Abstract
Homologous DNA recombination promotes genetic diversity and the maintenance of genome integrity, yet no enzymes with specificity for the Holliday junction (HJ)—a key DNA recombination intermediate—have been purified and characterized from metazoa or their viruses. Here we identify critical structural elements of RuvC, a bacterial HJ resolvase, in uncharacterized open reading frames from poxviruses and an iridovirus. The putative vaccinia virus resolvase was expressed as a recombinant protein, affinity purified, and shown to specifically bind and cleave a synthetic HJ to yield nicked duplex molecules. Mutation of either of two conserved acidic amino acids abrogated the catalytic activity of the A22R protein without affecting HJ binding. The presence of bacterial-type enzymes in metazoan viruses raises evolutionary questions.
Homologous DNA recombination is a ubiquitous process that promotes genetic diversity and the maintenance of genome integrity in prokaryotic and eukaryotic organisms. The central intermediate in the recombination process is a four-way DNA junction usually referred to as a Holliday junction (HJ), consisting of two homologous duplex DNA molecules joined by cross-over strands (1, 2). Although HJ-resolving activities have been found in extracts prepared from a wide variety of organisms, only enzymes from bacteria, bacteriophages, yeast mitochondria, and archaea have been genetically identified and characterized. The latter include Escherichia coli RuvC (3–5), bacteriophage T4 endonuclease VII (6), bacteriophage T7 endonuclease I (7, 8), lambdoid prophage RusA (9), Saccharomyces cerevisiae mitochondrial CCE1 (10), Schizosaccharomyces pombe mitochondrial YDC2 (11), and Pyrococcus furiosus Hjc protein (12). These enzymes can be divided into two functional groups. Members of the first have RuvC as their prototype, include the resolvases from bacteria, mitochondria, and archaea, have high selectivity for HJs, exhibit sequence specificity for cleavage, and are thought to have roles in recombination and DNA repair. Members of the second group, composed of the bacteriophage enzymes, cleave a variety of branched DNAs, exhibit low sequence specificity, and have roles in recombination and the processing of DNA before packaging. Absent from either group, however, is a well-characterized HJ endonuclease from metazoa or their viruses.
The poxviruses, of which vaccinia virus is the prototype, are large DNA viruses that replicate in the cytoplasm of infected cells and consequently provide a unique system for studying enzymes involved in RNA and DNA synthesis (13). Although the ability of poxviruses to undergo recombination has been long known (14), this process has not been separated from replication (15), and the viral proteins involved have remained elusive. An enzyme that cleaves HJs may participate in recombination as well as other steps in processing and packaging of poxvirus DNA. The genomes of poxviruses consist of a single linear double-stranded DNA molecule with covalently linked hairpin termini (16, 17) that are formed from long concatemeric intermediates (18, 19) by a sequence-specific process known as telomere resolution (20, 21). The target of the resolution reaction is the palindromic concatemer junction, which can adopt a cruciform HJ structure in supercoiled plasmids (22, 23). Concatemer resolution is independent of replication and requires late stage gene expression (24, 25). Activities that cleave supercoiled plasmids into linear molecules with cross-linked ends have been detected in extracts of purified virions or infected cells, but the genes encoding them have not been identified (23, 26, 27). Lately, attention has shifted to a possible role of the poxvirus-encoded topoisomerase I in recombination and resolution, and this enzyme has been shown to resolve synthetic HJs and cruciform structures in vitro (28, 29).
Several factors led us to renew the search for a poxvirus HJ endonuclease. First, the complete genome sequences of several poxviruses as well as those of a number of bacterial genomes encoding RuvC family resolvases have become known. Second, the crystal structure of RuvC and the structural requirements for HJ resolution by this enzyme have been determined (30, 31). We describe here how this information, together with improvements in sequence similarity search programs, allowed us to identify the critical motifs and structural elements of RuvC in yeast as well as previously uncharacterized open reading frames from all sequenced poxviruses and an iridovirus. These relationships cannot be detected by standard database searches, and the existence of RuvC homologs outside of bacteria was not reported previously. We have expressed the RuvC homolog encoded by vaccinia virus and demonstrated specific cleavage of a synthetic HJ. This endonuclease activity was lost upon mutation of conserved acidic amino acids predicted to form part of the metal binding site of RuvC.
Materials and Methods
Sequence and Structure Analysis.
