Abstract
An approach for purifying nuclear proteins that bind directly to the hyperphosphorylated C-terminal repeat domain (CTD) of RNA polymerase II was developed and used to identify one human phosphoCTD-associating protein as CA150. CA150 is a nuclear factor implicated in transcription elongation. Because the hyperphosphorylated CTD is a feature of actively transcribing RNA polymerase II (Pol II), phosphoCTD (PCTD) binding places CA150 in a location appropriate for performing a role in transcription elongation-related events. Several recombinant segments of CA150 bound the PCTD. Predominant binding is mediated by the portion of CA150 containing six FF domains, compact modules of previously unknown function. In fact, small recombinant proteins containing the fifth FF domain bound the PCTD. PCTD binding is the first specific function assigned to an FF domain. As FF domains are found in a variety of nuclear proteins, it is likely that some of these proteins are also PCTD-associating proteins. Thus FF domains appear to be compact protein-interaction modules that, like WW domains, can be evolutionarily shuffled to organize nuclear function.
The C-terminal repeat domain (CTD) of the largest subunit of RNA polymerase II is an unusual protein moiety that mediates critical interactions between the core transcription machinery and other nuclear factors. The mammalian CTD is a conserved Pol II-specific domain consisting of 52 repeats of the consensus sequence YSPTSPS (1). In its hypophosphorylated form, the CTD interacts with several factors as a central organizer of the “holoenzyme” involved in preinitiation complex formation and transcription initiation (2–5). Hyperphosphorylation of the CTD accompanies the transition into active elongation mode (6–9) and heralds dramatic changes in the properties of the transcription complex. Two alterations notable in the current context are an increase in Pol II's elongation competence (e.g., 10, 11–13) and a dramatic change in CTD-associated factors (14).
Our current knowledge of factors that associate with the CTD is fragmentary, and in particular proteins that bind the CTD or phosphoCTD (PCTD) directly have been identified in only a few instances. One example is capping enzyme from yeast and human cells, which binds to the phospho- but not the nonPCTD (15–18); incidentally, this discrimination can explain how capping enzymes are targeted specifically to elongating RNA polymerase II. Other examples of demonstrated CTD-binding proteins are the cleavage/polyadenylation factors CstF and CPSF (19). In the case of CstF, binding appears to be predominantly through the 50-kDa subunit. We recently demonstrated that the unusual peptidyl prolyl isomerase, ESS1/Pin1, binds tightly to the PCTD via its WW domain (20). This binding could facilitate isomerization of phosphoserine (PSer)–Pro bonds in the CTD by the peptidyl prolyl cis/trans isomerase domain, altering the conformation of the CTD repeats and modulating the binding of factors to the PCTD.
A few additional proteins that bind the CTD directly have been identified by yeast two-hybrid screens. Corden and colleagues uncovered two classes of mammalian CTD-binding proteins that resemble splicing components (21), and one of these has a yeast homolog identified through its effects on pre-mRNA metabolism (22, 23). Also using a yeast two-hybrid approach, Schaeffner and colleagues identified several interacting proteins (24, 25).
This paper describes a procedure for detecting and purifying HeLa PCTD-associating proteins (PCAPs) by using an assay exploiting radioactively labeled recombinant PCTD and interaction blot analysis (Far Western). The first human PCAP we purified and identified by using this procedure was CA150. CA150 was discovered as a component of a fraction required for HIV-1 Tat activation (26). Overexpression of CA150 leads to promoter-specific inhibition of elongation (27), and CA150 has been found associated with elongation factors Tat-SF1 and P-TEFb (A.C.G. and M.A.G.-B., unpublished results). The CA150 sequence contains several known or potential protein interaction motifs, including WW domains and FF motifs [putative protein–protein interaction domains named for two conserved phenylalanine (F) residues (28)]. Unexpectedly, we found that the principal PCTD-interacting portion of CA150 is the FF domain-containing segment. The results described here uncover a physical connection between the conserved nuclear factor CA150 and RNA polymerase II, providing a basis for attempts to understand CA150 function. More generally, the data reveal that a single FF domain can display PCTD binding and suggest a function for these recently described domains.
Materials and Methods
Suppliers.
