Abstract
Deletion of 5 residues (Δ5) from the central cytoplasmic loop of the lactose permease of Escherichia coli has no significant effect on expression or activity, whereas Δ12 leads to increased rates of permease turnover after membrane insertion and decreased transport activity, and Δ20 abolishes insertion and activity. By expressing Δ12 or Δ20 in two halves, both expression and activity are restored to levels approximating wild type. Replacing deleted residues with random hydrophilic amino acids also leads to full recovery. However, introduction of hydrophobic residues decreases expression and activity in a context-dependent manner. Thus, a minimum length of the central cytoplasmic loop is vital for proper insertion, stability, and efficient transport activity, because of constraints at the cytoplasmic ends of helices VI and VII. Furthermore, the results are consistent with the idea that the middle cytoplasmic loop provides a temporal delay between insertion of the first six helices into the membrane before insertion of the second six helices.
Keywords: insertion, membrane proteins, ribosomes, signal recognition particle
Many advances have been made with respect to knowledge of translocation of proteins across the plasma membrane of Escherichia coli, as well as integration of proteins into the membrane (1). Protein targeting to the E. coli inner membrane can occur via the secretion (Sec) pathway or the signal recognition particle (SRP) pathway. The extensively studied Sec pathway utilizes a cytosolic chaperone, SecB, that binds posttranslationally to the mature region of preproteins (2). The SecB/preprotein complex is targeted to the membrane, where SecA is activated for high-affinity recognition of the complex by the membrane-embedded bacterial translocon (SecYEG) (3). SecB is released from the preprotein SecA and drives preprotein translocation by mediating repeated cycles of ATP binding and hydrolysis (4).
The SRP pathway involves cytosolic factors homologous to components involved in protein targeting to the endoplasmic reticulum (ER) membrane in eukaryotes (5). Briefly, a hydrophobic signal sequence in a polytopic membrane protein (PMP) may serve as a signal for the bacterial SRP (Ffh protein + 4.5S RNA). Thereafter, the SRP/ribosome/nascent chain complex (SRP-RNC) docks with FtsY, the bacterial homologue of the mammalian SRP receptor, SRα, on the cytoplasmic face of the inner membrane. Subsequently, binding of GTP by FtsY and Ffh triggers the RNC transfer to the SecYEG translocon complex (6). Thereafter, each transmembrane domain (TMD) is translated on the ribosome and may be inserted cotranslationally into the SecYEG complex. Concurrently and/or subsequently, one or more TMDs at a time presumably move laterally to the periphery of the translocon and into the bilayer (7).
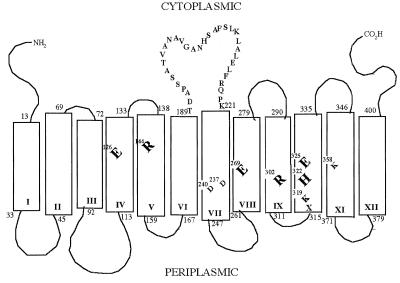
The lactose permease (lac permease) (8), encoded by the lacY gene of E. coli, catalyzes galactoside/H+ symport and is an important model for secondary transport proteins in organisms from Archaea to the mammalian central nervous system that transduce free energy stored in electrochemical ion gradients into solute concentration gradients (8, 9). The permease has been solubilized and purified in a completely active state (10) and shown to function as a monomer (11). The protein contains 12 α-helices that traverse the membrane in zigzag fashion, connected by relatively hydrophilic loops with both the N and C termini on the cytoplasmic face (Fig. 1A) (12, 13). In a functional mutant devoid of native Cys residues, each residue has been replaced with Cys (14). Analysis of the mutant library has led to the following developments (12–16): (i) The great majority of the mutants are expressed normally in the membrane and exhibit significant activity, and only six side chains are clearly irreplaceable for active transport—Glu-126 (helix IV) and Arg-144 (helix V), which are indispensable for substrate binding, and Glu-269 (helix VIII), Arg-302 (helix IX), His-322, and Glu-325 (helix X), which are critical for H+ translocation and coupling with substrate translocation. (ii) Helix packing and tilts, as well as ligand-induced conformational changes, have been determined by using site-directed biochemical and biophysical techniques. (iii) Positions that are accessible to solvent have been revealed. (iv) Positions where the reactivity of the Cys replacement is increased or decreased by ligand binding have been identified. (v) The permease has been shown to be a highly flexible molecule. (vi) A working model for lactose/H+ symport has been formulated.
Figure 1.
Secondary structure of lac permease. The one-letter amino acid code is used, and putative transmembrane helices are shown in boxes. Residues in the central cytoplasmic loop are shown. Residues that are irreplaceable with respect to active transport are enlarged—Glu-126 (helix IV) and Arg-144 (helix V) are critical for substrate binding, and Glu-269 (helix VIII), Arg-302 (helix IX), His-322 (helix X), and Glu-325 (helix X) are essential for H+ translocation and coupling. The charge pairs Asp-237 (helix VII)/Lys-358 (helix XI) and Asp-240 (helix VII)/Lys-319 (helix X) are also shown.
