Abstract
The HIV regulatory proteins Tat and Rev have a nucleolar localization property in human cells. However, no functional role has been attributed to this localization. Recently it has been demonstrated that expression of Rev induces nucleolar relocalization of some protein factors involved in Rev export. Because the function of Rev is to bind HIV RNA and facilitate transport of singly spliced and unspliced RNA to the cytoplasm, it is likely that the nucleolus plays a critical role in HIV-1 RNA export. As a test for trafficking of HIV-1 RNAs into the nucleolus, a hammerhead ribozyme that specifically cleaves HIV-1 RNA was inserted into the body of the U16 small nucleolar RNA, resulting in accumulation of the ribozyme within the nucleoli of human cells. HeLa CD4+ and T cells expressing this nucleolar localized ribozyme exhibit dramatically suppressed HIV-1 replication. The results presented here suggest a trafficking of HIV-1 RNA through the nucleoli of human cells, thus posing a different paradigm for lentiviral RNA processing.
The expression of HIV type 1 (HIV-1) is controlled by a posttranscriptional mechanism. From a single primary transcript several mRNAs are generated. These RNAs can be divided into three main classes: unspliced 9-kb, singly spliced 4-kb, and the multiply spliced 2-kb RNAs. Each of these RNAs is exported to the cytoplasm for translation and, in the case of the 9-kb RNA, for packaging into virions (1). Normally, pre-mRNAs must undergo a splicing process to remove one or more introns before being exported to the cytoplasm. HIV-1 overcomes this limitation, allowing singly spliced and unspliced RNA to be exported via interaction with its own encoded Rev protein. This regulatory protein binds an RNA stem-loop structure termed the Rev response element located within the env coding region of singly spliced and unspliced HIV RNAs (2–5). Binding of Rev to this element promotes the export, stability, and translation of these HIV-1 RNAs (6–15). The export process is mediated by the nuclear export signal of Rev, which binds the receptor exportin 1/CRM1. It is believed that CRM1 bridges the interaction of Rev with the nucleoporins required for export to the cytoplasm (16).
When Rev and Tat are expressed independently of other HIV transcripts, these proteins localize within the nucleolus of human cells (17–22). The simultaneous presence of a nuclear export signal as well as a nuclear import/localization signal confers upon Rev the ability to shuttle between the nucleus and the cytoplasm (16). It has recently been reported that in HeLa cells, the expression of Rev induces the relocalization of the nucleoporins Nup98 and Nup214, along with a significant fraction of CRM1, into the nucleolus (23). This result has led to the hypothesis that formation of the Rev-CRM1-nucleoporin complex targeted to the nuclear pore complex occurs in the nucleolus. It can be similarly hypothesized that HIV RNAs are also relocalized to the nucleolus before cytoplasmic export. Previous studies, which used in situ hybridization assays to define the subcellular localization of HIV RNAs, failed to detect these RNAs in the nucleoli (24–27). This failure to detect these RNAs is most likely due to the dynamic process of RNA transport, making it difficult to identify discrete nucleolar localization. Therefore we have investigated the same problem, using an alternative strategy based on the use of nucleolar localized ribozymes.
Ribozymes are RNAs with catalytic activity (28). The hammerhead ribozyme is the simplest in terms of size and structure and can readily be engineered to perform intermolecular cleavage on targeted RNA molecules. These properties make this ribozyme a useful tool for inactivating gene expression and a potential therapeutic agent. Moreover, ribozymes can be very effective inhibitors of gene expression when they are colocalized with their target RNAs (29, 30). We have taken advantage of ribozyme-mediated inactivation of targeted RNAs to investigate whether there is nucleolar trafficking of HIV RNA. To determine whether this trafficking takes place, a hammerhead ribozyme targeted to a highly conserved sequence in the U5 region of HIV-1 was delivered into the nucleoli of human cells. The ability of this nucleolar-localized ribozyme to inhibit the replication of HIV-1 was demonstrated in HeLa CD4+ and CEM T lymphocytes, which are markedly resistant to HIV-1 infection as a consequence of ribozyme-mediated cleavage of HIV-1. These results can be best explained by nucleolar trafficking of HIV RNA, making them susceptible targets for ribozyme-mediated inhibition. The inhibition of HIV by a nucleolar localized ribozyme has important implications for the biology and pathogenesis of HIV-1 infection, as well as therapeutic potential.
Materials and Methods
Plasmid Constructs.
The U16Rz was prepared synthetically by PCR (31) using the primers A, B, C, D, E, and F:
A: 5′-CTTGCAATGATGTCGTAATTTGCGTCTTACTCTGTTCTCAGCGACAGTTGAA-3′
B: 5′-TGTGCCCGTTTCGTCCTCACGGACTCATCA(C/G)TGTTGTGTGATTTTCAACTGTCGCTGAG-3′
C: 5′-GGACGAAACGGGCACACAAAACCTGCTGTCAGTAAGCTGGTACAGAAGGTTG-3′
D: 5′-TTTCTTGCTCAGTAAGAATTTTCGTCAACCTTCTGTACCAGCTTACTGAC-3′
E: 5′-CCCCCGAGCTCCTTGCAATGATGTCGTAA-3′
F: 5′-CCCCCCAAGCTTTTTCTTGCTCAGTAAGAA-3′
The B primer contains an equimolar mixture of C and G nucleotides at the position indicated to simultaneously generate by PCR the wild-type (wt) and mutant versions of U16Rz. The PCR product was subcloned in the SacI and HindIII sites of the pGEM 9zf(−) vector (Promega), giving rise to the pGEM/U16Rz wt and mutant constructs. These constructs were linearized with HindIII and transcribed in vitro to test their catalytic activity against an RNA oligonucleotide containing the HIV-1 RNA target (data not shown).
