Abstract
The anaphase-promoting complex (APC) is a cell cycle-regulated ubiquitin-protein ligase that targets cyclin B, securin and other destruction box containing proteins for proteolysis. Nine APC subunits have been identified in vertebrates and eleven in yeast, but for none of them it is known how they contribute to the catalysis of ubiquitination reactions. Here we report the mass spectrometric identification of CDC26 and of the RING-H2 finger protein APC11 in the human APC. We have expressed these proteins and several other APC subunits in Escherichia coli and have tested their activities in vitro. We find that APC11 alone is sufficient to allow the synthesis of multiubiquitin chains in the presence of E1 and UBC4. These multiubiquitin chains are partly unanchored and partly bound to APC11 itself. APC11 and UBC4 are also able to ubiquitinate securin and cyclin B, but these reactions show a decreased dependency on the destruction box. The integrity of the putative zinc binding RING-H2 finger is required for the ability of APC11 to support ubiquitination reactions. These results suggest that APC11 and UBC4 catalyze the formation of isopeptide bonds in APC-mediated ubiquitination reactions.
Keywords: cyclin B, cyclosome, mechanism, proteolysis, securin
The anaphase-promoting complex (APC) or cyclosome is a cell cycle-regulated ubiquitin-protein ligase that regulates important events in mitosis such as the initiation of anaphase and exit from telophase (reviewed in refs. 1 and 2). Together with the ubiquitin-activating enzyme E1 and ubiquitin-conjugating enzymes (E2s), the APC assembles multiubiquitin chains on a variety of regulatory proteins and thus targets them for proteolysis by the 26S proteasome. To initiate sister chromatid separation, the APC has to ubiquitinate the anaphase inhibitor securin, whereas exit from mitosis requires the APC-mediated ubiquitination of B-type cyclins. These reactions depend on activation of the APC by CDC20 and CDH1, which transiently associate with the APC at the end of mitosis and in G1, respectively. APC substrates contain either a destruction (D) or a KEN box sequence whose presence is required for recognition by the APC (3).
In ubiquitination reactions, the APC fulfills the role of a ubiquitin-protein ligase (E3); i.e., it allows the transfer of ubiquitin residues from E2 enzymes to substrate proteins. This reaction results in the formation of an isopeptide bond between the C terminus of ubiquitin and the epsilon amino group of a lysine residue in the substrate. In subsequent reaction cycles, also lysine residues within the attached ubiquitin residues can serve as acceptor sites, resulting in the assembly of a multiubiquitin chain, which is thought to function as a recognition signal for the 26S proteasome. How the APC mediates these ubiquitination reactions is not known. So far, 9 subunits have been identified in the vertebrate APC, whereas 11 are known in budding yeast (2, 4). Initial analyses did not reveal any homologies between APC subunits and two previously identified ubiquitin-protein ligases, Ubr1 and E6-AP, suggesting that E3s may represent a structurally and mechanistically diverse class of enzymes. Recent results suggest that the APC belongs to a family of E3s that comprises at least two other multisubunit complexes, the Skp1-cullin-F box protein complex and the von Hippel Lindau complex (reviewed in refs. 5–7). Both the Skp1-cullin-F box protein and von Hippel Lindau complexes are characterized by the presence of a cullin subunit whose conserved C-terminal domain interacts with a protein called Rbx1/Hrt1/Roc1 in budding yeast and ROC1 in humans (8–12). This protein may recruit the E2 enzyme CDC34 to the Skp1-cullin-F box protein complex and is characterized by the presence of a RING-H2 finger consensus, a subtype of the RING finger motif, which is known to coordinate two atoms of zinc (13). The APC subunit APC2 is a distant member of the cullin family (14, 15), and a RING-H2 finger protein related to Rbx1/Hrt1/Roc1, called Apc11, has been identified in the budding yeast APC (14).
Although various functions have been proposed for RING finger motifs such as representing nucleic acid or protein binding domains, numerous recent observations suggest a prominent role of these domains in ubiquitination reactions. The analysis of rbx1/hrt1/roc1 and apc11 mutants in budding yeast indicates that these proteins are essential for Skp1-cullin-F box protein complex and APC activity, respectively (8, 9, 11, 12, 14). A dimeric complex of CUL1 and ROC1 obtained by transient transfection into human cells and partial purification has been reported to catalyze the assembly of unanchored multiubiquitin chains in the presence of E1 and CDC34, suggesting that a cullin and a RING-H2 finger subunit may represent the essential catalytic subunits of this type of E3 (10). Also, immunoprecipitates from transiently transfected cells containing APC11, APC2, and some unidentified proteins have been reported to contain ubiquitination activity (8). Intriguingly, RING-H2 finger domains have also been identified in several E3s that do not contain cullin subunits. For example, RING-H2 finger sequences are required for the activity of Ubr1, an E3 that ubiquitinates proteins with destabilizing N-terminal amino acid residues (16), for the activity of MDM2, a protein that can ubiquitinate the tumor suppressor p53 (17, 18), and for the activity of c-CBL, a protein that can mediate the ubiquitination of the platelet-derived growth factor receptor (19–21). When expressed in Escherichia coli, several other RING-H2 finger proteins have been shown to support the synthesis of multiubiquitin chains, suggesting that many if not all proteins with this motif may have this ability (22).