Sequence similarity searches were performed with the gapped blastp program (32) and the Non-Redundant Protein Database at the National Center for Biotechnology Information (National Institutes of Health, Bethesda). Iterative database searches were performed with the Position-Specific Iterating (psi)-blast program with position-specific scoring matrices (PSSMs) and a cutoff expectation (E) value of 0.01 (32). Multiple alignments of protein sequences were constructed by using the clustal_x program (33) or the Multiple Alignment Construction and Analysis Workbench (macaw) program (34) and adjusted manually on the basis of psi-blast results. Secondary structure prediction and secondary-structure-based threading were carried out by using the phd (35, 36) and psi-pred programs (37).
Plasmid Construction.
The A22R ORF was amplified by polymerase chain reaction (PCR) using vaccinia virus strain WR genomic DNA as template and oligonucleotide primers 5′-GGGCGGTACCACCATGGCTGAAACTTTAACCAGTTCGTCTCAATC (KpnI and NcoI restriction sites underlined, initiating codon in boldface letters) and 5′-GGGGGGATCCTTAGTGATGGTGATGGTGATGCATTTTTTTTATGTAATTTCTAGATTTAC (BamHI restriction site underlined, the six consecutive histidine codons in italics). The PCR product was digested with NcoI and BamHI, gel purified, and inserted into pET11d (38), downstream of the bacteriophage T7 RNA polymerase promoter, to create pET11- A22-his. A second plasmid was constructed by inserting the A22-his PCR fragment into the KpnI and BamHI site of pcDNA 3 (Invitrogen) to create pcDNA-A22-his which was used as a template for PCR-mediated constructions. Mutated A22R ORFs A22D30N and A22E81Q were constructed by site-directed mutagenesis using a two-step recombinant PCR procedure. All plasmids were grown in E. coli DH5α, and the structures of relevant portions were confirmed by DNA sequencing.
Bacterial Expression and Metal-Affinity Purification of Recombinant A22R Protein (rA22).
Bacterial synthesis of rA22, rA22D30N, and rA22E81Q was carried out in the T7 RNA polymerase expression system (38). E. coli strain BL-21(DE3) pLysS was transformed with pET11-A22-his, pET11-A22D30N-his, and pET11-A22E81Q-his, and transformants were typically grown in 1 liter of LB broth plus 150 μg of ampicillin per ml. Expression of recombinant proteins was induced with 0.4 mM isopropyl thiogalactoside for 2–3 h. Induced bacteria were centrifuged and the pellet was frozen and then resuspended in 40 ml of buffer-200 (25 mM Tris⋅HCl, pH 7.5/200 mM NaCl/10% glycerol/10 mM 2mercaptoethanol/0.2% Nonidet P-40 detergent/2.5 mM benzamidine/0.2 mM phenylmethylsulfonyl fluoride/25 mM imidazole). The suspension was passed through a 19-gauge needle to disrupt the bacterial pellet and frozen and thawed once to facilitate lysis. The resulting lysate was treated on ice with 10 μg of micrococcal nuclease per ml plus 1 mM CaCl2 to digest the bacterial DNA. The lysate was centrifuged at 26,000 × g for 20 min to remove insoluble debris. The clarified bacterial extract (40 ml) was mixed with 2 ml of Ni-nitrilotriacetate (NTA) metal affinity resin (Qiagen), previously equilibrated in buffer-200, and rA22 and mutant proteins were bound to the resin in a batch procedure for 3–4 h. Afterward, the resin was pelleted, the supernatant was discarded, and the resin was resuspended in buffer-500, which contains the same components as buffer-200 except that the NaCl and imidazole concentrations were 500 mM and 40 mM, respectively. The resin suspension was transferred to a column, and the resin was washed successively with 10 column-volumes of buffer-500 followed by 10 column-volumes of buffer-200 containing 40 mM imidazole without Nonidet P-40. The immobilized proteins were eluted in 2 ml of buffer-200 containing 200 mM imidazole without Nonidet P-40. The eluted proteins were dialyzed against 20 mM Tris⋅HCl, pH 7.5/100 mM NaCl/1 mM DTT/0.2 mM EDTA/10% glycerol/0.2 mM phenylmethylsulfonyl fluoride. Mock-affinity-purified protein from bacterial extract was prepared as described above except that BL-21 pLysS cells transformed with the parental pET11d vector were used.
Preparation of DNA Substrates.