MacroPrep High S and MacroPrep High Q resins were from Bio-Rad. O-phospho-l-serine, O-phospho-l-serine agarose, and reduced glutathione were from Sigma. Hybond-C Extra and Glutathione Sepharose 4B were from Amersham Pharmacia Biotech. Primers were from GIBCO/BRL and the Duke University core DNA facility. pGEX-2TK was from Pharmacia. 32P-ATP was from ICN. Centricons were from Amicon.
Extract (0.5 M) of HeLa Nuclear Proteins.
HeLa nuclei were prepared and extracted with 0.15 M NaCl as described (29). Residual pellet (25 ml) was homogenized on ice in a final volume of 200 ml of 0.5 M NaCl in HGE (25 mM Hepes, pH 7.6/15% glycerol/0.1 mM EDTA) + PMSF (1:1,000 dilution of saturated solution in isopropanol) + 10 mM NaF. The homogenate was centrifuged at 149,000 × g for 60 min at 4°C. The supernatant was termed the 0.5 M NaCl extract of the nuclear pellet.
High S Column of 0.5 M NaCl Extract of Nuclear Pellet.
All steps were at 4°C. An aliquot [140 mg (33 ml)] of the 0.5 M NaCl extract of the nuclear pellet was diluted with HGE + PMSF + 10 mM NaF to a final volume of 165 ml. The sample was centrifuged at 20,000 × g for 20 min to pellet precipitated proteins. The supernatant was applied to a 5-ml High S resin equilibrated in 0.1 M NaCl in HGE. The column was washed with 0.1 M NaCl in HGE, and proteins were then eluted with 3 × 5-ml steps of each of the following NaCl concentrations in HGE: 0.25 M; 0.5 M; 0.75 M; 1 M; and 2 M. Samples were analyzed by SDS/PAGE with Coomassie staining and Far Western blotting.
Purification and Identification of PCAP150.
Five milliliters of the 0.5 M NaCl High S eluate was diluted to 0.0165 M with HGE and applied to a 0.5 ml PSer column equilibrated in 0.0165 M NaCl, HGE. The column was eluted, stepwise, with 3 × 0.5 ml of 0.01 M, 0.05 M, 0.1 M, 0.2 M, 0.4 M, 0.6 M, and 1 M PSer. Samples were analyzed on duplicate 4–15% SDS gels; one was Coomassie stained and one was Far Western blotted with PCTD. A prominent 150 kDa PCAP was present in the 0.1 M PSer step fraction. To resolve this protein for identification, the 0.1 M PSer fraction was diluted to 0.075 M in HGE and applied to a 0.5-ml Q column equilibrated with 0.075 M NaCl, HGE. Proteins were eluted stepwise with 3 × 0.5 ml of 0.075 M, 0.1 M, 0.2 M, 0.3 M, 0.4 M, and 1 M NaCl. Fractions were gel analyzed as before. The PCAP of 150 kDa was found in the 0.2 M NaCl step fraction and was sufficiently pure for identification. The protein band was excised and subjected to in-gel trypsinization and MALDI mass spectrometry analysis (John Leszyk, Laboratory for Protein Microsequencing and Proteomic Mass Spectrometry, University of Massachusetts Medical School, Worcester Foundation Campus, Shrewsbury, MA). The masses of the tryptic peptides revealed the 150 kDa PCAP to be CA150.
Far Western Analysis.
Proteins were separated on duplicate 4–15% precast SDS gels (Bio-Rad) and either stained with Coomassie blue or electroblotted to nitrocellulose. Membranes were then incubated overnight at 4°C in PBS/0.2% Tween20/3% dry milk/10 mM NaF/PMSF/2 mM DTT. Blots were probed in the above solution with 32P-βgalyCTD (20, 30) for 4 h at 4°C. Membranes were then washed in PBS/0.2% Tween20 for 4 × 10 min at 4°C. Membranes were then exposed overnight to film or PhosphorImager screen.
Construction and Expression of CA150 Plasmids.