To obtain insight into the mechanism of the APC, we have reinvestigated the subunit composition of the human APC by mass spectrometry. We report the identification of two subunits previously only identified in yeast, CDC26 and APC11. We show that APC11 alone is sufficient to catalyze the ubiquitination of APC substrates in the presence of E1 and the E2 enzyme UBC4.
Materials and Methods
Mass Spectrometry.
p10 and p14 were excised from silver stained gels and were processed as described (23, 24). Tryptic digestion was carried out for 3 h at 37°C. After the digestion, the supernatant was loaded on reversed phase resin (POROS R2, PerSeptive Biosystems, Framingham, MA) in a capillary, was washed with acidified water, and was eluted with organic phase directly into a nanoelectrospray needle (25) (Protana, Odense, Denmark). Peptide sequencing by tandem mass spectrometry was performed on a quadrupole time of flight instrument (QSTAR, Sciex, Toronto). Peptide sequence tags (26) were assigned in the spectra resulting from peptide fragmentation. Calculated fragments from the peptides corresponding to the retrieved expressed sequence tag (EST) database hits were compared with the complete fragmentation spectrum to verify the complete peptide sequence. Protana's pepsea software was used for database searches.
DNA Constructs.
Fusion proteins between glutathione S-transferase (GST) and APC10, APC11, and CDC26 were generated by PCR-based amplification of individual open reading frames, starting from amino acid 2 until the stop codon and subsequent blunt end ligation into the pGex4T-1 plasmid (Amersham Pharmacia) cut with BamHI and filled in with T4 DNA-polymerase. The APC10 coding region was amplified from a cDNA-containing plasmid, whereas APC11 and CDC26 were amplified from single-stranded cDNA prepared from 3 μg of total HeLa RNA. Human securin was amplified from HeLa single-stranded cDNA and was cloned into pTrcHis2A (Invitrogen), creating a fusion with a C-terminal myc-His tag. Mutagenesis of Cys23, Cys26, Cys51, and Cys54 of APC11 and Arg61 and Leu64 of human securin to alanine was performed by using standard PCR-based technology. All mutations were evaluated by DNA sequencing. His-tagged UBC4 was generated by recloning the ORF into the pQE30 vector (Qiagen, Chatsworth, CA).
Protein Expression and Purification.
E.coli DH5α carrying the individual expression plasmids were grown overnight, were induced with 0.3 mM isopropyl-β-thiogalactopyranoside for 2–3 h, and were harvested by centrifugation. Lysis was performed either under native (GST-APC10, GST-APC11 wild type and mutants, GST-CDC26, His-UBC4) or denaturing conditions (securin-myc-His and securin mutant). In brief, securin was purified on nickel-nitrilotriacetic acid agarose (Qiagen) by using 8 M urea as denaturing agent. Subsequently, the eluted protein was dialyzed against buffer QA (27) containing decreasing amounts of urea. Purification of GST-fusion proteins was performed by lysis in Tris⋅KCl buffer (TK) (50 mM Tris, pH 8.0/150 mM KCl), plus 2 mM DTT and 1% Triton X-100 followed by affinity purification on glutathione Sepharose (Pharmacia). Bound proteins were eluted in TK, 2 mM DTT, and 10 mM glutathione. Free glutathione was removed by repeated concentration and redilution in TK and 0.5 mM DTT by using Centriprep 10 concentrators (Amicon). In some experiments, GST-tagged APC subunits bound to glutathione beads were used instead of soluble, eluted proteins. His-UBC4 was purified on nickel-nitrilotriacetic acid agarose in TK, 1 mM β-mercaptoethanol, and 1% Triton X-100. Protein was eluted in TK, 1 mM β-mercaptoethanol, and 250 mM imidazol and was concentrated/rediluted on Centriprep10 using TK and 1 mM β-mercaptoethanol. UBA1 (E1) and UBCx were expressed and purified as described (27). UBC2 was a gift from M. Rolffe (Mitotix, Cambridge, MA). CDC34 was a gift from R. Yew (Harvard Medical School, Boston). APC was immunopurified from HeLa log phase cells using a polyclonal anti-CDC27 antibody (28). CDH1 was purified as described (28).
Ubiquitination Assays.
