Abstract
Cryptococcus neoformans, which causes fatal infection in immunocompromised individuals, has an elaborate polysaccharide capsule surrounding its cell wall. The cryptococcal capsule is the major virulence factor of this fungal organism, but its biosynthetic pathways are virtually unknown. Extracellular polysaccharides of eukaryotes may be made at the cell membrane or within the secretory pathway. To test these possibilities for cryptococcal capsule synthesis, we generated a secretion mutant in C. neoformans by mutating a Sec4/Rab8 GTPase homolog. At a restrictive temperature, the mutant displayed reduced growth and protein secretion, and accumulated ∼100-nm vesicles in a polarized manner. These vesicles were not endocytic, as shown by their continued accumulation in the absence of polymerized actin, and could be labeled with anti-capsular antibodies as visualized by immunoelectron microscopy. These results indicate that glucuronoxylomannan, the major cryptococcal capsule polysaccharide, is trafficked within post-Golgi secretory vesicles. This strongly supports the conclusion that cryptococcal capsule is synthesized intracellularly and secreted via exocytosis.
INTRODUCTION
Naturally occurring glycoconjugates are incredibly diverse and are generally found outside of cells. In bacteria, these molecules include peptidoglycan, lipopolysaccharides (LPS), and capsular polysaccharides. These compounds are synthesized on either peptide (peptidoglycans) or lipid (LPS and capsular polysaccharides) primers by bacterial glycosyltransferases. These enzymes are usually localized in the membrane, thereby coupling the synthesis and export of extracellular polysaccharides (van Heijenoort, 2001; Raetz and Whitfield, 2002; Whitfield, 2006).
In fungi, most of the cell wall is composed of polysaccharides, including α- and/or β-glucans, chitin, and some mannans (Bose et al., 2003; Latge et al., 2005; Klis et al., 2006; Lesage and Bussey, 2006; Ruiz-Herrera et al., 2006). Glucans and chitin are polymers of glucose and N-acetylglucosamine (GlcNAc), respectively, and the corresponding synthases are thought to function at the plasma membrane (Leal-Morales et al., 1988; Cortes et al., 2002; Konomi et al., 2003; Valdivia and Schekman, 2003; Ortiz and Novick, 2006). It is unclear how synthesis of these polymers is initiated. Mannans are highly branched mannose polymers (100–300 mannoses in Saccharomyces cerevisiae) that are assembled in the secretory pathway as components of protein N-glycan moieties (Dean, 1999; Munro, 2001). The polypeptides that will bear mannan are synthesized in the endoplasmic reticulum (ER) and receive core N-glycans before moving to the Golgi apparatus. There they encounter glycosyltransferases that extend the N-glycan chains, and the completed mannan-bearing proteins progress to the cell surface.
The major cell wall component of plants is cellulose, an α-1,4–linked glucose polymer. Like fungal wall components, it is made at the cell surface (Somerville, 2006). Two other categories of plant cell wall polymers are pectin and hemicellulose, which provide rigidity and flexibility to the cell wall by interconnecting cellulose fibers (Reiter, 2002; Scheible and Pauly, 2004). These complex polymers are made in the secretory pathway, but elucidating their biosynthetic pathways has been extremely challenging (Dhugga, 2005; Bacic, 2006).
In animals, most cell-associated extracellular glycoconjugates are protein- or lipid- linked molecules that are made in the secretory pathway and displayed on the cell surface. One exception is hyaluronan, a linear polymer of disaccharide units of glucuronic acid and GlcNAc, which is synthesized at the plasma membrane (Weigel et al., 1997; Itano and Kimata, 2002). In general, therefore, extracellular polysaccharide synthesis in eukaryotic cells occurs either at the cell membrane, as in bacteria, or in the secretory pathway.
We are interested in the synthesis of polysaccharide molecules on the surface of Cryptococcus neoformans. C. neoformans is an environmental basidiomycetous yeast that causes fatal infections in mammals. The two most widespread serotypes, A and D, are a leading cause of fungal meningoencephalitis in immunocompromised individuals, with the majority of cases caused by serotype A strains. The characteristic of C. neoformans that is unique among pathogenic fungi is the polysaccharide capsule that surrounds its cell wall (Bose et al., 2003; Janbon, 2004). Similar to bacterial capsules, the cryptococcal capsule is important for immune evasion in the host (Kozel and Gotschlich, 1982; Vecchiarelli, 2005), but its composition and structure differ greatly from its bacterial counterparts. The cryptococcal capsule consists mainly of two polysaccharide species, glucuronoxylomannan (GXM) and galactoxylomannan (GalXM). GXM is a 1.7-7-MDa polymer (McFadden et al., 2006) composed of α-1,3-linked mannose which is O-acetylated and substituted with xylose and glucuronic acid residues (Cherniak and Sundstrom, 1994). GalXM is more complex, with an α-1,6-galactan backbone and xylosylated side chains of mannose and galactose (Vaishnav et al., 1998). GXM comprises ∼90% of the capsule mass and is considered to be the major virulence factor of C. neoformans. Strains lacking GXM are avirulent in animal models (Chang and Kwon-Chung, 1994, 1998, 1999; Chang et al., 1996), and GXM itself has immunosuppressive effects (Monari et al., 2006). A critical question about capsule polymers is how and where they are synthesized. The complex structures of GXM and GalXM suggest they might be made in the secretory pathway. On the other hand, the strong precedent from other extracellular polymers outlined above suggests they might be synthesized at the cell membrane.
A location for capsule synthesis was first hypothesized in 1967, when Edwards et al. (1967) observed a halo between the cell wall and the capsule using conventional transmission electron microscopy (TEM) and suggested it was a “zone of capsule synthesis.” Several years later, Takeo et al. (1973a, 1973b) demonstrated that this halo corresponded to a layer of 20-nm “granules,” which they speculated were involved in polymerization of capsule precursors. These investigators used a freeze-etching technique to compare C. neoformans grown in vitro to cells grown in a mouse model of infection, which have more extensive capsule. They noted that the number of intracellular vesicles in the latter cells was greater and hypothesized that the capsule was synthesized inside these vesicles. In both cell populations they also observed unusual plasma membrane invaginations containing small vesicles, which they termed the “baglike paramural bodies.” This observation led them to alternatively speculate that capsule was made in the vesicles of the paramural body.
Twenty years later, Sakaguchi et al. (1993) used the quick-freeze, deep-etch technique to improve image resolution in the comparison of in vivo- and in vitro-grown C. neoformans cells and observed that a “particle-accumulating layer” between the cell wall and the capsule was thicker in cells grown in vivo. They proposed that the accumulated particles represented capsule precursors that were then polymerized into fibrils, but did not indicate a model for overall capsule synthesis. More recently, two other reports touched on the location of capsule synthesis in C. neoformans. As part of a morphological study of cryptococcal cells during the course of mouse infection, Feldmesser et al. (2001) performed immunoelectron microscopy (immuno-EM) using anti-GXM antibodies, biotin-conjugated secondary antibodies, and gold-conjugated streptavidin and observed 200–300-nm aggregations of label within the cytosol, which they termed “vacuolar structures.” In another report, this group used the same immuno-EM method and noted “clusters” of gold particles in the cytoplasm and cell wall of C. neoformans (Garcia-Rivera et al., 2004).
The reports summarized above differ in the proposed location of capsule synthesis, with some investigators suggesting it happens inside the cell and others suggesting this process occurs outside the cell wall. They also propose that a variety of structures are involved with capsule synthesis, ranging from intracellular vacuoles, vesicles, or paramural bodies to extracellular particles. We sought to definitively address these fundamental questions of capsule synthesis, by testing the hypothesis that the major capsular polysaccharide, GXM, is made within the cell and secreted via exocytosis.
