Abstract
Each of the trypanosome small nuclear ribonucleoproteins (snRNPs) U2, U4/U6, and U5, as well as the spliced leader (SL) RNP, contains a core of common proteins, which we have previously identified. This core is unusual because it is not recognized by anti-Sm Abs and it associates with an Sm-related sequence in the trypanosome small nuclear RNAs (snRNAs). Using peptide sequences derived from affinity-purified U2 snRNP proteins, we have cloned cDNAs for five common proteins of 8.5, 10, 12.5, 14, and 15 kDa of Trypanosoma brucei and identified them as Sm proteins SmF (8.5 kDa), -E (10 kDa), -D1 (12.5 kDa), -G (14 kDa), and -D2 (15 kDa), respectively. Furthermore, we found the trypanosome SmB (T. brucei) and SmD3 (Trypanosoma cruzi) homologues through database searches, thus completing a set of seven canonical Sm proteins. Sequence comparisons of the trypanosome proteins revealed several deviations in highly conserved positions from the Sm consensus motif. We have identified a network of specific heterodimeric and -trimeric Sm protein interactions in vitro. These results are summarized in a model of the trypanosome Sm core, which argues for a strong conservation of the Sm particle structure. The conservation extends also to the functional level, because at least one trypanosome Sm protein, SmG, was able to specifically complement a corresponding mutation in yeast.
Trans splicing in trypanosomes is an essential step in the expression of all mRNAs and results in joining of a short, noncoding miniexon sequence [spliced leader (SL)] to each of the protein-coding sequences that are part of long polycistronic precursors (reviewed in ref. 1). As in the cis-spliceosome, small nuclear RNAs (snRNAs) U2, U4, and U6, in addition to the SL RNA, are essential cofactors for trans splicing (2). In addition, the trypanosomatid U5 snRNA has been identified (3–5). Surprisingly, Schnare and Gray (6) recently discovered also a U1-like small RNA in the trypanosomatid species Crithidia fasciculata and Leishmania tarentolae, which may be required for cis splicing of internal introns such as the one of the poly(A) polymerase gene (7).
We had previously established affinity purification procedures that allowed the identification of protein components in the trans-spliceosomal small nuclear ribonucleoproteins (snRNPs) from Trypanosoma brucei. A set of at least five polypeptides of 8.5, 10, 12.5, 14, and 15 kDa, which we have called common proteins, was detected originally in the SL RNP, the U2 snRNP, and the U4/U6 snRNP (8). Common proteins were localized by immunofluorescence predominantly in the nucleoplasm of trypanosomes (9). They make up a stable core shared between these snRNPs and bind to an snRNA region resembling the Sm sequence of cis-spliceosomal snRNPs (10). Using polyclonal Abs that we generated against a mixture of four of these proteins (8.5, 10, 12.5, and 14 kDa), we showed later that these core proteins are present in the U5 snRNP (4) and the SLA (spliced leader-associated) RNP (11). For the trypanosomal U4 and U5 snRNAs, we have recently demonstrated by mutational analysis that the Sm-like sequence is essential for core snRNP assembly in vivo and that it is important for nuclear localization of these snRNPs (12, 13).
One of the common proteins has been cloned from the trypanosomatid species Leptomonas collosoma and identified as a member of the Sm protein family, specifically as an SmE homologue, suggesting that core complexes from trypanosomes share some similarities with those from other organisms (14). In addition to the common proteins, trypanosome snRNPs contain specific components. So far, only two of them are cloned: first, a 40-kDa U2 snRNP-specific protein of T. brucei, which only in its N-terminal half is homologous to the mammalian U2 A′ protein (15), and second, a 277-kDa U5-specific protein with extensive homology to the cis-spliceosomal PRP8/p220 factors (4).
The assembly of the seven canonical Sm polypeptides (SmB/B′, -D1, -D2, -D3, -E, -F, and -G) into core snRNPs and the interactions involved have been investigated in particular in the mammalian and yeast systems (16–22). A heptameric, ring-like structure has been proposed for the mammalian Sm core protein complex, based on crystal structures for the B-D3 and D1-D2 heterodimers (23) and consistent with earlier electron-microscopic images (24, 25). For the trypanosome system important questions remained open: How do the mammalian and the trypanosome core complex compare with each other, in particular because no immunological crossreactivity could be established?
Here we report on the protein sequences of seven common proteins from T. brucei and Trypanosoma cruzi, based on cDNA cloning and database searches. We establish that each of these proteins carries a bipartite Sm motif, including interesting deviations from the Sm consensus and from cis-spliceosomal counterparts. A thorough analysis of protein–protein interactions in vitro and heterologous studies on the Sm protein function in vivo revealed that most of the interactions previously identified in the mammalian and yeast Sm cores are conserved in the trypanosome counterpart. In sum, structural and functional conservation of the Sm complex appears to be surprisingly high, although the Sm binding site on the snRNAs is rather degenerate in comparison with other eukaryotes.
Materials and Methods
Cell Culture and Extract Preparation.
The procyclic form of T. brucei brucei strain 427 was grown at 28°C in SDM-79 medium as described (26). The preparation of total cell extract and DEAE chromatography were carried out according to ref. 27.