Standard ubiquitination reactions were carried out in 10 μl of QA for various time periods and contained 0.5 μg GST-fusion protein, either bound to glutathione-Sepharose and washed with QA or added as purified, soluble protein, 0.5 μg E1, 0.5 μg E2, 20 μg ubiquitin, or, where indicated iodinated ubiquitin, 0.5 μl of an ATP regenerating system, and, where indicated, 20 or 50 ng of substrate [securin-myc-His, maltose binding protein (MBP), GST-p53]. Reactions were stopped by addition of sodium-dodecyl-sulfate (SDS) containing sample buffer. Ubiquitination assays in Fig. 3C were incubated for 30 min at 37°C and were diluted 20-fold with cold QA containing 0.5% Triton X-100. After addition of 5 μl of glutathione agarose, the reaction was incubated for 60 min at 4°C with agitation. Before centrifugation, an aliquot of the reaction was removed (total, Fig. 3C). A second aliquot was removed after pelleting the agarose beads (supernatant, Fig. 3C). The remaining bead fraction was washed twice with QA and 0.5% Triton X-100, and a final aliquot was removed (beads, Fig. 3C). Thioester formation experiments were performed as described (27).
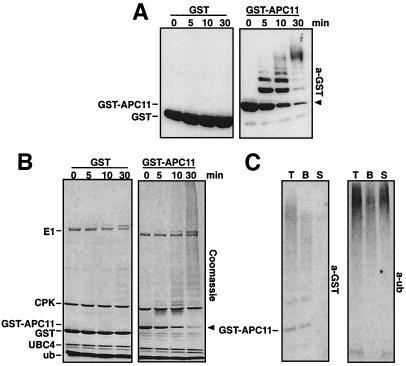
Figure 3.
Characterization of ubiquitin conjugates formed in the presence of GST-APC11. (A) Purified GST (Left) or GST-APC11 (Right) were incubated for the indicated time with E1, UBC4, an ATP regenerating system, and ubiquitin as in Fig. 2B. Samples were separated by 12% SDS/PAGE. GST and GST-APC11 were detected by immunodecoration with anti-GST antibodies. The position of unmodified GST-APC11 is indicated by an arrowhead. (B) Identical samples as in A were separated by 15 + 8% SDS/PAGE and were stained with Coomassie blue. Identified proteins are indicated on the left (CPK, creatine phosphokinase; for all others, see text). The arrowhead indicates the position of GST-APC11. (C) A GST-APC11-mediated ubiquitination reaction as described in Fig. 2B was split into three aliquots as described in Materials and Methods. Identical amounts of total (T), beads (B), and supernatant (S) fractions were loaded on a 15 + 8% polyacrylamide gel and were transferred to poly(vinylidene difluoride) membrane. One part (Left) was decorated with anti-GST antibodies, the other part (Right) with anti-ubiquitin antibodies.
Antibodies.
Mouse polyclonal antibodies against CDC26 were generated with a peptide encoding the 35 N-terminal amino acids. Antibodies against GST, MBP, ubiquitin, and p53 were obtained from Santa Cruz Biotechnology. All other antibodies used are described (29).
Results
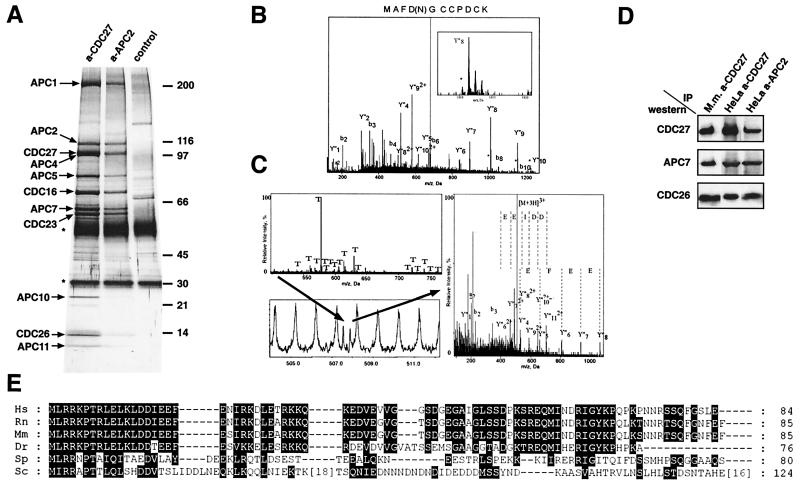
Our recent characterization of APC10/DOC1 suggested that additional subunits only identified in the yeast APC so far may also exist in higher eukaryotes (24). We therefore reinvestigated the subunit composition of human APC. SDS/PAGE and silver staining revealed two proteins of 14 and 10 kDa (p14 and p10) that specifically coimmunoprecipitated with antibodies to the APC subunits CDC27 and APC2 (Fig. 1A). We sequenced tryptic peptides from p14 and p10 by nanoelectrospray tandem mass spectrometry. A p10-derived peptide with the sequence MAFDGCCPDCK did not match any sequence in the more than one million human ESTs in the public database. Close inspection of the obtained spectrum showed asparagine deamidation by the presence of small accompanying peaks corresponding to the sequence MAFNGCCPDCK (Fig. 1B). This sequence retrieved more than 100 overlapping ESTs. All of these encode sequences highly homologous to budding yeast Apc11 (14). This indicates that p10 is the human ortholog of Apc11 and that this protein, which we call APC11, can be modified posttranslationally by deamidation on residue 31.
