Abstract
The procyclic form of Trypanosoma brucei exists in the midgut of the tsetse fly. The current model of its surface glycocalyx is an array of rod-like procyclin glycoproteins with glycosylphosphatidylinositol (GPI) anchors carrying sialylated poly-N-acetyllactosamine side chains interspersed with smaller sialylated poly-N-acetyllactosamine–containing free GPI glycolipids. Mutants for TbGPI12, deficient in the second step of GPI biosynthesis, were devoid of cell surface procyclins and poly-N-acetyllactosamine–containing free GPI glycolipids. This major disruption to their surface architecture severely impaired their ability to colonize tsetse fly midguts but, surprisingly, had no effect on their morphology and growth characteristics in vitro. Transmission electron microscopy showed that the mutants retained a cell surface glycocalyx. This structure, and the viability of the mutants in vitro, prompted us to look for non-GPI–anchored parasite molecules and/or the adsorption of serum components. Neither were apparent from cell surface biotinylation experiments but [3H]glucosamine biosynthetic labeling revealed a group of previously unidentified high apparent molecular weight glycoconjugates that might contribute to the surface coat. While characterizing GlcNAc-PI that accumulates in the TbGPI12 mutant, we observed inositolphosphoceramides for the first time in this organism.
INTRODUCTION
The tsetse fly–transmitted protozoan parasite Trypanosoma brucei causes human sleeping sickness and the cattle disease Nagana in sub-Saharan Africa. The organism undergoes a complex life cycle between the mammalian host and the insect, tsetse, vector. The bloodstream (trypomastigote) form of the parasite avoids the host's innate immune system through the expression of a dense monolayer of 5 × 106 glycosylphosphatidylinositol (GPI)-anchored variant surface glycoprotein (VSG) dimers and avoids specific immune responses through antigenic variation (Cross, 1996; Vanhamme et al., 2001). The bloodstream-form parasites exist as dividing “slender” forms and nondividing “stumpy” forms that are preadapted for survival in the tsetse fly. After ingestion in a blood meal, the stumpy trypomastigote-form parasites differentiate into dividing procyclic-form parasites that colonizes the tsetse midgut. The procyclic trypanosomes express a radically different cell surface coat, thought to be made up ∼3 × 106 procyclin glycoproteins (Roditi et al., 1987; Mowatt et al., 1987; Richardson et al., 1988) and a smaller number (∼1 × 106) of poly-N-acetyllactosamine–containing free GPIs (Lillico et al., 2003; Vassella et al., 2003; Nagamune et al., 2004; Roper et al., 2005). The procyclins are polyanionic, rod-like (Roditi et al., 1989; Treumann et al., 1997), proteins encoded by procyclin genes (Roditi and Clayton, 1999). In T. brucei strain 427, used in this study, the parasites contain (per diploid genome) two copies of the GPEET1 gene encoding 6 Gly-Pro-Glu-Glu-Thr repeats, one copy each of the EP1-1 and EP1-2 genes, encoding EP1 procyclins with 30 and 25 Glu-Pro repeats, respectively, two copies of the EP2-1 gene, encoding EP2 procyclin with 25 Glu-Pro repeats and two copies of the EP3-1 gene, encoding EP3 procyclin with 22 Glu-Pro repeats (Acosta-Serrano et al., 1999). The EP1 and EP3 procyclins contain a single N-glycosylation site, occupied exclusively by a conventional Man5GlcNAc2 oligosaccharide, at the N-terminal side of the Glu-Pro repeat domain (Treumann et al., 1997; Acosta-Serrano et al., 1999). Whereas neither EP2 nor GPEET procyclin is N-glycosylated, GPEET1 procyclin is phosphorylated on six of seven Thr residues (Butikofer et al., 1999; Mehlert et al., 1999; Schlaeppi et al., 2003). In culture, the procyclin expression profile depends on the carbon source (Vassella et al., 2000) and metabolic state of the cells (Morris et al., 2002; Vassella et al., 2004) and in the tsetse fly there appears to be a program of procyclin expression such that GPEET procyclin is expressed early, giving way to EP1 and EP3 procyclin expression (Acosta-Serrano et al., 2001; Vassella et al., 2001). GPEET and EP procyclins contain similar GPI membrane anchors. These are based on the ubiquitous ethanolamine-P-6Manα1–2Manα1-6Manα1-4GlcNα1-6PI core (where, in this case, the phosphatidylinositol [PI] lipid is a 2-O-acyl-lyso-PI structure; Treumann et al., 1997), but they also contain the largest and most complex known GPI side chains. These side chains are large poly-disperse branched poly-N-acetyllactosamine structures (with an average of ∼8–12 repeats, depending on the preparation) that can terminate with α2-3–linked sialic acid residues (Ferguson et al., 1993; Treumann et al., 1997). Sialic acid is transferred from serum sialoglycoconjugates to terminal βGal residues by the action of a cell surface GPI-anchored trans-sialidase enzyme (Engstler et al., 1993; Pontes de Carvalho et al., 1993; Montagna et al., 2002). Trans-sialylation of surface components plays a role in the successful colonization of the tsetse fly (Nagamune et al., 2004). In vivo, the N-termini of the procyclins are removed by tsetse fly gut proteases (Acosta-Serrano et al., 2001), though the role of this event is unclear (Liniger et al., 2004), and it is thought that the underlying (protease resistant) anionic repeat units and associated GPI anchor side chains might protect the parasite from the approach tsetse fly gut hydrolases (Acosta-Serrano et al., 2001).
In previous studies, the cell surface architecture of procyclic trypanosomes has been manipulated by gene knockout of the procyclin genes themselves (Vassella et al., 2003) or genes encoding enzymes that act in the later parts of the GPI biosynthetic pathway, i.e., TbGPI10 and TbGPI8 (Nagamune et al., 2000, 2004; Lillico et al., 2003), or by galactose-starvation (Roper et al., 2005). The procyclin, TbGPI10 and TbGPI8 knockouts all resulted in parasites devoid of GPI-anchored procyclins, but this was apparently compensated for by an up-regulation in free GPI expression.
In this article, we describe the phenotype of procyclic trypanosome TbGPI12 null mutants that cannot synthesize GPI structures beyond GlcNAc-PI. To our surprise, these mutants are viable in culture, though unable to colonize the tsetse midgut. The mutant also revealed that the procyclic trypanosome surface coat contains molecules other than GPI-anchored proteins and free GPIs. Adsorbed serum components were excluded as coat components and [3H]glucosamine labeling revealed a hereto unidentified high-molecular-weight glycoconjugates that may constitute some or all of the residual surface coat.
MATERIALS AND METHODS
Cell Culture and Generation of Procyclic-form T. brucei TbGPI12 Null and Conditional Null Mutants
Puromycin (PAC) and blasticydin (BSD) antibiotic resistance genes were cloned into TbGPI12-targeted gene replacement plasmids and the tetracycline-inducible expression plasmid (pLew100) containing the TbGPI12 ORF with a C-terminal myc-tag were prepared as described in (Chang et al., 2002). Plasmid maxi-preps were prepared using a Qiagen kit (Qiagen, Crawley, West Sussex, United Kingdom), digested with NotI, heated at 65°C for 30 min, precipitated with ethanol, redissolved in sterile water, and used for electroporation of procyclic forms of T. brucei strain 427 clone 29.13, referred in this article as wild-type (WT) cells. These cells were grown in SDM-79 (Brun and Schonenberger, 1979) in the presence of 15% fetal bovine serum (FBS), haemin 7.5 mg/L at 28°C, containing the appropriate antibiotics for selection (G418 at 15 μg/ml to maintain the T7 RNA polymerase; hygromycin at 50 μg/ml to maintain the tetracycline repressor protein; phleomycin at 2.5 μg/ml for pLew100; puromycin at 1 μg/ml; and blasticydin at 10 μg/ml). Cell densities were measured using a hemocytometer and plotted as the product of cell density and dilution.
