basal bodies and centrosomes. Bottom, merged red and green fluorescence images demonstrate colocalization of basal bodies/centrosomes and GFP-tagged IFTA1 (yellow, bracket).
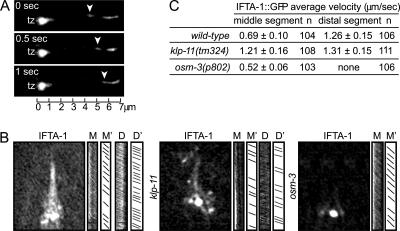
Using time-lapse microscopy, we observed that GFP-tagged IFTA-1 undergoes bidirectional movement along C. elegans sensory cilia (Figure 3A and Supplemental Video 1). In wild-type animals, IFTA-1::GFP moves along the ciliary middle segments at ∼0.7 μm/s and accelerates to ∼1.3 μm/s along distal segments (Figure 3, B and C). To more closely examine IFTA-1 motility, we analyzed the rate of IFTA-1 transport in two kinesin-2 mutants, klp-11 (subunit of heterotrimeric kinesin-II) and osm-3 (homodimeric OSM-3-kinesin), both of which drive anterograde IFT in the nematode. Using this scheme, we found that GFP-tagged IFTA-1 moves at a unitary fast speed (∼1.3 μm/s) along both the middle and distal segments of klp-11(tm324) mutants, whereas in osm-3 mutants it moves more slowly (∼0.5 μm/s) along the remaining middle segments (Figure 3, B and C). Our transport data for IFTA-1 are essentially identical to those previously found for known BBS/IFT-particle components, namely, that they are transported anterogradely in a biphasic manner in wild-type animals and in a monophasic manner in kinesin-II (klp-11) or OSM-3-kinesin (osm-3) single mutants (Snow et al., 2004; Ou et al., 2005a). Together, the above-mentioned data indicate that IFTA-1 is a novel, bona fide IFT-associated protein.
Figure 3.
C. elegans IFTA-1 undergoes biphasic IFT. (A) GFP-tagged IFTA-1 undergoes IFT. Shown is a kymograph (3 consecutive 0.5-s time points) depicting the anterograde motility (away from the transition zone, tz) of a GFP-tagged IFTA-1–associated particle (arrowhead) along the ciliary axoneme of one phasmid neuron. Scale bar is shown at the bottom of the kymograph. (B and C) The anterograde IFT motility of IFTA-1 is biphasic. Shown in B are representative “still” fluorescence images and corresponding kymographs (M and D) and kymograph schematics (M′ and D′), obtained from the analysis of IFTA-1::GFP transport (using IFT motility assays) along amphid sensory cilia. Note that the individual diagonal lines of the kymographs correspond to motility rates for individual fluorescent particles. Images (B) and table (C) show that in wild-type cilia, GFP-tagged IFTA-1 displays biphasic IFT, with intermediate velocities along middle segments (M and M′) and fast velocities along distal segments (D and D′). Also shown is the monophasic IFT behavior of IFTA-1::GFP along the ciliary axonemes of animals with loss-of-function mutations in a heterotrimeric kinesin-II subunit gene, klp-11, and a homodimeric OSM-3-kinesin subunit gene, osm-3.
IFTA-1 Is Required for the Proper Functioning and Assembly of Cilia
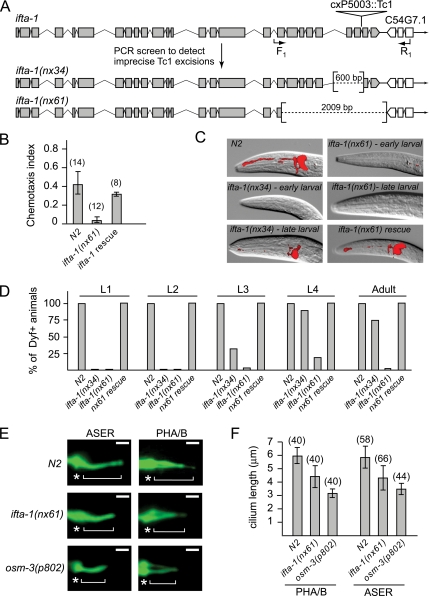
To further investigate the cilia-related function of IFTA-1, we generated mutant alleles of C. elegans ifta-1 by taking advantage of a previously isolated mutator strain, mut-7(cxP5003::Tc1), which contains a Tc1 transposable element within the 24th intron of ifta-1 (Figure 4A; Korswagen et al., 1996). Using a PCR-based strategy, mut-7(cxP5003::Tc1) animals were screened for imprecise Tc1 excision events, and two deletion alleles of ifta-1 were isolated (Figure 4A). The ifta-1(nx34) allele is defined by a 600-base pair deletion spanning introns 22–25, and the ifta-1(nx61) allele possesses a 2009-base pair deletion that extends from exon 18 onward.