DNA substrates were prepared from the following oligonucleotides:
HJ-1, 5′-GGTAGGACGGCCTCGCAATCGGCTTTGACCGAGCACGCGAGATGTCAACG;
HJ-2, 5′-CGTTGACATCTCGCGTGCTCGGTCAATCGGCAGATGCGGAGTGAAGTTCC;
HJ-3, 5′-GGAACTTCACTCCGCATCTGCCGATTCTGGCTGTGGCGTGTTTCTGGTGG;
HJ-4, 5′-CCACCAGAAACACGCCACAGCCAGAAAGCCGATTGCGAGGCCGTCCTACC;
D-5, 5′-GGTAGGACGGCCTCGCAATCGGCTTTCTGGCTGTGGCGTGTTTCTGGTGG;
YJ-1, 5′-GGTAGGACGGCCTCGCAATCGGCTTATCGGCAGATGCGGAGTGAAGTTCC.
Each oligonucleotide was gel purified by electrophoresis through a 12% (20:1, acrylamide to bisacrylamide) polyacrylamide gel containing 8.0 M urea and 1× TBE (89 mM Tris–borate/2 mM EDTA, pH 8.3). The bands were excised and the oligonucleotides were extracted by fragmenting and soaking the polyacrylamide gel in 4 ml of TE (10 mM Tris⋅HCl/1 mM EDTA, pH 8.0) overnight. The oligonucleotides were purified and concentrated by using Sep-Pak C18 cartridges (Waters). The substrates were prepared by annealing the following oligonucleotides: HJ (HJ-1, HJ-2, HJ-3, and HJ-4); duplex DNA (HJ-4 and D-5); Y-junction (HJ-3, HJ-4, and YJ-1); and three-way junction (HJ-1, HJ-3, and HJ-4). Before annealing the oligonucleotides, the HJ-4 strand was usually 5′-end labeled by using T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol; 1 Ci = 37 GBq). The kinase reaction was terminated by extraction with phenol and the unincorporated label was removed by passing the aqueous layer through a G-50 spin column (Pharmacia).
Cleavage Assay.
Reaction mixtures (20 μl) contained indicated amounts of rA22 or mutated forms, indicated amounts of 32P-labeled DNA substrate, 20 mM Tris⋅HCl (pH 8.0), 0.5 mM MgCl2, 1 mM DTT, 100 μg/ml BSA, 5% glycerol, and 50 mM NaCl. After 25 min at 37°C, the reactions were terminated by the addition of a stop solution to make final concentrations of 20 mM EDTA, 0.2% SDS, 4% glycerol, and 0.02% bromophenol blue. The duplex products were resolved by electrophoresis in a nondenaturing 10% (20:1) polyacrylamide gel containing 1× TBE. The gel was dried on DE-81 paper (Whatman) and autoradiography was performed to visualize the bands. To examine single-stranded cleavage products, 5 μl of the reaction mixture was mixed with 10 μl of 90% formamide/10 mM EDTA/0.02% bromophenol blue. After heating at 95°C for 3 min, the mixture was placed on ice and then analyzed by electrophoresis through a 12% (20:1) polyacrylamide gel containing 8.0 M urea and 1× TBE. The wet gel was then exposed to x-ray film for autoradiography.
Mobility-Shift Assay.
DNA binding reaction mixtures (20 μl) contained 0.5 μl of rA22 or mutated forms, indicated amounts of 32P-labeled DNA, 20 mM Tris⋅HCl (pH 8.0), 1 mM DTT, 100 μg/ml BSA, 5% glycerol, 50 mM NaCl, and 1 mM EDTA. DNA binding mixtures were incubated for 20 min at room temperature, and 0.5 μl of tetrahistidine antibody (0.2 mg/ml; Qiagen) was added to designated reactions and the incubation was continued for 20 min. Competition assays were done with 25 μg/ml poly(dI-dC)⋅(dI-dC) for 5 min at room temperature before the addition of 32P-labeled HJ. Afterward, a loading dye solution was added to give final concentrations of 20 mM Hepes (pH 7.5), 5% glycerol, and 0.02% bromophenol blue, and the mixture was placed on ice. The DNA binding reaction products were analyzed by using a 4% (30:1) polyacrylamide gel containing 0.5× TBE. Electrophoresis was performed at 4°C at 150–200 V until the bromophenol blue dye marker migrated toward the bottom of gel. The gel was dried on DE-81 paper and the bands were visualized by autoradiography.
Results
Poxvirus, Iridovirus, and Yeast Mitochondrial Homologs of the Bacterial HJ Resolvase RuvC.