Secretion is a highly conserved biological pathway that has been well studied in the model yeast S. cerevisiae. Pioneering work by Schekman and colleagues identified multiple yeast genes involved in secretion (Novick and Schekman, 1979; Novick et al., 1980), which are now known to have homologues in other eukaryotic systems. Secretion mutants in other fungal species have been generated, typically by mutating highly conserved small GTPases (Craighead et al., 1993; Mao et al., 1999; Lee et al., 2001; Punt et al., 2001; Liu et al., 2005; Siriputthaiwan et al., 2005). One such protein is Sec4p, which regulates exocytosis (Guo et al., 1999). In S. cerevisiae exocytosis mutants, vesicles of ∼100 nm in diameter accumulate upon shift to a restrictive temperature, and these “post-Golgi” vesicles have been shown to contain secreted proteins (Novick et al., 1980; Holcomb et al., 1987; Harsay and Bretscher, 1995). In this study, we mutated a C. neoformans Sec4p homolog to address whether GXM uses the secretory pathway to exit the cell.
MATERIALS AND METHODS
Strains and Growth Conditions
C. neoformans strains stored in 25% glycerol at −80°C were streaked onto yeast extract peptone dextrose (YPD) plates for growth at room temperature (RT) or at 30°C. Where indicated, plates were supplemented with nourseothricin (100 μg/ml) or G418 (100 μg/ml). Liquid cultures were grown in YPD medium with shaking (230 rpm) at RT or at 30°C. Serotype A strain H99 was provided by G. Cox (Duke University) and serotype D strain JEC21 was from J. Lodge (St. Louis University). Generation of other C. neoformans strains is described below.
Identification of a Cryptococcal Homolog of the Sec4/Rab8 Subfamily of Rab GTPases
BLAST was used to query the Institute of Genome Research (TIGR) annotations of the C. neoformans serotype D strain JEC21 (http://www.tigr.org/tdb/e2k1/cna1/) with the amino acid sequence of S. cerevisiae Sec4p (NP_116650). The closest homolog (E-value of 6 × 10−61) was CNC04340 (GenBank XP_569689), which showed 53% identity and 67% similarity to Sec4p when whole sequences were compared using AlignX (VectorNTI Suite, Invitrogen, Carlsbad, CA). The genomic sequence of XP_569689, termed SAV1, was then used to identify the corresponding locus in the serotype A strain H99 sequence in the Broad Institute C. neoformans database (http://www.broad.mit.edu/annotation/genome/cryptococcus_neoformans/Home.html); this was identified as Supercontig 5: 681180–681966. The amino acid sequence encoded by this serotype A protein predicted by TWINSCAN (Tenney et al., 2004; protein 57_15; http://genome.slu.edu/) was shown to be identical to that predicted for serotype D Sav1p.
Plasmid Construction
To clone the complete SAV1 gene with its untranslated regions (UTRs), we PCR-amplified H99 genomic DNA (prepared as in Nelson et al., 2001) with primers AYP72 and AYP73 (Table 1). The resulting fragment, consisting of SAV1 with 1 kb of 5′UTR and 1.5 kb of 3′UTR sequence, was cloned into the pCR2.1-TOPO plasmid (Invitrogen) to form pAY19. SAV1 was sequenced to ensure that no errors were introduced during PCR. To obtain the desired Val23Asn mutation, we performed PCR-based site-directed mutagenesis of pAY19 using a QuikChange kit (Stratagene, La Jolla, CA) with primers AYP78 and AYP79. This mutated plasmid was confirmed by sequencing and termed pAY20.
Table 1.
Primers used for PCR in this study
| Primer name | Sequence (5′-3′) |
|---|---|
| Cloning of SAV1 with UTRs | |
| AYP72 | GCATCGACAATACC TGCAAGCTCATCATCG |
| AYP73 | CCTACTGCCCAGGAATTCCTCTTGACC |
| Site-directed mutagenesis of SAV1 | |
| AYP78 | GCAACTCTTGATAGGT AAC GGCAAGTCATGTCTCCTCTTGC |
| AYP79 | GCAAGAGGAGACATGACTTGCC GTT ACCTATCAAGAGTTGC |
| Confirmation of downstream integration | |
| AYP30 | CGAATCCGAGACAGACAT |
| AYP108 | CGTCTCTCATATGCTGCAGC |
| Confirmation of upstream integration | |
| AYP31 | CTGCATGCTTATGTGAGTCC |
| AYP109 | CCGTCTTGTTA GCTCCAAGG |
Bold residues indicate the engineered mutation in SAV1 (see text).
For the next step, we used a modified form of pGMC200 (McDade and Cox, 2001). pGMC200, provided by G. Cox, contains a nourseothricin acetyl transferase cassette (NAT) for drug selection in C. neoformans. The modified form of pGMC200, termed pGMC200-MCS, contains an additional multiple cloning site upstream of the actin promoter of NAT (H. Liu and T. L. Doering, unpublished results). We inserted the 2.7-kb BamHI fragment of pAY20 into a BglII-digested pGMC200-MCS. The resulting plasmid, pAY25, contains 1 kb of the 5′UTR, the sav1 gene bearing a mutation encoding the desired Val23Asn amino acid change, and 0.5 kb of the SAV1 3′UTR, inserted upstream of NAT. pAY25 was next modified by insertion of an additional 1 kb of the 3′UTR of SAV1 (excised from pAY20 with BamHI and XbaI; the unlabeled gray region in Figure 1) downstream of NAT to form pAY30 (Figure 1). Finally, we replaced the pAY30 segment consisting of the mutated sav1 sequence followed by NAT with the wild-type SAV1 gene in tandem with a neomycin-resistant cassette (Neo) from plasmid pMH12-T (provided by J. Lodge). The resulting plasmid was termed pAY51 (Figure 1). The same constructs were made for the serotype D strains using sequences obtained from JEC21 genomic DNA. Additional cloning details are available from the authors.
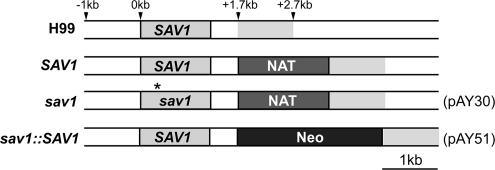
Figure 1.
Chromosomal position of the SAV1 gene and drug markers in the cryptococcal strains used in this work. H99, the serotype A parent strain; SAV1, a nourseothricin-resistant wild-type control strain; sav1, the sav1 mutant strain with the same marker; sav1::SAV1, a restored strain in which sav1 has been replaced with the wild-type SAV1 gene and the NAT marker with Neo. The drug resistance cassettes were inserted 0.5 kb downstream of the SAV1 stop codon in each case, and each construct used for biolistic transformation contained 1-kb flanking regions for homologous recombination. The asterisk in sav1 indicates the position of the sequence encoding the Val23Asn mutation. The gray region 3′ to the labeled genes designates corresponding regions of the 3′UTR. Plasmid designations in parentheses refer to constructs described in Materials and Methods.
Generation of C. neoformans Strains
To replace the chromosomal SAV1 gene with a mutated copy, strain H99 was subjected to biolistic transformation (Toffaletti et al., 1993) with linearized pAY30, allowed to recover on YPD plates at RT for 24 h, replated onto nourseothricin/YPD plates, and incubated at RT until colonies appeared. Genomic DNA from 48 colonies was screened for the integration of NAT by PCR with primers shown in Table 1: AYP30 and AYP108 for confirmation of downstream integration and AYP31 and AYP109 for upstream integration. A SAV1 gene fragment spanning the mutation was sequenced from all 10 colonies with correctly integrated NAT, and two strains, one with the desired mutation (GTG to AAC) and one with wild-type sequence, were chosen for study. The former strain was termed sav1, and the latter strain (a matched wild-type control with NAT inserted at the same genomic site) was termed SAV1 (see Figure 1).
To restore wild-type SAV1, the sav1 strain was biolistically transformed with BstXI-digested pAY51. Transformants were allowed to recover as above, replated onto G418-containing YPD plates to select for cells harboring Neo, and incubated at 30°C. They were then screened for loss of nourseothricin resistance, as well as for the ability to grow at 37°C (see Results). SAV1 and sav1 strains were confirmed to have the single desired insertion in the genome by DNA blotting (unpublished data). A set of serotype D strains was similarly generated from the parent strain JEC21, except that sav1 candidate colonies were screened for lethality at 37°C and for morphological change and dull appearance of colonies at 30°C before PCR, sequencing, and DNA blotting for confirmation (unpublished data). The sav1::SAV1 strain was generated by biolistic cotransformation (Goins et al., 2006) of the sav1 strain with two DNA fragments: one with the wild-type SAV1 with 5′ and 3′ 1-kb flanking sequences and the other containing Neo. PCR and DNA blotting showed that this strain contains one replacement at the desired genomic site and one additional ectopic insertion of Neo (unpublished data).