PCR-generated fragments of CA150 were inserted into pGEX-2TK. The 5′ primers used were GGCCAGATCTGACGATAATAAAGACATTGAC; GGCCAGATCTGAAGAACGCAGGGAAAAGAAAAAT; GGCCAGATCTTCGAAGACCAGAGGTGAGAAG; GGCCAGATCTCGAGAAAGGGAGGTTCAAAAG; GGCCAGATCTGCACTTACCAAAAAAAAGAGAGAG; GGCCAGATCTGAAGAATATATCAGAGACAAA; GGCCAGATCTGAATCGGATCAGCACCTGAAA. The 3′ primers used were: GGCCGAATTCTTATTTTGTTGATCGTCTCGT; GGCCGAATTCTTACTCTGCCCTGGTCTTTACATA; GGCCGAATTCTTAATCTTCTTTCTCTTTCTTCCT; GGCCGAATTCTTATTCTCGAAGGCTTGCCTCAAT; GGCCGAATTCTTATTCAATGTGTTCATTAAAAAG; GGCCGAATTCTTAAAATTCTCTTTGTTTTTTCCT; GGCCGAATTCTTATTTGGTCTCTTTCAAAAGCGT; GGCCGAATTCTTACACAATCAGTTTACGCCTCTC. By using these primers and pEFBOST7CA150 (27) as the template, constructs described in Figs. 2 and 4 and Table 1 were amplified by using Pfu DNA polymerase according to the manufacturer's instructions with the following cycling conditions: the DNA was melted at 94°C for 5 min, cycled 35 times through melting at 94°C for 30 seconds, annealing at 50°C for 1 min, and extending at 72°C for 3.5 min; followed by extension at 72°C for 10 min. The primers were constructed with a BglII restriction site in the 5′ primer and a EcoRI restriction site in the 3′ primer. PCR products were separated by gel electrophoresis, excised, and purified by using QIAEX II Gel Extraction Kit (Qiagen, Chatsworth, CA), and then digested with BglII and EcoRI. These products were then ligated into gel-purified pGEX-2TK digested with BamHI and EcoRI. The constructs were verified by DNA sequencing. Plasmids were transformed into DH5α for selection and maintenance and then into BLR21/DE3 for expression. The fusion proteins were purified by batch adsorption to Glutathione Sepharose 4B according to manufacturer's instructions.
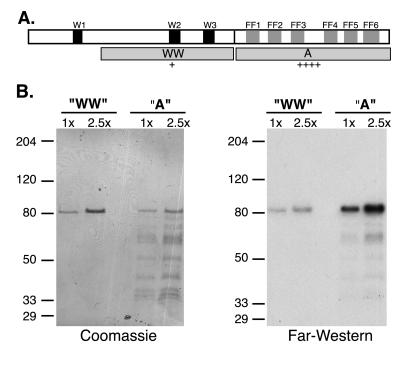
Figure 2.
Domains and PCTD binding of CA150. (A) Upper open box represents the 1,098-aa CA150 protein, with WW domains shaded black and FF motifs, numbered sequentially, shaded gray. Gray boxes in second row labeled WW and A represent segments of protein expressed in bacteria and tested for PCTD binding in B. Construct names and amino acid numbers: WW (); A (631–1098). Relative binding affinities to the PCTD, as judged by eye from Far Western analyses, are indicated below each construct. (B) GST fusion proteins carrying segments WW and A were run in two different amounts (1× = ≈100 ng for WW and ≈50 ng for A) on duplicate SDS gels that were either stained (Coomassie) or blotted to nitrocellulose and probed with 32P-labeled PCTD (Far Western). Prestained marker positions are indicated at left (apparent Mr). The amino terminal-most segment, containing WW1, was not successfully expressed in bacteria.
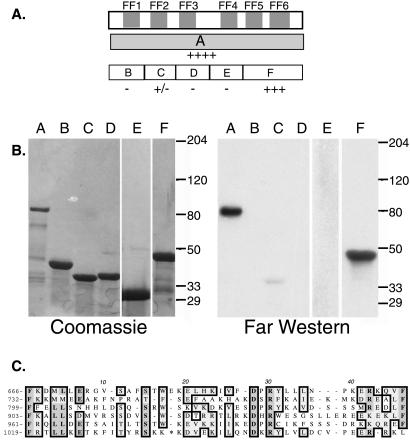
Figure 4.
CA150 fragments bind PCTD by Far Western. (A) The upper box represents the C-terminal region of CA150 with the FF domains shaded gray. The gray box on the next line represents construct A. The open boxes on the third line represent constructs B–F. Construct names and amino acid numbers: A (631–1098); B (); C (); D (); E (); F (952–1098). Relative binding affinities to PCTD are indicated below each construct. (B) Constructs A–F were separated on duplicate 4–15% SDS gels. One gel was stained with Coomassie, whereas the duplicate was transferred to Nitrocellulose and probed with 32PCTD (Far Western). (C) Alignment of the FF motifs of CA150. The starting amino acid of each motif is indicated at left. Homologous amino acids are boxed and * in FF6 = LIQESDQHL.