Southern Blotting
Genomic DNA was prepared using DNAzol (Helena Biosciences, Gateshead, United Kingdom). Probes were amplified by PCR, gel-purified, and dUTP-fluorescein–labeled by random priming (Gene Images Kit, GE Healthcare, Little Chalfont, United Kingdom).
Cell-free System Experiment
Procyclic-form T. brucei membranes were prepared (Masterson et al., 1989) from wild-type cells, TbGPI12 null mutant, and TbGPI12 conditional null mutant grown with daily addition of 1 μg/ml tetracycline, or without tetracycline for 16 d, in media containing 15% FBS certified tetracycline-free (Clontech, Palo Alto, CA). Trypanosome membranes were washed twice and resuspended at 1 × 10−9/ml in 2× incorporation buffer (Güther and Ferguson, 1995). Aliquots (2 × 107 cells) were added to equal volume of water containing 2 μCi of UDP[3H]GlcNAc (41.6 Ci/mmol, Perkin Elmer-Cetus) and 2 mM GDP-Man and labeled for 20 min at 30°C. Subsequent chloroform/methanol/water and butan-1-ol extractions were performed as described before (Güther et al., 1994). Samples and glycolipid standards were run on aluminum-backed silica gel-60 HPTLC (Merck, Rahway, NJ) and developed using chloroform/methanol/1 M ammonium acetate/13 M ammonia/water (180:140:9:9:23, vol/vol). Radiolabeled compounds were detected by fluorography at −80°C after spraying with En3Hance (Perkin-Elmer Cetus, Norwalk, CT) using Kodak XAR-5 film (Eastman Kodak, Rochester, NY) and intensifying screens.
Anti-myc Western Blotting
Washed parasites were lysed in 20 mM Tris-HCl, pH 7.2, 2% SDS, boiled and adjusted to 0.3% SDS, and 1% Triton X-100 in 20 mM Tris-HCl, pH 7.2, and aliquots equivalent to 2 × 108 cells were immunoprecipitated with 1 μg anti-myc mAb (Upstate Biotechnology, Lake Placid, NY) and protein G agarose (Sigma, Poole, Dorset, United Kingdom). The washed beads were boiled in SDS sample buffer and applied to an Invitrogen gel (Paisley, United Kingdom; 4–12%, NuPage with MOPS buffer). Proteins were transferred to nitrocellulose, and the blocked filter was probed with anti-myc mAb (10 ng/ml) and developed with an anti-mouse secondary antibody conjugated to horseradish peroxidase (Sigma; diluted 1:10,000) and ECL reagent (Amersham).
Mass Spectrometry of Lipid Extracts
Wild-type procyclic-form T. brucei, TbGPI12 null mutant cells (5 × 108 total) were washed and resuspended in 0.1 ml phosphate-buffered saline and extracted with chloroform/methanol/water (10:10:3 vol/vol/vol) overnight at 4°C and then sonicated for 15 min in a sonicating water bath. The extract was centrifuged for 5 min at full speed in an Eppendorf microfuge, and the supernatant was transferred to a fresh Eppendorf tube. After drying under a stream of nitrogen, the products were partitioned between butan-1-ol and water (0.2 ml each). The aqueous phase was extracted twice more with 0.2 ml water-saturated butan-1-ol. The combined butan-1-ol phases were back-washed three times with 0.4 ml butanol-saturated water. The washed butan-1-ol phases were dried under nitrogen and dissolved in 200 μl chloroform/methanol/water (10:10:3 vol/vol/vol). Small aliquots were transferred to Waters nanotips (Millipore, Milford, MA; type F) and analyzed by electrospray ionization–mass spectrometry (ES-MS) in negative ion mode on an ABI Q-StarXL mass spectrometer (Surrey, United Kingdom). Individual ions were subjected to collision induced dissociation (CID) and tandem mass spectrometry (ES-MS/MS). The product ion spectra were used to identify phospholipid type and molecular species.
FACS Analysis
For FACS analysis, an aliquot of wild-type and TbGPI12 null mutant cells were centrifuged, resuspended in 1 ml of fresh SDM-79 media, and incubated with anti-EP mAb 247 (Richardson et al., 1988) diluted 1:100 (30 min, room temperature). Parasites were washed with cold PBS and incubated with FITC anti-mouse (Sigma; diluted 1:10000) for 1 h at room temperature. After incubation, parasites (at the concentration of 5 × 106 cells/ml) were analyzed in a Becton Dickinson FACSCalibur (Cockeysville, MD) using detector FL1-A.
Metabolic Labeling of Live T. brucei Procyclic Forms
T. brucei procyclic forms (5 ml at 107/ml) were washed twice with SDM-79 media depleted of l-proline, d-mannose, d-glucose, d-glucosamine, l-serine, l-ethanolamine and containing 2% FBS and resuspended in 10 ml of the same media and split into 2-ml aliquots. One aliquot was labeled with 5 μCi/ml [14C]proline for 2 h at 28°C. Another aliquot, after addition of l-proline and l-hydroxy-proline, was labeled either with [3H]ethanolamine (50 μCi/ml) or [3H]glucosamine (200 μCi/ml) for 20 h at 28°C, in the latter case with the further addition of l-serine. The final concentrations of all supplements were as described for SDM-79 (Brun and Schönenberger, 1979). After labeling, cells were washed and resuspended in PBS. An equal volume of 2× SDS-sample buffer containing 0.2 M DTT was added, and the samples were boiled and applied into 4–12% Nupage BisTris polyacrylamide gels and run using MOPS running buffer (Invitrogen). Gels were stained with Coomassie blue, soaked in En3Hance (Perkin Elmer-Cetus), dried, and placed in contact with Kodak XAR-5 film and intensifying screen at −80°C.
Ruthenium Red Stain and Ultramicroscopy
Parasites in culture medium were washed twice in cold PBS and fixed at 4°C for 1 h in 2.5% glutaraldehyde, 0.1 M cacodylate buffer containing 0.15% ruthenium red, 5 mM CaCl2, and 5% sucrose and processed according to Zufferey et al., (2003) through 1% osmium tetroxide/ruthenium red. Further processing included 30-min en bloc staining of the cells with 1% aqueous uranyl acetate before a graded ethanol dehydration, rinsing with propylene oxide (2 × 5 min), and embedding in Epon-Araldite resin. Ultrathin sections were prepared on 200-mesh grids, stained in Reynold's lead citrate for 5 min, and viewed at 120 kV on a Zeiss 912 Omega transmission electron microscope (Thornwood, NY), recording zero-loss images on a 2K Proscan digital camera system (Proscan, Lagerlechfeld, Germany).
Surface Biotinylation of Procyclic Form T. brucei
Wild-type and TbGPI12 null mutant procyclic cells (10 ml cultures at 3 × 107/ml for each) were washed three times with 20 ml PBS and resuspended in 20 ml of EZ-link sulfo-NHS-biotin (Pinpoint Cell Surface Protein Isolation Kit, Pierce, Rockford, IL) in PBS and rotated at 4°C for 30 min. After quenching, lysate preparation, and centrifugation, the supernatant was purified using Neutravidin beads. Proteins were eluted by boiling the beads with SDS-sample buffer containing DTT and were applied into 4–12% Nupage BisTris polyacrylamide gels run with MOPS buffer (Invitrogen). The gel was stained with Coomassie blue, and the more abundant bands in the null mutant lane were cut out of the gel, alkylated, digested with trypsin, and processed for mass-fingerprinting. (Fingerprints Proteomic Facility, University of Dundee, Scotland). Identical samples were applied into a duplicate gel to develop with silver to visualize procyclins.
Tsetse Fly Infections
Pupae of Glossina morsitans morsitants were obtained from the Institute of Zoology, Slovak Academy of Science (Bratislava, Slovakia). Newly hatched (teneral) flies were fed with an infected bloodmeal, which consisted of 107 parasites mixed with washed defribinated horse blood (containing 10% FBS). In the case of flies infected with TbGPI12 conditional null mutant cells, the bloodmeal also contained 25 μg/ml tetracycline. The latter was always included, at the same concentration, in successive bloodmeals until flies were dissected. Tetracycline is not harmful for tsetse flies, and it has been successfully used in the past to induce trypanosome gene expression within tsetse compartments (Peacock et al., 2005). Infected flies were fed with bloodmeals every 2–3 d. After 2 wk, midguts were isolated from infected flies and disrupted by mechanical force in cold SDM-79 containing 10% FBS. Isolated parasites from individual midguts were kept on ice until counted on a hemocytometer.