Figure 1.
Characterization of CDC26 and APC11 as subunits of the vertebrate APC. (A) Identification of APC subunits by silver staining. Immunoprecipitates obtained from logarithmically growing HeLa cells with CDC27 or APC2 antibodies were analyzed by SDS/PAGE and silver staining. The positions of APC subunits are indicated. The IgG light and heavy chains are marked by stars. (B and C) Identification of APC11 and CDC26 by nanoelectrospray tandem mass spectrometry. (B) Mass spectrometric sequencing of APC11. Tandem mass spectrum of a tryptic peptide (m/z 680.9) obtained after in-gel digestion of p10. The presence of peaks shifted by 1 Da (marked by asterisks) allowed to localize the site of deamidation. (C) Mass spectrometric sequencing of CDC26. Mass spectrum of in-gel digested human p14 (Upper left). The peaks marked with T correspond to trypsin autolysis products. The peak marked by an arrow (Lower left) was sequenced by nanoelectrospray mass spectrometry. The right panel shows the tandem mass spectrum of the selected peak. (D) APC was immunoprecipitated from logarithmically growing HeLa cells (HeLa) and from mouse myeloma Ag8.653 cells (Mm) with CDC27 or APC2 antibodies. Precipitates were subsequently analyzed by SDS/PAGE and immunoblotting by using CDC27, APC7, and CDC26 antibodies. (E) Alignment of the amino acid sequences of CDC26 orthologs from various species. CDC26 orthologs were identified by psi-blast by using the human CDC26 sequence as a query. Amino acid residues that are identical or similar (according to the Blosum 62 substitution matrix) are highlighted by black boxes. Amino acid residues are given in the one-letter code. For Sc Cdc26 insertions of 18 and 16 amino acids are indicated. Ce, Caenorhabditis elegans; Dr, Danio rerio; Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norwegicus; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe.
For p14 peptides, only few ions defining the peaks were detected because of the low protein amounts (Fig. 1C Left). Only one peptide was detected with a signal to noise ratio of 1:2, and the 1/3-Da spacing between the peaks revealed it to be triply charged. The high resolution and the mass accuracy of the quadrupole time of flight mass spectrometer allowed recognition of this peptide and its selection for MS/MS analysis. The resulting tandem mass spectrum, acquired by using the long analysis time of the nanoelectrospray ion source, is shown in Fig. 1C Right). The sequence tag (530.28)EFEELDD(1407.58) was assigned, leading to unambiguous identification of a single sequence LDDIEEFENIRK in a total of 25 overlapping ESTs. Pairwise comparison of the contiq from these ESTs with the sequence of known yeast APC subunits revealed low homology to budding yeast Cdc26 (14, 30) and fission yeast Hcn1 (31) (Fig. 1E). Coimmunoprecipitation and immunoblotting experiments confirmed that this protein is associated with the human APC and showed that a crossreacting protein also exists in the APC isolated from mouse cells, cow brain, and Xenopus eggs (Fig. 1D; data not shown). We conclude that this protein, which we call CDC26, is a constitutive subunit of the vertebrate APC.