Figure 4.
ifta-1 is a ciliogenic gene required for the normal structure and function of sensory cilia. (A) Generation of two mutant alleles of ifta-1 by transposon excision. Shown is the genomic position of cxP5003::Tc1 within ifta-1, and the resultant 600- and 2009-base pair deletions obtained after excision. (B) ifta-1(nx61) mutants are defective in chemotaxis toward iso-amyl alcohol compared with wild-type (N2) animals. Rescue of the chemotaxis defect was observed in ifta-1(nx61) worms expressing a wild-type ifta-1::gfp transgene. (C) ifta-1(nx34) and ifta-1(nx61) animals are defective in their ability to uptake the fluorescent dye DiI. Shown are DIC-fluorescence merged images of the head region of wild-type and ifta-1 mutant worms after a dye-fill assay. Note that the dye-fill defective phenotype (Dyf) of ifta-1 mutants is fully rescued in ifta-1 mutants expressing a wild-type ifta-1::gfp transgene. (D) The Dyf phenotype of ifta-1(nx61) mutants is fully penetrant at all larval and adult stages, whereas ifta-1(nx34) animals have a stage-specific Dyf phenotype. Note that each data point represents the analysis of 50 worms. (E and F) The cilia length of ifta-1(nx61) mutants is shorter than wild-type worms but longer than osm-3(p802) mutants. Presented are fluorescence images (E) and a graph (F) showing the cilia lengths obtained for PHA/B phasmid and ASER amphid neurons, by using srb-6p::gfp and gcy-5p::gfp reporters, respectively. For E: Bar, 2 μm; star, transition zone; bracket, cilia. The n values for F are presented in brackets.
Both ifta-1 mutant strains are homozygous viable and display no obvious morphological, locomotory, or behavioral phenotypes; however, the animals possess various ciliary abnormalities. First, ifta-1 mutants display cilia-related chemosensory (Che) defects, as evidenced by a significantly reduced ability to chemotax toward iso-amyl alcohol (Figure 4B), a behavior that is mediated by AWA and AWC neuronal sensory cilia (Bargmann et al., 1993).
Second, similar to known IFT machinery mutants in C. elegans, ifta-1 mutant cilia have lost the ability to take up the fluorescent dye DiI (Figure 4, C and D). This dye-fill defective (Dyf) phenotype indicates that ifta-1 mutant cilia may be structurally defective, because in wild-type worms, dye filling of six amphid and both phasmid sensory neurons normally occurs via environmentally exposed and fully intact ciliary endings. Interestingly, although the Dyf phenotype of ifta-1(nx61) mutants (with the larger deletion) is almost fully penetrant at all larval stages and at adulthood, ifta-1(nx34) mutants (with a smaller deletion) display only a partial Dyf phenotype. ifta-1(nx34) animals are fully Dyf at the L1 and L2 stages, partially Dyf at the L3 stage, and almost wild-type for dye-fill at the L4 and adult stages (Figure 4, C and D). Based on the Dyf phenotype data, the nx61 deletion is likely a severe loss-of-function allele, which may be a null, whereas the nx34 allele seems to retain some functionality.
It is important to note that the Che and Dyf phenotypes of ifta-1 mutants are directly attributable to loss of ifta-1 gene function, because expression of a wild-type ifta-1::gfp transgene in the ifta-1 mutant backgrounds fully rescues these defects (Figure 4, B–D). In addition, these rescue experiments verify the functionality of the ifta-1::gfp transgene, thereby validating the ciliary localization and IFT motility for GFP-tagged IFTA-1, described above (Figures 2, A and B, and 3, A–C).
Finally, using cell-specific transcriptional GFP reporters to directly visualize the ASER (gcy-5p::gfp) and PHA/B (srb-6p::gfp) neuronal cilia, ifta-1(nx61) mutant animals were observed to possess abnormal ciliary structures, a finding that is consistent with the Dyf phenotype discussed above. Specifically, we determined that the ciliary axonemes of ifta-1(nx61) mutants are shorter (PHA/B, 4.4 ± 0.9 μm and ASER, 4.2 ± 0.8 μm) than those of N2 control animals (PHA/B, 5.8 ± 0.8 μm and ASER, 5.9 ± 0.6 μm), but they are longer than those of osm-3(p802) mutants (PHA/B, 3.1 ± 0.4 μm and ASER, 3.5 ± 0.3 μm), which are composed only of middle segments (Figure 4, E and F). These observations indicate that ifta-1(nx61) mutant sensory cilia consist of an intact middle segment and part of the distal segment, similar to that observed in mutant animals defective for IFT subcomplex A components (Perkins et al., 1986).