Antisense Affinity Selection of T. brucei U2 snRNPs and Protein Microsequencing.
For affinity selection of U2 snRNPs on a preparative scale, 100 μg of a biotinylated antisense 2′-O-methyl RNA oligonucleotide, complementary to nucleotides 1–15 of T. brucei U2 snRNA (8), and 20 ml DEAE fraction were used.
For microsequence analysis, proteins from a preparative 15% SDS/polyacrylamide gel were transferred to nitrocellulose by the method of Towbin et al. (28). After Ponceau S staining, strips of nitrocellulose containing individual U2 snRNP proteins (8.5 kDa, 10 kDa, 12.5 kDa, and 14 kDa) or a mixture of proteins in the 15-kDa region (15K-mix) were cut out from the blot (for the peptide sequences, see Fig. 7, which is published as supplemental material on the PNAS web site, www.pnas.org).
cDNA and Genomic Cloning of the T. brucei/T.cruzi Genes for Sm Proteins.
Total poly(A+) mRNA was isolated from procyclic T. brucei cells. cDNA was prepared by reverse transcription (RT) and with an N(dT)18 primer (N, G/A/C); for PCR amplification, SL1–25 (specific for nucleotides 1–25 of the T. brucei SL RNA) and a degenerate oligonucleotide (see supplemental material) were used. PCR products were cloned and sequenced, partial sequences were completed by 3′ rapid amplification of cDNA ends (RACE) procedures, and the assembled cDNA sequences were confirmed by genomic cloning.
The T. brucei SmB sequence was cloned by genomic PCR, based on sequence information obtained by database searches (no. AQ652994). As a result, a 490-nt genomic DNA fragment containing the ORF and flanking sequences was cloned and sequenced.
For the T. cruzi gene for SmD3 a partial genomic sequence (AQ444257) was initially identified by database searches (T. cruzi Genome Project, Sanger Center, Cambridge, U.K.), using the human SmD3 protein sequence. The missing N-terminal sequence was added by RT-PCR reactions from T. cruzi total RNA, using SL- and gene-specific primers, followed by cloning and sequencing of the product. The cDNA sequence was confirmed by PCR amplifying, cloning, and sequencing a genomic fragment carrying the ORF and flanking sequences.
T. brucei and T. cruzi Recombinant Proteins.
Glutathione S-transferase (GST) derivatives.
The ORFs of T. brucei Sm proteins B, D1, D2, E, F, and G and of T. cruzi SmD3 were PCR amplified from genomic DNA and cloned into pGEX-2TK vector. Constructs were expressed in Escherichia coli BL 21 (DE3) pLysS cells.
His tag derivatives.
Similarly the ORFs were cloned into pQE30 vector. Constructs were expressed in E. coli M15 [pREP4], and proteins were purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography.
Analysis of Protein–Protein Interactions.
T7-transcribed mRNA was translated in rabbit reticulocyte lysate in the presence of 25 μCi of l-[35S]methionine. After a 2-h translation at 30°C, 35S-labeled products were analyzed by SDS/PAGE and fluorography.
For studying in vitro protein–protein interactions (heterodimer formation), 1 μg of a GST-Sm protein or GST alone (as negative control) was immobilized on 25 μl of glutathione agarose beads and incubated with in vitro-translated 35S-labeled Sm protein (20 μl of translation reaction) in 200 μl of binding buffer (50 mM Tris⋅HCl, pH 7.5/150 mM NaCl/5 mM MgCl2/0.05% Nonidet P-40/0.5 mM DTT). After a 1-h incubation at 25°C, the beads were washed five times in the same buffer. Selected proteins were analyzed on a 12.5% polyacrylamide tricine/SDS gel (29) by fluorography (Amersham).
For characterizing SmF/E/G and SmG/E/F heterotrimeric interactions, GST-SmF (or GST-SmG) was immobilized on glutathione agarose (see above) and incubated with in vitro-translated, unlabeled second Sm protein or mock in binding buffer for 1 h at 30°C. After washing in binding buffer, a third, 35S-labeled Sm protein was incubated for 1 h at 30°C with the agarose-bound proteins. Then beads were washed extensively in binding buffer, and 35S-labeled proteins were analyzed as described above.
In the case of the SmF/D2/D1 heterotrimeric interaction, GST-SmF (or GST as a control) were first immobilized on glutathione agarose. Second, the other two Sm proteins were incubated in 200 μl of binding buffer, one of them in His-tagged recombinant form (1 μg), the other one 35S-labeled in vitro. Third, after a 1-h incubation at 30°C, this protein mixture was combined with the glutathione–agarose bound GST(-Sm) protein and incubated for another hour at 30°C with constant rotation. Protein analysis was performed as described above.
Complementation Assays in Yeast.
The ORFs of T. brucei Sm proteins D1, D2, E, F, and G were PCR amplified from genomic DNA and cloned into the yeast vector pYX142 (R&D Systems) under the control of the strong triose phosphate isomerase (TPI) promoter. Standard yeast media, techniques, and plasmid shuffling strategies were applied (30). The following yeast strains were used during this study: BSY729 and BSY795 (22) and YRB10 (31). New yeast strains BSY456 and BSY860, carrying disruptions of the yeast SmG and SmD1 genes, respectively, complemented URA-marked plasmid and were constructed to test complementation of SmG and SmD1.