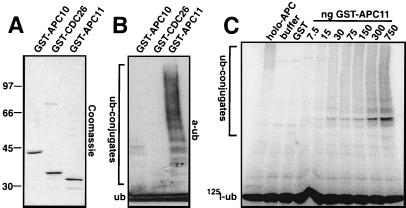
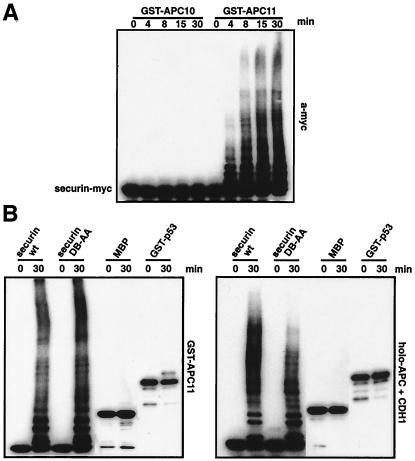
Our identification of CDC26 and APC11 increased the number of known human APC subunits to 11, but for none of these is it known how they function in ubiquitination reactions. To address this question, we expressed nine human APC subunits as recombinant proteins containing an N-terminal GST tag (Fig. 2A; data not shown). To avoid possible heterooligomerization problems with endogenous APC subunits, we expressed all proteins in E. coli whose genome does not contain obvious APC homologs. To analyze their activities, we incubated the purified GST-fusion proteins with E1, UBC4, ubiquitin, and an ATP-regenerating system and measured the formation of multiubiquitin chains by immunoblotting with ubiquitin antibodies. Remarkably, under these conditions, multiubiquitin chains were formed in the presence of GST-APC11 but not in the presence of any of the other APC subunits (Fig. 2B; data not shown). In dose-response experiments, the amount of ubiquitin conjugates formed depended on the concentration of GST-APC11 (Fig. 2C). Multiubiquitin chains could also be detected when radiolabeled ubiquitin (Fig. 1C), GST-, or myc-tagged ubiquitin were used (data not shown). Interestingly, similar multiubiquitin chains were generated by purified human APC under these conditions: i.e., in the absence of substrates (Fig. 1C, holo-APC).
Figure 2.
Formation of polyubiquitin chains in the presence of GST-APC11. (A) Bacterial lysates expressing GST-APC10, GST-CDC26, and GST-APC11 were incubated with glutathione beads, were washed, were separated by SDS/PAGE, and were stained with Coomassie blue. (B) Identical amounts of GST-APC10, GST-CDC26, and GST-APC11 lysates as in A were bound to glutathione beads, were washed, and were used for in vitro ubiquitination. Samples were separated by SDS/PAGE (15 + 8%), were transferred to poly(vinylidene difluoride) membrane, and were immunodecorated with anti-ubiquitin antibodies. (C) Purified holo-APC, buffer QA, GST (750 ng), or the indicated amounts of GST-APC11 were incubated with E1, UBC4, and ATP as in B in the presence of 125I-ubiquitin. Samples were incubated for 10 min at 37°C, were separated by SDS/PAGE, and were visualized by phosphorimaging.
When we analyzed the behavior of GST-APC11 and UBC4 in these reactions by immunoblotting, we observed that GST-APC11 itself was quantitatively converted into multiubiquitinated forms (Fig. 3A), whereas UBC4 was only monoubiquitinated (data not shown). We noticed that the formation of multiubiquitin chains could also be visualized by Coomassie blue staining (Fig. 3B). A comparison of the Coomassie-stained conjugates with the GST-APC11 containing conjugates detected by immunoblotting suggested that only a portion of the multiubiquitin chains are bound to GST-APC11. To test this possibility, GST-APC11 was removed at the end of the reaction by the addition of glutathione beads and centrifugation. Quantitative detection of the conjugates with radiolabeled ubiquitin antibodies showed that approximately 60% of these chains remained in the supernatant, although GST-APC11 was exclusively detected in the pellet (Fig. 3C), demonstrating that most multiubiquitin chains are not bound to GST-APC11. Because of the lack of suitable antibodies, we were unable to test directly whether these chains are anchored to E1 or the creatine phosphokinase, which is present in the ATP-regenerating system. However, this is unlikely because Coomassie staining indicated that creatine phosphokinase is not detectably modified during the course of the reaction, whereas about 50% of the E1 in the reaction mixture is converted into a discrete ubiquitinated form (Fig. 3B). Analysis of this E1 modification using GST-ubiquitin suggested that E1 is diubiquitinated under these conditions (data not shown).
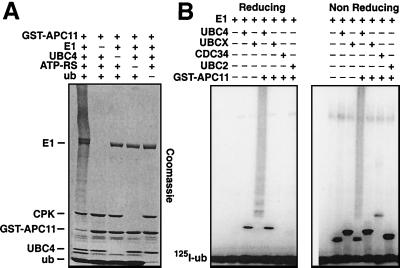
Omission of either E1, UBC4, the ATP-regenerating system, or ubiquitin from GST-APC11 containing reaction mixtures showed that the formation of multiubiquitin chains depended strictly on all of the above-listed components (Fig. 4A). This reaction is therefore attributable to a genuine E1- and E2-dependent ubiquitination reaction. APC-dependent ubiquitination reactions are supported by UBC4 and UBCx but not by other E2 enzymes (27, 32, 33). Likewise, we found that reactions occurring in the presence of GST-APC11 are not supported by UBC2 or CDC34, although these enzymes were active as judged by their ability to form thioesters with ubiquitin (Fig. 4B). Unexpectedly, UBCx also was unable to support GST-APC11-dependent ubiquitination reactions (Fig. 4B).