Together, the above-mentioned data show that loss of ifta-1 gene function leads to the disruption of sensory cilia structure and function. Considering our observation that the IFTA-1 protein undergoes IFT, our findings strongly suggest that IFTA-1 is a component of the core IFT machinery that is required to build and maintain ciliary structures.
IFTA-1 Functions Similarly to IFT Particle Subcomplex A Proteins
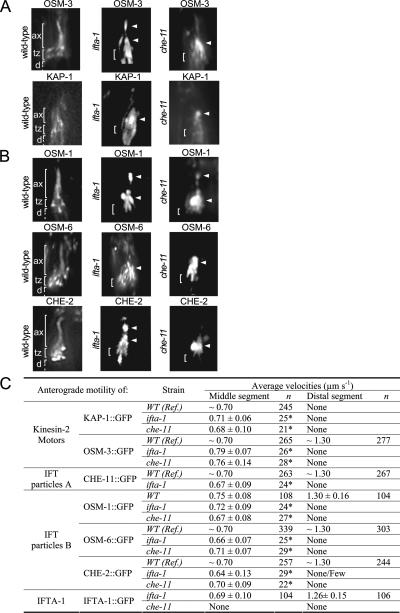
To decipher the role of IFTA-1 in the IFT process, IFT motility assays were used to determine transport profiles for IFTA-1 in various IFT mutant backgrounds as well as for other IFT proteins in the ifta-1 mutant background. First, the ciliary localization and IFT motility of GFP-tagged IFT components were assessed in ifta-1(nx61) mutants. Using this scheme, we found that anterograde IFT motor subunits KAP-1 (a component of heterotrimeric kinesin-II) and OSM-3 (homodimeric OSM-3-kinesin) as well as IFT-B subcomplex proteins (CHE-2, OSM-1, and OSM-6), all accumulate within the ciliary axonemes of ifta-1(nx61) mutants (Figure 5, A and B, and Supplemental Figure 2). In addition, IFT motility (as judged from the kymographs and visual inspection of the time-lapse recordings) of all examined GFP-tagged IFT proteins along ifta-1 mutant cilia was severely abrogated, with only infrequent anterograde transport events being detected in the middle segments and no transport events observed in the truncated distal segments (Figure 5C). Few or no retrograde transport events were detectable in our IFT assays. The ability of IFT proteins to enter ifta-1 mutant cilia, presumably via anterograde IFT and to subsequently form large ciliary accumulations or deposits that are essentially immotile, is strikingly similar to that observed for the IFT-A subcomplex gene mutant che-11(e1810) (Figure 5, A–C). In Chlamydomonas and C. elegans IFT-A subcomplex mutants, retrograde IFT motility is proposed to be defective, resulting in the ciliary axonemal accumulation of the IFT machinery and a disruption of the cycling IFT process (Perkins et al., 1986; Piperno et al., 1998; Schafer et al., 2003).
Figure 5.
ifta-1 mutants phenocopy the retrograde IFT defects of che-11 mutants. (A and B) The anterograde IFT machinery, namely, the kinesin-2 motors (A) and IFT-B subcomplex proteins (B), accumulates along ifta-1 and che-11 mutant cilia. Shown are representative fluorescence images of one set of amphid cilia from wild-type (N2), ifta-1(nx61), and che-11(e1810) animals, expressing the indicated GFP-tagged protein. Images demonstrate that in contrast to wild-type worms, the anterograde IFT motor subunits (KAP-1::GFP and OSM-3::GFP) and IFT-B subcomplex proteins (OSM-1::GFP, OSM-6::GFP, and CHE-2::GFP) accumulate (see arrowheads) within the ciliary axonemes of ifta-1 and che-11 mutants, indicating a retrograde IFT defect. Note that in wild-type panels, the ciliary axonemes (ax), transition zones (tz), and dendrites (d) are denoted. Note also that all images are similarly sized and orientated, with the transition zone region denoted in all panels by a bracket. (C) Anterograde IFT motility velocities of fluorescent IFT-proteins along the amphid cilia of WT, ifta-1(nx61) and che-11(e1810) mutants. The asterisk denotes that compared with wild-type cilia, the number of detectable anterograde transport events along ifta-1(nx61), and che-11(e1810) mutant cilia was low; hence, only a small n value could be obtained. References (Ref.) refer to Snow et al. (2004) for OSM-6::GFP, Ou et al. (2005a) for KAP-1::GFP, OSM-3::GFP, CHE-2::GFP, and CHE-11::GFP, and Ou and Scholey, unpublished data, for OSM-1::GFP.