Table 1.
PhosphoCTD binding to FF5
| Construct | Complete FF domain contained | P-CTD binding |
|---|---|---|
| F (952–1098)* | FF5, FF6 | +++ |
| I (952–1004) | FF5 | ++ |
| J (1005–1027) | − | − |
| K (1039–1072) | − | − |
| L (1005–1072) | FF6 | − |
| M (952–1027) | FF5 | ++ |
Amino acid numbers.
In-Solution Binding Assay.
Five micrograms each of proteins M and J were desalted by using Centricon filters. A 50% slurry of the glutathione beads was prepared according to manufacturer's instructions. The desalted protein was added to 20 μl 50% glutathione bead slurry, and PBS was added to a final volume of 100 μl. This solution was incubated with rocking at 4°C for 30 min, the sample centrifuged at 500 × g for 5 min, and the supernatant removed. Fifty microliters PCTD solution (≈5 μg protein/3,000 cpm) in PBS was added to the beads, and PBS was added to a total volume of 100 μl. After incubation with rocking at 4°C for 30 min, the beads were pelleted by centrifugation at 500 × g for 5 min. Ninety microliters of supernatant was collected from each sample. For washing, the beads were resuspended in 90 μl PBS for 5 min, incubated at 4°C with rocking, and pelleted as above. The wash supernatant was collected. Eighty microliters of PBS was added to the pelleted beads to ease handling and to give a final volume of 90 μl, as for the supernatant and wash samples. Twenty percent of each sample was boiled with Laemmli sample buffer containing SDS for 3 min. The samples were electrophoresed on a 6% SDS polyacrylamide gel, fixed in Coomassie stain, and exposed to a PhosphorImager screen.
Immunoprecipitations from HeLa Nuclear Extract.
CA150 antibodies were raised in rabbits against a GST fusion protein containing the C-terminal 772 amino acids of CA150. Anti-GST and anti-CA150 antibodies were antigen affinity purified from the same rabbit polyclonal sera (31). Fifty microliters of HeLa cell nuclear extract [15 mg/ml (32)] was diluted to 200 μl final volume with IP buffer (20 mM Hepes, pH 7.9/150 mM KCl/20% glycerol/1 mM DTT/1% Triton-X100/0.5% Nonidet P-40/0.2 mM EDTA). Fifteen micrograms of antigen-affinity purified antibody was added, followed by incubation with end-over-end rotation at 4°C for 4 h. Immune complexes were collected with 500–750 μg magnetic protein A beads (BioMag Protein A beads, PerSeptive Biosystems, Framingham, MA) and a magnet. The pellets were washed 4 times with 1 ml of IP buffer by rotating 5 min at 4°C. The pellets and supernatants were then electrophoresed on 7.5% SDS gels and analyzed by immunoblotting with enhanced chemiluminesence by using either anti-CA150, mAb 8WG16 [which recognizes the largest subunit of Pol II (Berkeley Antibody, Richmond, CA)], or anti-TBP (Promega). The equivalent of 5 μg IgG of each pellet was loaded per lane, and equal volumes of supernatants were loaded.
Expression and Purification of Epitope-Tagged CA150 Proteins in Vivo.
The plasmid pEFBOST7CA150 (27) expressed full length (amino acids 1–1098) or the C-terminal half of CA150 (amino acids 590-1098) with expression driven by the polypeptide chain elongation factor 1α promoter. Analysis of protein expression was carried out in whole-cell lysates from 293T cells transiently expressing the CA150 proteins with an N-terminal epitope tag from T7 phage capsid. Cell extracts for immunoprecipitation experiments were made by incubating cells on ice for 30 min in IP buffer containing PMSF at 50 μg/ml. Cell debris was removed by centrifugation at 10,000 × g for 5 minutes at 4°C in a microcentrifuge. Fifty microliters bed volume of monoclonal anti-T7 antibody covalently crosslinked to agarose beads (Novagen) were used to immunoprecipitate the T7 epitope tagged CA150 proteins. These pellets were washed 4 times with 1 ml of IP buffer by rotating 5 min per wash at 4°C. The immunoprecipitates were subjected to 10% SDS/PAGE and analyzed by immunoblotting with anti-T7 monoclonal antibody and the 8WG16 monoclonal antibody. Proteins were detected by using enhanced chemiluminescence.