The U16Rz wt and mutant sequences were amplified by PCR from the pGEM/U16Rz wt and mutant constructs, using the SalI 5′ and XbaI 3′ primers:
SalI 5′: 5′- CCCCCCCCGTCGACCTTGCAATGATGTCGTAATTTG-3′
XbaI 3′: 5′- CCCCTCTAGAAAAAATTTCTTGCTCAGTAAGAATTT-3′
The PCR products were ligated in the SalI and XbaI sites of the pTz/U6+1 expression cassette (32) generating the U6+1/U16Rz wt and mutant constructs. The pTz/U6+1 cassette contains the U6 promoter that allows transcription driven by RNA pol III but does not contain the sequence elements necessary for 5′ capping, which might interfere with the nucleolar localization encoded in the C/D motif of U16. Six thymidines were added at the 3′ end of the ribozyme coding sequence to terminate RNA polymerase III transcription.
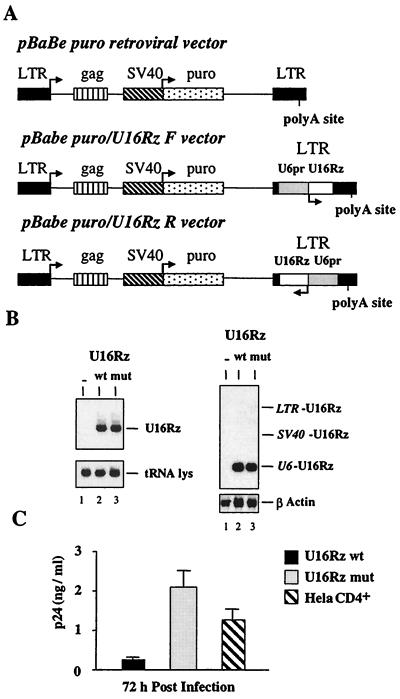
The BamHI–XbaI fragments from U6+1/U16Rz wt and mutant constructs (containing the U6 promoter) were filled in, and the resulting fragment was inserted in both orientations into the NheI site of the pBabe puro retroviral vector [U3 region of the 3′ long terminal repeat (LTR)], giving rise to the following constructs: pBabe puro/U16Rz F (wt and mutant) and pBabe puro/U16Rz R (wt and mutant) (see Fig. 3A).
Figure 3.
Delivery and in vivo activity of the U16Rz in the HeLa CD4+ cell line. (A) Schematic representation of pBabe puro retroviral constructs. The expression cassette U6+1/U16Rz wt and U16Rz mutant were cloned in both orientations in the 3′LTR (U3 region) of the pBabe puro parental vector (48), giving rise to the constructs pBabe puro/U16Rz wt or U16Rz mutant, F (forward) and pBabe puro/U16Rz wt or U16Rz mutant, R (reverse). (B) HeLa CD4+ cells were transfected with the pBabe puro retroviral vector constructs, and pooled populations were selected for puromycin resistance. Five micrograms of total RNAs was extracted from the different transfected HeLa CD4+ pooled populations and electrophoresed in a 6% polyacrylamide/7 M urea gel (Left) or in a 1% agarose/formaldehyde gel (Right), blotted onto nylon filters, and hybridized with specific probes as described in Materials and Methods. Lane 1 contains total RNA extracted from parental HeLa CD4+ cells. Lanes 2 and 3 contain total RNA extracted from the HeLa CD4+ pooled populations transfected with either the pBabe puro/U16Rz wt or pBabe puro/U16Rz mutant, respectively, both in the F orientation. (C) The pooled populations of HeLa CD4+ expressing U16Rz wt, U16Rz mutant, or the untransfected parental HeLa CD4+ cells were infected with HIV-1-IIIB at an moi of 0.001. The HIV-1 p24 antigen accumulation was determined at 72 h after infection. The data presented represent average values of four independent experiments, including the standard deviation.
Cell Culture.
293 (American Type Culture Collection: CRL 1573) and HeLa-CD4-LTR-β-gal cells (obtained from the AIDS Research and Reference Reagent Program no. 1470) were both propagated in Dulbecco's modified Eagle's medium with high glucose (Irvine Scientific) supplemented with 10% FBS (GIBCO/BRL), 200 mM l-glutamine (Irvine Scientific), 10 units/ml penicillin G (GIBCO/BRL), 10 mg/ml streptomycin (GIBCO/BRL), and 0.2 mg/ml G418 (GIBCO/BRL) and 0.1 mg/ml hygromycin (H-Sigma) only for the HeLa-CD4-LTR-β-gal cells. Transfection and infection of the cells were carried out in the absence of any antibiotics. Cells were plated at approximately 1 × 106 cells per 100-mm dish, 1 day before the transfection and then transiently transfected with 2–10 μg of DNA, using the calcium phosphate DNA precipitation method according to the manufacturer's instructions (GIBCO/BRL). Forty-eight hours after transfection, HeLa-CD4-LTR-β-gal cells were selected in 1.5 μg/ml puromycin (Sigma) to obtain uniformly selected pools. CEM cells were maintained in RPMI medium 1640 supplemented with 10% FCS (Irvine Scientific), penicillin (10 units/ml; Irvine Scientific), and streptomycin (100 μg/ml; Irvine Scientific).
Packaging Cell Line.