lac permease is a useful model for studying polytopic membrane protein (PMP) insertion. While the insertion of single transmembrane helix proteins is poorly understood (7), the process for PMPs is even more complicated (17, 18). In vivo studies indicate that lac permease is inserted cotranslationally (19), since the N-terminal 50 amino acid residues cause ribosomal attachment to the membrane during translation. Conditional mutants lacking 4.5S RNA, Ffh (20), or FtsY (21) do not insert permease into the membrane, whereas SecA mutants have no effect. Furthermore, a fundamental requirement for efficient permease insertion is a specific interaction between the N-terminal six helices and the C-terminal six helices that stabilizes the functional complex (22, 23). Additionally, interaction between Asp-237 (helix VII) and Lys-358 (helix XI) is required for efficient insertion, indicating that the formation of the salt bridge is important for insertion of the C-terminal half of the permease (24). In vitro studies suggest that the chaperone GroEL aids folding of the permease and prevents aggregation (25) and that phosphatidylethanolamine (PE) plays a role in late maturation of the polypeptide (26).
Alignment of the major facilitator superfamily (MFS) reveals that the central cytoplasmic loop is consistently longer than the other loops (27). Although this loop in lac permease contains a portion of the binding site for the regulatory protein IIAGlc (28–31), Cys-scanning (14) and insertional (32–34) mutagenesis indicate that this region does not play an important role in the transport mechanism. The findings presented in this paper demonstrate that the cytoplasmic loop between helices VI and VII provides a temporal delay between insertion of the first six helices into the translocon and their movement into the bilayer before insertion of the last six helices.
Materials and Methods
Materials.
[1-14C]Lactose and l-[35S]methionine were purchased from Amersham. Deoxynucleotides were synthesized on an Applied Biosystems 391 DNA synthesizer. Restriction endonucleases, T4 DNA ligase, and Vent polymerase were from New England Biolabs. Penta-His antibody was purchased from Qiagen (Valencia, CA).
Bacterial Strains and Growth.
E. coli HB101 [hsdS20 (rB−, mB−), recA13, ara-14, proA2, lacY1, galK2, rpsL20 (Smr), xyl-5, mtl-1, supE44, Δ−/F−] was used as a carrier for the plasmids described and qualitative assessment of downhill lactose transport on MacConkey indicator plates (Difco) containing 20 mM lactose. E. coli T184 [lacI+O+Z−Y−(A), rpsL, met−, thr−, recA, hsdM, hsdR/F′, lacIqO+ZD118(Y+ A+)] (35) expressing given permease mutants was grown aerobically at 37°C in Luria–Bertani (LB) broth with ampicillin (100 μg/ml). Fully grown cultures were diluted 10-fold and grown for 2 h at 37°C before induction with 1 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG). After additional growth for 2 h at 37°C, cells were harvested by centrifugation.
Construction of Permease Mutants.
By using plasmid pT7–5/cassette lacY encoding wild-type lac permease with six contiguous His residues at the C terminus (His6 tag), oligonucleotide-directed site-specific mutagenesis by inverse PCR was used to generate deletion mutants Δ5, Δ12, and Δ20. After restriction endonuclease digestion with PstI and SpeI, the PCR products were subcloned back into similarly treated parental vector. Construction of the Δ5, Δ12, and Δ20 mutants as two contiguous, nonoverlapping peptides corresponding to the N- and C-terminal six helices was carried out as described (36). Thereafter, restriction endonuclease digestion with PstI and SpeI was followed by subcloning back into the similarly treated parental vector. For insertion of stretches of random amino acids, a unique KasI restriction endonuclease site was introduced at the deletion site in the DNA encoding the Δ5, Δ12, and Δ20 mutants. Linkers were inserted into the unique KasI restriction endonuclease site of the Δ mutants as described in Table 1.
Table 1.
Genotypes and phenotypes of the mutants used in this study
| Mutant | Phenotype | Sequence |
|---|---|---|
| WT | Red | 189TDAPSSATVANAVGANHSAFSL––––––––––––––––––––––KLALELFRQPK221 |
| Δ5 | Red | 189TDAPSSATVANAVGANH–––––––––––––––––––––––––––KLALELFRQPK221 |
| Δ5-KasI | Red | 189TDAPSSATVANAVGANH–––––––––––––––GA––––––––––KLALELFRQPK221 |
| Δ5-split | Red | 189TDAPSSATVANAVGANH*–––––––––––––––––––––––––MKLALELFRQPK221 |
| Δ5 + 12 Hphil 1 | Red | 190TDAPSSATVANAVGANH––––––––––GAEDEDCPEDEHCA–––KLALELFRQPK221 |
| Δ5 + 12 Hphil 2 | Red | 190TDAPSSATVANAVGANH––––––––––GAEDEDEDEDEDCA–––KLALELFRQPK221 |