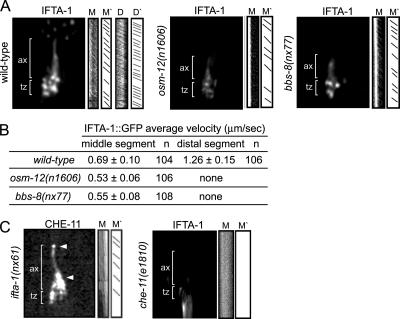
Next, we examined IFTA-1 anterograde motility along the ciliary axonemes of bbs mutants. Loss of C. elegans BBS protein function destabilizes anterograde motor–IFT particle assemblies within the middle segments, resulting in kinesin-II and OSM-3-kinesin motors that are uncoupled and separately transporting IFT-A and IFT-B subcomplex proteins, respectively (Ou et al., 2005a). In bbs-7 [osm-12(n1606)] and bbs-8(nx77) mutant cilia, we found that IFTA-1 has the same transport properties as kinesin-II motors and the IFT-A subcomplex component, CHE-11 (Figure 6, A and B; Ou et al., 2005a). Specifically, it was determined that GFP-tagged IFTA-1 particles are moved along bbs mutant middle segments at ∼0.5 μm/s, a rate characteristic of uncoupled kinesin-II/IFT-A assemblies, and it was not observed to enter the distal segments (Figure 6B). These data suggest that in the destabilized IFT assembly background of bbs mutants, IFTA-1 is closely associated with heterotrimeric kinesin-II and the IFT-A subcomplex protein CHE-11.
Figure 6.
IFTA-1 is an IFT particle subcomplex A-type protein. (A and B) Unlike wild-type (N2) cilia, in bbs mutant cilia, IFTA-1::GFP moves only along the middle segments (M and M′) at slow rates characteristic of uncoupled kinesin-II/IFT-A subcomplex assemblies and does not enter the distal segments. Shown in A are representative still fluorescence images and corresponding kymographs (M) and kymograph schematics (M′), obtained from the analysis of IFTA-1 transport (by using IFT motility assays) along wild-type (N2), bbs-8(nx77), and osm-12(n1606) mutant cilia. All images are similarly sized and orientated, with the transition zones and ciliary axonemes denoted in all panels. (C) CHE-11 protein function is required for the ciliary localization of IFTA-1 but not vice versa. Shown are representative still fluorescence images and corresponding kymographs (M) and kymograph schematics (M′), obtained from the analysis of IFT (by using IFT motility assays) of CHE-11::GFP in ifta-1(nx61) mutants, and IFTA-1::GFP in che-11(e1810) mutants. Note that CHE-11::GFP accumulates (see arrowheads) within the ciliary axonemes of ifta-1 mutants. In contrast, IFTA-1::GFP does not enter the ciliary axonemes of che-11 mutant cilia. Both images are similarly sized and orientated, with the transition zones and ciliary axonemes denoted in both images.
Finally, to further examine the potential association of IFTA-1 with the IFT-A subcomplex, we analyzed CHE-11:: GFP in ifta-1 mutants and in the reciprocal experiment, IFTA-1::GFP in che-11 mutants. In ifta-1(nx61) animals, GFP-tagged CHE-11 accumulates within the ciliary axonemes, and few transport events are detectable, similar to that described above for kinesin-II and IFT-B subcomplex components (Figure 6C). By contrast, in che-11(e1810) mutants, the transition zone (basal body) and ciliary axonemal signals for IFTA-1::GFP are barely detectable or absent (Figure 6C). This latter finding provides further support for the notion that IFTA-1 is associated with the IFT-A subcomplex, which includes CHE-11.