The Phoenix Packaging cell line (http://www.stanford.edu/nolan/NL phnxr.html) was cultured in Dulbecco's modified Eagle's medium (Irvine Scientific) containing 10% FCS (Irvine Scientific), penicillin (10 units/ml; Irvine Scientific), and streptomycin (100 μg/ml; Irvine Scientific). Retrovirus particles were collected from Phoenix cells transiently transfected with 6 μg of the various pBabe constructs and were used to transduce CEM cells as previously described (33). For puromycin-resistance selection, 1.5 μg/ml puromycin was added to the medium, and cells were incubated in the presence of this drug for 3 weeks to obtain pooled, drug-resistant populations of cells. Single stable clones were obtained from the pools by limiting dilution.
HIV Infectious Assays.
Twenty-four hours before infection of the cells with HIV-I-IIIB, 1 × 105 HeLa-CD4-LTR-β-gal cells were plated in 3 ml of medium per well in a six-well plate. Cells were infected in triplicate with 5 μl of HIV-1-IIIB overnight in the presence of 4 μg/ml protamine sulfate (Elkins-Sinn, Cherry Hill, NJ). The HIV-1-IIIB viral stock had been propagated on peripheral blood lymphocytes and was determined to contain 1 × 104 TCID50/ml. After overnight incubation the cells were washed three times with Hanks' balanced salts solution and refed with medium. Aliquots of medium for HIV-1 p24 antigen analysis were collected when cells reached confluency, 72 h after infection. p24 values were determined with the HIV-1 p24 antigen capture assay (Science Applications International Corp., Frederick, MD) according to the manufacturer's instructions. CEM cells (2.5 × 105) derived from stable clones were infected with HIV-1NL4–3 at a multiplicity of infection (moi) of 0.0002 (low moi experiment, Fig. 4B) and 0.002 (high moi experiment, Fig. 5). Infections were performed in duplicate. After infection, the cells were resuspended in 12 ml of complete medium, and p24 accumulation in the supernatant was monitored over time. The cells were split and refed twice a week. The p24 analyses were performed using the HIV-1 p24 antigen capture assay kit (Science Applications International Corp. Frederick). For the high moi experiment the p24 analyses were performed on days 7 and 11 (data not shown), after which the pathogenic effects of the virus on the control cells made p24 determinations irrelevant.
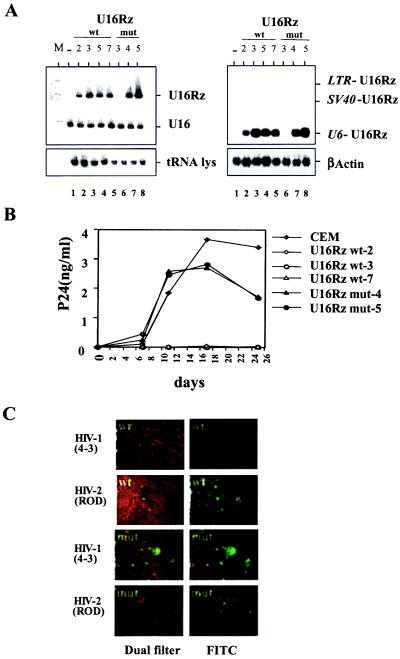
Figure 4.
Delivery and intracellular activity of the U16Rz in the human T lymphoblastoid CEM cell line. (A) Human T lymphoblastoid CEM cells were transduced with the different pBabe puro retroviral constructs (Fig. 3A), and pooled populations were selected for puromycin resistance. Single stable clones were selected by limiting dilution from the pooled clones. Total RNAs (5 μg) from the single stable clones were electrophoresed in a denaturing polyacrylamide (Left) or on an agarose-formaldehyde gel (Right), blotted onto nylon filters, and hybridized with the specific probes as described in Materials and Methods. Lane M contains the labeled HpaII-digested Bluescript KS (+) DNA used as a size marker. Lane 1 contains total RNA extracted from untransduced CEM cells. Lanes 2–5 contain total RNA extracted from single CEM stable clones 2, 3, 5, and 7, which were cloned from the CEM cell pool transduced with the pBabe puro/U16Rz wt, in the F orientation. Lanes 6–8 contain total RNA extracted from single CEM stable clones 3, 4, and 5 cloned from the CEM cell pool transduced with the pBabe puro/U16Rz mutant, in the F orientation. A low level of expression from the U16Rz mutant clone 3 can be observed after prolonged exposure of the hybridized blot. (B) Three single, stably transduced CEM clones (2, 3, and 7) expressing the wt U16Rz, and clones 4 and 5 expressing the U16Rz mutant along with the parental CEM cells were infected with HIV-1NL4–3 at an moi of 0.0002. The HIV-1-encoded p24 accumulation was determined at days 7, 11, 17, and 25 after infection. (C) HIV indirect immunofluorescence assays performed, as described previously (34), using heat-inactivated HIV-1 seropositive human serum, on CEM clones expressing the U16Rz wt (clone 3) (upper four panels) and U16Rz mutant (clone 4) (lower four panels) infected with HIV-1NL4–3 or HIV-2ROD, at days 17 and 7 after infection, respectively. The infected cells are FITC-stained and fluoresce green. The cells were counterstained in 1% trypan blue dye in PBS. The images on the Left were obtained with a dual filter (FITC/rhodamine). The uninfected cells appear red under the rhodamine filter, and infected cells exhibit a green fluorescence. The images on the Right were acquired with just the FITC filter, revealing only the green fluorescence of the infected cells.