RESULTS
Creation and Growth Phenotype of TbGPI12 Null Mutant Procyclic T. brucei
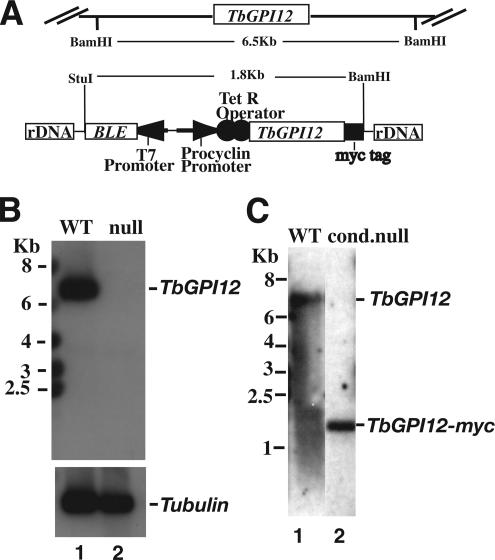
TbGPI12 is present as a single copy per haploid genome (Chang et al., 2002). We replaced one allele with puromycin acetyltransferase (PAC) and, after antibiotic selection, we replaced the second allele with blasticydin deaminase (BSD). After dual antibiotic selection, a Southern blot with a TbGPI12 ORF probe showed that both alleles were replaced and that we had created a ΔTbGPI12::PAC/ΔTbGPI12::BSD null mutant (Figure 1B). A tetracycline-inducible TbGPI12 conditional null mutant was also created by introducing an ectopic C-terminally myc-tagged version of TbGPI12, targeted to the ribosomal DNA locus of the null mutant using the pLew100 vector (Wirtz et al., 1999). A Southern blot with a TbGPI12 ORF probe showed the presence of the ectopic gene and the absence of endogenous copies, i.e., a genotype of TiTbGPI12myc/ΔTbGPI12::PAC/ΔTbGPI12::BSD (Figure 1C).
Figure 1.
Creation of TbGPI12 null and conditional null procyclic trypanosomes. (A) Schematic of the endogenous TbGPI12 locus and the ectopic TbGPI12myc construct incorporated into the rDNA locus by homologous recombination, indicating the positions of BamHI and StuI restriction sites and the sizes of the TbGPI12-containing fragments. (B) Southern blot of genomic DNA from wild-type (lane 1) and TbGPI12 null (lane 2) cells digested with BamHI and StuI and probed with the TbGPI12 ORF. The same blot was reprobed with a β-tubulin probe (bottom) to confirm equal loadings of DNA. (C) Southern blot of genomic DNA from wild-type (lane 1) and the TbGPI12 conditional null (cond. null) mutant clone used in this study (lane 2) digested with BamHI and StuI and probed with the TbGPI12 ORF.
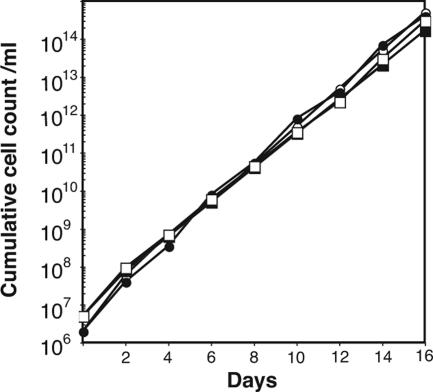
The TbGPI12 null mutant and the conditional null mutant (with and without tetracycline induction) had identical growth phenotypes in culture to the wild-type cells (Figure 2), demonstrating that, unlike the situation in bloodstream-form trypanosomes (Chang et al., 2002), TbGPI12 is a nonessential gene in cultured procyclic T. brucei. The mutant cells also had normal morphology, as judged by light and scanning electron microscopy (data not shown).
Figure 2.
Growth phenotypes of the wild-type and TbGPI12 null and conditional null mutants. Cells were inoculated into culture, counted and diluted fivefold with fresh media every 2 d. The cumulative cell counts allow for the fivefold dilutions. The growth characteristics for wild-type (●), TbGPI12 null (○) and TbGPI12 conditional null cells under permissive (plus tetracycline, closed squares) and nonpermissive (minus tetracycline, □) conditions were indistinguishable.
TbGPI12 Null Mutants Lack GlcNAc-PI de-N-Acetylase Activity and Accumulate GlcNAc-PI
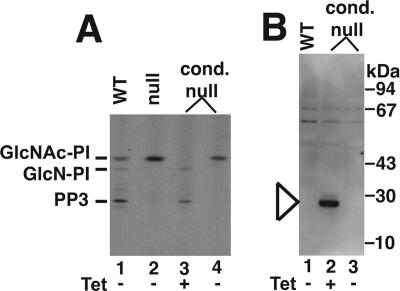
We were able to confirm that we had deleted all GlcNAc-PI de-N-acetylase activity in the null mutants in two ways.
First, cell-free systems of washed parasite membranes made from wild-type and TbGPI12 null and conditional null mutants (grown with and without tetracycline) were labeled with UDP-[3H]GlcNAc in the presence of excess unlabeled GDP-Man. Under these conditions, the wild-type cell-free system incorporates [3H]GlcNAc into [3H]GlcNAc-PI and downstream de-N-acetylated products such as GlcN-PI, with label accumulating in the GPI intermediate PP3, i.e., EtNP-Man3GlcN-(acyl)PI (Field et al., 1992; Güther and Ferguson, 1995 and Figure 3A, lane 1). In the TbGPI12 null mutant, the label accumulated only in GlcNAc-PI (Figure 3A, lane 2), suggesting that these membranes do not contain any residual GlcNAc-PI de-N-acetylase activity. The de-N-acetylation of GlcNAc-PI to GlcN-PI is an essential step in GPI biosynthesis, and no further reactions of GlcNAc-PI are expected without de-N-acetylation (Sharma et al., 1997). The same result was observed with the TbGPI12 conditional null mutant in the absence of tetracycline (Figure 3A, lane 4), indicating the complete absence of detectable TbGPI12 expression under nonpermissive conditions. On the other hand, the same mutant grown under permissive (plus tetracycline) conditions yielded a cell-free system that did not accumulate any GlcNAc-PI (Figure 3A, lane 3), suggesting some overexpression of GlcNAc-PI de-N-acetylase activity in these cells compared with wild type. The expression of the myc-tagged GlcNAc-PI de-N-acetylase protein under permissive conditions, but not under nonpermissive conditions, is shown by anti-myc Western blot (Figure 3B). The band with an apparent molecular weight of 28 kDa (Figure 3B, lane 2) is consistent with the predicted molecular weight of the TbGPI12-myc translation product (29.5 kDa). It is worth noting that the level of protein expression under permissive conditions was insufficient for direct Western blotting of whole cell lysates with anti-myc antibody and that immunoprecipitation of whole cell lysates from 2 × 108 cells with anti-myc was necessary to preconcentrate the target protein before Western blotting.
Figure 3.
Cell-free system GlcNAc-PI de-N-acetylase assays and anti-myc Western blots for wild-type and TbGPI12 mutants. (A) Cell-free systems were prepared from wild-type cells (lane 1), TbGPI12 null cells (lane 2), and TbGPI12 conditional null cells grown under permissive conditions (+Tet; lane 3) and nonpermissive conditions (−Tet; lane 4). Each was labeled with UDP-[3H]GlcNAc in the presence of GDP-Man, and extracted glycolipids were analyzed by HPTLC and fluorography. The positions of authentic standards of GlcNAc-PI, its de-N-acetylation product GlcN-PI and its major metabolite EtNP-Man3GlcN-(acyl)PI (PP3) are indicated. (B) Whole cell lysates from wild-type (lane 1) and TbGPI12 conditional null cells grown under permissive (+Tet; lane 2); and nonpermissive (−Tet) conditions were immunoprecipitated with anti-myc antibody, subjected to SDS-PAGE, transferred to nitrocellulose, and probed with anti-myc antibody. A band with an apparent molecular weight consistent with TbGPI12-myc can be seen in lane 2, marked by the arrowhead. The absence of any enzyme activity (A) and of any protein band (B) shows that tetracycline regulation of gene expression is very tight in this mutant.