The data presented above demonstrate that the functions of IFTA-1 and CHE-11 are very closely linked. ifta-1 mutants phenocopy che-11 mutants, with each possessing moderately truncated ciliary axonemes, a severely disrupted IFT process, and ciliary axoneme accumulations of IFT machinery. In addition, IFTA-1 and CHE-11 proteins are both transported in an identical manner along bbs mutant cilia, and CHE-11 is required for the proper incorporation of IFTA-1 into an IFT machinery assembly. Overall, our findings lead us to conclude that IFTA-1 is a bona fide IFT protein that is required for retrograde IFT, likely in close physical association with the IFT-A subcomplex (Figure 7).
Figure 7.
Model of IFTA-1 function as a component of retrograde IFT. Presented is an extension of previous models by Snow et al. (2004) and Ou et al. (2005a), where two kinesin-2 motors were shown to drive anterograde IFT (heterotrimeric kinesin-II, drawn in brown; and homodimeric OSM-3-kinesin, drawn in blue), and BBS-7 and BBS-8 proteins were found to stabilize the association of IFT-A and IFT-B subcomplexes. (A) Loss of IFTA-1 function causes a ciliary accumulation of IFT machinery components, including IFT-B subcomplexes and the IFT-A subcomplex protein (CHE-11), indicating that IFTA-1 function is not required for entry of IFT machinery assemblies into cilia via anterograde IFT but that it is required for recycling of the IFT machinery back to the base of cilia via retrograde IFT. Defective retrograde IFT results in a partial loss of the singlet-microtubule-derived distal segment. (B) The IFT-A subcomplex gene mutant che-11 closely phenocopies the retrograde IFT defects of ifta-1 mutants. In addition, CHE-11 function is required for the ciliary localization of IFTA-1 protein. (C) Together, the data presented in A and B indicate that in wild-type C. elegans sensory cilia, IFTA-1 is closely associated with IFT-A subcomplexes and functions within the retrograde arm of the IFT process.
DISCUSSION
Understanding the molecular basis of IFT is an important goal in our drive to elucidate how cilia and flagella are built, maintained, and function. Furthermore, the ever-increasing number of cilia-related diseases (Badano et al., 2006), most of which are directly linked to defects in IFT, highlights the medical importance of such research. To investigate the poorly understood ciliogenic mechanism of IFT, we are using C. elegans to identify and functionally characterize novel components of the core IFT machinery. Previous research by us and others using the nematode model has lead directly to the discovery of a number of novel IFT machinery components, including several BBS proteins, DYF-1, DYF-3, and DYF-13, and also has provided significant insight into how the bipartite structure of nematode sensory cilia is assembled (Snow et al., 2004; Blacque et al., 2005; Ou et al., 2005a, b.
In this study, we identified the protein product of C. elegans C54G7.4 as a strong candidate IFT machinery component, based on previous reports that this gene/protein is found in numerous data sets enriched for ciliary genes and proteins (Avidor-Reiss et al., 2004; Li et al., 2004; Blacque et al., 2005; Efimenko et al., 2005; Pazour et al., 2005) and that a Drosophila homologue (OSEG4) localizes to ciliary structures (Avidor-Reiss et al., 2004). We present an in-depth analysis of C54G7.4 function and find that the encoded protein (IFTA-1) localizes to ciliary structures and undergoes IFT. Furthermore, we demonstrate that loss of IFTA-1 function disrupts sensory cilia structure and function, including a severe abrogation of the IFT motility process. Based on phenotypic overlap between ifta-1 mutants and the IFT-A subcomplex mutant che-11 and the fact that IFTA-1 and CHE-11 proteins display identical transport profiles in the destabilized IFT background of bbs mutants, we conclude that IFTA-1 plays important roles in retrograde IFT.
Retrograde IFT is proposed to be driven by a minus-end–directed multisubunit motor, IFT-Dynein, which to date has been shown to comprise a cytoplasmic dynein heavy chain (DHC1b) (Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999b; Wicks et al., 2000), a dynein light chain (LC8) (Pazour et al., 1998), and a dynein light intermediate chain (D2LIC) (Perrone et al., 1999; Hou et al., 2002; Schafer et al., 2003). In the above-mentioned studies, a role for IFT-dynein in retrograde IFT was established from the observations that Chlamydomonas and C. elegans IFT-dynein subunit mutants assemble swollen truncated cilia and flagella, which contain large accumulations of IFT particle subunits that cannot be recycled to the base of the cilium. IFT-A subcomplex proteins are proposed to play roles in retrograde IFT based on a number of observations. First, loss of Chlamydomonas IFT-A subcomplex protein function causes a specific reduction of retrograde IFT motility, with anterograde IFT motility relatively unchanged (Piperno et al., 1998). Second, similar to that observed in Chlamydomonas IFT-dynein subunit mutants, the sensory cilia of C. elegans IFT-A mutants are moderately truncated and swollen, containing accumulations of electron dense material, including IFT particle subunits (Perkins et al., 1986; Schafer et al., 2003).