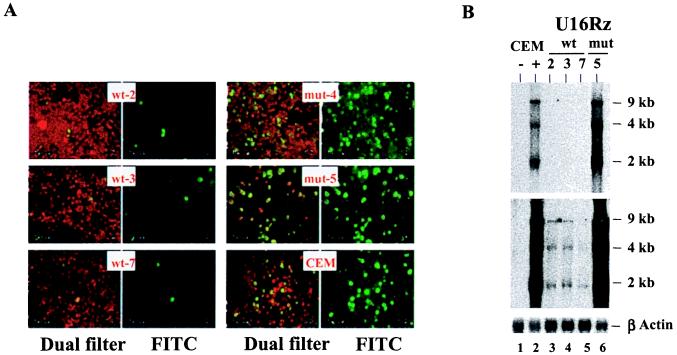
Figure 5.
CEM stable clones expressing the U16Rz wt are resistant to an elevated moi of HIV-1. (A) HIV indirect immunofluorescence assays were performed as described in Fig. 4C. CEM clones 2, 3, and 7 expressing the U16Rz wt, clones 4 and 5 expressing the U16Rz mutant, and parental CEM cells were infected with HIV-1NL4–3 at an moi of 0.002. Immunofluorescence monitoring of infection was performed at day 11 after infection. The infected cells are FITC-stained (Right). The dual filter (FITC/rhodamine, Left) shows uninfected cells (red) and infected cells (green). (B) Northern blot analysis was performed on 5-μg RNA samples electrophoresed in a 1% agarose/formaldehyde gel. Total RNAs were extracted from the above HIV-1-infected CEM stable clones. Hybridization was carried out with a Rev cDNA sequence. The signals obtained from the endogenous β-actin (Bottom) were used as loading controls. After overnight exposure the HIV-1 RNA was detected (Top) only in the parental CEM cells (lane 2) and in the CEM clone 5 expressing the U16Rz mutant (lane 6). RNA prepared from uninfected CEM cells (lane 1) was used as a negative control. HIV-1 RNAs from the U16Rz wt-expressing clones 2, 3, and 7 (lanes 3, 4, and 5, respectively) was detectable only after 3 days of prolonged exposure of the hybridized filter, as shown in Middle.
RNA Preparation and Northern Blot Analyses.
Total RNA was prepared using the RNA-STAT 60 reagent (Tel-Test “B”; Tel-Test, Friendswood, TX) according to the manufacturer's protocol. The RNA was electrophoresed in a 1% agarose/formaldehyde gel or a 6% polyacrylamide/7 M urea gel and blotted onto a nylon filter. To simultaneously detect the U16Rz RNA and the endogenous U16 small nucleolar RNA (snoRNA), we used a radiolabeled probe complementary to the 3′ end of U16 (Fig. 4A Left). To detect only the U16Rz RNA we used a probe complementary to the hammerhead ribozyme sequence. To detect the loading controls, we used probes specific for β-actin mRNA and tRNA3Lys.
In Situ Hybridization.
We performed in situ hybridizations as previously described (http://singerlab.aecom.yu.edu/protocols). For probes we used the following aminoallyl-T-modified primers:
U3: 5′-GT*TCTCTCCCTCT*CACTCCCCAAT*ACGGAGAGAAGAACGAT*CATCAATGGCT*G-3′
U16Rz: 5′-T*TTTGTGTGCCCGT*TTCGTCCTCACGGACT*CATCAGTGTTGT*GTGATTTTCAACT*G-3′
The T* indicates the aminoallyl-T nucleotides. The specific primer for U3 was chemically conjugated with the Cy3 fluorophore (CyTM3 monofunctional reactive dye; Amersham Pharmacia). The U16Rz probe was chemically conjugated with Oregon green 488 (Molecular Probes). Digital image processing was used to analyze the localization of U16Rz and U3 snoRNA within the 293 cells. Images were collected with an Olympus BX50 microscope (Fig. 2A) and a DEI-50 video camera (Optronics). A 60× objective and FITC and Cy3 filters were used to detect the U16Rz and U3 snoRNA signals, respectively. A dual filter for FITC + Cy3 was also used to show the yellow overlapping signals from the two RNAs. A 4′, 6-diamidino-2-phenylindole filter was used to identify the nucleus (blue signal). The image in Fig. 2B was collected with a Zeiss LSM310 laser scanning microscope at 6,400-fold magnification. HIV indirect immunofluorescence assays were performed as described previously (34) on CEM-derived clones infected with HIV-1NL4–3 or HIV-2ROD. The images were collected with an Olympus BX50 microscope and a DEI-50 video camera (Optronics) with a 40× objective (Figs. 4C and 5A).
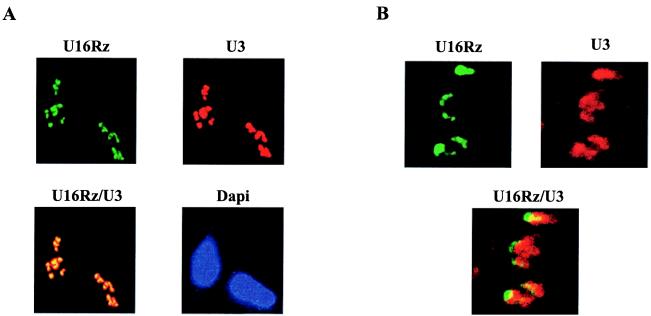
Figure 2.
In vivo intracellular localization of U16Rz. (A) U16Rz localizes in the nucleolus. 293 cells were grown on coverslips and transiently transfected with 2 μg of the U6+1/U16Rz wt and U16Rz mutant (data not shown) constructs. After 48 h the cells were fixed in 4% paraformaldehyde and the in situ hybridization was performed. (Upper Left) Hybridization with the Oregon green 488 probe specific for the U16Rz. (Upper right) Hybridization with the Cy3 red probe specific for the U3 snoRNA, used to detect the nucleoli. (Lower Left) The overlap between the red signal of U3 and the green signal of U16Rz appears as a yellow signal. (Lower Right) 4′,6-Diamidino-2-phenylindole staining to delineate the nuclei. (B) High-resolution confocal microscope analysis of the U3 snoRNA and U16Rz distribution in the nucleoli. This analysis shows partial overlap between the U3 (red signal) and the U16Rz (green signal) in the nucleoli.