Results
cDNA Cloning of Common Protein Components of T. brucei snRNPs: Sequence Analysis Reveals Five Sm Polypeptides Homologous to SmD1, -D2, -E, -F, and -G.
To obtain peptide information, we purified the U2 snRNP from T. brucei extracts on a preparative scale, based on affinity selection with an antisense 2′-O-methyl RNA oligonucleotide (Fig. 1); polypeptides of 8.5, 10, 12.5, and 14 kDa, as well as a mixture of at least three proteins in the 15-kDa region (15K-mix), were eluted and subjected to microsequencing. For each of these proteins, internal peptide sequences were derived, as well as one N-terminal sequence for the 12.5-kDa protein. Suitable sequences were then selected for designing degenerate primers, resulting in the RT-PCR amplification of 5′ and 3′ halves of the corresponding cDNAs. We then used the new cDNA sequence information to PCR amplify from total T. brucei DNA the corresponding genomic DNA fragments containing the entire ORFs.
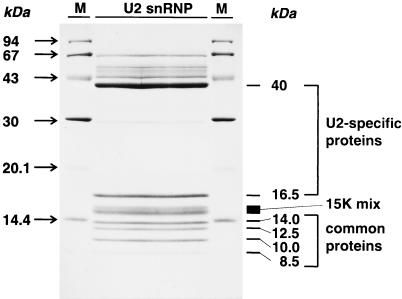
Figure 1.
Preparative affinity purification of U2 snRNPs from T. brucei. U2 snRNPs were affinity purified from T. brucei extract, separated on a preparative SDS/polyacrylamide gel (15%), transferred to nitrocellulose, and visualized by Ponceau S staining. The identification of U2-specific and common proteins is indicated on the right (compare with ref. 8), molecular mass markers on both sides (sizes in kDa). For microsequencing, the four common proteins 8.5 kDa, 10 kDa, 12.5 kDa, and 14 kDa were selected, and in addition, the region around 15 kDa containing both common and U2-specific proteins (15K-mix).
As Fig. 2 shows, each of these five sequences contains the bipartite Sm motif characteristic of a large family of proteins, most of which represent the core proteins of snRNPs U1, U2, U4, U5, and U6 (see supplementary material for a complete alignment). Sequence comparisons indicated that the trypanosome Sm proteins of 8.5, 10, 12.5, 14, and 15 kDa are homologues of the canonical Sm proteins F, E, D1, G, and D2, respectively. Additional confirmation that the cloned Sm protein genes encode the spliceosomal common proteins came from immunoprecipitation experiments, using polyclonal anti-common protein Abs, which had been raised against four polypeptides (9). From a mixture of five in vitro-translated and 35S-labeled polypeptides (SmD1, -D2, -E, -F, and -G), only the four that had been used for immunization (SmD1, -E, -F, and -G) reacted; SmD2 did not (data not shown).
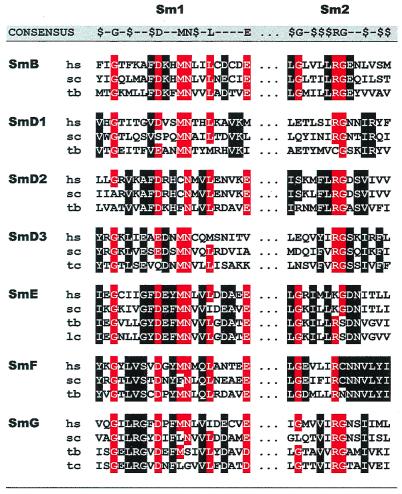
Figure 2.
Comparison of the seven trypanosome Sm proteins with their human and yeast homologues (hs, Homo sapiens; sc, Saccharomyces cerevisiae; tb, T. brucei; tc, T. cruzi; lc, L. collosoma). An alignment of the conserved Sm motifs 1 and 2 is presented with the consensus shown above ($, hydrophobic residue). Conserved amino acids are outlined by reverse print (red, overall conservation; black, subgroup conservation). Trypanosomal Sm proteins: T. cruzi SmG, putative (AA676151), L. collosoma SmE (AF126283). Human: SmD1 (P13641), SmD2 (P43330), SmD3 (P43331), SmE (P08578), SmF (X85372), SmG (S55054), SmB (P14678). Yeast: SmD1 (Q02260), SmD2 (Q06217), SmD3 (P43321), SmE (Q12330), SmF (P54999), SmG (P40204), and SmB (P40018)
Focusing on the characteristic Sm motif sequences (Fig. 2), we note several significant deviations from the overall consensus as well as from Sm homologues from other species that are–for all seven trypanosome Sm proteins–presented in Discussion. In sum, based on peptide sequence information, we have cloned five canonical Sm proteins that are common core components of the spliceosomal snRNPs, including the SL RNP.
Identification in Silico and Cloning of Trypanosome SmB and -D3 Protein Genes.