Our conclusion that IFTA-1 plays important roles in retrograde transport, likely via a close association with the IFT-A subcomplex, is based on three key findings. First, ifta-1 and the IFT-A subcomplex mutant che-11 possess similarly truncated ciliary structures, which are close to wild-type length, lacking only a small portion of the distal segment. Our structural analysis of ifta-1 mutant cilia by using transcriptional GFP markers did not detect any bulges along the ciliary axoneme, a defect that was previously observed in electron micrographs of IFT-A subcomplex mutants (Perkins et al., 1986). It is possible that GFP does not adequately illuminate ciliary axonemal bulges; it would therefore be interesting in the future to examine the ultrastructure of ifta-1 mutant cilia by using electron microscopy. Second, core IFT machinery components (kinesin-2 motors and IFT-A and IFT-B subcomplex proteins) were found to accumulate within ifta-1 mutant cilia, indicating that IFT assemblies are able to move into ifta-1 cilia, via anterograde IFT, but they are not able to recycle back to the ciliary base via retrograde IFT. Indeed, our analysis of anterograde IFT motility along che-11 and ifta-1 mutant cilia detected infrequent transport events (Figure 5), indicating that anterograde IFT can still operate in the absence of IFT-A subcomplex protein function. However, unlike the situation in wild-type animals, retrograde transport events in ifta-1 mutants could not be observed using our IFT assays. Third, using IFT motility assays, we found that IFTA-1 and CHE-11 are moved in an identical manner along bbs mutant cilia. In bbs mutants, the IFT particle subcomplexes and the kinesin-2 motors are uncoupled, resulting in two separately moving assemblies of kinesin-II/IFT-A subcomplex and OSM-3-kinesin/IFT-B subcomplex (Ou et al., 2005). Our data demonstrate that in the destabilized IFT assembly background of bbs mutants, IFTA-1 associates with kinesin-II/IFT-A, suggesting that IFTA-1 is closely linked to the IFT-A subcomplex.
In Chlamydomonas, biochemical techniques determined that the IFT-A subcomplex consists of at least six proteins: IFT144, IFT140, IFT139, IFT122A, IFT122B, and IFT43 (Cole et al., 1998). With the exception of IFT140 (CHE-11) and IFT122A (DAF-10), the protein sequences of the remaining IFT-A subcomplex proteins are unknown. Interestingly, the Chlamydomonas homologue of IFTA-1, the flagellar protein FAP118 (Pazour et al., 2005), displays striking similarities to the protein model presented previously for IFT122B in that both possess N-terminal WD repeat domains (Cole, 2003). In addition, the predicted molecular mass and theoretical isoelectric point (pI) of FAP118 is 136 kDa and 6.04, respectively (using www.expasy.org/tools/protparam). These values are remarkably close to the molecular mass (122 kDa) and pI (5.8–6.0) measurements that were determined biochemically for IFT122B (Cole et al., 1998). It is conceivable, therefore, that IFTA-1 may be the C. elegans homologue of IFT122B.
Further evidence that IFTA-1 is either closely associated with, or is a core component of, IFT-A subcomplexes comes from our observation that in a che-11 mutant, IFTA-1 fails to localize to ciliary structures (including the transition zones/basal bodies) (Figure 6C). Our findings indicate that CHE-11 function is required for the assembly of IFTA-1 into the IFT-A subcomplex, or an IFT-A-associated subcomplex, but not vice versa. Also arising from these data is the possible interpretation that IFT-A subcomplexes may be assembled in an ordered manner, similar to that previously shown for IFT-B subcomplexes (Haycraft et al., 2003; Lucker et al., 2005). In this scaffold model of protein–protein interactions, CHE-11 would occupy a more “central” or “core” position within the IFT-A subcomplex relative to IFTA-1, which could be positioned more “peripherally.” Accordingly, in the absence of CHE-11, the scaffolding requirement for IFTA-1 is lost, and IFTA-1 fails to be tethered to the IFT assembly.
Understanding the molecular mechanisms underlying IFT motility is of great interest, because IFT defects are now known to underlie numerous human ciliopathies such as polycystic kidney disease and Bardet–Biedl syndrome. Future research on identifying the full repertoire of IFT machinery components as well as uncovering how they are grouped and regulated in the context of moving multisubunit assemblies will be required to improve our understanding of how cilia are assembled and maintained. In addition, such work will help to elucidate emerging functions of cilia and IFT, which include exciting new roles in developmental signaling cascades (Davenport and Yoder, 2005).