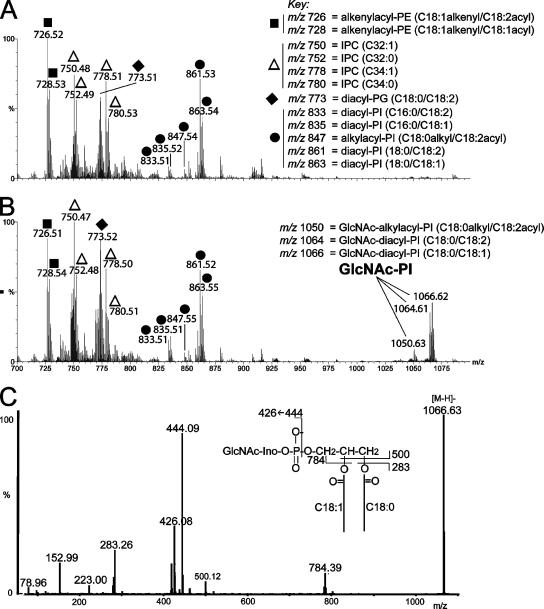
Second, negative ion ES-MS and ES-MS/MS of lipid extracts from wild-type and null mutant parasites showed the absence of detectable steady state levels of GlcNAc-PI in wild-type cells (Figure 4A), consistent with a very low steady state level of this biosynthetic intermediate, but the accumulation of considerable amounts of GlcNAc-PI in the null mutants (Figure 4, B and C).
Figure 4.
Negative ion electrospray mass spectrometry of lipid extracts from wild-type and TbGPI12 null mutant cells. Mass spectrum of lipid species from wild-type (A) and TbGPI12 null mutant cells (B) and a tandem (MS/MS) spectrum of the major GlcNAc-PI ion at m/z 1066.6 (C). The identities of the major ions are indicated. PE, phosphatidylethanolamine; IPC, inositolphosphoceramide; PG, phosphatidylglycerol; PI, phosphatidylinositol. The figures in brackets describe the carbon chain length and degree if unsaturation of the acyl, alkenyl, or alky chains or, for the IPCs, the ceramide moiety. The inset in C shows the product ion assignments for GlcNAc-PI. The ion at m/z 444 is [GlcNAc-myo-inositol-1,2-cyclic phosphate]−, and its dehydration product at m/z 426, are characteristic of negative ion GlcNAc-PI product ion spectra. The ions at m/z 79, 153, 223 and 283 are [PO3]−, [glycerol-cyclic phosphate]−, [inositol-1,2-cyclic phosphate–H2O]−, and [CH3(CH2)16CO2]−, respectively.
From these data we may conclude that TbGPI12 encodes all detectable GlcNAc-PI de-N-acetylase activity of procyclic trypanosomes and that, in the absence of this enzyme, procyclic trypanosomes can accumulate substantial amounts of the GlcNAc-PI precursor.
The negative ion ES-MS and ES-MS/MS analyses also identified the major phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and PI components of procyclic-form T. brucei (Figure 4, A and B), which appear to present in comparable relative amounts in the two samples. Of particular note is a cluster of ions at m/z 750, 752, 778, and 780. These produced intense inositol-1,2-cyclic phosphate and inositol-monophosphate product ions at (m/z 241 and 259) in ES-MS/MS (data not shown), characteristic of inositolphosphoceramide (IPC) ions. These are discussed later. Negative product ion spectra of IPCs do not provide fine detail on the long-chain base and fatty acid compositions of the ceramide portions of IPCs, other than total carbon numbers and degrees of unsaturation (which are C32:1, C32:0, C34:1, and C34:0 for the T. brucei IPCs). These are, on average, smaller than those found in T. cruzi epimastigotes IPCs, which are mainly C34:1, C34:0, C36:1, and C36:0 (Bertello et al., 1995), and Leishmania promastigote IPCs, which are mainly C34:1 and C36:1 (Zufferey et al., 2003, Denny et al., 2004). The ES-MS/MS product ion spectra of the two major PE ions at m/z 726 and 728 (data not shown) showed these to be alkenylacyl-PE species (also known as plasmenylethanolamines) that appear to be common in kinetoplastid organisms (Villas Boas et al., 1999; Zufferey et al., 2003).
TbGPI12 Null Mutants Lack Cell Surface Procyclin and Free GPIs
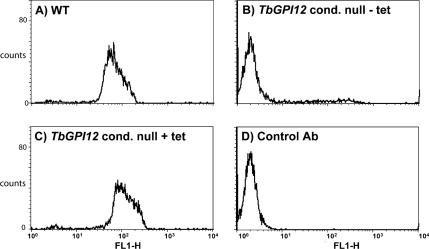
In the absence of a functional GPI anchor biosynthetic pathway, we would not expect the expression of cell surface procyclins, as has been described for TbGPI10 and TbGPI8 knockout procyclic trypanosomes (Nagamune et al., 2000, 2004; Lillico et al., 2003). Analysis of cells by flow cytometry using anti-procyclin antibodies and FITC-labeled secondary antibody revealed a strong signal for wild-type cells (Figure 5A) and only background labeling for the TbGPI12 conditional null mutant grown under nonpermissive conditions (Figure 5B). Strong labeling was restored in the TbGPI12 conditional null mutant cells under permissive (plus tetracycline) conditions (Figure 5C).
Figure 5.
Flow cytometric (FACS) analysis for surface procyclin in the wild-type and TbGPI12 conditional null mutants cells. (A) Wild-type cells stained with anti-procyclin antibody. (B) TbGPI12 conditional null mutants cells grown under nonpermissive (−tet) conditions. (C) TbGPI12 conditional null mutants cells grown under permissive (+tet) conditions. (D) No primary anti-procyclin antibody control.
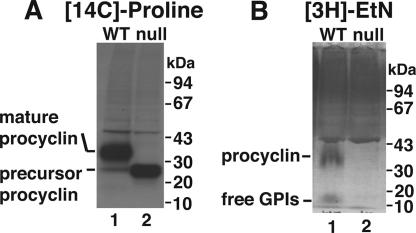
The absence of cell surface procyclin indicated by cytometry was not due to a lack of procyclin protein synthesis. SDS-PAGE and fluorography of anti-procyclin immunoprecipitated cell extracts labeled for 2 h with [14C]proline revealed low-molecular-weight procyclin precursor in the TbGPI12 mutants (Figure 6A, lane 2), whereas wild-type cells produced fully processed GPI-anchored procyclin (Figure 6A, lane 1). In pulse-chase experiments (2-h pulse and 18-h chase) the majority of [14C]proline-labeled procyclin made in the TbGPI12 mutants was found secreted into the medium as multiple degradation products (data not shown).
Figure 6.
Biosynthetic labeling of wild-type and TbGPI12 null cells with [14C]proline and [3H]ethanolamine. (A) Wild-type (lane 1) and TbGPI12 null cells (lane 2) were labeled with [14C]proline and lysed and anti-procyclin immunoprecipitates were subjected to SDS-PAGE and fluorography. (B) Wild-type (lane 1) and TbGPI12 null cells (lane 2) were labeled with [3H]ethanoamine and subjected to SDS-PAGE and fluorography.
Unlike the TbGPI10 and TbGPI8 null mutants, that can make poly-N-acetyllactosamine modified Man2GlcN-(acyl)lyso-PI and EtNP-Man3GlcN-(acyl)lyso-PI free GPIs, respectively (Lillico et al., 2003; Nagamune et al., 2004), TbGPI12 null mutants should not be able to synthesize any kind of GPI structure beyond GlcNAc-PI. Labeling of cells with [3H]ethanolamine confirmed this supposition. Thus, in wild-type cells, [3H]ethanolamine labeled procyclin and free GPIs (Figure 6B, lane 1), whereas these were absent in the TbGPI12 null mutants (Figure 6B, lane 2). The labeled band running above procyclin in both wild-type and mutant cells is most likely protein synthesis elongation factor 1a (Dever et al., 1989).