Results
We began a biochemical search in extracts of yeast and human cells for proteins that bind directly to the hyperphosphorylated CTD, on the premise that we currently are aware of only a fraction of nuclear factors with functionally important PCTD interactions. To generate PCTD probes, we used yeast CTD kinase I to efficiently hyperphosphorylate CTD-containing fusion proteins (30, 33). The first yeast phosphoCTD-associating protein (PCAP) we found was ESS1/Pin1, as mentioned. We now report the initial results of our search for PCAPs in extracts from human cell nuclei.
Purifying PCAPs from HeLa Nuclear Pellet.
As a source potentially enriched for PCAPs, we chose a nuclear pellet fraction from HeLa cells. This fraction is the centrifugally pelleted material from nuclei extracted with physiological levels of salt (see Methods), and it should contain the majority of template-engaged transcriptionally elongating Pol II in the cells (34). Although template-engaged Pol II is stably associated with the chromatin in the pellet, we expected that proteins in association with the hyperphosphorylated CTD of the elongating Pol II should be variably extractable by increasing salt concentrations. We therefore treated the nuclear pellet with buffers containing increasing levels of NaCl and tested the resulting fractions for PCAPs by interaction (Far Western) blotting, by using 32P-PCTD as a probe.
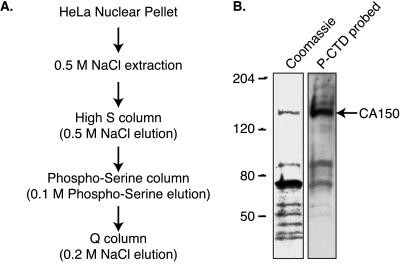
Indeed, the nuclear pellet proved to be a rich source of PCAPs. The majority were released from the pellet by a 0.5 M salt wash, and we therefore subjected this fraction to the purification scheme outlined in Fig. 1A. Proteins were first fractionated by ion-exchange chromatography by using a High-S resin, and PCAPs were detected by PCTD-binding in Far Western analysis. Because the majority of the PCAPs were found in the High-S 0.5 M step (unpublished data), we chose this fraction to apply to a PSer matrix. Salt-step elution of this material produced several PCAP-containing fractions. In particular, the 0.1 M PSer fraction contained a prominent band at 150 kDa that we purified by using a Q column. This 150-kDa protein (PCAP150) eluted in the 0.2 M salt step, and as seen in Fig. 1B, corresponded to a single Coomassie-stained band. The remainder of this paper describes studies of PCAP150. Note, however, that other PCAPs in the Q column fractions also corresponded to single Coomassie-stained bands (e.g., two flanking the 80 kDa marker in Fig. 1B). We have been able to identify several of these other PCAPs and to confirm their PCTD-binding capability by using recombinant proteins (S.M.C. and A.L.G., unpublished data). The approach therefore is an efficient method for identifying and purifying novel PCTD-associating proteins.
Figure 1.
Identification of CA150 as a PCTD associating protein. (A) To identify PCTD-binding proteins from HeLa cells, we purified HeLa nuclear proteins and assessed their PCTD-binding abilities by direct binding in a Far Western analysis. The flow chart indicates the steps in the purification of a PCAP of molecular weight 150,000. (B) The Coomassie-stained proteins and PCTD interaction of 0.2 M NaCl elution of Q column demonstrate that the protein of molecular weight 150,000 binds the PCTD. Mass spectrometric analysis revealed that this protein is CA150.
HeLa CA150 Is a PCAP with PCTD-Binding Domains in Both the N-Terminal and C-Terminal Segments.
We excised the 150-kDa band (Fig. 1B) and submitted it for identification by in-gel trypsinization and mass spectrometry. This analysis revealed it to be human CA150, a nuclear factor discovered by HIV Tat affinity chromatography and previously shown to affect transcription elongation in a promoter-specific fashion (see Introduction).