| Δ5 + 12 Hphob 1 | Halo | 190TDAPSSATVANAVGANH––––––––––GAVLIFWAIFVSCA–––KLALELFRQPK221 |
| Δ5 + 12 Hphob 2 | Halo | 190TDAPSSATVANAVGANH––––––––––GAIVIVIVIVIVCA–––KLALELFRQPK221 |
| Δ12 | Red | 189TDAPSSATVA––––––––––––––––––––––––––––––––––KLALELFRQPK221 |
| Δ12-KasI | Red | 189TDAPSSATVA––––––––––––––––––––––GA –––––––––KLALELFRQPK221 |
| Δ12-split | Red | 189TDAPSSATVA*––––––––––––––––––––––––––––––––MKLALELFRQPK221 |
| Δ12 + 12 Hphil 1 | Red | 190TDAPSSATVA–––––––––––––––––GAEDEDCPEDEHCA–––KLALELFRQPK221 |
| Δ12 + 12 Hphil 2 | Red | 190TDAPSSATVA–––––––––––––––––GAEDEDEDEDEDCA–––KLALELFRQPK221 |
| Δ12 + 12 Hphob 1 | White | 190TDAPSSATVA–––––––––––––––––GAVLIFWAIFVSCA–––KLALELFRQPK221 |
| Δ12 + 12 Hphob 2 | White | 190TDAPSSATVA–––––––––––––––––GAIVIVIVIVIVCA–––KLALELFRQPK221 |
| Δ20 | White | 189TDAPSS––––––––––––––––––––––––––––––––––––––––––ELFRQPK221 |
| Δ20-KasI | White | 189TDAPSS––––––––––––––––GA –––––––––––––––––––––––ELFRQPK221 |
| Δ20-split | Red | 189TDAPSS*––––––––––––––––––––––––––––––––––––––––MELFRQPK221 |
| Δ20 + 22 Hphil | Red | 190TDAPSS–––––––––GATSTATSTSCATSTATSTSCA––––––––––ELFRQPK221 |
| Δ20 + 29 Hphil | Red | 190TDAPSS–––––––GAGGGGGGGGAGGGGGGGGGAGGGGGGGGGA––––ELFRQPK221 |
| Δ20 + 40 Hphil | Red | 190TDAPSSGATSTATSTSCATSTATSTSCATSTATSTSCATSTATSTSCAELFRQPK221 |
Phenotype refers to color on MacConkey lactose indicator plates. Asterisks indicate a stop codon, and the italicized G and A indicate the amino acids glycine and alanine encoded by the unique KasI restriction enzyme site.
Lactose Transport.
For active transport, E. coli T184 was washed once with 100 mM KPi (pH 7.5)/10 mM MgSO4 and adjusted to an OD600 of 10.0 (0.7 mg of protein per ml). Transport was initiated by addition of [1-14C]lactose [5 mCi/mmol specific activity (1 mCi = 37 MBq), 0.4 mM or 2.0 mM final concentration], and samples were quenched at given times by 100 mM KPi (pH 5.5)/100 mM LiCl and assayed by rapid filtration (37).
Western Blots.
Crude membranes from the same cells used to assay active transport were prepared as described (38). Total membrane protein was assayed by a modified Lowry procedure (39). A sample containing 60 μg of membrane protein from each sample was subjected to NaDodSO4/12% PAGE (40). Proteins were electroblotted onto poly(vinylidene difluoride) membranes (Immobilon-PVDF; Millipore) and probed with a monoclonal antibody raised against a His5 epitope (Qiagen), followed by treatment with an anti-mouse IgG peroxidase-linked antibody (Amersham Pharmacia Biotech). The PVDF membrane was subsequently developed with fluorescent substrate (Renaissance, DuPont NEN) and exposed to film. Films were scanned with an imaging densitometer (Molecular Dynamics).
In Vivo Labeling with [35S]Methionine.
Cloned DNA was overexpressed by using the T7 RNA polymerase system (41–43). Briefly, plasmid DNA was transformed into E. coli T184/λDE3 (Novagen) bearing the T7 RNA polymerase gene and was grown at 37°C in LB broth supplemented with streptomycin (10 μg/ml) and ampicillin (100 μg/ml). Overnight cultures were diluted 1:10 with fresh medium at 37°C, and growth was continued for 3 h. The cells were washed twice in prewarmed M9 minimal medium containing 0.005% amino acids except methionine and supplemented with ampicillin (100 μg/ml). The cells were then resuspended in the minimal medium, and after growing under sulfur-starved conditions for 1 h at 37°C, 0.2 mM IPTG was added, and growth was continued for 12 min. Rifampicin was added to a final concentration of 0.2 mg/ml to inhibit the host cell RNA polymerase, and incubation was continued for an additional 40 min. Labeling was initiated by addition of [35S]methionine (1,000 Ci/mmol) to a final concentration of 2.5 nM. After 15-min incubation, cells were harvested and resuspended in 20 mM Tris⋅HCl (pH 7.5)/2 mM ethylenediaminetetraacetate (EDTA; potassium salt). Membranes were prepared by sonication as described (38), solubilized in 1% NaDodSO4/10% glycerol/1% 2-mercaptoethanol (vol/vol), and subjected to NaDodSO4/PAGE followed by autoradiography and visualization with a Storm 860 PhosphorImager (Molecular Dynamics).
Lifetime Studies.
Cloned DNA was overexpressed by using the T7 RNA polymerase system. E. coli T184 was transformed with a given plasmid and grown at 37°C in L-B broth supplemented with streptomycin (10 μg/ml) and ampicillin (100 μg/ml). Overnight cultures were diluted 1:10 with fresh medium at 37°C, grown for an additional 2 h, induced with 0.5 mM IPTG, and harvested after 2 h. After addition of chloramphenicol (34 μg/ml), aliquots were removed at 75-min intervals and flash frozen in liquid N2. Membranes were prepared by sonication and treated as described above.