After four iterations of a psi-blast search initiated with the E. coli RuvC sequence, protein MSV106 from Melanoplus sanguinipes entomopoxvirus was retrieved in addition to RuvC homologs from other bacteria and several bacteriophages. When a new blast search was initiated with MSV106, the sequences of orthologous proteins from other poxviruses, including the A22R protein of vaccinia virus, were retrieved at a statistically significant level (E < 10−6). Additional psi-blast searches initiated with the sequences of different RuvC-like proteins identified a homolog in the iridovirus Chilo iridescent virus, and also showed that the Sac. cerevisiae mitochondrial resolvase CCE1 and its ortholog from Sch. pombe belong to the same family (L.A. and E.V.K., unpublished results). A multiple alignment created with the macaw program resulted in the detection of five statistically significant motifs (P < 10−8) that are conserved throughout the RuvC family (Fig. 1). Mapping these conserved sequence motifs on the three-dimensional structure of RuvC (30), together with secondary structure predictions, suggested that the entire family preserves the core structural elements of RuvC separated by poorly conserved spacers in the yeast mitochondrial and viral forms (Fig. 1). The characteristic motifs of the RuvC family include acidic residues that are usually an aspartate near the end of strand 1, a glutamate near the end of the conserved strand 4, and two aspartates (DXXD) embedded in the C-terminal helix (Fig. 1). All of these residues are critical for the resolvase activity of RuvC; three of them form a spatially juxtaposed acidic triad that probably coordinates a metal ion (31). The catalytic residues and the adjacent secondary structure elements are perfectly conserved in the poxvirus A22R homologs (Fig. 1), which suggests that these proteins are active resolvases.
Figure 1.
Multiple sequence alignment of the RuvC family of HJ resolvases. The alignment was constructed by using the macaw program; only the five conserved motifs are shown. The lengths of the poorly conserved spacers between the motifs are indicated by italic numbers. The positions of the first and the last of the aligned amino acid residues in each sequence are also indicated. The protein designation consist of the gene, an abbreviated species name, and the GenBank identification number (separated by underlines). The consensus derived using 90% conservation is shown underneath the alignment; b indicates “big” residues (E, K, R, I, L, M, F, Y, W), h indicates hydrophobic residues (A, C, F, I, L, M, V, W, Y), s indicates small residues (A, C, S, T, D, N, V, G, P), u indicates “tiny” residues (G, A, S), and p indicates polar residues (D, E, H, K, N, Q, R, S, T). The conserved acidic residues constituting the catalytic triad of RuvC are highlighted by shading with reverse lettering. The multiple-alignment-based secondary structure prediction is shown on top of the alignment; E (e) indicates extended conformation (β-strand), and H (h) indicates α-helix (uppercase indicates the most confident prediction). Species abbreviations: Bacteria: Ct, Chlamydia trachomatis; Ec, E. coli; Hp, Helicobacter pylori; Mtu, Mycobacterium tuberculosis; Rp, Rickettsia prowazekii; Ssp, Synechocystis sp.; Tm, Thermotoga maritima; Tp, Treponema pallidum. Eukaryotic mitochondrial: Sc, Sac. cerevisiae; Sp, Sch. pombe. Viruses: biL66, LBPc2, Lactococcus lactis bacteriophages; CIV, Chilo iridescent virus; MCV, molluscum contagiosum virus; MSEPV, Melanoplus sanguinipes entomopoxvirus; Myx, myxoma virus; RFV, rabbit fibroma virus; VACC, vaccinia virus; YMV, Yaba monkey virus.
Expression and Purification of Recombinant Proteins.
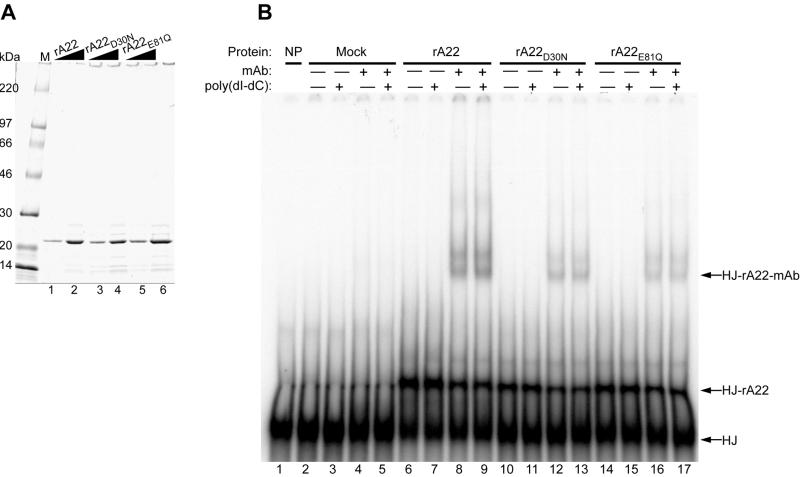
To determine whether the vaccinia virus RuvC homolog encodes a functional HJ resolvase, we expressed recombinant polyhistidine-tagged A22R protein (rA22) and mutated forms with the conserved aspartate-30 changed to asparagine (rA22D30N) or glutamate-81 changed to glutamine (rA22E81Q). On the basis of the properties of RuvC, these mutations were predicted to eliminate catalytic activity without affecting specific DNA binding (31). After affinity purification, wild-type and mutant recombinant proteins of 24 kDa were the major components visualized by SDS/PAGE and Coomassie blue staining (Fig. 2A).