Attempts To Find Molecules That Might Replace Procyclin and Free GPIs in TbGPI12 Null Mutants
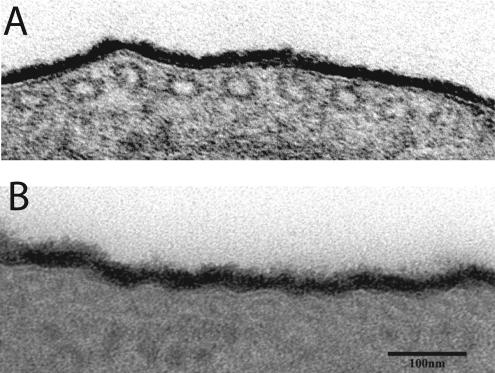
Transmission electron microscopy (TEM) of ultrathin sections of wild-type and TbGPI12 null procyclic trypanosomes stained with ruthenium red, a stain used to preserve prokaryote and eukaryote glycocalyx structure (Luft, 1966, 1971; Szubinska and Luft, 1971; Zufferey et al., 2003), revealed that the cell surface coats of these preparations were indistinguishable (Figure 7). This was a surprising finding because procyclins have been assumed to be the major component of the procyclic trypanosome surface coat (Roditi et al., 1989; Mehlert et al., 1998).
Figure 7.
Transmission electron microscopy of the surface glycocalyx of wild-type and TbGPI12 null cells. Ruthenium red–stained ultrathin sections of (A) wild-type and (B) TbGPI12 null cells. Bar, 100 nm.
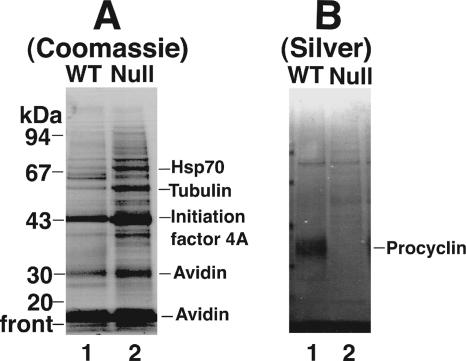
We considered that procyclins and free GPIs might have been replaced by adsorbed serum components from the medium, so we labeled the cell surface with a cleavable sulfo-NHS-biotin reagent. However, analysis of the neutravidin-purified surface-biotinylated components by SDS-PAGE and Coomassie blue staining did not reveal any obvious components unique to the null mutants (Figure 8A). Even those proteins that appeared more intense in the mutant compared with the wild-type cells proved to be intracellular T. brucei proteins, and not bovine serum proteins, by peptide mass fingerprinting. These same preparations were stained with silver, which then revealed the non-Coomassie–stainable cell-surface procyclins of the wild-type cells, as expected (Figure 8B, lane 1). However, no new biotin-labeled components were obvious in the TbGPI12 null cell extracts (Figure 8B, lane 2).
Figure 8.
Cell surface biotinylation of wild-type and TbGPI12 null cells. Wild-type cells (lane 1) and TbGPI12 null cells (lane 2) were surface-labeled with sulfo-NHS-SS-biotin, and labeled products were purified on neutravidin beads and analyzed by SDS-PAGE and Coomassie blue staining (A) or silver staining (B). The proteins indicated were identified by protein mass fingerprinting.
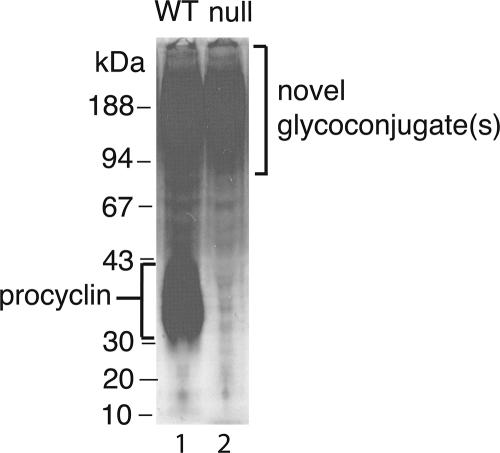
In an attempt to find nonbiotinylatable and/or non-silver–stainable surface molecules, we labeled the wild-type and TbGPI12 null mutant cells with [3H]glucosamine. We chose this label because glucosamine and/or its metabolite N-acetylglucosamine is/are common to all known trypanosomatid glycoconjugates (McConville and Ferguson, 1993; Guha-Niyogi and Turco, 2001; McConville et al., 2002; Atrih et al., 2005; Mendonca-Previato et al., 2005). Strong labeling of procyclin was apparent in the wild-type cells (Figure 9, lane 1), as expected, whereas this band was completely absent in the TbGPI12 null mutant, consistent with the cytometric and [14C]proline-labeling experiments described above (Figure 9, lane 2). Although [3H]glucosamine labeling did not reveal any obviously up-regulated glycoconjugates in the TbGPI12 null mutant, it did reveal a smear of labeled glycoconjugates, ranging in apparent molecular weight from ∼90 to >200 kDa, that were labeled in both the wild-type and TbGPI12 null mutant parasites.
Figure 9.
Biosynthetic labeling of wild-type and TbGPI12 null cells with [3H]glucosamine. Wild-type (lane 1) and TbGPI12 null cells (lane 2) were labeled with [3H]glucosamine and subjected to SDS-PAGE and fluorography.
TbGPI12 Null Mutant Procyclic Trypanosomes Fail To Infect the Tsetse Midgut
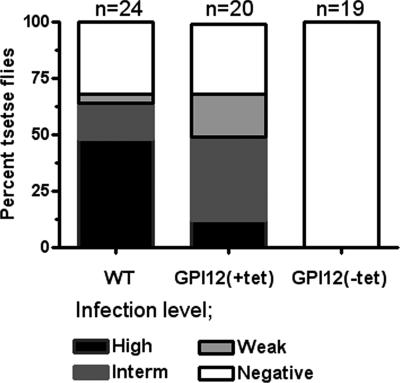
The discovery of a non-GPI surface glycocalyx in TbGPI12 null mutant parasites, which seem to be displayed with a similar thickness to that observed on the surface of WT cells (Figure 7), prompted us to determine its possible functional role in establishing a midgut infection in the tsetse fly. Thus, we infected teneral flies with WT and TbGPI12 conditional null cells cultured under permissive and nonpermissive (i.e., with and without tetracycline) conditions, and after 2 weeks their midguts were dissected and checked for the presence of parasites. As observed in (Figure 10), although flies infected with either WT or TbGPI12 conditional null cells grown under permissive conditions and also fed with bloodmeals containing 25 μg/ml tetracycline developed comparable infection indexes, no parasites were detected in flies infected with TbGPI12 conditional null cells (previously grown under nonpermissive conditions) and continuously fed with bloodmeals lacking tetracycline, suggesting that TbGPI12 expression is essential for the colonization of the tsetse fly.
Figure 10.
TbGPI12 null mutant cells are unable to establish midgut infections. Tsetse flies were fed with infected bloodmeals containing wild-type (WT) or TbGPI12 conditional null mutants under permissive (GPI12(+tet)) and nonpermissive (GPI12(−tet)) conditions. After 15 d, flies were dissected and infections scored as heavy (black; more than 100 parasites per field), intermediate (dark gray; 20–100 parasites), weak (light gray; 1–19 parasites), and negative (no detectable parasites). The number of dissected flies is indicated on top of each group.
Taken together, although the components and the chemical nature of the second (non-GPI) surface glycocalyx from procyclic trypanosomes remain to be determined, these experiments suggest that GPI-anchored proteins and free GPIs are probably the most functionally important components of the surface glycocalyx of procyclic T. brucei with respect to colonization of the tsetse vector.