Results
Membrane Expression of lac Permease Is Sensitive to the Length of the Middle Cytoplasmic Loop.
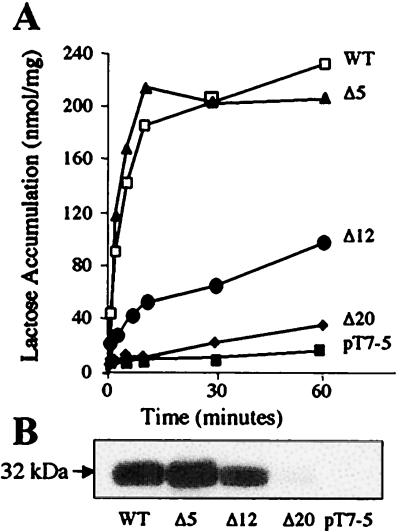
Deletion analysis (44) indicates that the central loop of lac permease is approximately 33 residues in length, extending from Thr-189 to Lys-221 (Fig. 1 and Table 1). Deletions were generated outwards from Ala-205, which is equidistant from the ends of helices VI and VII. E. coli HB101 (lacZ+Y−) expressing wild-type lac permease with six His residues at the C terminus grows as red colonies on MacConkey indicator plates (Table 1) and E. coli T184 (lacZ−Y−) expressing the same construct catalyzes rapid lactose accumulation to a steady-state level of approximately 240 nmol per mg of protein (Fig. 2A) and is expressed well in the membrane (Fig. 2B).
Figure 2.
Effect of progressive deletion of the central cytoplasmic loop. (A) Time course of lactose transport by E. coli T184 expressing the given mutants. (B) Permease expression. Membranes were prepared as described in the text, and 100 μg of protein was subjected to NaDodSO4/PAGE and electroblotting. The blot was incubated with an anti-His5 antibody, followed by horseradish peroxidase-linked protein A and luminescent substrate before exposure to film. WT, wild type; Δ5, deletion of residues 206–210; Δ12, deletion of residues 199–210; Δ20, deletion of residues 195–214; pT7–5, plasmid pT7–5 with no lacY insert.
Deletion of residues 206–210 (Δ5) results in properties identical to those of wild-type permease (Fig. 2). Although deletion of residues 199–210 (Δ12) leads to a red phenotype with E. coli HB101 on MacConkey plates, a significant decrease in the rate and steady-state level of lactose accumulation is observed (100 nmol per mg of protein) with almost no reduction in the steady-state level of protein expression in the membrane. Dramatically, deletion of residues 195–214 (Δ20) leads to white colonies on indicator plates (Table 1), a very low steady-state level of accumulation (35 nmol per mg of protein), and almost undetectable levels of expression (Fig. 2; Table 1).
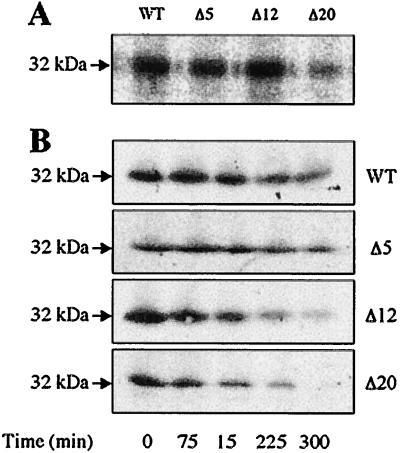
Progressive Deletion of the Central Cytoplasmic Loop Increases the Rate of Degradation and Ultimately Decreases Insertion.
There are two classes of mutants that cause low levels of lac permease in the membrane: (i) truncation mutants at the cytoplasmic end of helix XII in which the permease is inserted but subsequently degraded rapidly (42, 43, 45) and (ii) point mutants in which the permease does not enter the membrane (24, 46). [35S]Met labeling for 15 min shows that the wild type and the Δ5 and Δ12 mutants are inserted into the membrane at comparable rates, whereas the Δ20 mutant is inserted at a highly reduced rate (Fig. 3A). Lifetime experiments with chloramphenicol to block permease synthesis demonstrate that both the wild type and the Δ5 mutant are stable for at least 5 h. In contrast, the Δ12 mutant is degraded at a significantly increased rate (Fig. 3B). By overexposing immunoblots of the Δ20 mutant, which is expressed at much lower levels, it is apparent that the Δ20 mutant is also degraded at an increased rate (Fig. 3B).
Figure 3.
Insertion and stability of deletion mutants in the membrane. (A) [35S]Met labeling. Membranes prepared from cells expressing wild-type permease or a given mutant were labeled for 15 min with [35S]methionine, subjected to NaDodSO4/PAGE, and autoradiographed. Aliquots containing the same amount of membrane protein (30 μg) were applied to each lane, and prestained molecular size markers were used as indicated by the arrow at the left. (B) Lifetime studies. E. coli T184 harboring a plasmid encoding a given mutant was diluted 1:10 from overnight cultures and grown for 2 h at 37°C. IPTG was then added, and growth was continued for 2 h. Chloramphenicol (34 μg/ml) was added, and samples were collected at given times and flash frozen in liquid N2. Membranes were prepared, subjected to NaDodSO4/PAGE, and autoradiographed. WT, wild type; Δ5, deletion of residues 206–210; Δ12, deletion of residues 199–210; Δ20, deletion of residues 195–214.