Figure 4.
Ubiquitination reactions occurring in the presence of APC11 depend on E1, UBC4, ATP, and ubiquitin. (A) Coomassie-stained gel of reactions containing (+) or missing (−) the indicated components. All reactions were incubated for 30 min at 37°C, and missing components were substituted with buffer. (B) Autoradiogram of reactions separated under reducing (Left) or nonreducing (Right) conditions. All reactions were performed in the presence of E1, ATP, 125I-ubiquitin, and with either UBC4, UBCx, CDC34, UBC2, GST-APC11, or combinations of these. Reactions were incubated for 5 min at room temperature, were stopped by addition of SDS/sample buffer with or without DTT, and were analyzed by SDS/PAGE.
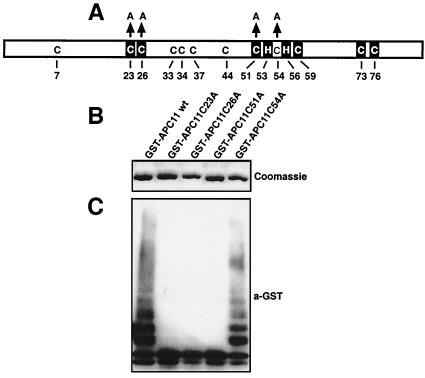
Based on analogy with RING fingers whose atomic structure is known (34), the sequence of APC11 is predicted to coordinate two atoms of zinc. We found that ubiquitination reactions did not occur in the presence of APC11 mutants in which conserved cysteine residues of the RING-H2 finger motif are changed to alanine, whereas mutation of a cysteine residue outside the RING-H2 finger consensus had no effect (Fig. 5). The integrity of the RING-H2 finger therefore appears to be required for APC11 to support the formation of polyubiquitin chains.
Figure 5.
The formation of multiubiquitin chains depends on the integrity of the APC11 RING-H2 finger. (A) Schematic representation of APC11. Cysteines (C) and two histidine residues (H) are indicated. Boxed residues represent the predicted Zn coordinating amino acids. The cysteine residues that were changed to alanine (A) in this study are indicated by arrows. (B) Bacterial lysates containing GST-APC11 wild type and the indicated GST-APC11 mutants were immobilized on glutathione-beads, were washed, and were eluted with SDS/sample buffer. Shown is a 12% polyacrylamide gel stained with Coomassie blue. (C) Identical amounts of bacterial lysates as in B were bound to glutathione beads, were washed, and were incubated with a reaction mixture as in Fig. 2B for 30 min at 37°C. Proteins were visualized by immunodecoration with anti-GST antibodies followed by enhanced chemiluminescence.
To test whether bona fide substrates of the APC can be ubiquitinated by APC11 and UBC4, we added purified myc-tagged human securin to APC11 reaction mixtures. Securin was rapidly multiubiquitinated in reactions containing GST-APC11, whereas no ubiquitination was observed in the presence of other APC subunits (Fig. 6A). APC11 could also ubiquitinate cyclin B (data not shown) but not MBP and only very poorly GST-p53 (Fig. 6B). A D box mutant of securin was ubiquitinated similarly well as wild-type securin in the presence of GST-APC11, whereas holo-APC bound to CDH1 showed decreased activity toward the D box mutant (Fig. 6B).
Figure 6.
GST-APC11 mediates the ubiquitination of the APC substrate securin. (A) Time course of reactions containing either purified GST-APC10 (Left) or GST-APC11 (Right), E1 and UBC4, an ATP regenerating system, ubiquitin and securin-myc-His. At the indicated time points, reactions were stopped by addition of SDS/sample buffer. Separation on 15 + 8% SDS/PAGE was followed by Western blotting and immunodecoration with an anti-myc antibody (9E10). (B) Purified GST-APC11 (Left) or holo-APC plus CDH1 (Right) were incubated with E1, UBC4, ATP, and ubiquitin as in A for 0 and 30 min at 37°C in the presence of either 20 ng securin-myc-His wild type, 20 ng of securin-myc-His D box mutant, 50 ng MBP, or 50 ng of GST-p53. After separation on SDS/PAGE and transfer to poly(vinylidene difluoride) membranes, proteins were detected by using anti-myc (9E10) antibodies (securin), anti-MBP antibodies, or anti p53 antibodies.