Electron Microscopy
Strains were grown in 50 ml of YPD medium at RT overnight, diluted to 2 × 106 cells/ml in fresh YPD medium, and grown at RT for 6 h. The cultures were divided into two flasks and grown at RT or at 30°C with shaking for 3 h. The cultures were then centrifuged in 50-ml conical tubes, the medium was discarded, and the cell pellets were quickly resuspended in 1 ml of primary fixation mix (0.1 M sorbitol, 1 mM MgCl2, 1 mM CaCl2, 2% glutaraldehyde in 0.1 M PIPES buffer, pH 6.8; Wright, 2000). The cell suspension was transferred into 1.5-ml microcentrifuge tubes and sedimented in a tabletop microcentrifuge (0.5 min, RT, top speed). Cell pellets were resuspended in 1 ml of primary fixation mix and fixed overnight at 4°C.
For optimal visualization of membrane structure, cells were postfixed with potassium permanganate. Cells were sedimented and washed three times in water (each wash involved resuspension of the pellet followed by a 10-min incubation at RT) and transferred into 13 × 100 mm borosilicate glass tubes (Fisher Scientific, Pittsburgh, PA). Cells were again sedimented to remove water and resuspended in 2% KMnO4 aqueous solution. The final centrifugation and resuspension steps were repeated, and the cells were postfixed for 45 min and then washed in water as above until a purple color was no longer visible. Cells were next transferred to microcentrifuge tubes and dehydrated by successive 10-min incubations in water–ethanol mixtures with increasing ethanol concentrations. The ethanol-substituted samples were then substituted in propylene oxide (twice for 30 min each) and infiltrated and embedded in Eponate 12 resin (Ted Pella, Redding, CA). Blocks were polymerized for at least 24 h at 65°C. The samples were cut into 60–80-nm sections with a Leica Ultracut UCT ultramicrotome (Leica Microsystems, Bannockburn, IL) and were viewed with a JEM 1200EX transmission electron microscope (JEOL USA, Peabody, MA) after uranyl acetate-lead citrate staining.
For immuno-EM, cells were fixed in glutaraldehyde as above, washed in 50 mM citrate buffer, pH 6.0, three times, and resuspended in 1 ml of 40 mg/ml Lysing enzymes (L1412, Sigma, St. Louis, MO) in the same buffer. After 30 min of partial cell wall digestion at 30°C, cells were washed in 0.1 M phosphate buffer, pH 7.0, three times and postfixed in 1% OsO4 in the same buffer for 1 h at RT. Blocks were prepared as described above. Sections were blocked with a solution of 5% FBS in 100 mM PIPES buffer (pH 7.0) for 30 min, labeled with an anti-GXM mAb (either 3C2 from T. Kozel, University of Nevada at Reno, or 2H1 from A. Casadevall, Albert Einstein College of Medicine) for 1 h, washed in blocking buffer, and incubated with 12-nm gold-conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch, West Grove, PA). Sections were then washed once in 100 mM PIPES buffer, pH 7.0, and twice in water, stained with uranyl acetate and lead citrate, and viewed as above.
Acid Phosphatase Secretion Assay
Acid phosphatase secretion was assayed using intact cells (To-e et al., 1973). Overnight cultures of C. neoformans strains grown at RT in a minimal medium supplemented with phosphate (0.5% KH2PO4; Cerdá-Olmedo and Lipson, 1987) were washed in a minimal medium without phosphate (0.5% KCl instead of KH2PO4) and resuspended in 25 ml of the same medium at 1 × 107 cells/ml. From this, 12.5 ml was then incubated with shaking either at RT or at 30°C. After 6 h of incubation, the cells from 1-ml aliquots of the cultures were harvested, resuspended in 400 μl of substrate buffer (2.5 mM para-nitrophenylphosphate in 50 mM acetate buffer, pH 4.0), and incubated at 37°C for 5 min. The reactions were stopped by adding 800 μl of saturated Na2CO3, and absorbance was read in a spectrophotometer. The difference in A420 between the final cell suspension and a blank reaction without cells was calculated, and results were normalized by cell density (A600).
Statistics
Statistical significance was assessed with Microsoft Excel X for Mac (Redmond, WA) using either Student's t test, to compare pairs of samples, or z-test, for comparison to the normalized wild-type value of 100 in acid phosphatase assays.
RESULTS
Sav1p Is A Cryptococcal Homolog of the Sec4/Rab8 Subfamily of Rab GTPases
To generate a late secretion mutant in C. neoformans, we targeted a homolog of Sec4p. Sec4p is a small GTPase of the Rab subfamily that acts as a “master regulator” of exocytosis (Novick and Zerial, 1997; Guo et al., 1999; Segev, 2001), and conditional sec4 mutants in S. cerevisiae accumulate post-Golgi vesicles under restrictive conditions (Novick et al., 1980; Mulholland et al., 1997). We find at least seven Rab GTPases in C. neoformans, using the Institute of Genome Research (TIGR) annotations of the C. neoformans serotype D strain JEC21 (http://www.tigr.org/tdb/e2k1/cna1/) and TWINSCAN (Tenney et al., 2004) predicted proteins of serotype A strain H99 (http://genome.slu.edu/). The two of these that are closest to the S. cerevisiae Sec4p sequence, XP_568088 and XP_569689, are very similar (54% identity and 70% similarity). However, we noted that the former contains a Ypt1/Rab1-specific stretch of sequence (amino acids 33–44), whereas the latter contains Sec4/Rab8-specific sequence (amino acids 39–44) (Nussbaum and Collins, 2005). This indicated that XP_569689 is a member of the Sec4/Rab8 subfamily. We termed this gene SAV1, because cells with a mutant copy were secretion-deficient and accumulated vesicles (see below).
Extending the homology to Sec4-related proteins, Sav1p is closely related to the Colletotrichum lindemuthianum Clpt1p (Dumas et al., 2001; Siriputthaiwan et al., 2005; 72% identity, 79% similarity), which was identified as a filamentous fungal homolog of Sec4p. Notably, like the Schizosaccharomyces pombe Sec4p homolog Ypt2p (Haubruck et al., 1990), Sav1p is more closely related to the higher eukaryotic Rab8 (58% identity and 71% similarity to human Rab8) than to the S. cerevisiae Sec4p. The sequences of these proteins and other fungal homologues are shown in Figure 2. Ypt2p and Clpt1p were both able to complement a temperature-sensitive mutant of SEC4 in S. cerevisiae, sec4-8 (Haubruck et al., 1990; Dumas et al., 2001), although the Aspergillus niger homolog, SrgAp, was not (Punt et al., 2001). Expression of the cryptococcal homolog, Sav1p, also did not complement sec4-8 (Supplementary Figure S1).
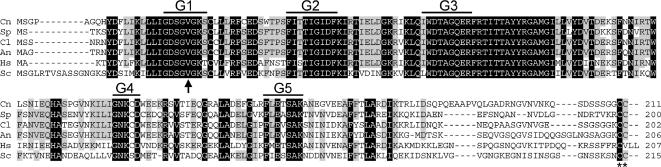
Figure 2.
Sequence comparison of selected Sec4p homologues. Cn, C. neoformans Sav1p; Sp, S. pombe Ypt2p; Cl, C. lindemuthianum Clpt1p; An, Aspergillus niger SrgAp; Hs, Homo sapiens Rab8p; Sc, S. cerevisiae Sec4p. Residues that are identical in all sequences are shaded black; those conserved in four or five of six sequences are in gray. G1–G5 indicate conserved GTP binding domains, the vertical arrow indicates the conserved Val residue that was mutagenized to Asn in S. pombe (Craighead et al., 1993), and a conserved prenylation site at the C-terminus is marked with asterisks.