To identify the PCTD-interacting domain(s) of CA150, we tested the PCTD-binding properties of recombinant CA150 protein segments purified from Escherichia coli. In the Far Western interaction assay, the WW domain-containing portion of CA150 showed weak binding (“WW” in Fig. 2), whereas the C-terminal segment displayed strong binding (“A” in Fig. 2), even though more protein was loaded on the gel for construct WW (Coomassie). The segment of the protein represented by construct A contains the five FF motifs described by Bedford and Leder (28). We noticed a sixth apparent FF motif C-terminal to these sequences (Fig. 4, below).
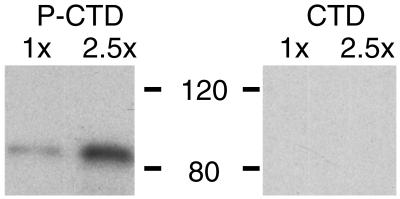
To check whether the binding displayed by fragment A depended on CTD phosphorylation, we tested in parallel its binding to a CTD probe and to a PCTD probe. As shown in Fig. 3, the binding observed in the Far Western assay is specific for the hyperphosphorylated CTD.
Figure 3.
The FF region binding is specific for the phosphorylated CTD. Construct A of Fig. 2 was run in duplicate in two different amounts on an SDS gel, and the proteins were electroblotted to nitrocellulose strips. One strip was reacted with hyperphosphorylated βgal-yCTD fusion protein (PCTD) and the other with nonphosphorylated βgal-yCTD fusion protein (CTD). Bound fusion protein was visualized by using anti-βgal antibodies and chemiluminescence.
Individual FF Repeats Bind the PCTD.
We sectioned the C-terminal half of CA150 into several pieces based on potential domain structure (Figs. 2 and 4) and tested PCTD-binding of each piece. The segment containing FF domain 5 plus the remaining C-terminal residues of the protein (construct F) bound the PCTD well, although somewhat less strongly than the entire C-terminal half of CA150 (Fig. 4B, compare lanes A and F), whereas three other FF-containing segments displayed no binding (Fig. 4B, lanes B, D, and E). A second segment, carrying FF 2, showed very weak but reproducible binding (Fig. 4B, lane C). These results demonstrate that a small terminal portion of the 150-kDA protein, consisting of only 147 amino acids, carries the major PCTD-binding activity of CA150, as judged by Far Western. Note that the binding by each fragment (F and C) was weaker than the binding by the segment carrying both of these (A), suggesting that CA150 carries multiple PCTD-interacting domains with different PCTD interaction properties.
To determine which part of terminal segment F carries PCTD-binding activity, we divided it into subfragments and expressed them in bacteria. The resulting fusion proteins were tested for PCTD binding by interaction blotting as before. As shown in Table 1, we found that two fragments carrying a complete FF5 (I and M) bound the PCTD probe, whereas a fragment carrying a partial FF5 (J) did not. Finally, a protein segment carrying only FF6 (L) did not demonstrate binding, nor did one carrying only part of FF6 (K). These results suggest that FF5 is mainly responsible for the PCTD binding displayed by the terminal segment F.
As an additional test of FF5 binding to the PCTD, we carried out in-solution experiments. Fusion constructs containing either segment M or segment J were purified and bound to agarose beads (Methods). The ability of the proteins on the beads to bind the PCTD was then tested by pull-down experiments. Segment M, carrying a complete FF domain 5, clearly bound the labeled PCTD (Fig. 5), whereas segment J, representing principally an interFF domain region, did not. These results, carried out with presumed native fusion proteins, recapitulate the results from Far Western analyses that used proteins that had been denatured in SDS and renatured on the blotting membrane. Thus, by two complementary methods, we find that FF5 binds the PCTD whereas neighboring segments do not.
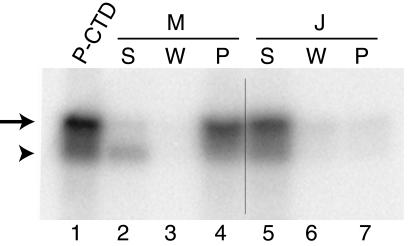
Figure 5.
FF5 (fragment M, amino acids 952-1027; see Table 1) binds the PCTD in solution. We bound the M (FF5) and the J (control, amino acids 1005–1027; Table 1) GST-fusion proteins to glutathione beads and then tested the ability of the bead-bound proteins to pull down 32PCTD (Methods). The 32PCTD in each sample was visualized by PhosphorImager. Lane 1 is the starting 32PCTD solution. Lanes 2–4 are the supernatant, wash, and pellet for sample M. Lanes 5–7 are the supernatant, wash, and pellet for sample J. Hyperphosphorylated CTD is indicated by the arrow and hypophosphorylated CTD is indicated by the arrowhead.