Figure 2.
Binding of affinity-purified rA22, rA22D30N, and rA22E81Q to a synthetic HJ. (A) Recombinant polyhistidine-tagged rA22, rA22D30N, and rA22E81Q were expressed in E. coli and purified by chromatography on a metal-affinity resin, analyzed on an SDS/4–20% polyacrylamide gel, and stained with Coomassie blue. Lanes: M, molecular mass markers; 1, 2.1 μg of rA22; 2, 4.2 μg of rA22; 3, 2.3 μg of rA22D30N; 4, 4.6 μg of rA22D30N; 5, 2.6 μg of rA22E81Q; and 6, 5.2 μg of rA22E81Q. Wedges indicate increasing amounts of protein. (B) Binding of rA22, rA22D30N, and rA22E81Q to the HJ. Affinity-purified recombinant proteins or mock-affinity-purified proteins from bacterial extracts were incubated with 0.1 pmol of HJ in the presence of EDTA, and with (+) or without (−) 75-fold excess poly(dI-dC)⋅poly(dI-dC) or anti-tetrahistidine monoclonal antibody (mAb). The native products were analyzed on a 4% polyacrylamide gel. Lanes: 1, no protein; 2–5, mock-affinity-purified proteins; 6–9, 0.42 μg of rA22; 10–13, 0.46 μg of rA22D30N; 14–17, 0.52 μg of rA22E81Q. Free HJ, HJ-rA22, and HJ-rA22-mAb complexes are indicated on the right side of the autoradiogram.
Binding of rA22 and Mutated Forms to a HJ Probe.
A synthetic HJ substrate for RuvC with a mobile central core and nonhomologous arms to prevent dissociation by branch migration (31) was prepared by annealing four partially complementary 50-mer oligonucleotides of which one was 5′-32P-labeled. When rA22 or the mutated forms were incubated with the HJ in the presence of EDTA to inhibit any nuclease activity, a discrete band migrating more slowly than the free probe was observed on nondenaturing polyacrylamide gels (Fig. 2B). A similar band was not detected when protein was omitted or when the probe was incubated with mock-affinity-purified proteins from a bacterial extract (Fig. 2B). The identities of the DNA–protein complexes were established by the formation of supershifted species when a monoclonal antibody that recognizes four or more consecutive histidines was added to the binding reaction mixture (Fig. 2B). The detection of a doublet could signify the binding of one or two antibody molecules. The intensities of the HJ complexes were not reduced by a ≈75-fold excess of poly(dI-dC)⋅poly(dI-dC) added as a nonspecific competitor.
Nicking of a Synthetic HJ by rA22.
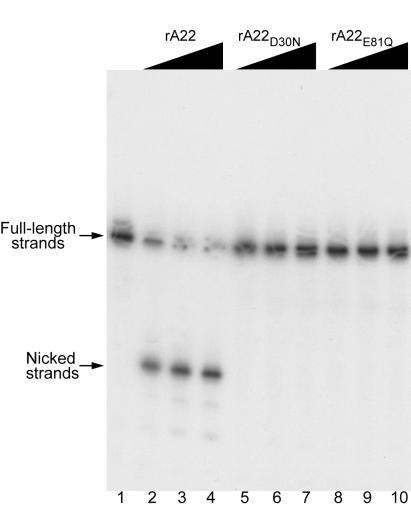
The ability of rA22 or the mutated forms to nick a HJ was examined next. Increasing amounts of rA22 or mutated proteins were incubated with the HJ containing a 50-nucleotide 32P-labeled strand. When analyzed on a denaturing polyacrylamide gel, a major band of approximately 24 nucleotides was detected, indicating endonuclease activity (Fig. 3). When analyzed on a nondenaturing gel, the HJ product was resolved into a 50-bp nicked duplex (shown later). Neither rA22D30N nor rA22E81Q cut the 32P-labeled strand of the HJ (Fig. 3) even though they could bind to the probe (Fig. 2B). Because all three recombinant proteins were prepared in the same manner and were of similar purity (Fig. 2A), we could exclude an activity produced by a contaminating E. coli endonuclease.