Because secretion is an essential process and there is only one homolog of Sec4p in C. neoformans (Sav1p), we sought to generate a conditional mutant of SAV1. In S. pombe, mutation of a conserved Val residue within the first GTP-binding domain of Ypt2p (Figure 2, arrow) produced a temperature-sensitive mutant (Craighead et al., 1993). We adopted this strategy to obtain a temperature-sensitive mutant of Sav1p, using site-directed mutagenesis to generate a mutation encoding the same Val-to-Asn change in the G1 region of SAV1. The mutated gene, in tandem with a drug resistance marker and flanked by endogenous UTR sequences, was introduced into serotype A C. neoformans strain H99 by biolistic transformation. Alternate cross-over events yielded both the desired mutant (sav1) and a matched drug-resistant control strain (SAV1), as shown in Figure 1 and detailed in the Materials and Methods. We also generated a strain in which sav1 was restored to wild type by replacement of the mutated gene, termed sav1::SAV1 (Figure 1).
The sav1 Mutation Confers Temperature-sensitive Growth and Defective Protein Secretion
Once sav1 and control strains were generated, we tested whether the Val23Asn mutation indeed conferred the expected temperature-sensitive growth phenotype. As shown in Figure 3, sav1 showed no growth defect as compared with the SAV1 and sav1::SAV1 strains at RT, but grew more slowly at 30°C. Furthermore, 37°C was completely lethal to the sav1 mutant, which under these conditions displayed massive cell wall damage (unpublished data). For further studies we chose the semipermissive temperature of 30°C, where cell growth was impaired but cell wall damage was minimal.
Figure 3.
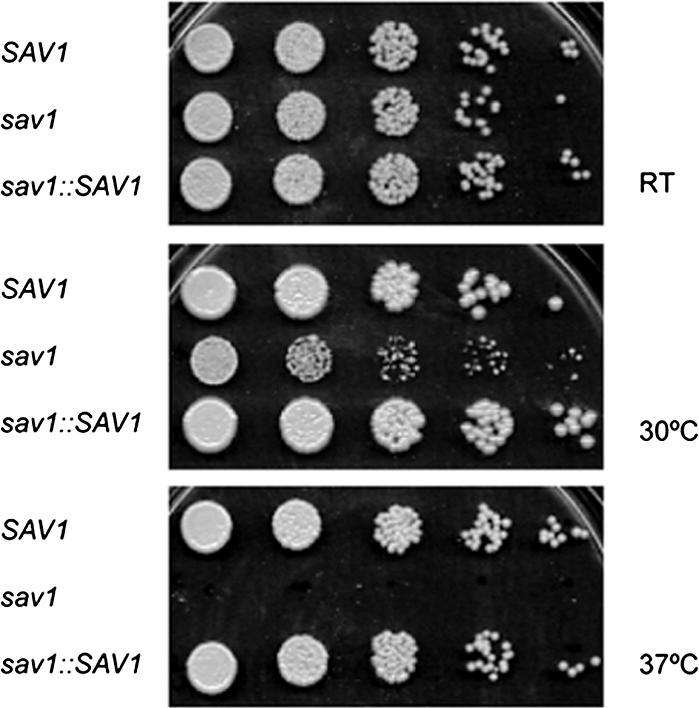
sav1 displays temperature sensitivity at 30°C. SAV1, sav1, and sav1::SAV1 strains grown on YPD plates at RT were suspended in sterile water and diluted to 6 × 105 cells/ml, and serial fivefold dilutions were spotted onto YPD plates. Plates were incubated at the indicated temperatures for 3 d.
The originally generated strains were of serotype A. To assess consistency of the sav1 phenotype, we generated a parallel set of strains in a serotype D background. The serotype D sav1 mutant was even more sensitive to high temperatures, exhibiting a slight growth defect at RT and very slow growth at 30°C (Supplementary Figure S2).
Next, we tested whether Sav1p is involved in protein secretion. The invertase assay, a commonly used secretion assay for S. cerevisiae, was not suitable for C. neoformans (unpublished data), probably because of the difficulty of generating protoplasts in this organism. We instead used an assay for acid phosphatase secretion (van Rijn et al., 1972; To-e et al., 1973; Novick and Schekman, 1979). This assay takes advantage of the localization of secreted acid phosphatase: this enzyme is retained in the cell wall after secretion, where it can act on an exogenously supplied colorimetric substrate. As shown in Figure 4, sav1 cells displayed a significant protein secretion defect both at RT and 30°C. The sav1::SAV1 strain secreted a normal level of acid phosphatase at both temperatures, confirming that the observed secretion defect is due to the mutation in SAV1. This result suggests that Sav1p is required for protein secretion, like other Sec4/Rab8 GTPases.
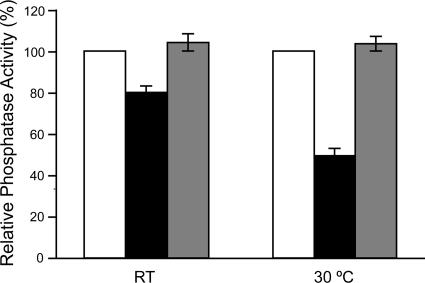
Figure 4.
The sav1 mutation results in decreased acid phosphatase secretion. Acid phosphatase secretion was induced by depletion of phosphate as described in Materials and Methods and is shown corrected for cell density and normalized to wild-type levels. □, SAV1; ■, sav1; ▩, sav1::SAV1; all strains are serotype A. The mean and SE values plotted were compiled from seven independent experiments performed in duplicate for the SAV1 and sav1 strains, and four independent experiments performed in duplicate for the sav1::SAV1 strain. Statistically significant differences were observed between the sav1 mutant and wild-type strains (p < 0.001 at both temperatures) and between the sav1 mutant and complemented strains (p = 0.003 at RT; p < 0.001 at 30°C). There was no significant difference between wild-type and complemented strains (p = 0.19 for RT; p = 0.21 for 30°C).
The sav1 Mutant Exhibits Separation Defects and Accumulates Vesicles at the Septum and the Bud
Defects in exocytosis often lead to alterations in morphology. We observed that when sav1 cells were grown at 30°C for more than 6 h, budding cells with abnormally thick necks (Figure 5A) became prevalent. This phenotype was reminiscent of S. pombe, where exocytosis mutants exhibit cell separation defects due to their inability to properly localize enzymes required to digest septa (Wang et al., 2002; Martin-Cuadrado et al., 2005). We suspected that this was also occurring in the cryptococcal sav1 cells and examined their morphology more closely by TEM. The preparation method we used (with KMnO4, see Materials and Methods) does not preserve capsule but is excellent for visualizing membrane structures and cell walls. These studies revealed abnormally wide and thick septa in dividing sav1 cells, with accumulation of ∼100-nm vesicles at the septum (Figure 5B). Cells undergoing septation were rarely observed in wild-type C. neoformans in the growth conditions used; cells were either engaged in budding or had already separated.
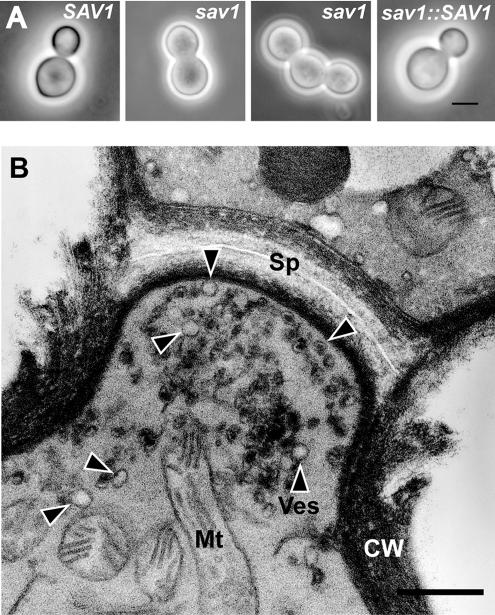
Figure 5.
sav1 exhibits a cell separation defect at 30°C. (A) Light micrographs of serotype A SAV1, sav1, and sav1::SAV1 strains grown in YPD for 10 h at 30°C. Scale bar, 5 μm. (B) Electron micrograph of a sav1 cell grown in the same way, showing an undigested septum and accumulation of ∼100-nm vesicles. Sp, septum; Ves or arrowheads, vesicles; Mt, mitochondria; CW, cell wall. Scale bar, 0.5 μm.