The phosphorylated CTD fusion protein used in the experiment of Fig. 5 contained some hypophosphorylated species (lower band in lane 1), and, compared with the hyperphosphorylated material, these species were poorly bound by construct M (compare lanes 2 and 4). This result is consistent with that of Fig. 3, which showed that CA150 does not bind the nonphosphorylated CTD. The relative enrichment for the hyperphosphorylated CTD in the bound fraction (Fig. 5, lane 4) shows that segment M binds PCTDs carrying multiple phosphates more effectively than those with few phosphates. A similar preference was seen for ESS1 (20). As for all PCAPs, it will be interesting to determine the specific binding determinants on the PCTD.
Coimmunoprecipitation of CA150 and RNA Polymerase II.
We sought to determine whether native CA150 is associated with Pol II in HeLa nuclear extracts by determining whether these proteins coimmunoprecipitate. CA150 and GST antibodies were affinity purified from rabbit polyclonal sera raised against a GST-CA150 fusion protein. The affinity-purified anti-CA150 IgGs recognize only CA150 in Western blots (not shown). The GST antibodies were used as a negative control for nonspecific interactions.
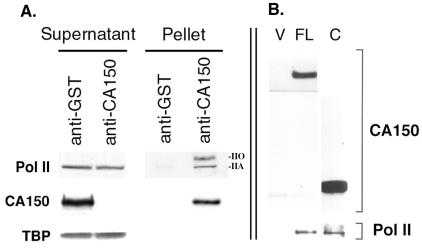
We used sufficient anti-CA150 to deplete CA150 from the nuclear extract, and we analyzed the resulting supernatants and pellets by Western blotting with antibodies against the Pol II large subunit, CA150, and TATA-binding protein (Fig. 6A). As seen in the “Pellet” anti-CA150 lane, both hyperphosphoryated and hypophosphorylated Pol II (II0 and IIA, respectively) coimmunoprecipitated with CA150. Because the nuclear extract contains little II0 relative to IIA (“Supernatant” lanes), this result indicates that CA150 selectively associated with Pol II0 in the nuclear extract, consistent with our finding that CA150 binds the PCTD. The TATA-binding protein (TBP) did not coprecipitate with CA150/Pol II, demonstrating the specificity of the immunoprecipitations.
Figure 6.
RNA polymerase II coimmunoprecipitates with CA150 from HeLa nuclear extracts. (A) CA150 was immunoprecipitated by using CA150 antibodies. GST antibodies were used as a negative control. Equal amounts of supernatants and pellets from the immunoprecipitations were separated by SDS/PAGE and subsequently analyzyed by immunoblotting with specific antibodies against RNA polymerase II (Pol II), CA150, and the TATA-binding protein (TBP). Both the hyperphosphorylated IIO form and the hypophosphorylated IIA form of Pol II largest subunit were detected in the anti-CA150 pellets. (B) The C-terminal half of CA150, containing the six FF repeats, is sufficient for binding to Pol II in vivo. Full length CA150 (FL) or the C-terminal half of CA150 (C) were expressed in 293T cells and immunoprecipitated via an amino-terminal T7 epitope tag. As a negative control, immunoprecipitates from 293T cells transfected with the expression vector alone (V) were also analyzed. The immunoprecipitates were separated by SDS/PAGE and immunoblotted with T7 tag antibodies, to detect CA150 proteins and with Pol II antibodies to detect subunits IIa/IIo (not resolved on this gel).
We also investigated the association of CA150 with Pol II in 293T cells by overexpressing epitope-tagged CA150. We performed immunoprecipitation experiments from human 293T cells that were transfected with either full length CA150 (amino acids 1–1098) or the FF-containing C-terminal half of CA150 (amino acids 590-1098). Both proteins had amino-terminal T7 epitope tags that allowed efficient immunoprecipitation of the proteins from whole cell lysates by using an anti-T7 monoclonal antibody. As a negative control, we used whole cell lysates from cells transfected with empty vector. Fig. 6B shows that both full length CA150 and its C-terminal half were able to coimmunoprecipitate Pol II. This result demonstrates that the C-terminal half of CA150, which contains the FF domains, is sufficient for association with Pol II.