Figure 3.
Nicking of a synthetic HJ by rA22. Recombinant proteins were incubated for 30 min at 37°C with 0.2 pmol of HJ that was 5′-32P-end-labeled on strand 4. Reactions were stopped by addition of SDS and EDTA, and the products were analyzed by electrophoresis on a denaturing 12% polyacrylamide gel and visualized by autoradiography. Lanes: 1, no protein; 2–4, 0.30 to 3.0 μg of rA22; 5–7, 0.43 to 4.3 μg of rA22D30N; 8–10, 0.43 to 4.3 μg of rA22E81Q.
Substrate Specificity of rA22.
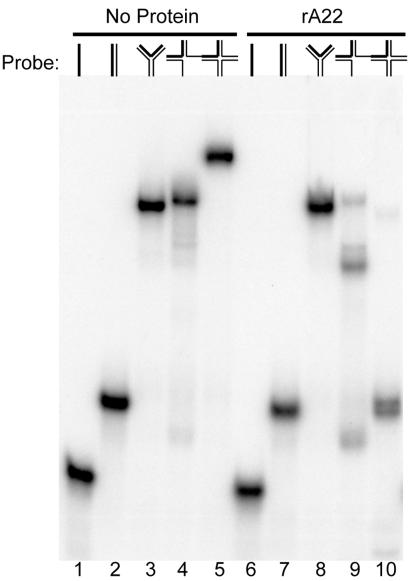
Resolvases of the same class as E. coli RuvC exhibit marked selectivity for synthetic DNA substrates that model four- or three-stranded junctions, whereas bacteriophage T4 endonuclease VII and bacteriophage T7 endonuclease I have broader activities (39). We therefore prepared several different synthetic probes from 50-mer oligonucleotides: single-stranded DNA, duplex DNA, a Y-junction DNA, a two-stranded branched DNA, a three-stranded junction, and a four-way HJ. The 5′-32P-labeled probes were incubated with rA22 and the products were analyzed by nondenaturing PAGE. rA22 converted the four-stranded HJ into a DNA product that comigrated with a 50-bp duplex and converted the three-stranded junction to products that migrated slower and faster than the duplex (Fig. 4). In contrast, rA22 did not cleave single-stranded DNA, double-stranded DNA, Y-junctions (Fig. 4) or double-stranded branched forms (data not shown). Neither rA22D30N nor rA22E81Q cleaved any of the substrates (data not shown).
Figure 4.
Substrate specificity of rA22. rA22 was incubated for 25 min at 37°C with 0.20 pmol of single-stranded, duplex, Y-junction, three-stranded junction, or four-stranded HJ that was 5′-32P-end-labeled on one strand. Reactions were stopped by addition of SDS and EDTA, and the products were analyzed by electrophoresis on a nondenaturing 10% polyacrylamide gel and visualized by autoradiography. Lanes: 1–5, no recombinant protein; 6–10, 3.2 μg of rA22. DNA substrates are shown diagrammatically above the autoradiograph with the 32P-end-labeled strand indicated in boldface.
To determine the binding specificity of rA22, incubations with the various DNA probes were repeated in the presence of EDTA to prevent cleavage and with or without anti-tetrahistidine monoclonal antibody to induce supershifting. rA22 formed complexes with the three- and four-stranded junctions, as anticipated from the resolution assays, and to a much lesser extent with Y-junctions and duplex molecules (data not shown).
Symmetrical Cleavage of HJs.
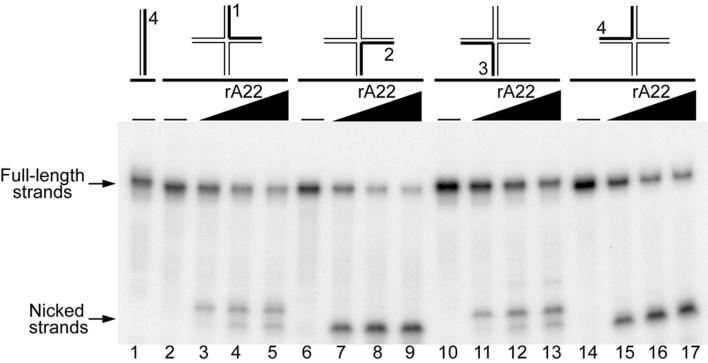
The pattern of cleavage in each strand of the HJ was examined. Four identical HJs differing only in the strand that was 5′-32P-labeled were incubated with increasing amounts of rA22 and the products were separated in a denaturing gel. In each reaction, full-length and faster migrating strands were detected. There were two predominant nicks in strand 1, one in strand 2, two in strand 3 corresponding to strand 1, and one in strand 4 corresponding to strand 1 (Fig. 5). In contrast, no nicking was observed when the same substrates were incubated with rA22D30N or rA22E81Q (data not shown). Thus, nicks in strands 1 and 3 and in 2 and 4 appeared to be symmetrically related as occurs with other RuvC-related resolvases. The sequence specificity of rA22, however, is likely to be different from that of RuvC, which preferentially nicks strands 1 and 3 of the HJ used here (31).