Discussion
Our identification of two low molecular weight proteins in the human APC as orthologs of yeast Apc11 and Cdc26/Hcn1 increases the number of known vertebrate APC subunits to 11. Surprisingly, our work shows that, of these, only APC11 is required to allow the synthesis of multiubiquitin chains in the presence of E1 and UBC4, although APC11 represents only about 1% of the total APC mass. Because APC11 contains a RING-H2 finger motif (14) whose integrity is required for APC11 activity (this work), our results support the recent notion that many if not all RING-H2 finger proteins may function as ubiquitin-protein ligases or subunits of such enzymes (22). It has recently been proposed that dimers of RING-H2 finger and cullin subunits represent the minimal catalytic unit of cullin containing E3 complexes (10, 11). Our results suggest that a RING-H2 finger protein alone can provide a minimal E3 activity, at least in the case of the APC, similar to what has been observed for MDM2 (18, 35–37). Like MDM2, APC11 can assemble ubiquitin chains on itself and on substrate proteins. In addition, our results suggest that chains not anchored to any substrate protein are generated in the presence of APC11. These results suggest an important role for APC11 in the catalysis of ubiquitination reactions. Based on these results and on the earlier observation that the budding yeast APC11 gene is essential for the in vivo function of the APC (14), we propose that APC11 also fulfills a catalytic role within the holo-APC.
To further characterize this role, we compared several aspects of APC11-mediated reactions with reactions catalyzed by holo-APC. Hallmarks of reactions mediated by holo-APC are that ubiquitination is strongly activated by either CDC20 or CDH1, that only substrates bearing a D or a KEN box are efficiently ubiquitinated, and that two distinct E2 enzymes can collaborate with the APC (1–4). In contrast, no stimulation was seen when CDH1 was added to APC11-dependent reactions (data not shown), indicating that additional APC subunits are required to transduce the activating effect of this protein to APC11. Because there are at least 10 other subunits in the APC core besides the small APC11 subunit, and because CDC20 and CDH1 appear to confer substrate specificity to holo-APC (reviewed in ref. 4), we did also not expect that APC11 would display substrate specificity. Consistently, we found that APC11 was able to ubiquitinate wild-type and D box mutants of APC substrates similarly well. However, APC11 showed a preference for substrates of holo-APC, such as securin and cyclin B, whereas other proteins were either ubiquitinated only poorly or not at all. We further observed that neither ROC1 nor ROC2 were able to efficiently ubiquitinate APC substrates (data not shown), suggesting that some substrate specificity already exists at the level of RING-H2 finger proteins.
Unexpectedly, we noticed that only UBC4 was able to support APC11-dependent ubiquitination reactions, although holo-APC is equally well supported by UBC4 and UBCx, at least in vitro (27, 33). It is unknown which of these E2s collaborates with the APC in vivo, but the observation that neither UBC4 nor UBCx orthologs are essential for viability in budding yeast is consistent with the possibility that multiple E2s function in the APC pathway (38, 39). Remarkably, the majority of ubiquitination reactions mediated by RING-H2 fingers in vitro depends on members of the UBC4/5 family (20, 22, 35, 37). Only human ROC1 has been shown to mediate the formation of ubiquitin chains in the presence of CDC34 (10), and in yeast Rbx1/Hrt1/Roc1 has been found to stimulate Cdc34 autoubiquitination (9, 11), but both reactions also required the presence of the cullin CUL1 or Cdc53. It is therefore conceivable that additional APC subunits such as APC2 have to bind to APC11 to allow a functional interaction with UBCx.
What could be the role of APC11 in the catalysis of ubiquitination reactions? The answer to this question is not known, but several possibilities can be envisioned. First, the finding that the integrity of the RING finger is essential for the ubiquitination activity of APC11 and of several other RING finger proteins raises the interesting possibility that zinc could directly participate in the catalysis of ubiquitination reactions, as it does in various other reactions mediated by metalloenzymes. However, it has been proposed that RING finger related proteins, so called U-box proteins, can also support ubiquitination reactions without being able to coordinate zinc (40), suggesting that zinc has a structural rather than a catalytic role. Second, the observation that many RING fingers physically interact with E2 enzymes suggests that the RING finger may simply represent an E2 binding domain (9, 11, 19, 20, 22, 41, 42). However, a recent analysis of Ubr1 has shown that the RING-H2 finger of this E3 is required for activity but not for binding of Ubc2 (16), and Lorick et al. (22) found that E2 binding is not sufficient for RING finger proteins to support ubiquitination reactions. RING finger domains must therefore have other roles in ubiquitination reactions than providing binding sites for E2s. Because RING fingers do not seem to form ubiquitin thioesters themselves (ref. 11; our unpublished results), they may instead function as allosteric activators of E2s. Further mechanistic studies will be required to test this hypothesis.
Acknowledgments
We are grateful to F. Dupeux and K. Kashofer for help with the recombinant protein expression, to A. Schleiffer for sequence alignments, to I. Waizenegger for mutating securin, to E. Kramer for the CDH1 baculovirus, to B. Koch for GST-p53, to G. Schaffner for DNA-sequencing, to M. Glotzer for comments on the manuscript, to R. Schroeder for discussions, and to K. Wilgenbus for support. This work was supported by a grant from the Austrian Industrial Research Promotion Fund to J.-M.P.