Results
Ribozyme Construction.
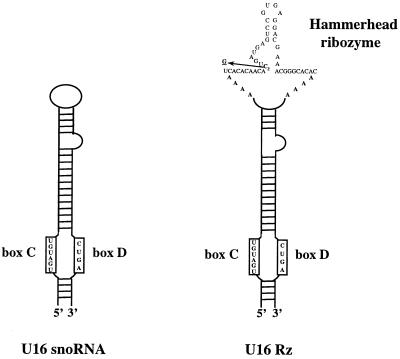
We chose the well-studied U16 snoRNA (35) as the RNA vector for nucleolar localization of a hammerhead ribozyme targeted against a conserved sequence in the 5′LTR of HIV-1 RNA (36). U16 is a member of the C/D box snoRNA class that primarily guides 2′-O-methylation of the ribose moiety of specific pre-rRNA nucleotides, although some members of this class are involved in pre-rRNA processing (37). Moreover, Buonomo et al. (38) have demonstrated that a chimeric U16 RNA harboring an HIV-1 Rev binding element localizes to the nucleolus after transcription from the human U6 small nuclear RNA promoter.
The apical loop of U16 snoRNA was used to insert the anti-HIV-1 hammerhead ribozyme (U16Rz, Fig. 1). A functionally disabled ribozyme was constructed by mutating the C3 nucleotide of the hammerhead catalytic core to a G (Fig. 1) (46) and was used as a negative control in these studies. The chimeric U16Rz was shown to site-specifically cleave an HIV-1 transcript in vitro, whereas the disabled ribozyme had no cleavage activity (data not presented).
Figure 1.
Schematic representation of U16 snoRNA and U16-hammerhead ribozyme (U16Rz). U16 snoRNA is folded in a stem-and-loop structure; the C and D boxes are highlighted. The C and D boxes are the nucleolar signal (39, 40) and are important for the stability and processing of U16 (41). The hammerhead ribozyme was designed to cleave at the highly conserved position +115 relative to the transcription initiation site of the HIV-1 (36, 42). The sequence of the hammerhead active core was derived from the studies of Uhlenbeck and Haseloff (43–45). A disabled version of the hammerhead ribozyme was obtained by single point mutation of the C3 nucleotide of the catalytic core to a G (indicated by an arrow) (46). Four adenosines were added to each side of the hammerhead ribozyme to enlarge the loop of U16 and facilitate interaction of the ribozyme with the HIV-1 target site.
In Vivo Expression and Intracellular Localization of the U16Rz.
The chimeric U16/ribozyme RNAs were expressed by the human U6 small nuclear RNA promoter (clone U6+1) (32) after transient transfection into human 293 cells, and in situ hybridization was carried out, using fluorescent probes to detect the U16Rz. A separate fluorescent probe was used to identify endogenous U3 snoRNA, which served as a nucleolar control. The pattern obtained demonstrated that the ribozyme and the U3 snoRNAs colocalize within the nucleoli (Fig. 2). Laser scanning confocal microscopy reveals partial overlap between the U3 snoRNA and U16Rz fluorescent signals within the nucleoli (Fig. 2B). Some of the observed localization of U3 and the U16Rz may be in coiled bodies as well as the nucleoli, inasmuch as the studies of Narayanan et al. (47) suggest that coiled bodies can play a role in the biogenesis and/or intranuclear transport of the C/D box snoRNAs.
U16Rz in Vivo Activity.
To test the effect of nucleolar localization of an anti-HIV-1 hammerhead ribozyme on HIV-1 infectivity, the chimeric U16Rz wt and the U16Rz mutant expression cassettes were inserted within the 3′LTR (U3 region) of the pBabe puro retroviral vector (48) in both orientations [forward (F) and reverse (R) with respect to the direction of transcription from the 5′LTR; Fig. 3A]. As a rapid preliminary test for efficacy of the U16Rz, HeLa CD4+ cells were transfected with the different retroviral constructs, and pooled populations were selected for puromycin resistance. RNAs were isolated from these cells and analyzed for ribozyme expression (Fig. 3B). Cells transfected with the pBabe/U16Rz clone in the F orientation expressed the highest levels of ribozymes, and therefore we used only the F orientation for subsequent analyses. Northern analyses demonstrate that the U16Rz is transcribed almost exclusively from the U6 small nuclear RNA promoter (Fig. 3B Right).
We assayed by HIV-1 infectious challenge the ribozyme-inhibitory effect of the pooled population of HeLa CD4+ cells expressing the U16Rz wt and U16Rz mutant. The results of HIV-1 p24 analyses demonstrate that the nucleolar U16Rz wt confers protection from infection, whereas both the pooled populations expressing the U16Rz mutant and the parental HeLa CD4+ cells are readily infectable by HIV-1 (Fig. 3C).