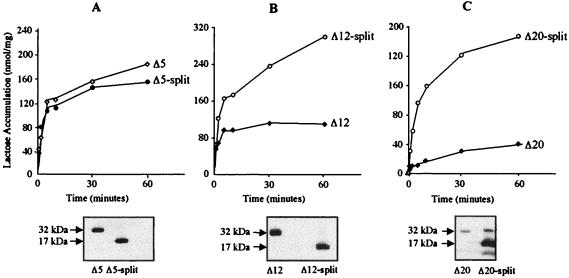
Expression of the Δ12 and Δ20 Mutants in Two Nonoverlapping Fragments Rescues Activity and Expression.
As described previously (22, 36, 47, 48), lac permease can be expressed in two fragments that complement functionally and stabilize each other against degradation (split permease). Expression of Δ5 in two fragments corresponding to the N-terminal six helices (N6) and the C-terminal six helices (C6) of the permease produces no significant effect on either activity or expression, whereas splitting Δ12 results in a very significant increase in activity with little or no effect on expression (Fig. 4 A and B). Most dramatically, when the poorly expressed and essentially inactive Δ20 mutant is expressed as a split construct, the phenotype on indicator plates becomes red (Table 1), a dramatic increase in expression is observed, and both the rate and steady-state level of lactose accumulation are rescued to levels approaching wild type (Fig. 4C).
Figure 4.
Effect of introducing a discontinuity on expression and activity. Time courses of lactose transport and levels of membrane expression of mutants Δ5 and Δ5-split (A), Δ12 and Δ12-split (B), and Δ20 and Δ20-split (C). E. coli T184 harboring a plasmid encoding a given mutant or its derivative split construct were assayed for lactose transport (Upper) and membrane expression of the permease (Lower) as described in the text.
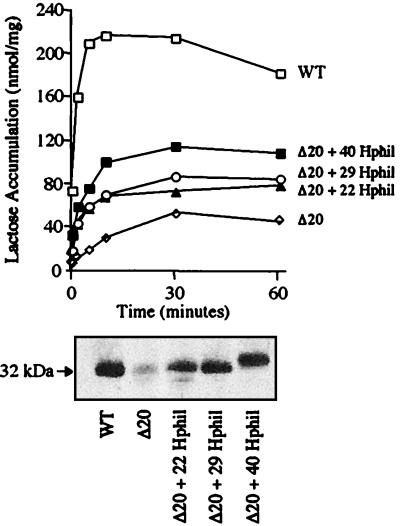
Effect of Inserting Random Amino Acid Stretches into the Deletion Mutants.
Insertion of the three progressively longer random hydrophilic amino acid sequences into the Δ20 mutant (Δ20 + 22 Hphil, Δ20 + 29 Hphil, and Δ20 + 40 Hphil; see Table 1) results in a progressive increase in expression paralleled by increases in transport activity (Fig. 5). Conversely, whereas the introduction of a hydrophilic sequence (Δ12 + 12 Hphil 1; Table 1) into the well-expressed deletion mutants Δ5 and Δ12 has little effect on expression or activity, insertion of a hydrophobic sequence (Δ12 + 12 Hphob 1; Table 1) into these mutants decreases expression and activity in a context-dependent manner. Thus, when the hydrophobic sequence is inserted at position 199, insertion and activity are abolished. However, insertion of the identical sequence at position 206 causes a significantly less drastic effect (Fig. 6). Although data are not shown, similar results were obtained with other hydrophilic and hydrophobic amino acid stretches (Δ12 + 12 Hphil 2 and Δ12 + 12 Hphob 2; Table 1).
Figure 5.
Insertion of random hydrophilic amino acid stretches into Δ20 rescues expression and activity. E. coli T184 harboring a plasmid encoding the Δ20 mutant with a given hydrophilic insertion in the middle cytoplasmic loop were assayed for lactose transport (Upper) and membrane expression of the permease (Lower) as described in the text. WT, wild type; Δ20, deletion of residues 195–214; Δ20 + 22 Hphil, Δ20 with 22 random hydrophilic amino acid residues (Table 1) inserted into the Δ20 mutant; Δ20 + 29 Hphil, Δ20 with 29 random hydrophilic amino acid residues (Table 1) inserted into the Δ20 mutant; Δ20 + 40 Hphil, Δ20 with 40 random hydrophilic amino acid residues (Table 1) inserted into the Δ20 mutant.
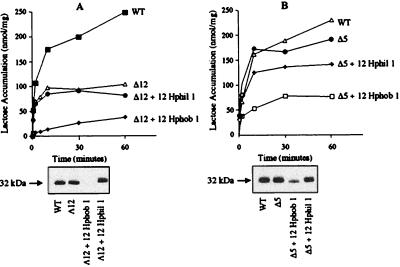
Figure 6.