Abbreviations
- APC
anaphase-promoting complex
- D box
destruction box
- MBP
maltose binding protein
- GST
glutathione S-transferase
- EST
expressed sequence tag
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Morgan D O. Nat Cell Biol. 1999;1:E47–E53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- 2.Zachariae W, Nasmyth K. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- 3.Pfleger C M, Kirschner M W. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 4.Peters J-M. Exp Cell Res. 1999;248:339–349. doi: 10.1006/excr.1999.4443. [DOI] [PubMed] [Google Scholar]
- 5.Gieffers C, Schleiffer A, Peters J M. Protoplasma. 2000;211:20–28. [Google Scholar]
- 6.Deshaies R J. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 7.Tyers M, Jorgensen P. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 8.Ohta T, Michel J J, Schottelius A J, Xiong Y. Mol Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 9.Skowyra D, Koepp D M, Kamura T, Conrad M N, Conaway R C, Conaway J W, Elledge S J, Harper J W. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 10.Tan P, Fuchs S Y, Chen A, Wu K, Gomez C, Ronai Z, Pan Z Q. Mol Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 11.Seol J H, Feldman R M, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies R J. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamura T, Koepp D M, Conrad M N, Skowyra D, Moreland R J, Iliopoulos O, Lane W S, Kaelin W G, Jr, Elledge S J, Conaway R C, et al. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 13.Borden K L, Freemont P S. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 14.Zachariae W, Shevchenko A, Andrews P D, Ciosk R, Galova M, Stark M J, Mann M, Nasmyth K. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Peters J M, King R W, Page A M, Hieter P, Kirschner M W. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- 16.Xie Y, Varshavsky A. EMBO J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda R, Yasuda H. Oncogene. 2000;19:1473–1476. doi: 10.1038/sj.onc.1203464. [DOI] [PubMed] [Google Scholar]
- 18.Fang S, Jensen J P, Ludwig R L, Vousden K H, Weissman A M. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 19.Yokouchi M, Kondo T, Houghton A, Bartkiewicz M, Horne W C, Zhang H, Yoshimura A, Baron R. J Biol Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- 20.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 21.Levkowitz G, Waterman H, Ettenberg S A, Katz M, Tsygankov A Y, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 22.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevchenko A, Jensen O N, Podtelejnikov A V, Sagliocco F, Wilm M, Vorm O, Mortensen P, Boucherie H, Mann M. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossberger R, Gieffers C, Zachariae W, Podtelejnikov A V, Schleiffer A, Nasmyth K, Mann M, Peters J M. J Biol Chem. 1999;274:14500–14507. doi: 10.1074/jbc.274.20.14500. [DOI] [PubMed] [Google Scholar]
- 25.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Nature (London) 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 26.Wilm M, Mann M. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, King R W, Peters J M, Kirschner M W. Curr Biol. 1996;6:455–466. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- 28.Kramer E R, Scheuringer N, Podtelejnikov A V, Mann M, Peters J-M. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gieffers C, Peters B H, Kramer E R, Dotti C G, Peters J M. Proc Natl Acad Sci USA. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang L H, Murray A W. Mol Biol Cell. 1997;8:1877–1887. doi: 10.1091/mbc.8.10.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada H, Kumada K, Yanagida M. J Cell Sci. 1997;110:1793–1804. doi: 10.1242/jcs.110.15.1793. [DOI] [PubMed] [Google Scholar]
- 32.Aristarkhov A, Eytan E, Moghe A, Admon A, Hershko A, Ruderman J V. Proc Natl Acad Sci USA. 1996;93:4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King R W, Peters J M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 34.Borden K L, Lally J M, Martin S R, O'Reilly N J, Solomon E, Freemont P S. Proc Natl Acad Sci USA. 1996;93:1601–1606. doi: 10.1073/pnas.93.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda R, Tanaka H, Yasuda H. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 36.Honda R, Yasuda H. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez M S, Desterro J M, Lain S, Midgley C A, Lane D P, Hay R T. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seufert W, Jentsch S. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Townsley F M, Ruderman J V. Yeast. 1998;14:747–757. doi: 10.1002/(SICI)1097-0061(19980615)14:8<747::AID-YEA271>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 40.Aravind L, Koonin E V. Curr Biol. 2000;10:R132–R134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 41.Bailly V, Lauder S, Prakash S, Prakash L. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 42.Moynihan T P, Ardley H C, Nuber U, Rose S A, Jones P F, Markham A F, Scheffner M, Robinson P A. J Biol Chem. 1999;274:30963–30968. doi: 10.1074/jbc.274.43.30963. [DOI] [PubMed] [Google Scholar]