To confirm the inhibitory effect of the U16Rz in a T cell model, the CEM T lymphoblastoid cell line was transduced with the same retroviral constructs (Fig. 3A). Single stable clones were selected from the pooled population of puromycin-resistant cells, and the steady-state accumulation of ribozyme transcripts was analyzed by Northern blotting and hybridization (Fig. 4A). In situ hybridization analysis performed on the stably transduced CEM cells confirmed the colocalization between U3 snoRNA and U16Rz (data not shown). Three different clones expressing the U16Rz wt (2, 3, and 7) and two different clones expressing the U16Rz mutant (4 and 5) along with the untransduced parental CEM cells were used in HIV-1 challenge assays (Fig. 4A). The results from these viral challenges showed no measurable HIV-1 p24 in the supernatants of the U16Rz wt clones during the 25-day period of analysis, whereas the untransduced CEM cells and U16Rz mutant clones gave rise to nanogram levels of p24 antigen (Fig. 4B). The results of the p24 analysis were confirmed by an HIV indirect immunofluorescence assay using serum from an HIV-1-infected patient (Fig. 4C). No HIV antigen staining was detected in the CEM stable clones expressing the U16Rz wt, whereas intense staining was detected in the U16Rz mutant clones, indicating viral production and spread in these cell cultures. The levels of CD4 expression for all of the cell lines used in the challenge assays were monitored, and no differences were observed (data not presented).
To verify that the ribozyme-expressing CEM cells were resistant to HIV-1 as a consequence of ribozyme function, we challenged the U16Rz wt, U16Rz mutant, and untransduced CEM cells with HIV-2, which is CD4-tropic but does not harbor the ribozyme target site. All of the cell lines tested were readily infected by HIV-2, as shown by HIV-2 reverse transcriptase analyses (data not shown) and HIV indirect immunofluorescence analyses (Fig. 4C). We next evaluated the extent of ribozyme-mediated inhibition of HIV-1 in the stably transduced CEM cell lines by using a 10-fold higher moi. Despite the higher moi used in this challenge, CEM clones expressing the U16Rz wt still showed a dramatic resistance to HIV-1 infection as monitored by HIV immunostaining and HIV-1 RNA levels (Fig. 5). Measurements of p24 antigen demonstrated that some U16Rz wt-expressing cells had in fact been infected at this higher moi (data not shown), but the immunofluorescence staining revealed that the extent of viral spread was dramatically reduced in comparison to the U16Rz mutant and CEM cell controls (Fig. 5A).
Discussion
We have taken advantage of the catalytic activity and nucleolar localization properties of a C/D box motif snoRNA fused to an anti-HIV-1 hammerhead ribozyme to address the question of whether HIV-1 RNAs pass through the nucleolus. The U16Rz constructs accumulate within the nucleoli of human cells, and very few if any of these constructs are found outside of the nucleolus (Fig. 2A).
Both a stably transfected, pooled population of HeLa CD4+ cells and stably transduced lymphoblastoid CEM cells expressing the U16Rz wt are highly resistant to the spread of HIV-1 infection (Figs. 3C and 4B). In contrast, cells expressing the U16Rz mutant are readily infected by HIV-1 (Figs. 3C and 4B). Both U16Rz wt and U16Rz mutant CEM cells are infectable by HIV-2, which does not harbor the cleavage site for the ribozyme (Fig. 4C). The extent of inhibition of HIV-1 replication mediated by the nucleolar localized U16Rz is dramatic and suggests that one or more classes of HIV-1 RNAs pass through the nucleoli before nuclear export.
Several RNAs have been reported to pass through the nucleolus for processing, particle assembly, or other modification (49). These include c-myc, N-myc, and myoD1 mRNAs (50); the signal recognition particle RNA (51, 52); U6 small nuclear RNA (53); some pre-tRNAs in yeast (54); and the RNase P RNA (55). There is also evidence that telomerase RNA is processed within the nucleolus (56, 57). Transcription and replication of the Borna disease virus have also been shown to occur within the nucleolus (58). Importantly, the HTLV-I env RNAs have been demonstrated to be partially localized in the nucleolus (59). HTLV-I and HIV-1 have a similar posttranscriptional regulation mechanism, and the Rex protein, a functional homolog of HIV-1 Rev, also has nucleolar localization properties. To date, published data concerning nucleolar localization of HIV-1 RNAs are inconclusive. Using electron microscopy and in situ hybridization, Romanov et al. (60) detected a subgenomic mRNA expressing the HIV-1 p37gag (containing the Rev response element) in all of the subcellular compartments (including the nucleoli) of HL Tat cells. Interestingly, they observed that expression of Rev induced relocalization of HIV RNAs into two nonrandom patterns. One of these, the long track in the nucleoplasm, was radially organized around and in contact with the nucleoli. Other investigators using in situ hybridization analyses performed on mammalian cell lines transfected with different HIV-1 subgenomic or genomic constructs failed to detect HIV-1 RNA in the nucleolus (24–27). The discrepancy in these results might be due to the different HIV-1 constructs, cell lines, and in situ hybridization protocols used by the various investigators. Furthermore, it should be taken into consideration that RNA export is a dynamic process; the rate of export as well as the amount of the HIV-1 RNA passing through the nucleolus can be limiting factors for in situ hybridization-mediated detection of nucleolar localized transcripts.
The use of an anti-HIV-1 nucleolar-localized ribozyme represents a valid approach to the problem of HIV-1 nucleolar trafficking.
Colocalization of ribozyme and target RNAs has been shown to markedly enhance ribozyme efficacy (29, 30). Studies by Samarsky et al. (30) have provided a definitive demonstration that nucleolar colocalization of a snoRNA-appended ribozyme and snoRNA target resulted in a nearly complete cleavage of the target.