Insertion of random hydrophilic or hydrophobic amino acid stretches into the Δ12 or Δ5 mutant. E. coli T184 harboring a plasmid encoding either Δ12 (A) or Δ5 (B) with a given hydrophilic or hydrophobic insertion in the middle cytoplasmic loop were assayed for lactose transport (Upper) and membrane expression of the permease (Lower) as described in the text. WT, wild type; Δ12, deletion of residues 199–210; Δ12 + 12 Hphob 1, Δ12 with 12 random hydrophobic amino acid residues (Table 1) inserted into the Δ12 mutant; Δ12 + 12 Hphil 1, Δ12 with 12 random hydrophilic amino acid residues (Table 1) inserted into the Δ12 mutant; Δ5, deletion of residues 206–210; Δ5 + 12 Hphob 1, Δ5 with 12 random hydrophobic amino acid residues (Table 1) inserted into the Δ5 mutant; Δ5 + 12 Hphil 1, Δ5 with 12 random hydrophilic amino acid residues (Table 1) inserted into the Δ5 mutant.
Discussion
Since the long central cytoplasmic loop in the lac permease and presumably the other members of the major facilitator superfamily plays no direct role in the transport mechanism, conservation of length in this loop presents an interesting evolutionary quandary. Sequence alignments indicate that residues in this region are very poorly conserved, and apart from the regulation of certain members of the oligosaccharide/H+-linked cluster by IIAGlc (28–31), it has not been possible to assign a role for this region of the protein. It is demonstrated here with lac permease that a minimum length is necessary for activity, stability, and insertion into the membrane.
The Central Cytoplasmic Loop Plays No Direct Role in the Transport Mechanism.
Extensive site-directed mutagenesis has shown that no residue in the central cytoplasmic loop is essential for active transport (14). Likewise, the ability to insert entire proteins [cytochrome b562 and HisP (49) or the biotin acceptor domain from a Klebsiella carboxylase (33)] into this region without a significant effect on activity supports this idea. The work presented here provides additional support in two respects: (i) split permease generated by introducing a discontinuity into the Δ20 mutant (i.e., devoid of the central loop) exhibits expression and activity comparable to wild type (Fig. 4C); and (ii) introduction of random hydrophilic amino acid stretches into the poorly expressed and almost inactive Δ20 mutant restores highly significant expression and activity (Fig. 5).
Shortening the Central Cytoplasmic Loop Constrains the lac Permease.
Although the Δ12 mutant is expressed at levels similar to wild type, a lower initial rate and steady-state level of lactose accumulation are observed (Fig. 2 A and B, respectively). By introducing a discontinuity into the mutant, there is a significant increase in the rate and steady-state level of accumulation without altering expression levels (Fig. 4B). Labeling studies with [35S]methionine demonstrate that Δ12 is inserted into the membrane at a rate comparable to that of wild type (Fig. 3A). However, the Δ12 mutant is clearly significantly less stable than the wild type, as demonstrated by an enhanced rate of degradation (Fig. 3B). Moreover, the rate of degradation in the Δ20 mutant is also increased (Fig. 3B). Therefore, shortening the central cytoplasmic loop appears to introduce a steric constraint between helices VI and VII that decreases activity and also destabilizes the permease in the membrane.
The Length and Hydrophilic Nature of the Middle Loop Are Critical for Efficient Insertion.
Studies on insertion of P-glycoprotein (P-gp) (50) show that up to six N-terminal helices can be extracted from the membrane with urea during translation, suggesting that the intermediate is stored in the aqueous channel of the mammalian translocon. The finding led to the suggestion that the long central cytoplasmic loop of P-gp may serve as a signal integration sequence that triggers the translocon to open laterally, allowing protein integration into the bilayer before translocation and insertion of the C-terminal six-helix bundle. Further evidence for this concept is provided by the observation that the internal diameter of the active translocon is between 40 and 60 Å (51–53), which is large enough to accommodate up to six helices. Given this hypothesis, in lac permease, as in P-gp and the entire major facilitator superfamily, the central cytoplasmic loop might contain a signal integration sequence. However, introduction of a covalent discontinuity into the Δ20 mutant leads to complete recovery of expression and activity under conditions where the central loop is deleted. Although it could be argued that the N- and C-terminal helices are inserted into the membrane via different translocons, insertion of random hydrophilic amino acid stretches into the Δ20 mutant also rescues expression and activity (Figs. 4C and 5). Thus, it is highly unlikely that the middle loop contains a signal integration sequence. Rather, it seems clear that a minimum length of polypeptide is required for efficient insertion and activity.
To define the nature of the polypeptide best suited for efficient insertion and activity, two different random 12-amino acid hydrophilic or hydrophobic stretches were inserted into Δ12 or Δ5. Whereas insertion of random hydrophilic amino acid residues has no effect on expression or activity (Fig. 6 A and B), insertion of the same length of random hydrophobic amino acid residues abolishes expression and activity of the Δ12 mutant and compromises Δ5 expression and activity. In other words, the site of insertion of the hydrophobic stretches is context dependent in such a fashion that the closer the insertion to the end of helix VI, the more severe the defect.