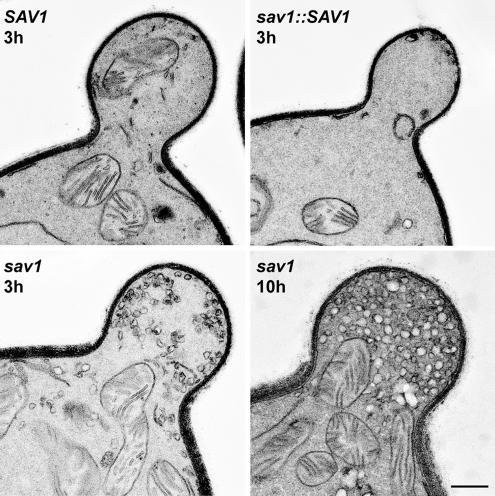
The classic manifestation of exocytic defects in budding yeast is accumulation of post-Golgi vesicles, particularly in the growing bud itself (Novick and Schekman, 1979). When sav1 cells were shifted to the restrictive temperature of 30°C for 3 h, we observed striking accumulation of vesicles at the bud (Figure 6). This phenotype was absent in the two control strains and was enhanced with increased time at 30°C (Figure 6). The accumulated vesicles were ∼100 nm in size, consistent with their identification as post-Golgi secretory vesicles.
Figure 6.
Vesicles accumulate in the bud of sav1 cells. 3 h after temperature shift to 30°C, putative post-Golgi vesicles are accumulated in the bud of sav1 cells, but not in SAV1 or sav1::SAV1. All strains are serotype A. This vesicle accumulation becomes more dramatic upon longer incubation (10 h, right lower panel). Scale bar, 0.4 μm.
We observe more vesicles in the serotype D wild-type strain JEC21 than in the serotype A wild-type strain H99. The sav1 mutation in a JEC21 background also resulted in more extensive accumulation of vesicles at 30°C in a shorter period (unpublished data), consistent with our preliminary observation that protein secretion is more active in serotype D (Supplementary Figure S3).
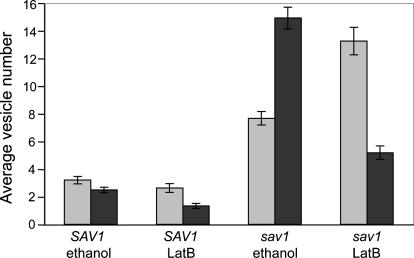
Because of the homology between Sec4p and Sav1p, and the morphological and secretion defects observed, we believe the vesicles that accumulate in sav1 cells are exocytic. However, we tested the formal possibility that these vesicles could be endocytic, by using the actin-depolymerizing drug, latrunculin B (LatB). Endocytosis is inhibited when cells are treated with actin-depolymerizing drugs, because internalization of endocytic vesicles requires actin patches (Ayscough, 2000, 2005; Huckaba et al., 2004; Kaksonen et al., 2005). In contrast, such drugs should not interfere with generation of secretory vesicles, although it will affect their polarization because this process depends on F-actin cables (Karpova et al., 2000). We therefore expected that cells grown with LatB should lack endocytic vesicles, but would generate post-Golgi vesicles that become dispersed throughout the cell. When C. neoformans strains were treated with 400 μM LatB, no F-actin structures were detected (Supplementary Figure S4). Despite this perturbation, there was no change in the total accumulation of 100-nm vesicles in sav1 cells. This remained at a level of fourfold over wild type (vesicles per EM section), with or without LatB (Figure 7). The LatB vesicles, however, were no longer polarized (Figure 7; electron micrograph in Supplementary Figure S5).
Figure 7.
sav1 cells contain more total vesicles than SAV1 cells, with or without latrunculin B (LatB), but in LatB conditions the vesicles are no longer polarized to the bud. For each strain, cells were shifted to 30°C for 3 h concurrent with LatB or ethanol (control) treatment and examined by EM. Vesicles present in sections of 60 budding cells were counted, and the average numbers of vesicles per section in the mother and bud were plotted. ▩, vesicles in a mother cell; ■, vesicles in a bud/daughter cell. Mean and SE are plotted. The fraction of vesicles observed in the bud decreased significantly in each strain upon LatB treatment [45.6 to 33.6% (p = 0.02) for SAV1; 66.2 to 28.8% (p < 0.001 for sav1 cells)].
The Accumulated Vesicles in sav1 Contain GXM
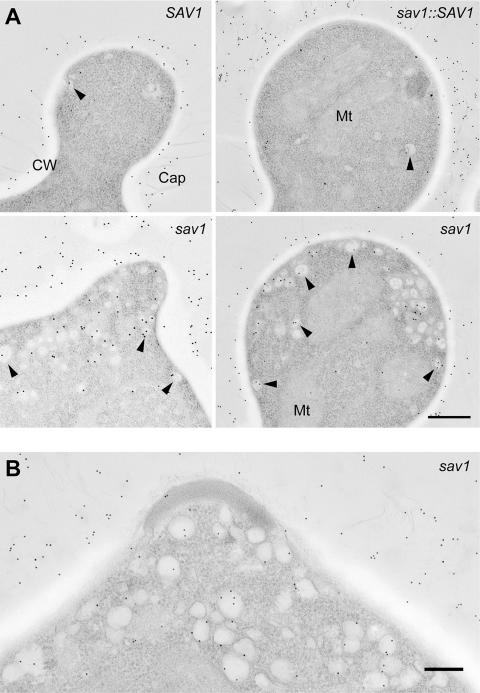
Previous studies of C. neoformans have shown that the capsule enveloping daughter cells consists of newly synthesized material (Pierini and Doering, 2001; Zaragoza et al., 2006). If this material is synthesized intracellularly and exits via the secretory pathway, we expect to detect GXM in the post-Golgi vesicles that accumulate in sav1 buds under restrictive conditions. To test this, we used anti-GXM antibodies and immuno-EM. The EM method used above (Figures 5 and 6) is not suitable for immuno-EM, because the strong oxidizing reagent used in the preparation does not preserve the antigenicity of carbohydrates, specifically capsule in this case. Further, en bloc staining with uranyl acetate alters the ultrastructure of the capsule (A. Yoneda and T. L. Doering, unpublished observation). To avoid these technical problems, we instead prepared samples using glutaraldehyde and osmium tetroxide fixations followed by embedding in plastic resin (Materials and Methods), and then performed immuno-EM using two independently derived anti-GXM monoclonal antibodies and gold-conjugated secondary antibody. In both SAV1 and sav1::SAV1 cells, the extracellular capsule was abundantly labeled with gold particles, whereas there were only a few gold particles visible within the cell. Although extracellular capsule was also labeled in sav1 cells grown at 30°C for 3 h, significant numbers of gold particles were visible inside the cell, and the majority of them were observed within vesicles (Figure 8).
Figure 8.
savl cells accumulate vesicles that contain GXM. Strains were grown in YPD at RT for 6 h and then shifted to 30°C for 3 h (for serotype A strains) or for 1.5 h (for serotype D strains). Ultrathin sections were labeled with the anti-GXM mAb 3C2 and 12-nm gold-conjugated anti-mouse antibody as described in Materials and Methods, and images of buds are shown. Similar results were obtained using the anti-GXM mAb 2H1 (unpublished data). (A) Serotype A strains; scale bar, 0.4 μm. (B) The serotype D sav1 strain; scale bar, 0.2 μm. Arrowheads, labeled vesicles; Cap, capsule; other abbreviations as in Figure 5. Note that when ∼100-nm vesicles are cut into 80-nm sections, in many cases only part of the vesicle is visible. Such portions may appear as small areas devoid of ribosomes, but may still be labeled with anti-GXM antibodies.
The buds of sav1 cells grown at 30°C do bear newly synthesized cell wall and capsule material (unpublished data; studies performed as in Pierini and Doering, 2001), due to the incomplete nature of the secretory block. This indicates that no unusual capsule rearrangement occurs under these conditions.