If core particles are conserved in their structural organization from human to trypanosomes, we would expect seven Sm constituents. In fact, we succeeded in identifying the two missing components, SmB and SmD3, by database searching, using the known cis-spliceosomal homologues.
The SmB gene of T. brucei was found in a database search using the human SmB protein sequence. The short genomic sequence that we found (no. AQ652994; TIGR T. brucei Genome Project) encodes a small polypeptide of 109 aa, which shares significant homology with known SmB proteins (Fig. 2; for a complete alignment, see supplementary material).
Through searches with the human SmD3 sequence, we succeeded to identify a homologous gene from T. cruzi, a species closely related to T. brucei. The short genomic sequence (no. AQ444257, Sanger Center T. cruzi Genome Project) contains a partial ORF. The missing N-terminal sequence was obtained by RT-PCR from total T. cruzi RNA and used to amplify a genomic fragment covering the complete coding sequence. The newly identified ORF encodes a polypeptide of 115 aa, which displays significant homology to the known SmD3 proteins (Fig. 2; supplementary material). Phylogenetic analysis indicates that the trypanosome SmB and -D3 proteins are more closely related to human SmB and -D3 than to the human Lsm8 and Lsm4 proteins, respectively.
Analysis of Protein–Protein Interactions in the Trypanosome Sm Core Complex: Heterodimeric and -trimeric Subcomplexes.
Considering the rather low degree of homology between the trypanosome Sm proteins and counterparts from other eukaryotes, we were interested to determine whether protein–protein contacts are conserved in the trypanosome system. We first characterized the interactions between individual Sm proteins, by using GST derivatives and in vitro-translated, 35S-labeled proteins (Fig. 3). Testing all pairwise combinations clearly demonstrated that there are strong and specific dimeric interactions between individual Sm proteins. Only SmD2 showed no apparent specificity, because it was bound by each of the GST-Sm derivatives with similar efficiency. In each case, binding of 35S-labeled Sm proteins to GST alone as a control was undetectable (Fig. 3A, lanes GST), implying that the Sm protein interactions measured do not depend on the GST portion. Except for SmD3, dimeric interactions were tested by using either of the two partners as GST derivative to determine whether interactions are reciprocal. The following interactions were detected (Fig. 3 A and B; summarized in Fig. 3C):
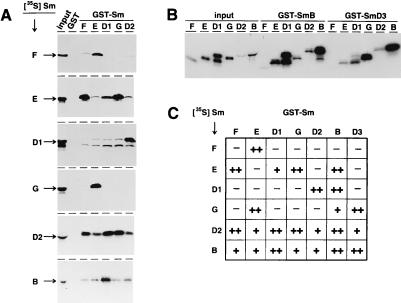
Figure 3.
Identification of specific protein–protein interactions in the Sm core. (A) In vitro-translated, 35S-labeled proteins (SmF, -E, -D1, -G, -D2, and -B, as indicated on the left) were incubated with immobilized GST-Sm proteins (lanes GST-SmF, -E, -D1, -G, and -D2) or, as a control, with GST alone (lanes GST); after washing, bound material was analyzed by SDS/PAGE and fluorography. In addition, 10% of the radiolabeled input of each reaction is shown (lanes input). Note that in vitro translation of 35S-labeled SmD1 consistently produced a doublet (see, for example, lane input/D1). The aberrant mobility of 35S-labeled SmD2 (lane input) is probably caused by comigrating abundant globin protein. (B) As described for A, the GST-SmB and GST-SmD3 interactions with each of six 35S-labeled Sm proteins (F, E, D1, G, D2, and B; as indicated) were characterized. (C) Summary of the Sm protein interactions. The relative strength of the hetero- and homodimeric interactions is indicated as + + (strongest interaction; signal greater than 10% of the input), + (intermediate; less than 10% of the input), and − (not significant; below 1% of the input).
We found four strong reciprocal interactions: D1/D2, F/E, E/G, and B/D1. In addition, strong but nonreciprocal binding was observed between GST-SmF and -SmD2 as well as between GST-SmB and -SmE. GST-SmD3 strongly interacted with both SmG and SmB (the reciprocal interactions with 35S-labeled SmD3 could not be tested; see above). Significantly, we found a strong homotypic interaction only for SmB.
In addition to these dimeric Sm protein interactions, we identified several heterotrimeric Sm protein complexes, by using GST derivatives and in vitro-translated proteins (Fig. 4). In principle, a GST-Sm protein was immobilized, followed by incubations with two other Sm proteins: first, with unlabeled Sm protein; second, with 35S-labeled Sm protein.
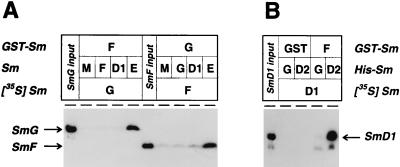
Figure 4.