DISCUSSION
As for bloodstream-form trypanosomes (Chang et al., 2002), the TbGPI12 gene clearly encodes all the GlcNAc-PI de-N-acetylase activity of procyclic-form trypanosome and its deletion prevents GPI synthesis beyond the early intermediate GlcNAc-PI (Figure 3). The accumulation of substantial quantities of GlcNAc-PI in the TbGPI12 null mutants, detected by ES-MS (Figure 4), indicates that the first step of GPI biosynthesis (i.e., the transfer of GlcNAc from UDP-GlcNAc to PI) is not sensitive to product inhibition. In the cell-free system and living cells, GlcNAc-PI de-N-acetylase activity may be a rate-limiting step in GPI biosynthesis and procyclin surface expression, respectively, because both are slightly higher in the TbGPI12 conditional null mutant when the expression of the ectopic copy of TbGPI12 is fully induced by tetracycline (Figures 3 and 5).
The molecular species of GlcNAc-PI that accumulate in the TbGPI12 null mutant mostly contain C18:0/C18:1 and C18:0/C18:2 diacyl-PI lipids, together with a small amount of C18:0/C18:2 alkylacyl-PI (Figure 4B). Interestingly, the relationship of C18:0/C18:2 (m/z 861) > C18:0/C18:1 (m/z 863) for the major diacyl-PI species is reversed for the GlcNAc-PIs that accumulate, where C18:0/C18:2 (m/z 1064) < C18:0/C18:1 (m/z 1066). In addition, the C16:0/C18:2 and C16:0/C18:1 diacyl-PI species (m/z 833 and 835) do not appear to be converted to GlcNAc-PI species, whereas the less-abundant C18:0/C18:2 alkylacyl-PI is converted to the corresponding GlcNAc-PI at m/z 1050. This apparent selection of particular PI species for GlcNAc addition may be consistent with a recent report (Martin and Smith, 2006b) that suggests that, at least in bloodstream-form T. brucei, two distinct pools of PI are synthesized, one for bulk membrane lipid and one for GPI anchor biosynthesis.
An unexpected finding from the lipid mass spectrometry data are the identification of IPC in T. brucei, although these are clearly not converted to GlcNAc-IPC in the TbGPI12 null mutant. To our knowledge, this is the first evidence for the presence of IPCs in T. brucei, but it is consistent with the very recent discovery of novel IPC synthase genes in this and other kinetoplastid organisms (Denny et al., 2006). In T. brucei, IPCs appear to be specific to the procyclic form because mass spectrometric analyses of bloodstream-form T. brucei lipids show them to be mostly C18:0/C18:2 and C18:0/C22:4 diacyl-PIs (Martin and Smith, 2006a, 2006b).
Thus far, the molecular architecture of procyclic-form T. brucei has been manipulated in four ways: 1) Knockout of the multiple procyclin genes themselves (Vassella et al., 2003); 2) Knockout of TbGPI10, encoding the third α-mannosyltransferase of GPI biosynthesis (Nagamune et al., 2000, 2004); 3) Knockout of TbGPI8, encoding the catalytic subunit of the GPI transamidase complex (Lillico et al., 2003); and 4) Conditional knockout of TbgalE, encoding UDP-Glc 4′-epimerase, producing galactose starvation under nonpermissive conditions (Roper et al., 2005). In this article, we describe altering the cell surface in a distinct way by knocking out the TbGPI12 gene that, uniquely, simultaneously prevents the cell surface expression of procyclins and poly-N-acetyllactosamine–containing free GPI glycolipids (Figures 5 and 6).
The procyclin and TbGPI10 and TbGPI8 knockouts all result in the complete loss of cell surface procyclins, but this is compensated for by an up-regulation in the expression of wild-type (procyclin and TbGPI8 knockout) or slightly modified (TbGPI10 knockout) poly-N-acetyllactosamine–containing free GPI glycolipids. In the TbGPI12 knockout, reported here, poly-N-acetyllactosamine–containing free GPI glycolipids cannot be made and, therefore, cannot compensate for the loss of surface procyclins. We were surprised that the TbGPI12 gene was not essential for the growth of procyclic-form T. brucei, because deletion of this gene simultaneously wipes out both surface procyclin and poly-N-acetyllactosamine–containing free GPI expression. Currently, these molecules are assumed to represent the majority of the cell surface coat glycocalyx of this lifecycle stage of the organism (Mowatt and Clayton, 1987; Roditi et al., 1987, 1989; Richardson et al., 1988; Mehlert et al., 1998; Lillico et al., 2003; Vassella et al., 2003, Roper et al., 2005). It seems unlikely that the minimal glycolipid GlcNAc-PI, which accumulates in the mutant, could compensate for loss of these much larger surface molecules. Curiously, whereas the TbGPI10 and TbGPI8 knockouts have significantly and slightly, respectively, impaired cell culture growth kinetics compared with wild-type cells (Nagamune et al., 2004), the TbGPI12 knockout cells have perfectly normal growth characteristics (Figure 2). We have no obvious explanation for these differences between the mutants.
Analysis of the cell surface by ruthenium red staining and TEM showed that the density and depth of the cell surface glycocalyx did not appear to be significantly altered in the TbGPI12 null mutants compared with wild type (Figure 7). This, and the excellent growth characteristics of the TbGPI12 null mutants, prompted us to look for the possible up-regulation of other parasite surface molecules and/or the adsorption of serum components from the medium to compensate for the loss of procyclins and poly-N-acetyllactosamine–containing free GPIs. However, neither of these were apparent from surface biotinylation, followed by isolation on neutravidin and detection with Coomassie blue or silver stain (Figure 8). Because carbohydrate-rich proteoglycan- or mucin-like molecules might contain few primary amine groups for biotinylation, we took a different approach, namely biosynthetic radiolabeling with the ubiquitous trypanosomatid glycoconjugate precursor glucosamine. This revealed polydisperse glycoconjugates with high apparent molecular weights (90 to >200 kDa) on SDS-PAGE that were present in similar levels in TbGPI12 null and wild-type cells (Figure 9). It remains to be seen whether the majority of the surface coat observed by TEM is constituted by the novel glycoconjugates revealed by [3H]glucosamine labeling. This material is certainly a candidate, but the answer will have to await its isolation and characterization and the generation of mono-specific reagents that recognize its principal component(s).
TbGPI10 and TbGPI8 knockouts are partially and severely compromised, respectively, with respect to their ability to colonize the tsetse midgut (Nagamune et al., 2004). The difference in tsetse infectivity is thought to correlate with surface sialic acid content, governed by the status of the normally GPI-anchored transialidase (TS) enzyme (Nagamune et al., 2004). Thus, although TbGPI10 knockout parasites secrete anchorless TS and, therefore, still acquire surface sialic acid, the GPI-transamidase null TbGPI8 knockout parasites fail to remove the GPI-addition signal peptide from the TS precursor, to degrade it intracellularly, and to sialylate their surface. In the case of the TbGPI12 knockout cells, we observe normal growth kinetics but severely reduced tsetse infectivity (Figure 10). These cells will be able to secrete TS (like the TbGPI10 knockouts) but cannot express cell surface procyclin or free GPI sialic acid acceptors. The TbGPI12 null phenotype with respect to tsetse colonization is, therefore, like that of TbGPI8 null parasites, supporting the hypothesis that sialylation of procyclin GPI anchor side chains and/or free GPI side chains (that, uniquely, are both are absent in TbGPI12 mutant) are essential for tsetse midgut infection.
Although deletion of surface free GPIs and/or procyclins does not prevent procyclic trypanosome cell division in culture, galactose starvation is cytostatic (Roper et al., 2005). We previously suggested that galactose starvation might exert these cytostatic effects via inhibition of poly-N-acetyllactosamine side chain addition to procyclin GPI anchors and/or free GPIs, an idea that was consistent with the apparently compensatory 10-fold up-regulation in side chain–free procyclins before the cessation of cell division. However, this notion may need to be revised in the light of the healthy growth of the TbGPI12 knockout because this mutant has no GPI structures that can be modified with poly-N-acetyllactosamine side chains. Thus, it is possible that it is the effects of galactose starvation on one or more non-GPI–anchored glycoprotein(s), possibly the novel high apparent molecular weight molecules identified in this article, that cause cell division to cease. Alternatively, one could argue that the accumulation of procyclins without GPI side chains under galactose starvation might be toxic to the cells and that because this cannot occur in the TbGPI12 knockout cells, the latter grow normally.