Figure 5.
DNA strands of HJ are nicked symmetrically by rA22. A set of four HJs, each with a different 5′-32P-end-labeled strand indicated in bold (0.1 to 0.14 pmol) was incubated with rA22 for 30 min and analyzed by electrophoresis on a denaturing polyacrylamide gel as in Fig. 3. Lanes 1, 2, 6, 10, and 14 show the mobilities of duplex or HJ probes without added proteins. The wedges above the other lanes indicate 0.34 to 3.4 μg of rA22 added to the reaction mixtures. Upper and lower arrows point to full-length and nicked strands, respectively.
Discussion
The present finding of a HJ endonuclease encoded by all sequenced poxviruses culminates a long search for such an enzyme, which could have multiple roles. Branched structures are likely to arise during DNA replication or homologous recombination, and a HJ endonuclease might be required for their resolution before packaging in virus particles. Such a role has been demonstrated for bacteriophage T4 endonuclease VII (40). A HJ resolvase has been postulated to play a role in converting concatemeric poxviral genomes into unit-length molecules with hairpin termini. In one model, the palindromic concatemer junctions are extruded as cruciforms, which have the same topology as HJs (22, 23). Resolution would occur by cleavage across the base of the junction and religation. Poxvirus topoisomerases can carry out such reactions, although with low efficiency in vitro (28, 29). The A22R protein can also resolve cruciform structures (unpublished data of A.D.G and B.M.), and genetic studies are needed to sort out the in vivo roles of the poxvirus topoisomerase and HJ endonuclease.
The finding that the HJ resolvase in poxviruses and iridoviruses belongs to the RuvC family presents an evolutionary puzzle, since these are typical bacterial enzymes. In standard database searches, the poxvirus A22R orthologs show statistically significant similarity to each other (probability of occurrence by chance <10−5) but not to any other proteins, which strongly suggests their monophyletic origin. So far, only homologs of eukaryotic genes with various levels of conservation have been detected in poxviruses, leading to the common notion of evolution of these viruses by incremental acquisition of host genes. The only detected eukaryotic representatives of the RuvC family are the mitochondrial resolvases CCE1 from Sac. cerevisiae and its ortholog from Sch. pombe (10, 11, 41). Poxvirus resolvases could have evolved from a prokaryotic source via a fungal intermediary; this makes an interesting parallel to the apparent common origin of the poxvirus and fungal RNA 5′-triphosphatases (42) and the specific relationships that are observed between poxvirus nucleoside triphosphate phosphohydrolase I and capping enzyme and the corresponding enzymes encoded in linear plasmids from yeast mitochondria (refs. 43 and 44; unpublished data of L.A. and E.V.K.).