In vitro assembly of heterotrimeric Sm protein complexes. (A) Heterotrimeric SmF/E/G and SmG/E/F complexes were assembled by incubating immobilized GST-SmF or -SmG with an in vitro-translated unlabeled protein (SmF, SmD1, SmE, or mock lysate, as indicated), and, after washing, with 35S-labeled SmG or SmF. Bound proteins were detected by SDS/PAGE and fluorography. For comparison, 10% of the radiolabeled SmG and SmF proteins is shown in the input lanes. The arrows on the left mark the positions of SmG and SmF. (B) A heterotrimeric SmF/D2/D1 complex was formed by incubating immobilized GST-SmF (or GST as a control) with preassembled His-SmD2 and 35S-labeled SmD1 (or His-SmG and 35S-labeled SmD1). Binding of radiolabeled SmD1 was detected by SDS/PAGE and fluorography. The position of SmD1 is marked by an arrow on the right.
Using this assay we demonstrated that SmF, -E, and -G efficiently form a heterotrimeric complex (Fig. 4A). Immobilized GST-SmF was incubated in four separate reactions with unlabeled in vitro-translated SmF, -D1, -E protein, or with control lysate. After washing, a second incubation followed with 35S-labeled SmG protein. As a result, [35S]SmG was bound only when the second incubation contained SmE; with the other Sm proteins (SmF or D1) and with control lysate (mock reaction), binding was at background levels. We conclude that SmF, -E, and -G form a specific heterotrimeric complex. This result could also be confirmed when the components were used in the opposite order, i.e., glutathione-agarose-bound SmG, unlabeled in vitro-translated SmE, and 35S-labeled SmF (Fig. 4A). Only in this combination, efficient heterotrimer formation was achieved, but not with unlabeled SmG or SmD1 as the bridging partner or with control lysate.
We further demonstrated the specific assembly of a heterotrimeric complex SmF/D2/D1, by using GST-SmF, His-tagged SmD2, and 35S-labeled SmD1 (Fig. 4B). Immobilized GST-SmF bound 35S-labeled SmD1 efficiently only after preincubation with His-tagged SmD2; if His-tagged SmG was used instead, very weak background levels of SmD1 binding were detected. This interaction strictly depended on immobilized SmF, because no binding was observed with GST alone.
In sum, we have obtained strong evidence for the specific formation of two heterotrimeric complexes: first, GST-SmF/E/G and GST-SmG/E/F; second, GST-SmF/D2/D1.
Functional Complementation of Yeast Mutants with T. brucei Sm Proteins.
Sequence analyses indicate that the trypanosomal Sm proteins are very distant from their counterparts of other eukaryotes (Fig. 2). To determine whether they are more closely related with respect to their function, we attempted to complement disruptions of the essential SMD1, -D2, -E, -F, and -G genes in yeast (reviewed in ref. 22) with some of the trypanosomal Sm proteins. The coding sequences of the trypanosomal Sm proteins E, F, G, D1, and D2 were placed downstream of a yeast promoter in a centromeric LEU2 vector. The resulting plasmids were introduced in diploid strains carrying a disruption of a yeast Sm protein, and tetrads were dissected. Using this strategy, we demonstrated that the trypanosomal SmD2 homologue was unable to complement a disruption of its yeast counterpart (data not shown). Similarly, the trypanosomal SmF and SmE homologues could not rescue a yeast SMF disruption. However, the trypanosomal 14-kDa protein (SmG homologue) functionally complemented a yeast SmG disruption, although the slow-growth phenotype indicated a partial effect. To confirm the specificity of this complementation, a second test was used. In this case, each of the five trypanosomal constructs was introduced in haploid yeast strains carrying a chromosomal deletion of one of either the SMD1, SME1, or SMG genes, which was complemented by a plasmid-derived copy linked to the URA3 marker. Transformants were transferred to fluoroorotic acid (FOA)-containing plates that prevent growth of cells carrying a URA3 marker. Therefore, in this assay, growth on FOA-containing media indicates that the trypanosomal gene is able to complement the corresponding yeast mutant. None of the trypanosomal genes was able to rescue inactivation of the yeast SME1 or SMD1 mutants (data not shown). However, consistent with the previous tests, the trypanosomal SmG protein was able to suppress a deletion of the yeast SMG gene, albeit again at low efficiency (Fig. 5). This complementation was highly specific because the trypanosome SmE, -F, -D1, and -D2 genes did not complement the same mutation (Fig. 5) and because the SmG construct was not able to complement disruption of the yeast SME1 and SMD1 genes (data not shown). In sum, these results indicate that the T. brucei 14-kDa protein is indeed a functional homologue of the yeast SmG protein.
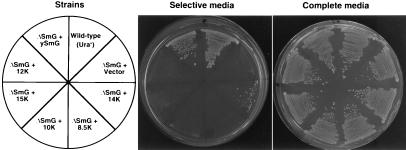
Figure 5.
The trypanosomal 14-kDa protein complements specifically a yeast SMG disruption. A haploid yeast strain carrying a chromosomal disruption of the SMG gene complemented by a wild-type copy of the gene on a URA3-marked plasmid was transformed with LEU2-marked plasmids expressing the various trypanosomal proteins or, as a positive control, the yeast SmG protein in a LEU2-marked plasmid. As a negative control, the same strain was transformed with the expression vector only. Transformants were then subcloned on complete media (Right) or on selective media (Center) that contains FOA, a drug preventing growth of cells expressing the URA3 marker. A wild-type ura3− strain was used as a positive control for FOA selection. (Left) Relevant characteristics of the strains present in the different sectors. The various strains grew equally well on complete media (Right). Cells expressing the trypanosomal 14-kDa protein were able to grow on selective media, indicating that it complements the SMG gene disruption.