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Stiernagle and the C. elegans gene-knockout consortium for providing strains. This work was supported by the March of Dimes and Canadian Institutes of Health Research (CIHR) Grant CBM134736 (to M.R.L.; approximately two-thirds and one-third funding, respectively), Genome BC/Canada (to D.L.B.), and National Institutes of Health Grant GM-50718 (to J.M.S.). M.R.L. holds Michael Smith Foundation for Health Research (MSFHR) and CIHR scholar awards, D.L.B. holds a Canada Research Chair, O.E.B. is supported by an MSFHR fellowship, and A.K.M. is supported by a Natural Sciences and Engineering Research Council scholarship.
Abbreviations used:
- BBS
Bardet–Biedl syndrome
- IFT
intraflagellar transport
- IFT-A
intraflagellar transport particle A
- IFT-B
intraflagellar transport particle B.
Footnotes

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-06-0571 on October 4, 2006.
REFERENCES
- Afzelius B. A. Cilia-related diseases. J. Pathol. 2004;204:470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansley S. J., et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C. S. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006 doi: 10.1146/annurev.genom.7.080505.115610. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., Hartwieg E., Horvitz H. R. Odorant selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Blacque O. E., et al. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Blacque O. E., et al. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 2004;18:1630–1642. doi: 10.1101/gad.1194004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton J. B., Amundsen C. D., Silflow C. D., Lefebvre P. A. The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr. Biol. 2001;11:1591–1594. doi: 10.1016/s0960-9822(01)00485-7. [DOI] [PubMed] [Google Scholar]
- Cole D. G. Intraflagellar transport in the unicellular green alga, Chlamydomonas reinhardtii. Protist. 2003;154:181–191. doi: 10.1078/143446103322166491. [DOI] [PubMed] [Google Scholar]
- Cole D. G., Chinn S. W., Wedaman K. P., Hall K., Vuong T., Scholey J. M. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J., Spike C. A., Lundquist E. A., Shaw J. E., Herman R. K. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport J. R., Yoder B. K. An incredible decade for the primary cilium: a once forgotten organelle. Am. J. Renal Physiol. 2005;289:1159–1169. doi: 10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- Efimenko E., Bubb K., Mak H. Y., Holzman T., Leroux M. R., Ruvkun G., Thomas J. H., Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- Fujiwara M., Ishihara T., Katsura I. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. eleganssensory cilia. Development. 1999;126:4839–4848. doi: 10.1242/dev.126.21.4839. [DOI] [PubMed] [Google Scholar]
- Haycraft C. J., Schafer J. C., Zhang Q., Taulman P. D., Yoder B. K. Identification of CHE-13, a novel intraflagellar transport protein required for cilia formation. Exp. Cell Res. 2003;284:251–263. doi: 10.1016/s0014-4827(02)00089-7. [DOI] [PubMed] [Google Scholar]
- Haycraft C. J., Swoboda P., Taulman P. D., Thomas J. H., Yoder B. K. The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development. 2001;128:1493–1505. doi: 10.1242/dev.128.9.1493. [DOI] [PubMed] [Google Scholar]
- Hearn T., Spalluto C., Phillips V. J., Renforth G. L., Copin N., Hanley N. A., Wilson D. I. Subcellular localization of ALMS1 supports involvement of centrosome and basal body dysfunction in the pathogenesis of obesity, insulin resistance, and type 2 diabetes. Diabetes. 2005;54:1581–1587. doi: 10.2337/diabetes.54.5.1581. [DOI] [PubMed] [Google Scholar]
- Hou Y., Cole D. G., Dentler W., Pazour G., Witman G. A dynein light intermediate chain is required for retrograde intraflagellar transport (IFT) Mol. Biol. Cell. 2002;13:41a. doi: 10.1091/mbc.E04-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korswagen H. C., Durbin R. M., Smits M. T., Plasterk R. H. Transposon Tc1-derived, sequence-tagged sites in Caenorhabditis elegans as markers for gene mapping. Proc. Natl. Acad. Sci. USA. 1996;93:14680–14685. doi: 10.1073/pnas.93.25.14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttala M., Tallila J., Salonen R., Kopra O., Kohlschmidt N., Paavola-Sakki P., Peltonen L., Kestila M. MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat. Genet. 2006;38:155–157. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- Li J. B., et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Lucker B. F., Behal R. H., Qin H., Siron L. C., Taggart W. D., Rosenbaum J. L., Cole D. G. Characterization of the intraflagellar transport complex B core: direct interaction of the IFT81 and IFT74/72 subunits. J. Biol. Chem. 2005;280:27688–27696. doi: 10.1074/jbc.M505062200. [DOI] [PubMed] [Google Scholar]