DISCUSSION
Understanding the synthesis of extracellular glycoconjugates poses unique challenges. Unlike polymerization of other cellular macromolecules, there are few “rules” governing the initiation of glycan synthesis, where this synthesis occurs, or how the products associate with the cell. In this study, we have addressed a long-standing question regarding the location of synthesis and mechanism of export of GXM, the major capsule polysaccharide of C. neoformans.
Early studies in C. neoformans suggested either intracellular or cell surface synthesis of capsule, but settling this question has remained elusive (Edwards et al., 1967; Takeo et al., 1973a, 1973b; Sakaguchi et al., 1993; Feldmesser et al., 2001; Garcia-Rivera et al., 2004). The major conclusion from our study is that GXM is trafficked within secretory vesicles, supporting a model that capsular materials are synthesized in the Golgi and targeted to the plasma membrane for exocytosis. Although our experiments do not distinguish between direct movement of GXM-containing post-Golgi vesicles to the plasma membrane and movement to that site via recycling endosomes (Gurunathan et al., 2002; Harsay and Schekman, 2002; Ang et al., 2004), they clearly indicate that capsule synthesis is intracellular.
Our studies used anti-GXM monoclonal antibodies to probe vesicle contents. They do not directly address the biosynthesis of the second capsule polysaccharide, GalXM, although we speculate that this polymer is similarly made in the secretory pathway. To prove this would require either antibodies of appropriate specificity or the identification of enzymes involved in GalXM synthesis, which could then be localized within the cell. These studies must therefore await the development of such reagents.
The primary GXM-containing structures we observed were secretory vesicles. In early ultrastructural studies, Takeo et al. (Takeo et al., 1973a, 1973b) hypothesized that 30–70-nm intracellular vesicles and/or ∼50-nm vesicles in paramural bodies were involved in capsule synthesis. The typical intracellular vesicle size we observed was 100 nm, although it ranged from 60 to 130 nm (Figures 6 and 8). This difference could be due to sample preparation, because the previous study used a freeze-etching technique, and we used plastic embedding and thin-sectioning. It is also possible that the previously observed small (30–50 nm) vesicles were endocytic (Prescianotto-Baschong and Riezman, 1998), in contrast to those that we observed. Structures similar to the paramural body are occasionally observed in C. neoformans strains grown in vitro (A. Yoneda and T. L. Doering, unpublished TEM observation), although their significance is not known. They could potentially be related to mammalian exosomes (Thery et al., 2002) or plant paramural bodies (An et al., 2006).
In more recent studies using anti-GXM antibodies, Feldmesser et al. (2001) showed immuno-gold labeling of 200–300-nm electron-lucent areas in the cytosol, and Garcia-Riviera et al. (2004) noted “clusters” of antibody labeling in the cytosol and in the cell wall. The size of the former entities and cell wall location of the latter suggest these do not correspond to post-Golgi vesicles.
In other ultrastructural studies, Sakaguchi et al. (1993) proposed that the “particle-accumulating layer” between the cell wall and the capsule represented capsule precursors. Our laboratory has shown that both α-1,3 glucan and proteins are required for capsule association with the cell wall (Reese and Doering, 2003, and A. J. Reese and T. L. Doering, unpublished results). Thus, an alternate model is that the observed particles are proteinaceous and are involved in linking the capsule and α-1,3-glucan or perhaps in the assembly of capsule fibers.
Mutation of the gene encoding a C. neoformans homolog of the Sec4/Rab8 GTPases, SAV1, yielded a mutant that accumulates exocytic vesicles. Interestingly, this gene does not itself complement the S. cerevisiae sec4-8 mutant (Supplementary Figure S1). This is the second reported fungal Sec4p that is unable to complement sec4-8, the first being one of the two Sec4p homologues present in Aspergillus niger (SrgAp, Figure 2; Punt et al., 2001).
We observed another interesting difference between S. cerevisiae and C. neoformans when C. neoformans cells were treated with LatB, which compromises vesicle polarization in both species by depolymerization of F-actin (Karpova et al., 2000; Figure 7 and Supplementary Figure S5). It has been reported that disassembly of F-actin by LatA caused vesicle accumulation in wild-type S. cerevisiae, although secretion per se was not blocked (Karpova et al., 2000). In contrast, total vesicle numbers in C. neoformans treated with LatB were slightly lower than control, whether in SAV1 or sav1 strains (Figure 7).
These differences between C. neoformans and S. cerevisiae are noteworthy, particularly because exocytosis in C. neoformans has not been specifically addressed before this work. One earlier study did suggest that the C. neoformans Cap59 protein is involved in trafficking of capsule and of proteins (Garcia-Rivera et al., 2004). In our studies of cap59 cells, we have not observed specific accumulation of secretory vesicles or other structures containing capsule material (A. Yoneda and T. L. Doering, unpublished results), or any defect in protein secretion (Supplementary Figure S3). It is possible that Cap59p, given its weak homology to glycosyltransferases, is involved in GXM synthesis itself.
C. neoformans constitutively produces capsule in laboratory media, but this structure enlarges when cells are grown under conditions that mimic in vivo growth, such as low iron or 5% CO2 (Granger et al., 1985; Vartivarian et al., 1993; Zaragoza et al., 2003). It has been assumed that these “in-ducing” conditions cause the cell to up-regulate capsule synthesis. However, the presence of larger capsules could also reflect upregulation of secretion or assembly of extracellular capsule polymers that have already been synthesized, down-regulation of capsule degradation or release, or production of longer polymers. Further investigation is needed to establish the underlying mechanism of capsule induction.
Although capsule is known to be shed from living cells, the mechanism and rate of release have not been characterized. For this reason quantifying shed GXM would not distinguish altered capsule secretion from changed levels of GXM release from the existing capsule. It would add to our understanding of capsule export if quantification of newly synthesized extracellular GXM were possible. However, there is no procedure available to completely remove pre-existing capsule while maintaining cell viability (Gates et al., 2004; Bryan et al., 2005). Our electron micrographs do show extracellular capsule on the surface of sav1 cells (Figure 8). This may be due to either the continued presence of capsule materials that were secreted before the temperature shift or the incomplete nature of the secretion block (Figure 4).
We have established the site of GXM synthesis and its manner of export. Despite these advances, however, many unanswered questions of cryptococcal capsule biosynthesis remain. These include the mechanisms of translocation of capsular materials across the cell wall after secretion, assembly of capsule fibrils, attachment of capsule to the cell wall, enlargement of the capsule structure upon environmental stimuli, and shedding of capsule into the extracellular milieu. In addition to these events downstream of export, we have not yet defined the initial stages of capsule biosynthesis, including the nature of any primer molecules and of the intracellular forms of GXM. Future studies, potentially employing the sav1 strain, should shed light on these puzzles of cryptococcal capsule biogenesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Randy Schekman, Patrick Brennwald, Wandy Beatty, and members of the Doering lab for helpful discussions; Dr. Michael Brent for advice on statistical analysis; and Dr. Indrani Bose and Morgann Reilly for comments on the manuscript. We are grateful to Drs. Thomas Kozel and Arturo Casadevall for generously providing anti-GXM monoclonal antibodies and Drs. Gary Cox and Jennifer Lodge for C. neoformans strains and plasmids. We also thank Drs. Vladimir Lupashin and Heather True-Krob for S. cerevisiae strains and plasmids. Work on capsule synthesis in the Doering lab is supported by National Institutes of Health R01 Grants GM71007 and GM66303 to T.L.D.
Abbreviations used:
- GXM
glucuronoxylomannan
- GalXM
galactoxylomannan.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0701) on October 4, 2006.