Discussion
Identification and Assignment of Trypanosome Sm Proteins.
Based on peptide sequence information, we have amplified cDNAs coding for five low-molecular weight snRNP polypeptides from T. brucei. The sequence of each of the isolated cDNAs was confirmed by PCR amplification and analysis of the corresponding genomic DNA (data not shown). In addition, we probed high-density filters of a genomic bacteriophage P1 library of T. brucei strain TREU927/4 individually with each of these five T. brucei Sm genes. Four of them (SmD2, -E, -F, and -G) could be detected, each of them in different P1 clones, indicating that at least these four Sm genes map to different genomic loci (data not shown). In sum, it appears that the Sm protein genes are not clustered in a single polycistronic transcription unit.
Originally we had identified this set of proteins as common components of the trypanosomal snRNPs, both by direct protein analysis of affinity-purified snRNPs (8) and by immunoprecipitation (9). Cloning and sequence analysis presented here revealed that they are members of the large family of Sm proteins and suggested homologies of each of them to known canonical Sm polypeptides from other eukaryotes.
Identifying the common proteins as classical Sm polypeptides was surprising for several reasons. First, no immunological relationship could previously be established between the trypanosomal common proteins and the Sm proteins of other eukaryotes (9, 32). Second, the binding sites in trypanosome snRNAs are relatively degenerate in comparison to the well-conserved Sm site of other eukaryotes (see below). Third, in contrast to the established role of the Sm proteins as a determinant of cap trimethylation and nuclear localization, the 5′ end structures of trypanosomal snRNAs are very diverse (spliced leader RNA, cap4; U2 and U4 snRNAs, m3G; U1, modified methyl guanosine cap; U5 snRNAs, no cap). This phenomenon raises the question how—despite a common Sm core—these different 5′ end modifications are specified.
Given the classical set of seven Sm proteins, why did we not detect SmB and -D3 in our initial protein analysis? Perhaps those two components are less stably bound than the others, similarly to the yeast SmB (33, 34), resulting in their loss during stringent affinity selections. This possibility would imply that a subcore lacking SmB and -D3 remains stably associated with the snRNA, consistent with data from Raker et al. (19) in the mammalian system. Alternatively, these two polypeptides may comigrate with the other Sm proteins, and we did not obtain peptide sequences from them. Several additional internal peptide sequences from proteins in the 15-kDa region we determined are not represented in any of the seven Sm proteins described here. This finding suggests that these additional peptide sequences are derived from other, most likely U2 snRNP-specific proteins, because affinity-purified U2 snRNPs were used as starting material. Finally, we cannot rule out the possibility that the trypanosome U2 snRNP lacks canonical SmB and -D3 proteins; however, in light of the strong and specific interactions that we have demonstrated (see below), we consider it unlikely.
Except for SmG, the other Sm polypeptides could be assigned to known homologues with high confidence, based on searches of the entire database and on comparing each trypanosome sequence with individual Sm proteins from different species (the variable amino acid positions within Sm motifs 1 and 2 are often conserved within individual Sm proteins; for example, see ref. 18).
Although assigning the identity of the trypanosome 14-kDa protein was difficult at first, its ability to specifically complement an SmG mutation in yeast strongly argued for its representing the trypanosome SmG orthologue. It is surprising that only one trypanosome Sm protein, SmG, complemented in yeast, in particular because it is one of the least conserved trypanosome Sm proteins. Additional confirmatory evidence came from protein interaction studies that demonstrated specific, strong, and reciprocal interaction with SmE, as well as strong D3/G binding (this study), consistent with the current model for the mammalian Sm core complex.
Comparison of the Trypanosome Sm Proteins with Those from Other Systems: Immunological Reactivity with Sm Abs.
The trypanosome Sm proteins appear to represent minimal versions of the canonical Sm polypeptides, because three of the seven, in particular SmB, are smaller than homologues from other species (see SmD1, -D3, and -B in Fig. 2 and supplementary material); the other four trypanosome proteins (SmD2, -E, -F, and -G) are of similar length as in other systems. It is of particular interest that both the trypanosome SmD1 and -D3 proteins are lacking the C-terminal RG dipeptide repeats of the human homologues. The arginines in this domain are methylated, constituting an important determinant of the Sm epitope (35); the lack of the RG-rich domain and its modification by arginine dimethylation in the trypanosome proteins may therefore explain why the trypanosome Sm proteins are not recognized by Sm Abs (9, 32).