- Mesland D. A., Hoffman J. L., Caligor E., Goodenough U. W. Flagellar tip activation stimulated by membrane adhesions in Chlamydomonas gametes. J. Cell Biol. 1980;84:599–617. doi: 10.1083/jcb.84.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama T., Toh Y., Ohshima Y., Koga M. The dyf-3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. J. Mol. Biol. 2005;346:677–687. doi: 10.1016/j.jmb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Orozco J. T., Wedaman K. P., Signor D., Brown H., Rose L., Scholey J. M. Movement of motor and cargo along cilia. Nature. 1999;398:674. doi: 10.1038/19448. [DOI] [PubMed] [Google Scholar]
- Ostrowski L. E., Blackburn K., Radde K. M., Moyer M. B., Schlatzer D. M., Moseley A., Boucher R. C. A proteomic analysis of human cilia: identification of novel components. Mol. Cell. Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- Otto E. A. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat. Genet. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Blacque O. E., Snow J. J., Leroux M. R., Scholey J. M. Functional coordination of intraflagellar transport motors. Nature. 2005a;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- Ou G., Qin H., Rosenbaum J. L., Scholey J. M. The PKD protein qilin undergoes intraflagellar transport. Curr. Biol. 2005b;15:R410–R411. doi: 10.1016/j.cub.2005.05.044. [DOI] [PubMed] [Google Scholar]
- Pazour G. J. Intraflagellar transport and cilia-dependent renal disease: the ciliary hypothesis of polycystic kidney disease. J. Am. Soc. Nephrol. 2004;15:2528–2536. doi: 10.1097/01.ASN.0000141055.57643.E0. [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Dickert B. L., Vucica Y., Seeley E. S., Rosenbaum J. L., Witman G. B., Cole D. G. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Dickert B. L., Witman G. B. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Wilkerson C. G., Witman G. B. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT) J. Cell Biol. 1998;141:979–992. doi: 10.1083/jcb.141.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Perrone C. A., Tritschler D., Taulman P., Bower R., Yoder B. K., Porter M. E. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol. Biol. Cell. 1999;14:2041–2056. doi: 10.1091/mbc.E02-10-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Siuda E., Henderson S., Segil M., Vaananen H., Sassaroli M. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J. Cell Biol. 1998;143:1591–1601. doi: 10.1083/jcb.143.6.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M. E., Bower R., Knott J. A., Byrd P., Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese T. S. Olfactory cilia in the frog. J. Cell Biol. 1965;25:209–230. doi: 10.1083/jcb.25.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J. L., Witman G. B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Schafer J. C., Haycraft C. J., Thomas J. H., Yoder B. K., Swoboda P. XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol. Biol. Cell. 2003;14:2057–2070. doi: 10.1091/mbc.E02-10-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J. M. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Signor D., Wedaman K. P., Orozco J. T., Dwyer N. D., Bargmann C. I., Rose L. S., Scholey J. M. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 1999b;147:519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor D., Wedaman K. P., Rose L. S., Scholey J. M. Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans. Mol. Biol. Cell. 1999a;10:345–360. doi: 10.1091/mbc.10.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. C., Northey J. G., Garg J., Pearlman R. E., Siu K. W. Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J. Proteome Res. 2005;4:909–919. doi: 10.1021/pr050013h. [DOI] [PubMed] [Google Scholar]
- Smith U. M. The transmembrane protein meckelin (MKS3) is mutated in Meckel-Gruber syndrome and the wpk rat. Nat. Genet. 2006;38:191–196. doi: 10.1038/ng1713. [DOI] [PubMed] [Google Scholar]
- Snow J. J., Ou G., Gunnarson A. L., Walker M. R., Zhou H. M., Brust-Mascher I., Scholey J. M. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- Stolc V., Samanta M. P., Tongprasit W., Marshall W. F. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. USA. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P., Adler H. T., Thomas J. H. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Wicks S. R., de Vries C. J., van Luenen H. G., Plasterk R. H. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev. Biol. 2000;221:295–307. doi: 10.1006/dbio.2000.9686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.