REFERENCES
- An Q., Huckelhoven R., Kogel K. H., van Bel A. J. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol. 2006;8:1009–1019. doi: 10.1111/j.1462-5822.2006.00683.x. [DOI] [PubMed] [Google Scholar]
- Ang A. L., Taguchi T., Francis S., Folsch H., Murrells L. J., Pypaert M., Warren G., Mellman I. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 2004;167:531–543. doi: 10.1083/jcb.200408165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough K. R. Endocytosis and the development of cell polarity in yeast require a dynamic F-actin cytoskeleton. Curr. Biol. 2000;10:1587–1590. doi: 10.1016/s0960-9822(00)00859-9. [DOI] [PubMed] [Google Scholar]
- Ayscough K. R. Defining protein modules for endocytosis. Cell. 2005;123:188–190. doi: 10.1016/j.cell.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Bacic A. Breaking an impasse in pectin biosynthesis. Proc. Natl. Acad. Sci. USA. 2006;103:5639–5640. doi: 10.1073/pnas.0601297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose I., Reese A. J., Ory J. J., Janbon G., Doering T. L. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell. 2003;2:655–663. doi: 10.1128/EC.2.4.655-663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan R. A., Zaragoza O., Zhang T., Ortiz G., Casadevall A., Dadachova E. Radiological studies reveal radial differences in the architecture of the polysaccharide capsule of Cryptococcus neoformans. Eukaryot. Cell. 2005;4:465–475. doi: 10.1128/EC.4.2.465-475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdá-Olmedo E., Lipson E. D. Phycomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1987. [Google Scholar]
- Chang Y. C., Kwon-Chung K. J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Kwon-Chung K. J. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 1998;66:2230–2236. doi: 10.1128/iai.66.5.2230-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Kwon-Chung K. J. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 1999;181:5636–5643. doi: 10.1128/jb.181.18.5636-5643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C., Penoyer L. A., Kwon-Chung K. J. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 1996;64:1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherniak R., Sundstrom J. B. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 1994;62:1507–1512. doi: 10.1128/iai.62.5.1507-1512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes J. C., Ishiguro J., Duran A., Ribas J. C. Localization of the (1,3)beta-D-glucan synthase catalytic subunit homologue Bgs1p/Cps1p from fission yeast suggests that it is involved in septation, polarized growth, mating, spore wall formation and spore germination. J. Cell Sci. 2002;115:4081–4096. doi: 10.1242/jcs.00085. [DOI] [PubMed] [Google Scholar]
- Craighead M. W., Bowden S., Watson R., Armstrong J. Function of the ypt2 gene in the exocytic pathway of Schizosaccharomyces pombe. Mol. Biol. Cell. 1993;4:1069–1076. doi: 10.1091/mbc.4.10.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta. 1999;1426:309–322. doi: 10.1016/s0304-4165(98)00132-9. [DOI] [PubMed] [Google Scholar]
- Dhugga K. S. Plant Golgi cell wall synthesis: from genes to enzyme activities. Proc. Natl. Acad. Sci. USA. 2005;102:1815–1816. doi: 10.1073/pnas.0409649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas B., Borel C., Herbert C., Maury J., Jacquet C., Balsse R., Esquerre-Tugaye M. T. Molecular characterization of CLPT1, a SEC4-like Rab/GTPase of the phytopathogenic fungus Colletotrichum lindemuthianum which is regulated by the carbon source. Gene. 2001;272:219–225. doi: 10.1016/s0378-1119(01)00536-4. [DOI] [PubMed] [Google Scholar]
- Edwards M. R., Gordon M. A., Lapa E. W., Ghiorse W. C. Micromorphology of Cryptococcus neoformans. J. Bacteriol. 1967;94:766–777. doi: 10.1128/jb.94.3.766-777.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmesser M., Kress Y., Casadevall A. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology. 2001;147:2355–2365. doi: 10.1099/00221287-147-8-2355. [DOI] [PubMed] [Google Scholar]
- Garcia-Rivera J., Chang Y. C., Kwon-Chung K. J., Casadevall A. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell. 2004;3:385–392. doi: 10.1128/EC.3.2.385-392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates M. A., Thorkildson P., Kozel T. R. Molecular architecture of the Cryptococcus neoformans capsule. Mol. Microbiol. 2004;52:13–24. doi: 10.1111/j.1365-2958.2003.03957.x. [DOI] [PubMed] [Google Scholar]
- Goins C. L., Gerik K. J., Lodge J. K. Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet. Biol. 2006;43:531–544. doi: 10.1016/j.fgb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Granger D. L., Perfect J. R., Durack D. T. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J. Clin. Invest. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Roth D., Walch-Solimena C., Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S., David D., Gerst J. E. Dynamin and clathrin are required for the biogenesis of a distinct class of secretory vesicles in yeast. EMBO J. 2002;21:602–614. doi: 10.1093/emboj/21.4.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E., Bretscher A. Parallel secretory pathways to the cell surface in yeast. J. Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsay E., Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J. Cell Biol. 2002;156:271–285. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubruck H., Engelke U., Mertins P., Gallwitz D. Structural and functional analysis of ypt2, an essential ras-related gene in the fission yeast Schizosaccharomyces pombe encoding a Sec4 protein homologue. EMBO J. 1990;9:1957–1962. doi: 10.1002/j.1460-2075.1990.tb08323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcomb C. L., Etcheverry T., Schekman R. Isolation of secretory vesicles from Saccharomyces cerevisiae. Anal. Biochem. 1987;166:328–334. doi: 10.1016/0003-2697(87)90581-1. [DOI] [PubMed] [Google Scholar]
- Huckaba T. M., Gay A. C., Pantalena L. F., Yang H. C., Pon L. A. Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 2004;167:519–530. doi: 10.1083/jcb.200404173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itano N., Kimata K. Mammalian hyaluronan synthases. IUBMB Life. 2002;54:195–199. doi: 10.1080/15216540214929. [DOI] [PubMed] [Google Scholar]
- Janbon G. Cryptococcus neoformans capsule biosynthesis and regulation. FEMS Yeast Res. 2004;4:765–771. doi: 10.1016/j.femsyr.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C. P., Drubin D. G. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Karpova T. S., Reck-Peterson S. L., Elkind N. B., Mooseker M. S., Novick P. J., Cooper J. A. Role of actin and Myo2p in polarized secretion and growth of Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:1727–1737. doi: 10.1091/mbc.11.5.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis F. M., Boorsma A., De Groot P. W. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202. doi: 10.1002/yea.1349. [DOI] [PubMed] [Google Scholar]
- Konomi M., Fujimoto K., Toda T., Osumi M. Characterization and behaviour of alpha-glucan synthase in Schizosaccharomyces pombe as revealed by electron microscopy. Yeast. 2003;20:427–438. doi: 10.1002/yea.974. [DOI] [PubMed] [Google Scholar]
- Kozel T. R., Gotschlich E. C. The capsule of Cryptococcus neoformans passively inhibits phagocytosis of the yeast by macrophages. J. Immunol. 1982;129:1675–1680. [PubMed] [Google Scholar]
- Latge J. P., Mouyna I., Tekaia F., Beauvais A., Debeaupuis J. P., Nierman W. Specific molecular features in the organization and biosynthesis of the cell wall of Aspergillus fumigatus. Med. Mycol. 2005;43(Suppl 1):S15–S22. doi: 10.1080/13693780400029155. [DOI] [PubMed] [Google Scholar]
- Leal-Morales C. A., Bracker C. E., Bartnicki-Garcia S. Localization of chitin synthetase in cell-free homogenates of Saccharomyces cerevisiae: chitosomes and plasma membrane. Proc. Natl. Acad. Sci. USA. 1988;85:8516–8520. doi: 10.1073/pnas.85.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. A., Mao Y., Zhang Z., Wong B. Overexpression of a dominant-negative allele of YPT1 inhibits growth and aspartyl protease secretion in Candida albicans. Microbiology. 2001;147:1961–1970. doi: 10.1099/00221287-147-7-1961. [DOI] [PubMed] [Google Scholar]
- Lesage G., Bussey H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006;70:317–343. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. H., Chou W. I., Lin S. C., Sheu C. C., Chang M. D. Molecular genetic manipulation of Pichia pastoris SEC4 governs cell growth and glucoamylase secretion. Biochem. Biophys. Res. Commun. 2005;336:1172–1180. doi: 10.1016/j.bbrc.2005.08.234. [DOI] [PubMed] [Google Scholar]