Closer inspection of the overall and subgroup sequences revealed several significant deviations (Fig. 2). The Sm motifs 1 and 2 are relatively degenerate and contain only a few highly conserved positions, such as the methionine-asparagine (MN) positions in Sm motif 1 or arginine-glycine (RG) in Sm motif 2. Many of the changes that we found in the trypanosome Sm proteins are conservative replacements by similar amino acids (Fig. 2). Some of the interesting differences in positions of the trypanosome proteins that are conserved in most or all other known Sm proteins are listed in the following. Only SmB conforms very well to the Sm consensus and to homologues from other species: (1) In SmD1 of T. brucei the conserved RG of motif 2 is altered to the unusual CG. (2) SmD2 usually contains in motif 1 a CN in place of the conserved MN; the T. brucei SmD2, however, an FN. At the second position of motif 2 (consensus glycine; as shown in Fig. 2), T. brucei contains an arginine instead of the serine in all other known species. (3) In SmD3 there is a characteristic replacement of aspartate by glutamate in the middle of motif 1, whereas T. cruzi contains glutamine instead. (4) The SmE-specific KG in motif 2 instead of the consensus RG is altered to an RS in T. brucei and L. collosoma. (5) Similarly, for SmF sequences known to date, there is a characteristic RC in place of the consensus RG in motif 2; in contrast, T. brucei SmF contains RN. (6) Finally, in SmG, the characteristic MN of motif 1 is an MS or LG in T. brucei and T. cruzi, respectively.
In contrast, there is almost no deviation in the 11 conserved hydrophobic positions of motifs 1 and 2, underlining their functional importance and consistent with a recent mutational analysis in yeast (21). In sum, we find in the trypanosome Sm proteins a multitude of alterations in conserved positions. In the future, this finding promises to yield interesting insights into conservation of structural features of the Sm core, depending on further crystallographic data on the mammalian Sm complex and modeling studies of the trypanosome core.
A Model of the Trypanosome Sm Protein Core: Structural and Functional Conservation?
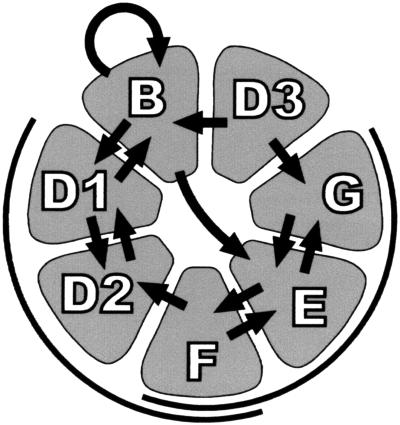
The recently proposed model for the mammalian core snRNP domain (23) raises the question how our data in the trypanosome system fit into such an arrangement of the seven Sm polypeptides. In sum, the combined data from our protein interaction assays are consistent with a heptameric ring-like structure of the seven trypanosome Sm proteins arranged in the same order (Fig. 6). Strong evidence for this model comes (i) from the specificity and reciprocal nature of dimeric interactions between Sm proteins and (ii) from our demonstration of two trimeric complexes, SmF/E/G and SmF/D2/D1, which overlap with each other. We note that SmD3 from T. cruzi interacts specifically with the other Sm proteins from T. brucei, indicating functional conservation within different trypanosomatid species. Taken together, these data strongly suggest that also in the trypanosome system a heptameric core complex of Sm polypeptides forms the common structural basis of snRNPs.
Figure 6.
Model of the trypanosome Sm core complex. The specific interactions between the trypanosome Sm proteins are schematically represented. Only the strong interactions (thick arrows) are shown; for 35S-labeled SmD2, only some of the strong interactions are represented because of an apparent lack of specificity (see Results). Because these data are based on GST precipitation assays (see Fig. 3 and 4), the arrow points from the GST fusion to the in vitro-translated protein. In addition, the heterotrimeric complexes F/D2/D1 and F/E/G are indicated. Most of these contacts can be fitted into the seven-membered-ring model for the mammalian Sm core (23).
We have detected several additional interactions, which had not been seen with the yeast proteins in the two-hybrid study (21), for example strong GST-SmB/E binding; furthermore, we observed a strong homotypic interaction for SmB. The functional significance of these interactions, which cannot be accounted for by the seven-membered-ring model, is currently unclear. They may be due to differences in the experimental approach (yeast two-hybrid versus in vitro binding assays) or to distortions by the GST portion of one binding partner (in our assay); more interestingly, they may reflect alternative interactions occurring during the multiple stages of snRNP assembly, but not present in the final assembly product.
On the basis of the in vitro interaction studies described here, it will be interesting to work on the reconstitution of a full trypanosome Sm core, with the aim to investigate its specific recognition of the rather degenerate Sm sequence.
Supplementary Material
Acknowledgments
We thank J. Hoheisel (Heidelberg) for T. cruzi genomic DNA and A. Ouaissi (Montpellier) for T. cruzi total RNA. We gratefully acknowledge the help of Sarah Melville and Vanessa Leech (T. brucei Genome Project, Cambridge, U.K.) with high-density filters and P1 clones, as well as Reinhard Lührmann (Marburg) and Shula Michaeli (Weizmann Institute, Israel) for discussions and unpublished data. This work was supported by the Deutsche Forschungsgemeinschaft and the German-Israeli Foundation.