The results taken as a whole are consistent with the model presented in Fig. 7. The open translocon/ribosome complex allows cotranslational insertion of the N-terminal six helices of lac permease. In the wild type, during translation of the central cytoplasmic loop, the N-terminal six helices move laterally from the translocon into the bilayer (Fig. 7A). Subsequently, the C-terminal six helices are inserted into the translocon and then into the bilayer. In the Δ mutants, as the length of the hydrophilic loop is progressively shortened, efficient insertion and activity are decreased appropriately. Likewise, artificially shortening the length of the hydrophilic segment between helices VI and VII by inserting hydrophobic stretches also reduces insertion and activity in a context-dependent fashion [i.e., hydrophobic insertion at position 199 (site I in Fig. 7A) is more deleterious than insertion of the identical sequence at position 206 (site II in Fig. 7A)]. In the Δ20 mutant (Fig. 7B), a likely scenario is that helix VII enters the translocon before clearance of the N-terminal six helices, thereby blocking further insertion and leading to degradation of the permease before insertion into the bilayer. This phenotype is reversed either by insertion of a stretch of random hydrophilic amino acid residues or by introducing a split (site III in Fig. 7B). By this means, the middle hydrophilic loop in the permease, which is clearly devoid of a signal integration sequence, acts as a temporal delay that allows insertion of the N-terminal half of the molecule into the bilayer before insertion of the C-terminal half into the translocon.
Figure 7.
A temporal role for the central cytoplasmic loop in membrane insertion. The open translocon/ribosome complex allows cotranslational insertion of the N-terminal six helices of lac permease. In the wild type, during translation of the central cytoplasmic loop, the N-terminal six helices move laterally from the translocon into the bilayer (A). Subsequently, the C-terminal six helices are translocated across the bilayer and inserted into the membrane. In the Δ mutants, as the length of the hydrophilic loop is progressively shortened, efficient insertion and activity are decreased appropriately. Likewise, by artificially shortening the length of the hydrophilic segment between helices VI and VII by inserting hydrophobic stretches (sites I and II), insertion and activity are also reduced in a context-dependent fashion. In the Δ20 mutant (B), helix VII enters the translocon before clearance of the N-terminal six helices, thereby blocking further insertion and leading to degradation of the permease before insertion into the bilayer. This phenotype is reversed either by insertion of a stretch of random hydrophilic amino acid residues or by introducing a split (site III).
Acknowledgments
We thank Miklos Sahin-Tóth for suggesting the lifetime experiments and both Eitan Bibi and Hajime Tokuda for critical reading of the manuscript. This work was supported in part by National Institutes of Health Grant DK51131 to H.R.K.
Abbreviations
- SRP
signal recognition particle
- IPTG
isopropyl 1-thio-β-d-galactopyranoside
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140224497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140224497
References
- 1.Fekkes P, Driessen A J. Microbiol Mol Biol Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumamoto C A, Frascetic O. J Bacteriol. 1993;175:2184–2188. doi: 10.1128/jb.175.8.2184-2188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Driessen A J, Fekkes P, van der Wolk J P. Curr Opin Microbiol. 1998;1:216–222. doi: 10.1016/s1369-5274(98)80014-3. [DOI] [PubMed] [Google Scholar]
- 4.Schiebel E, Driessen A J M, Hartl F-U, Wickner W. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 5.Walter P, Johnson A E. Ann Rev Cell Dev Biol. 1994;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 6.Valent Q A, Scotti P A, High S, de Gier J W, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson A E, van Waes M A. Ann Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 8.Kaback H R. J Membr Biol. 1983;76:95–112. doi: 10.1007/BF02000610. [DOI] [PubMed] [Google Scholar]
- 9.Kaback H R. Harvey Lect. 1989;83:77–103. [PubMed] [Google Scholar]
- 10.Viitanen P, Newman M J, Foster D L, Wilson T H, Kaback H R. Methods Enzymol. 1986;125:429–452. doi: 10.1016/s0076-6879(86)25034-x. [DOI] [PubMed] [Google Scholar]
- 11.Sahin-Tóth M, Lawrence M C, Kaback H R. Proc Natl Acad Sci USA. 1994;91:5421–5425. doi: 10.1073/pnas.91.12.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaback H R, Wu J. Q Rev Biophys. 1997;30:333–364. doi: 10.1017/s0033583597003387. [DOI] [PubMed] [Google Scholar]
- 13.Kaback H R, Voss J, Wu J. Curr Opin Struct Biol. 1997;7:537–542. doi: 10.1016/s0959-440x(97)80119-4. [DOI] [PubMed] [Google Scholar]
- 14.Frillingos S, Sahin-Tóth M, Wu J, Kaback H R. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 15.Kaback H R, Wu J. Acc Chem Res. 1999;32:805–813. [Google Scholar]