- Mao Y., Kalb V. F., Wong B. Overexpression of a dominant-negative allele of SEC4 inhibits growth and protein secretion in Candida albicans. J. Bacteriol. 1999;181:7235–7242. doi: 10.1128/jb.181.23.7235-7242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado A. B., Morrell J. L., Konomi M., An H., Petit C., Osumi M., Balasubramanian M., Gould K. L., Del Rey F., de Aldana C. R. Role of septins and the exocyst complex in the function of hydrolytic enzymes responsible for fission yeast cell separation. Mol. Biol. Cell. 2005;16:4867–4881. doi: 10.1091/mbc.E04-12-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade H. C., Cox G. M. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 2001;39:151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]
- McFadden D. C., De Jesus M., Casadevall A. The physical properties of the capsular polysaccharides from Cryptococcus neoformans suggest features for capsule construction. J. Biol. Chem. 2006;281:1868–1875. doi: 10.1074/jbc.M509465200. [DOI] [PubMed] [Google Scholar]
- Monari C., Bistoni F., Vecchiarelli A. Glucuronoxylomannan exhibits potent immunosuppressive properties. FEMS Yeast Res. 2006;6:537–542. doi: 10.1111/j.1567-1364.2006.00072.x. [DOI] [PubMed] [Google Scholar]
- Mulholland J., Wesp A., Riezman H., Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretary vesicle. Mol. Biol. Cell. 1997;8:1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. What can yeast tell us about N-linked glycosylation in the Golgi apparatus? FEBS Lett. 2001;498:223–227. doi: 10.1016/s0014-5793(01)02488-7. [DOI] [PubMed] [Google Scholar]
- Nelson R. T., Hua J., Pryor B., Lodge J. K. Identification of virulence mutants of the fungal pathogen Cryptococcus neoformans using signature-tagged mutagenesis. Genetics. 2001;157:935–947. doi: 10.1093/genetics/157.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Novick P., Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Zerial M. The diversity of Rab proteins in vesicle transport. Curr. Opin. Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- Nussbaum M., Collins R. N. Use of search algorithms to define specificity in Rab GTPase domain function. Methods Enzymol. 2005;403:10–19. doi: 10.1016/S0076-6879(05)03002-8. [DOI] [PubMed] [Google Scholar]
- Ortiz D., Novick P. J. Ypt32p regulates the translocation of Chs3p from an internal pool to the plasma membrane. Eur J. Cell Biol. 2006;85:107–116. doi: 10.1016/j.ejcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Pierini L. M., Doering T. L. Spatial and temporal sequence of capsule construction in Cryptococcus neoformans. Mol. Microbiol. 2001;41:105–115. doi: 10.1046/j.1365-2958.2001.02504.x. [DOI] [PubMed] [Google Scholar]
- Prescianotto-Baschong C., Riezman H. Morphology of the yeast endocytic pathway. Mol. Biol. Cell. 1998;9:173–189. doi: 10.1091/mbc.9.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt P. J., Seiboth B., Weenink X. O., van Zeijl C., Lenders M., Konetschny C., Ram A. F., Montijn R., Kubicek C. P., van den Hondel C. A. Identification and characterization of a family of secretion-related small GTPase-encoding genes from the filamentous fungus Aspergillus niger: a putative SEC4 homologue is not essential for growth. Mol. Microbiol. 2001;41:513–525. doi: 10.1046/j.1365-2958.2001.02541.x. [DOI] [PubMed] [Google Scholar]
- Raetz C. R., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese A. J., Doering T. L. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 2003;50:1401–1409. doi: 10.1046/j.1365-2958.2003.03780.x. [DOI] [PubMed] [Google Scholar]
- Reiter W. D. Biosynthesis and properties of the plant cell wall. Curr. Opin. Plant Biol. 2002;5:536–542. doi: 10.1016/s1369-5266(02)00306-0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Elorza M. V., Valentin E., Sentandreu R. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 2006;6:14–29. doi: 10.1111/j.1567-1364.2005.00017.x. [DOI] [PubMed] [Google Scholar]
- Sakaguchi N., Baba T., Fukuzawa M., Ohno S. Ultrastructural study of Cryptococcus neoformans by quick-freezing and deep-etching method. Mycopathologia. 1993;121:133–141. doi: 10.1007/BF01104068. [DOI] [PubMed] [Google Scholar]
- Scheible W. R., Pauly M. Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr. Opin. Plant Biol. 2004;7:285–295. doi: 10.1016/j.pbi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Segev N. Ypt/Rab GTPases: regulators of protein trafficking. Sci STKE. 2001;2001:RE11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- Siriputthaiwan P., Jauneau A., Herbert C., Garcin D., Dumas B. Functional analysis of CLPT1, a Rab/GTPase required for protein secretion and pathogenesis in the plant fungal pathogen Colletotrichum lindemuthianum. J. Cell Sci. 2005;118:323–329. doi: 10.1242/jcs.01616. [DOI] [PubMed] [Google Scholar]
- Somerville C. Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- Takeo K., Uesaka I., Uehira K., Nishiura M. Fine structure of Cryptococcus neoformans grown in vitro as observed by freeze-etching. J. Bacteriol. 1973a;113:1442–1448. doi: 10.1128/jb.113.3.1442-1448.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeo K., Uesaka I., Uehira K., Nishiura M. Fine structure of Cryptococcus neoformans grown in vivo as observed by freeze-etching. J. Bacteriol. 1973b;113:1449–1454. doi: 10.1128/jb.113.3.1449-1454.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney A. E., Brown R. H., Vaske C., Lodge J. K., Doering T. L., Brent M. R. Gene prediction and verification in a compact genome with numerous small introns. Genome Res. 2004;14:2330–2335. doi: 10.1101/gr.2816704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C., Zitvogel L., Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- To-e A., Ueda Y., Kakimoto S. I., Oshima Y. Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J. Bacteriol. 1973;113:727–738. doi: 10.1128/jb.113.2.727-738.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti D. L., Rude T. H., Johnston S. A., Durack D. T., Perfect J. R. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnav V. V., Bacon B. E., O'Neill M., Cherniak R. Structural characterization of the galactoxylomannan of Cryptococcus neoformans Cap67. Carbohydr. Res. 1998;306:315–330. doi: 10.1016/s0008-6215(97)10058-1. [DOI] [PubMed] [Google Scholar]
- Valdivia R. H., Schekman R. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA. 2003;100:10287–10292. doi: 10.1073/pnas.1834246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heijenoort J. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology. 2001;11:25R–36R. doi: 10.1093/glycob/11.3.25r. [DOI] [PubMed] [Google Scholar]
- van Rijn H. J., Boer P., Steyn-Parve E. P. Biosynthesis of acid phosphatase of baker's yeast. Factors influencing its production by protoplasts and characterization of the secreted enzyme. Biochim. Biophys. Acta. 1972;268:431–441. doi: 10.1016/0005-2744(72)90339-7. [DOI] [PubMed] [Google Scholar]
- Vartivarian S. E., Anaissie E. J., Cowart R. E., Sprigg H. A., Tingler M. J., Jacobson E. S. Regulation of cryptococcal capsular polysaccharide by iron. J. Infect. Dis. 1993;167:186–190. doi: 10.1093/infdis/167.1.186. [DOI] [PubMed] [Google Scholar]
- Vecchiarelli A. The cellular responses induced by the capsular polysaccharide of Cryptococcus neoformans differ depending on the presence or absence of specific protective antibodies. Curr. Mol. Med. 2005;5:413–420. doi: 10.2174/1566524054022585. [DOI] [PubMed] [Google Scholar]
- Wang H., Tang X., Liu J., Trautmann S., Balasundaram D., McCollum D., Balasubramanian M. K. The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell. 2002;13:515–529. doi: 10.1091/mbc.01-11-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel P. H., Hascall V. C., Tammi M. Hyaluronan synthases. J. Biol. Chem. 1997;272:13997–14000. doi: 10.1074/jbc.272.22.13997. [DOI] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Wright R. Transmission electron microscopy of yeast. Microsc. Res. Tech. 2000;51:496–510. doi: 10.1002/1097-0029(20001215)51:6<496::AID-JEMT2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Zaragoza O., Fries B. C., Casadevall A. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO(2) Infect. Immun. 2003;71:6155–6164. doi: 10.1128/IAI.71.11.6155-6164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O., Telzak A., Bryan R. A., Dadachova E., Casadevall A. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol. Microbiol. 2006;59:67–83. doi: 10.1111/j.1365-2958.2005.04928.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.