Abbreviations
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein
- SL
spliced leader
- RT
reverse transcription
- GST
glutathione S-transferase
- FOA
fluoroorotic acid
Footnotes
Data Deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF280390 (Sm-D3), AF280391 (Sm-F), AF280392 (Sm-G), AF280393 (Sm-D1), AF280394 (Sm-D2), and AF280395 (Sm-E)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150236097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150236097
References
- 1.Ullu E, Tschudi C, Günzl A. In: Molecular Biology of Parasitic Protozoa. Smith D F, Parson M, editors. Oxford: IRL; 1996. pp. pp.115–133. [Google Scholar]
- 2.Tschudi C, Ullu E. Cell. 1990;61:459–466. doi: 10.1016/0092-8674(90)90527-l. [DOI] [PubMed] [Google Scholar]
- 3.Dungan J M, Watkins K P, Agabian N. EMBO J. 1996;15:4016–4029. [PMC free article] [PubMed] [Google Scholar]
- 4.Lücke S, Klöckner T, Palfi Z, Boshart M, Bindereif A. EMBO J. 1997;16:4433–4440. doi: 10.1093/emboj/16.14.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y-X, Ben-Shlomo H, Michaeli S. Proc Natl Acad Sci USA. 1997;94:8473–8478. doi: 10.1073/pnas.94.16.8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnare M N, Gray M W. J Biol Chem. 1999;274:23691–23694. doi: 10.1074/jbc.274.34.23691. [DOI] [PubMed] [Google Scholar]
- 7.Mair G, Shi H, Li H, Djikeng A, Aviles H O, Bishop J R, Falcone F H, Gavrilescu C, Montgomery J L, Santori M I, et al. RNA. 2000;6:163–169. doi: 10.1017/s135583820099229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palfi Z, Günzl A, Cross M, Bindereif A. Proc Natl Acad Sci USA. 1991;88:9097–9101. doi: 10.1073/pnas.88.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palfi Z, Bindereif A. J Biol Chem. 1992;267:20159–20163. [PubMed] [Google Scholar]
- 10.Günzl A, Cross M, Bindereif A. Mol Cell Biol. 1992;12:468–479. doi: 10.1128/mcb.12.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palfi Z, Xu G-L, Bindereif A. J Biol Chem. 1994;269:30620–30625. [PubMed] [Google Scholar]
- 12.Bell M, Bindereif A. Nucleic Acids Res. 1999;27:3986–3994. doi: 10.1093/nar/27.20.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell M, Wöhner R-V, Bindereif A. Gene. 2000;247:77–86. doi: 10.1016/s0378-1119(00)00118-9. [DOI] [PubMed] [Google Scholar]
- 14.Goncharov I, Palfi Z, Bindereif A, Michaeli S. J Biol Chem. 1999;274:12217–12221. doi: 10.1074/jbc.274.18.12217. [DOI] [PubMed] [Google Scholar]
- 15.Cross M, Wieland B, Palfi Z, Günzl A, Röthlisberger U, Lahm H-W, Bindereif A. EMBO J. 1993;12:1239–1248. doi: 10.1002/j.1460-2075.1993.tb05765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehmeier T, Raker V, Hermann H, Lührmann R. Proc Natl Acad Sci USA. 1994;91:12317–12321. doi: 10.1073/pnas.91.25.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Lührmann R. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Séraphin B. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raker V A, Plessel G, Lührmann R. EMBO J. 1996;15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- 20.Fury M G, Zhang W, Christodoulopoulos I, Zieve G W. Exp Cell Res. 1997;237:63–69. doi: 10.1006/excr.1997.3750. [DOI] [PubMed] [Google Scholar]
- 21.Camasses A, Bragado-Nilsson E, Martin R, Séraphin B, Bordonné R. Mol Cell Biol. 1998;18:1956–1966. doi: 10.1128/mcb.18.4.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Séraphin B. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kambach C, Walke S, Young R, Avis J M, de la Fortelle E, Raker V A, Lührmann R, Li J, Nagai K. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 24.Kastner B, Bach M, Lührmann R. Proc Natl Acad Sci USA. 1990;87:1710–1714. doi: 10.1073/pnas.87.5.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plessel G, Lührmann R, Kastner B. J Mol Biol. 1997;265:87–94. doi: 10.1006/jmbi.1996.0713. [DOI] [PubMed] [Google Scholar]
- 26.Brun R, Schönenberger M. Acta Tropica. 1979;36:289–292. [PubMed] [Google Scholar]
- 27.Cross M, Günzl A, Palfi Z, Bindereif A. Mol Cell Biol. 1991;11:5516–5526. doi: 10.1128/mcb.11.11.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schägger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 30.Sherman F. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink G R, editors. San Diego, CA: Academic; 1991. pp. 3–21. [Google Scholar]
- 31.Bordonné R, Tarassov I. Gene. 1996;176:111–117. doi: 10.1016/0378-1119(96)00230-2. [DOI] [PubMed] [Google Scholar]
- 32.Michaeli S, Roberts T G, Watkins K P, Agabian N. J Biol Chem. 1990;265:10582–10588. [PubMed] [Google Scholar]
- 33.Neubauer G, Gottschalk A, Fabrizio P, Séraphin B, Lührmann R, Mann M. Proc Natl Acad Sci USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot H V, Mann M, Séraphin B, Rosbash M, Lührmann R, Fabrizio P. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- 35.Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Lührmann R. J Biol Chem. 2000;275:17122–17129. doi: 10.1074/jbc.M000300200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.