The marked inhibition of HIV-1 replication in cell lines expressing a nucleolar-localized U16Rz are best explained by nucleolar passage and colocalization of HIV-1 RNA with the ribozyme construct. We cannot exclude the possibility that a small fraction of non-nucleolar-localized U16Rz (undetectable by in situ hybridization; Fig. 2A) might be responsible for the observed functional inhibition of HIV-1. The small quantities of nonnucleolar U16Rz would have to be highly colocalized with the HIV RNA targets at other sites in the nucleus to provide the levels of inhibition observed in our studies, which is an unlikely possibility. The C/D box snoRNAs are transiently associated with the nuclear coiled bodies (47) before they accumulate in the nucleoli, whereas HIV-1 RNAs have never been observed to be associated with the coiled bodies (24–27), making it unlikely that the ribozyme-mediated cleavage events are occurring in these bodies. We therefore believe that the major inhibitory activity of U16Rz against HIV-1 RNAs is taking place within the nucleoli.
It is premature to assign a functional role for HIV-1 nucleolar trafficking or to conclude that all or a subset of HIV-1 transcripts participate in nucleolar trafficking. Importantly, the potent ribozyme-mediated inhibition of HIV-1 replication in our studies clearly demonstrates that a critical step in viral maturation is blocked by this approach.
Since the nucleolar localization properties of Rev and Tat have been demonstrated (17–22) it has been tempting to speculate that the assembly of a ribonucleoprotein particle containing the HIV-1 RNA and the two HIV-1 regulatory proteins along with other cellular factors takes place in the nucleolus. This ribonucleoprotein particle could be involved in the export and subsequent translation or packaging of the HIV-1 RNA in the cytoplasm. Alternatively, posttranscriptional 2′-O-methylation and/or pseudouridylation mediated by small nucleolar RNAs (37) could mark the HIV RNAs passing through the nucleoli before cytoplasmic export for a specific process, such as translation versus packaging into virions. If such modification of HIV-1 RNAs does occur, the virus could usurp one of a multitude of snoRNAs involved in modifying cellular RNAs. The results presented here warrant further investigation into the functional role(s) of HIV-1 RNA nucleolar trafficking.
Acknowledgments
We thank Dr. R. Barber for the confocal microscopy, Drs. J. Zaia and S. Li for assisting with the HIV-2 reverse transcriptase testing, Drs. Irene Bozzoni and Daniela Castanotto for critical reading of the manuscript, and W. Fitzgerald for technical assistance. I.B. is a recipient of National Institutes of Health Fellowship 5 F32 GM18898-02. This research was supported by National Institutes of Health Grants AI29329 and AI42552 to J.J.R.
Abbreviations
- wt
wild type
- LTR
long terminal repeat
- snoRNA
small nucleolar RNA
- moi
multiplicity of infection
References
- 1.Kingsman S M, Kingsman A J. Eur J Biochem. 1996;240:491–507. doi: 10.1111/j.1432-1033.1996.0491h.x. [DOI] [PubMed] [Google Scholar]
- 2.Zapp M L, Green M. Nature (London) 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- 3.Cochrane A W, Chen C-H, Rosen C. Proc Natl Acad Sci USA. 1990;87:1198–1202. doi: 10.1073/pnas.87.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Agostino D, Ciminale V, Pavlakis G, Chiecho-Bianchi L. AIDS Res Hum Retroviruses. 1995;11:1063–1071. doi: 10.1089/aid.1995.11.1063. [DOI] [PubMed] [Google Scholar]
- 5.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 6.Arrigo S J, Chen I S Y. Genes Dev. 1991;5:808–819. doi: 10.1101/gad.5.5.808. [DOI] [PubMed] [Google Scholar]
- 7.D'Agostino D M, Felber B K, Harrison J E, Pavlakis G N. Mol Cell Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emerman M, Vazeux R, Peden K. Cell. 1989;57:1155–1165. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg M B, Jarrett R F, Aldovini A, Gallo R C, Wong-Staal F. Cell. 1986;46:807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- 10.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammarskjold M L, Heimer J, Hammarskjold B, Sangwan I, Albert L, Rekosh D. J Virol. 1989;63:1959–1966. doi: 10.1128/jvi.63.5.1959-1966.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence J B, Cochrane A W, Johnson C V, Perkins A, Rosen C A. New Biol. 1991;3:1220–1232. [PubMed] [Google Scholar]
- 13.Schwartz S, Felber B K, Pavlakis G N. J Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malim M H, Hauber J, Lee S-Y, Maizel J V, Cullen B R. Nature (London) 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 15.Favaro J P, Maldarelli F, Arrigo S J, Schmidt M G. Virology. 1999;255:237–249. doi: 10.1006/viro.1998.9584. [DOI] [PubMed] [Google Scholar]
- 16.Hope T J. Arch Biochem Biophys. 1999;365:186–191. doi: 10.1006/abbi.1999.1207. [DOI] [PubMed] [Google Scholar]
- 17.Cullen B R, Hauber J, Campbell K, Sodrosky J P, Haseltine W A, Rosen C A. J Virol. 1988;62:2498–2501. doi: 10.1128/jvi.62.7.2498-2501.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luznik L, Martone M E, Kraus G, Zhang Y, Xu Y, Ellisman M H, Wong-Staal F. AIDS Res Hum Retroviruses. 1995;7:795–804. doi: 10.1089/aid.1995.11.795. [DOI] [PubMed] [Google Scholar]
- 19.Dundr M, Leno G H, Hammarskjold M-L, Rekosh D, Helga-Maria C, Olson M M O J. J Cell Sci. 1995;108:2811–2823. doi: 10.1242/jcs.108.8.2811. [DOI] [PubMed] [Google Scholar]