- 16.Weinglass A B, Kaback H R. Proc Natl Acad Sci USA. 1999;96:11178–11182. doi: 10.1073/pnas.96.20.11178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bibi E. Trends Biochem Sci. 1998;23:51–55. doi: 10.1016/s0968-0004(97)01134-1. [DOI] [PubMed] [Google Scholar]
- 18.Zen K, Consler T G, Kaback H R. Biochemistry. 1995;34:3430–3437. doi: 10.1021/bi00010a035. [DOI] [PubMed] [Google Scholar]
- 19.Stochaj U, Ehring R. Eur J Biochem. 1987;163:653–658. doi: 10.1111/j.1432-1033.1987.tb10914.x. [DOI] [PubMed] [Google Scholar]
- 20.MacFarlane J, Müller M. Biochem Soc Trans. 1995;23:560S. doi: 10.1042/bst023560s. [DOI] [PubMed] [Google Scholar]
- 21.Seluanov A, Bibi E. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 22.Bibi E, Kaback H R. Proc Natl Acad Sci USA. 1990;87:4325–4329. doi: 10.1073/pnas.87.11.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahin-Tóth M, Kaback H R, Friedlander M. Biochemistry. 1996;35:2016–2021. doi: 10.1021/bi952496g. [DOI] [PubMed] [Google Scholar]
- 24.Dunten R L, Sahin-Tóth M, Kaback H R. Biochemistry. 1993;32:3139–3145. doi: 10.1021/bi00063a028. [DOI] [PubMed] [Google Scholar]
- 25.Bochkareva E, Seluanov A, Bibi E, Girshovich A. J Biol Chem. 1996;271:22256–61. doi: 10.1074/jbc.271.36.22256. [DOI] [PubMed] [Google Scholar]
- 26.Bogdanov M, Dowhan W. J Biol Chem. 1999;274:36827–36830. doi: 10.1074/jbc.274.52.36827. [DOI] [PubMed] [Google Scholar]
- 27.Pao S S, Paulsen I T, Saier M H., Jr Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson T H, Yunker P L, Hansen C L. Biochim Biophys Acta. 1990;1029:113–116. doi: 10.1016/0005-2736(90)90443-r. [DOI] [PubMed] [Google Scholar]
- 29.Hoischen C, Levin J, Pitaknarongphorn S, Reizer J, Saier M H., Jr J Bacteriol. 1996;178:6082–6086. doi: 10.1128/jb.178.20.6082-6086.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seok Y J, Sun J, Kaback H R, Peterkofsky A. Proc Natl Acad Sci USA. 1997;94:13515–13519. doi: 10.1073/pnas.94.25.13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sondej M, Sun J, Seok Y J, Kaback H R, Peterkofsky A. Proc Natl Acad Sci USA. 1999;96:3525–3530. doi: 10.1073/pnas.96.7.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna E, Hardy D, Kaback H R. Proc Natl Acad Sci USA. 1992;89:11954–11958. doi: 10.1073/pnas.89.24.11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consler T G, Persson B L, Jung H, Zen K H, Jung K, Privé G G, Verner G E, Kaback H R. Proc Natl Acad Sci USA. 1993;90:6934–6938. doi: 10.1073/pnas.90.15.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Privé G G, Verner G E, Weitzman C, Zen K H, Eisenberg D, Kaback H R. Acta Crystallogr D. 1994;50:375–379. doi: 10.1107/S0907444993014301. [DOI] [PubMed] [Google Scholar]
- 35.Teather R M, Bramhall J, Riede I, Wright J K, Furst M, Aichele G, Wilhelm V, Overath P. Eur J Biochem. 1980;108:223–231. doi: 10.1111/j.1432-1033.1980.tb04715.x. [DOI] [PubMed] [Google Scholar]
- 36.Zen K H, McKenna E, Bibi E, Hardy D, Kaback H R. Biochemistry. 1994;33:8198–8206. doi: 10.1021/bi00193a005. [DOI] [PubMed] [Google Scholar]
- 37.Kaback H R. Methods Enzymol. 1974;31:698–709. doi: 10.1016/0076-6879(74)31075-0. [DOI] [PubMed] [Google Scholar]
- 38.Sahin-Tóth M, Kaback H R. Protein Sci. 1993;2:1024–1033. doi: 10.1002/pro.5560020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson G L. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 40.Newman M J, Foster D L, Wilson T H, Kaback H R. J Biol Chem. 1981;256:11804–11808. [PubMed] [Google Scholar]
- 41.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roepe P D, Zbar R I, Sarkar H K, Kaback H R. Proc Natl Acad Sci USA. 1989;86:3992–3996. doi: 10.1073/pnas.86.11.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna E, Hardy D, Pastore J C, Kaback H R. Proc Natl Acad Sci USA. 1991;88:2969–2973. doi: 10.1073/pnas.88.8.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolin C, Kaback H R. Biochemistry. 1999;38:8590–8597. doi: 10.1021/bi990650j. [DOI] [PubMed] [Google Scholar]
- 45.McKenna E, Hardy D, Kaback H R. J Biol Chem. 1992;267:6471–6474. [PubMed] [Google Scholar]
- 46.Jung K, Jung H, Colacurcio P, Kaback H R. Biochemistry. 1995;34:1030–1039. doi: 10.1021/bi00003a038. [DOI] [PubMed] [Google Scholar]
- 47.Wrubel W, Stochaj U, Sonnewald U, Theres C, Ehring R. J Bacteriol. 1990;172:5374–5381. doi: 10.1128/jb.172.9.5374-5381.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wrubel W, Stochaj U, Ehring R. FEBS Lett. 1994;349:433–438. doi: 10.1016/0014-5793(94)00719-5. [DOI] [PubMed] [Google Scholar]
- 49.Privé G G, Kaback H R. J Bioenerg Biomembr. 1996;28:29–34. [PubMed] [Google Scholar]
- 50.Borel A C, Simon S M. Cell. 1996;85:379–389. doi: 10.1016/s0092-8674(00)81116-2. [DOI] [PubMed] [Google Scholar]
- 51.Hanein D, Matlack K E, Jungnickel B, Plath K, Kalies K U, Miller K R, Rapoport T A, Akey C W. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 52.Hamman B D, Chen J C, Johnson E E, Johnson A E. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- 53.Hamman B D, Hendershot L M, Johnson A E. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]