Abbreviations
- HJ
Holliday junction
- rA22
recombinant A22R protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150238697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150238697
References
- 1.Holliday R. Genet Res. 1964;5:282–304. [Google Scholar]
- 2.Ortiz-Lombardia M, Gonzalez A, Eritja R, Aymami J, Azorin F, Coll M. Nat Struct Biol. 1999;6:913–917. doi: 10.1038/13277. [DOI] [PubMed] [Google Scholar]
- 3.Dunderdale H J, Benson F E, Parsons C A, Sharples G J, Lloyd R G, West S C. Nature (London) 1991;354:506–510. doi: 10.1038/354506a0. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connolly B, Parsons C A, Benson F E, Dunderdale H J, Sharples G J, Lloyd R G, West S C. Proc Natl Acad Sci USA. 1991;88:6063–6067. doi: 10.1073/pnas.88.14.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizuuchi K, Kemper B, Hays J, Weisberg R A. Cell. 1982;29:357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- 7.de Massy B, Weisberg R A, Studier F W. J Mol Biol. 1987;193:359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]
- 8.Dickie P, McFadden G, Morgan A R. J Biol Chem. 1987;262:14826–14836. [PubMed] [Google Scholar]
- 9.Sharples G J, Chan S N, Mahdi A A, Whitby M C, Lloyd R G. EMBO J. 1994;13:6133–6142. doi: 10.1002/j.1460-2075.1994.tb06960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleff S, Kemper B, Sternglanz R. EMBO J. 1992;11:699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White M F, Lilley D M. Mol Cell Biol. 1997;17:6465–6471. doi: 10.1128/mcb.17.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komori K, Sakae S, Shinagawa H, Morikawa K, Ishino Y. Proc Natl Acad Sci USA. 1999;96:8873–8878. doi: 10.1073/pnas.96.16.8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moss B. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 14.Fenner F, Comben B M. Virology. 1958;5:530–548. doi: 10.1016/0042-6822(58)90043-6. [DOI] [PubMed] [Google Scholar]
- 15.Merchlinsky M. J Virol. 1989;63:2030–2035. doi: 10.1128/jvi.63.5.2030-2035.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geshelin P, Berns K I. J Mol Biol. 1974;88:785–796. doi: 10.1016/0022-2836(74)90399-4. [DOI] [PubMed] [Google Scholar]
- 17.Baroudy B M, Venkatesan S, Moss B. Cell. 1982;28:315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- 18.Moyer R W, Graves R L. Cell. 1981;27:391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- 19.Baroudy B M, Venkatesan S, Moss B. Cold Spring Harbor Symp Quant Biol. 1982;47:723–729. doi: 10.1101/sqb.1983.047.01.083. [DOI] [PubMed] [Google Scholar]
- 20.Merchlinsky M, Moss B. Cell. 1986;45:879–884. doi: 10.1016/0092-8674(86)90562-3. [DOI] [PubMed] [Google Scholar]
- 21.Delange A M, Reddy M, Scraba D, Upton C, McFadden G. J Virol. 1986;59:249–259. doi: 10.1128/jvi.59.2.249-259.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickie P, Morgan A R, McFadden G. J Mol Biol. 1987;196:541–558. doi: 10.1016/0022-2836(87)90031-3. [DOI] [PubMed] [Google Scholar]
- 23.Merchlinsky M, Garon C, Moss B. J Mol Biol. 1988;199:399–413. doi: 10.1016/0022-2836(88)90613-4. [DOI] [PubMed] [Google Scholar]
- 24.Merchlinsky M, Moss B. J Virol. 1989;63:1595–1603. doi: 10.1128/jvi.63.4.1595-1603.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLange A M. J Virol. 1989;63:2437–2444. doi: 10.1128/jvi.63.6.2437-2444.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakritz N, Fogelsong P D, Reddy M, Baum S, Hurwitz J, Bauer W R. J Virol. 1985;53:935–943. doi: 10.1128/jvi.53.3.935-943.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuart D, Ellison K, Graham K, McFadden G. J Virol. 1992;66:1551–1563. doi: 10.1128/jvi.66.3.1551-1563.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekiguchi J, Seeman N C, Shuman S. Proc Natl Acad Sci USA. 1996;93:785–789. doi: 10.1073/pnas.93.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palaniyar N, Gerasimopoulos E, Evans D H. J Mol Biol. 1999;287:9–20. doi: 10.1006/jmbi.1999.2586. [DOI] [PubMed] [Google Scholar]
- 30.Ariyoshi M, Vassylyev D G, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 31.Saito A, Iwasaki H, Ariyoshi M, Morikawa K, Shinagawa H. Proc Natl Acad Sci USA. 1995;92:7470–7474. doi: 10.1073/pnas.92.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuler G D, Altschul S F, Lipman D J. Proteins Struct Funct Genet. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 35.Rost B, Sander C. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 36.Rost B, Schneider R, Sander C. J Mol Biol. 1997;270:471–480. doi: 10.1006/jmbi.1997.1101. [DOI] [PubMed] [Google Scholar]
- 37.Jones D T. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg A H, Lade B N, Chui D O, Lin S W, Dunn J J, Studier F W. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 39.Benson F E, West S C. J Biol Chem. 1994;269:5195–5201. [PubMed] [Google Scholar]
- 40.Kemper B, Brown D T. J Virol. 1976;18:1000–1015. doi: 10.1128/jvi.18.3.1000-1015.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitby M C, Dixon J. J Mol Biol. 1997;272:509–522. doi: 10.1006/jmbi.1997.1286. [DOI] [PubMed] [Google Scholar]
- 42.Ho C K, Pei Y, Shuman S. J Biol Chem. 1998;273:34151–34156. doi: 10.1074/jbc.273.51.34151. [DOI] [PubMed] [Google Scholar]
- 43.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aravind L, Koonin E V. J Mol Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]