- 20.Endo S, Kubota S, Siomi H, Adachi A, Oroszlan S, Maki M, Hatanaka M. Virus Genes. 1989;3:99–110. doi: 10.1007/BF00125123. [DOI] [PubMed] [Google Scholar]
- 21.Siomi H, Shida H, Maki M, Hatanaka M. J Virol. 1990;64:1803–1807. doi: 10.1128/jvi.64.4.1803-1807.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stauber R M, Pavlakis G N. Virology. 1998;252:126–136. doi: 10.1006/viro.1998.9400. [DOI] [PubMed] [Google Scholar]
- 23.Zolotukhin A S, Felber B K. J Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang G, Zapp M L, Yan G, Green M R. J Cell Biol. 1996;135:9–17. doi: 10.1083/jcb.135.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boe S-O, Bjorndal B, Rosok B, Szilvay A M, Kalland K-H. Virology. 1998;244:473–482. doi: 10.1006/viro.1998.9110. [DOI] [PubMed] [Google Scholar]
- 26.Favaro J P, Borg K T, Arrigo S J, Schmidt M G. Virology. 1998;249:286–296. doi: 10.1006/viro.1998.9312. [DOI] [PubMed] [Google Scholar]
- 27.Favaro J P, Maldarelli F, Arrigo S J, Schmidt M G. Virology. 1999;255:237–249. doi: 10.1006/viro.1998.9584. [DOI] [PubMed] [Google Scholar]
- 28.Rossi J J. Chem Biol. 1999;6:R33–R37. doi: 10.1016/S1074-5521(99)80001-5. [DOI] [PubMed] [Google Scholar]
- 29.Sullenger B A, Cech T R. Science. 1993;262:1566–1569. doi: 10.1126/science.8248806. [DOI] [PubMed] [Google Scholar]
- 30.Samarsky D A, Ferbeyre G, Bertrand E, Singer R H, Cedergren R, Fournier M J. Proc Natl Acad Sci USA. 1999;96:6609–6614. doi: 10.1073/pnas.96.12.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dillon P J, Rosen C A. BioTechniques. 1990;9:298–300. [PubMed] [Google Scholar]
- 32.Good P D, Krikos A J, Li S X L, Bertrand E, Lee N S, Giver L, Ellington A, Zaia J A, Rossi J J, Engelke D R. Gene Ther. 1997;4:45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
- 33.Scherr, M., Reed, M., Huang, C.-F., Riggs, A. D. & Rossi, J. J. (2000) Mol. Ther., in press. [DOI] [PubMed]
- 34.Sandstrom E G, Schooley R T, Ho D D, Byington R, Sarngadharan M G, MacLane M E, Essex M, Gallo R C, Hirsch M S. Transfusion. 1985;25:308–312. doi: 10.1046/j.1537-2995.1985.25485273806.x. [DOI] [PubMed] [Google Scholar]
- 35.Fragapane P, Prislei S, Michienzi A, Caffarelli E, Bozzoni I. EMBO J. 1993;12:2921–2928. doi: 10.1002/j.1460-2075.1993.tb05954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ojwang J O, Hampel A, Looney D J, Wong-Staal F. Proc Natl Acad Sci USA. 1992;89:10802–10806. doi: 10.1073/pnas.89.22.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinstein L B, Steitz J A. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 38.Buonomo S B C, Michienzi A, De Angelis F G, Bozzoni I. RNA. 1999;5:993–1002. doi: 10.1017/s1355838299990064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lange T S, Borovjagin A, Maxwell E S, Gerbi S A. EMBO J. 1998;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samarsky D A, Fournier M J, Singer R H, Bertrand E. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caffarelli E, Losito M, Giorgi C, Fatica A, Bozzoni I. Mol Cell Biol. 1998;18:1023–1028. doi: 10.1128/mcb.18.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K. Nature (London) 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 43.Uhlenbeck O C. Nature (London) 1987;328:596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- 44.Haseloff J, Gerlach W L. Nature (London) 1988;334:585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- 45.Ruffner D E, Stormo G D, Uhlenbeck O C. Biochemistry. 1990;29:10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- 46.Hertel K J, Pardi A, Uhlenbeck O C, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach W L, Hodgson R, Symons R H. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Narayanan A, Speckmann W, Terns R, Terns M P. Mol Biol Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgestern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pederson T. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bond V C, Wold B. Mol Cell Biol. 1993;13:3221–3230. doi: 10.1128/mcb.13.6.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobson M R, Pederson T. Proc Natl Acad Sci USA. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Politz J C, Yarovoi S, Kilroy S M, Gowda K, Zwieb C, Pederson T. Proc Natl Acad Sci USA. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tycowski K T, You Z H, Graham P J, Steitz J A. Mol Cell. 1998;2:629–638. doi: 10.1016/s1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 54.Bertrand E, Houser-Scott F, Kendall A, Singer R H, Engelke D R. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacobson M R, Cao L G, Taneja K, Singer R H, Wang Y L, Pederson T. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- 56.Mitchell J R, Cheng J, Collins K. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Narayanan A, Lukowiak A, Jady B E, Dragon F, Kiss T, Terns R M, Terns M P. EMBO J. 1999;18:5120–5130. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pyper J M, Clements J E, Zink M C. J Virol. 1998;72:7697–7702. doi: 10.1128/jvi.72.9.7697-7702.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalland K-H, Langhoff E, Bos H J, Gottlinger H, Haseltine W A. New Biol. 1991;3:389–397. [PubMed] [Google Scholar]
- 60.Romanov V I, Zolotukhin A S, Aleksandroff N A, Da Silva P P, Felber B K. Virology. 1997;228:360–370. doi: 10.1006/viro.1996.8398. [DOI] [PubMed] [Google Scholar]