ACKNOWLEDGMENTS
We thank Margaret Mullin and Craig Lapsley for their excellent technical assistance in processing the samples for electron microscopy and dissecting and maintaining tsetse flies, respectively; Jeremy Mottram for supplying the BSD resistance gene, the Proteomics Facility, Dundee, for mass fingerprinting; and Mick Urbaniak for helpful discussions. M.L.S.G. and M.A.J.F. dedicate this article to the memory of Dr. Anthony J.A. Ferguson. This work was supported by a Wellcome Trust Programme Grant to M.A.J.F. (071463). A.A.S. was supported by a Wellcome Trust Research Career Development Fellowship.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0702) on October 11, 2006.
REFERENCES
- Acosta-Serrano A., Cole R. N., Mehlert A., Lee M. G., Ferguson M. A., Englund P. T. The procyclin repertoire of Trypanosoma brucei. Identification and structural characterization of the Glu-Pro-rich polypeptides. J. Biol. Chem. 1999;274:29763–29771. doi: 10.1074/jbc.274.42.29763. [DOI] [PubMed] [Google Scholar]
- Acosta-Serrano A., Vassella E., Liniger M., Kunz Renggli C., Brun R., Roditi I., Englund P. T. The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc. Natl. Acad. Sci. USA. 2001;98:1513–1518. doi: 10.1073/pnas.041611698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atrih A., Richardson J. M., Prescott A. R., Ferguson M. A. Trypanosoma brucei glycoproteins contain novel giant poly-N-acetyllactosamine carbohydrate chains. J. Biol. Chem. 2005;280:865–871. doi: 10.1074/jbc.M411061200. [DOI] [PubMed] [Google Scholar]
- Bertello L. E., Goncalvez M. F., Colli W., de Lederkremer R. M. Structural analysis of inositol phospholipids from Trypanosoma cruzi epimastigote forms. Biochem. J. 1995;310(Pt 1):255–261. doi: 10.1042/bj3100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R., Schönenberger M. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- Butikofer P., Vassella E., Ruepp S., Boschung M., Civenni G., Seebeck T., Hemphill A., Mookherjee N., Pearson T. W., Roditi I. Phosphorylation of a major GPI-anchored surface protein of Trypanosoma brucei during transport to the plasma membrane. J. Cell Sci. 1999;112(Pt 11):1785–1795. doi: 10.1242/jcs.112.11.1785. [DOI] [PubMed] [Google Scholar]
- Chang T., Milne K. G., Guther M. L., Smith T. K., Ferguson M. A. Cloning of Trypanosoma brucei and Leishmania major genes encoding the GlcNAc-phosphatidylinositol de-N-acetylase of glycosylphosphatidylinositol biosynthesis that is essential to the African sleeping sickness parasite. J. Biol. Chem. 2002;277:50176–50182. doi: 10.1074/jbc.M208374200. [DOI] [PubMed] [Google Scholar]
- Cross G. A. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays. 1996;18:283–291. doi: 10.1002/bies.950180406. [DOI] [PubMed] [Google Scholar]
- Denny P. W., Goulding D., Ferguson M. A., Smith D. F. Sphingolipid-free Leishmania are defective in membrane trafficking, differentiation and infectivity. Mol. Microbiol. 2004;52:313–327. doi: 10.1111/j.1365-2958.2003.03975.x. [DOI] [PubMed] [Google Scholar]
- Denny P. W., Shams-Eldin H., Price H. P., Smith D. F., Schwarz R. T. The protozoan inositol phosphorylceramide synthase: a novel drug target which defines a new class of sphingolipid synthase. J. Biol. Chem. 2006;281:28200–28209. doi: 10.1074/jbc.M600796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Costello C. E., Owens C. L., Rosenberry T. L., Merrick W. C. Location of seven post-translational modifications in rabbit elongation factor 1 alpha including dimethyllysine, trimethyllysine, and glycerylphosphorylethanolamine. J. Biol. Chem. 1989;264:20518–20525. [PubMed] [Google Scholar]
- Engstler M., Reuter G., Schauer R. The developmentally regulated trans-sialidase from Trypanosoma brucei sialylates the procyclic acidic repetitive protein. Mol. Biochem. Parasitol. 1993;61:1–13. doi: 10.1016/0166-6851(93)90153-o. [DOI] [PubMed] [Google Scholar]
- Ferguson M. A., Murray P., Rutherford H., McConville M. J. A simple purification of procyclic acidic repetitive protein and demonstration of a sialylated glycosyl-phosphatidylinositol membrane anchor. Biochem. J. 1993;291:51–55. doi: 10.1042/bj2910051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field M. C., Menon A. K., Cross G. A. Developmental variation of glycosylphosphatidylinositol membrane anchors in Trypanosoma brucei. In vitro biosynthesis of intermediates in the construction of the GPI anchor of the major procyclic surface glycoprotein. J. Biol. Chem. 1992;267:5324–5329. [PubMed] [Google Scholar]
- Guha-Niyogi A., Sullivan D. R., Turco S. J. Glycoconjugate structures of parasitic protozoa. Glycobiology. 2001;11:45R–59R. doi: 10.1093/glycob/11.4.45r. [DOI] [PubMed] [Google Scholar]
- Güther M. L., Ferguson M. A. The role of inositol acylation and inositol deacylation in GPI biosynthesis in Trypanosoma brucei. EMBO J. 1995;14:3080–3093. doi: 10.1002/j.1460-2075.1995.tb07311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güther M. L., Masterson W. J., Ferguson M. A. The effects of phenylmethylsulfonyl fluoride on inositol-acylation and fatty acid remodeling in African trypanosomes. J. Biol. Chem. 1994;269:18694–18701. [PubMed] [Google Scholar]
- Lillico S., Field M. C., Blundell P., Coombs G. H., Mottram J. C. Essential roles for GPI-anchored proteins in African trypanosomes revealed using mutants deficient in GPI8. Mol. Biol. Cell. 2003;14:1182–1194. doi: 10.1091/mbc.E02-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liniger M., Urwyler S., Studer E., Oberle M., Renggli C. K., Roditi I. Role of the N-terminal domains of EP and GPEET procyclins in membrane targeting and the establishment of midgut infections by Trypanosoma brucei. Mol Biochem. Parasitol. 2004;137:247–251. doi: 10.1016/j.molbiopara.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Luft J. H. Fine structures of capillary and endocapillary layer as revealed by ruthenium red; Fed. Proc; 1966. pp. 1773–1783. [PubMed] [Google Scholar]
- Luft J. H. Ruthenium red and violet. II. Fine structural localization in animal tissues. Anat. Rec. 1971;171:369–415. doi: 10.1002/ar.1091710303. [DOI] [PubMed] [Google Scholar]
- Martin K. L., Smith T. K. Phosphatidylinositol synthesis is essential in bloodstream form Trypanosoma brucei. Biochem. J. 2006a;396:287–295. doi: 10.1042/BJ20051825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. L., Smith T. K. The glycosylphosphatidylinositol (GPI) biosynthetic pathway of bloodstream form Trypanosoma brucei is dependent on de novo synthesis of inositol. Mol. Microbiol. 2006b;61:89–105. doi: 10.1111/j.1365-2958.2006.05216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson W. J., Doering T. L., Hart G. W., Englund P. T. A novel pathway for glycan assembly: biosynthesis of the glycosyl-phosphatidylinositol anchor of the trypanosome variant surface glycoprotein. Cell. 1989;56:793–800. doi: 10.1016/0092-8674(89)90684-3. [DOI] [PubMed] [Google Scholar]
- McConville M. J., Ferguson M. A. The structure, biosynthesis and function of glycosylated phosphatidylinositols in the parasitic protozoa and higher eukaryotes. Biochem. J. 1993;294(Pt 2):305–324. doi: 10.1042/bj2940305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville M. J., Mullin K. A., Ilgoutz S. C., Teasdale R. D. Secretory pathway of trypanosomatid parasites. Microbiol. Mol. Biol. Rev. 2002;66:122–154. doi: 10.1128/MMBR.66.1.122-154.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlert A., Treumann A., Ferguson M. A. Trypanosoma brucei GPEET-PARP is phosphorylated on six out of seven threonine residues. Mol. Biochem. Parasitol. 1999;98:291–296. doi: 10.1016/s0166-6851(98)00168-6. [DOI] [PubMed] [Google Scholar]
- Mehlert A., Zitzmann N., Richardson J. M., Treumann A., Ferguson M. A. The glycosylation of the variant surface glycoproteins and procyclic acidic repetitive proteins of Trypanosoma brucei. Mol. Biochem. Parasitol. 1998;91:145–152. doi: 10.1016/s0166-6851(97)00187-4. [DOI] [PubMed] [Google Scholar]
- Mendonca-Previato L., Todeschini A. R., Heise N., Previato J. O. Protozoan parasite-specific carbohydrate structures. Curr. Opin. Struct. Biol. 2005;15:499–505. doi: 10.1016/j.sbi.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Montagna G., Cremona M. L., Paris G., Amaya M. F., Buschiazzo A., Alzari P. M., Frasch A. C. The trans-sialidase from the African trypanosome Trypanosoma brucei. Eur. J. Biochem. 2002;269:2941–2950. doi: 10.1046/j.1432-1033.2002.02968.x. [DOI] [PubMed] [Google Scholar]
- Morris J. C., Wang Z., Drew M. E., Englund P. T. Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J. 2002;21:4429–4438. doi: 10.1093/emboj/cdf474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowatt M. R., Clayton C. E. Developmental regulation of a novel repetitive protein of Trypanosoma brucei. Mol. Cell. Biol. 1987;7:2838–2844. doi: 10.1128/mcb.7.8.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamune K., Acosta-Serrano A., Uemura H., Brun R., Kunz-Renggli C., Maeda Y., Ferguson M. A., Kinoshita T. Surface sialic acids taken from the host allow trypanosome survival in tsetse fly vectors. J. Exp. Med. 2004;199:1445–1450. doi: 10.1084/jem.20030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamune K., Nozaki T., Maeda Y., Ohishi K., Fukuma T., Hara T., Schwarz R. T., Sutterlin C., Brun R., Riezman H., Kinoshita T. Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 2000;97:10336–10341. doi: 10.1073/pnas.180230697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock L., Bailey M., Gibson W. Tetracycline induction of gene expression in Trypanosoma brucei within the tsetse fly vector. Mol. Biochem. Parasitol. 2005;140:247–249. doi: 10.1016/j.molbiopara.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Pontes de Carvalho L. C., Tomlinson S., Vandekerckhove F., Bienen E. J., Clarkson A. B., Jiang M. S., Hart G. W., Nussenzweig V. Characterization of a novel trans-sialidase of Trypanosoma brucei procyclic trypomastigotes and identification of procyclin as the main sialic acid acceptor. J. Exp. Med. 1993;177:465–474. doi: 10.1084/jem.177.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. P., Beecroft R. P., Tolson D. L., Liu M. K., Pearson T. W. Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol. Biochem. Parasitol. 1988;31:203–216. doi: 10.1016/0166-6851(88)90150-8. [DOI] [PubMed] [Google Scholar]
- Roditi I., Carrington M., Turner M. Expression of a polypeptide containing a dipeptide repeat is confined to the insect stage of Trypanosoma brucei. Nature. 1987;325:272–274. doi: 10.1038/325272a0. [DOI] [PubMed] [Google Scholar]
- Roditi I., Clayton C. An unambiguous nomenclature for the major surface glycoproteins of the procyclic form of Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;103:99–100. doi: 10.1016/s0166-6851(99)00124-3. [DOI] [PubMed] [Google Scholar]
- Roditi I. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J. Cell Biol. 1989;108:737–746. doi: 10.1083/jcb.108.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper J. R., Guther M. L., Macrae J. I., Prescott A. R., Hallyburton I., Acosta-Serrano A., Ferguson M. A. The suppression of galactose metabolism in procyclic form Trypanosoma brucei causes cessation of cell growth and alters procyclin glycoprotein structure and copy number. J. Biol. Chem. 2005;280:19728–19736. doi: 10.1074/jbc.M502370200. [DOI] [PubMed] [Google Scholar]
- Schlaeppi A. C., Malherbe T., Butikofer P. Coordinate expression of GPEET procyclin and its membrane-associated kinase in Trypanosoma brucei procyclic forms. J. Biol. Chem. 2003;278:49980–49987. doi: 10.1074/jbc.M309548200. [DOI] [PubMed] [Google Scholar]
- Sharma D. K., Smith T. K., Crossman A., Brimacombe J. S., Ferguson M. A. Substrate specificity of the N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase of glycosylphosphatidylinositol membrane anchor biosynthesis in African trypanosomes and human cells. Biochem. J. 1997;328(Pt 1):171–177. doi: 10.1042/bj3280171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szubinska B., Luft J. H. Ruthenium red and violet. 3. Fine structure of the plasma membrane and extraneous coats in amoebae (A. proteus and Chaos chaos) Anat. Rec. 1971;171:417–441. doi: 10.1002/ar.1091710304. [DOI] [PubMed] [Google Scholar]
- Treumann A., Zitzmann N., Hulsmeier A., Prescott A. R., Almond A., Sheehan J., Ferguson M. A. Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J. Mol. Biol. 1997;269:529–547. doi: 10.1006/jmbi.1997.1066. [DOI] [PubMed] [Google Scholar]
- Vanhamme L., Pays E., McCulloch R., Barry J. D. An update on antigenic variation in African trypanosomes. Trends Parasitol. 2001;17:338–343. doi: 10.1016/s1471-4922(01)01922-5. [DOI] [PubMed] [Google Scholar]
- Vassella E., Acosta-Serrano A., Studer E., Lee S. H., Englund P. T., Roditi I. Multiple procyclin isoforms are expressed differentially during the development of insect forms of Trypanosoma brucei. J. Mol. Biol. 2001;312:597–607. doi: 10.1006/jmbi.2001.5004. [DOI] [PubMed] [Google Scholar]
- Vassella E., Butikofer P., Engstler M., Jelk J., Roditi I. Procyclin null mutants of Trypanosoma brucei express free glycosylphosphatidylinositols on their surface. Mol. Biol. Cell. 2003;14:1308–1318. doi: 10.1091/mbc.E02-10-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassella E., Den Abbeele J. V., Butikofer P., Renggli C. K., Furger A., Brun R., Roditi I. A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev. 2000;14:615–626. [PMC free article] [PubMed] [Google Scholar]
- Vassella E., Probst M., Schneider A., Studer E., Renggli C. K., Roditi I. Expression of a major surface protein of Trypanosoma brucei insect forms is controlled by the activity of mitochondrial enzymes. Mol. Biol. Cell. 2004;15:3986–3993. doi: 10.1091/mbc.E04-04-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villas Boas M. H., Lara L. S., Wait R., Bergter E. B. Identification of plasmenylethanolamine as a major component of the phospholipids of strain DM 28c of Trypanosoma cruzi. Mol. Biochem. Parasitol. 1999;98:175–186. doi: 10.1016/s0166-6851(98)00165-0. [DOI] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C., Cross G. A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Zufferey R., Allen S., Barron T., Sullivan D. R., Denny P. W., Almeida I. C., Smith D. F., Turco S. J., Ferguson M. A., Beverley S. M. Ether phospholipids and glycosylinositolphospholipids are not required for amastigote virulence or for inhibition of macrophage activation by Leishmania major. J. Biol. Chem. 2003;278:44708–44718. doi: 10.1074/jbc.M308063200. [DOI] [PubMed] [Google Scholar]