Discussion
We have described a method for identifying human PCAPs and have used the method to discover that nuclear factor CA150 is a novel PCAP. This finding provides a basis for a physical connection between the core transcription machine, Pol II, and CA150, a protein implicated in transcription elongation (27). Further, although CA150 contains several PCTD-binding regions, its principal PCTD-binding activity is mediated by FF domains, modules of previously unknown function. Because the mechanism of CA150 action is incompletely understood, knowing its PCTD-binding capacity and the domains responsible for this property is critical to elucidating its function(s).
PCTD binding is the first function assigned to an FF domain. FF domains therefore represent another example of protein interaction modules that coordinate discrete nuclear functions. We find that under our experimental regimen, FF domains nos. 2 and 5 bind the PCTD. It is possible that future work will uncover PCTD binding by other of CA150's FF domains, as we do not have independent evidence that the other domains, as we expressed them in bacteria, were capable of folding properly. Our results, however, clearly indicate that different FF domains display discrete properties. In this respect they are like another prominent family of protein interaction module, the WW domain, certain members of which have been shown to bind the PCTD [e.g., ESS1/Pin1 (20)] or the CTD [e.g., Rsp5 (35, 36)]. In fact, CA150 contains three WW domains in its N-terminal half. However, the PCTD binding of the CA150 WW domain region appears weaker than that of ESS1/Pin1. If binding by the CA150 N-terminal region is mediated by the WW domains, it appears quantitatively different from that of ESS1.
Our results lead us to suggest that other FF domain-containing proteins will bind the PCTD. In fact, FF domain segments of yeast splicing factor Prp40 (37–40) have been found to bind the PCTD (D. P. Morris & A.L.G., unpublished data). We therefore predict that the apparent ortholog of PRP40, mammalian FBP11, and the related FBP21 will be PCTD-associating proteins (38, 41). We further propose, of course, that CA150 counterparts identified in sequence databases for Drosophila melanogaster, Caenorhabditis elegans, zebrafish, and mouse will all be PCAPs, as their FF domains are well conserved.
The current findings for FF motifs, along with previous results for WW and other phospoCTD-interacting domains, illustrate an emerging principle that during evolution PCTD-interaction modules can be shuffled among proteins with distinct functions. This module-mediated targeting to the transcription elongation machinery furnishes a versatile means for evolving the organization of nuclear function. We therefore expect PCTD targeting to play multiple roles in coordinating RNA and DNA transactions in the nucleus. Identification of the PCTD-interacting domains in additional proteins will reveal the extent to which known PCTD-interaction modules mediate these interactions.
Acknowledgments
We thank Paul Modrich (Duke University Medical Center, Durham, NC) for generous gifts of nuclear pellet. This research was supported by a National Institutes of Health (NIH) grant to A.L.G. and an NIH grant to M.A.G.-B.
Abbreviations
- CTD
C-terminal repeat domain
- PCTD
hyperphosphorylated CTD (phosphoCTD)
- PCAP
PCTD-associating protein
- PSer
phosphoserine
- Pol II
RNA polymerase II
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160266597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160266597
References
- 1.Corden J L. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 2.Lu H, Flores O, Weinmann R, Reinberg D. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chesnut J D, Stephens J H, Dahmus M E. J Biol Chem. 1992;267:10500–10506. [PubMed] [Google Scholar]
- 4.Hampsey M, Reinberg D. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 5.Myer V E, Young R A. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 6.Cadena D L, Dahmus M E. J Biol Chem. 1987;262:12468–12474. [PubMed] [Google Scholar]
- 7.Weeks J R, Hardin S E, Shen J, Lee J M, Greenleaf A L. Genes Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien T, Hardin S, Greenleaf A L, Lis J T. Nature (London) 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 9.Greenblatt J. Curr Opin Cell Biol. 1997;9:310–319. doi: 10.1016/s0955-0674(97)80002-6. [DOI] [PubMed] [Google Scholar]
- 10.Marshall N F, Peng J, Xie Z, Price D H. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mancebo H S, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J M, Greenleaf A L. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- 14.Bentley D. Curr Opinion Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 15.Cho E J, Takagi T, Moore C R, Buratowski S. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Program A E, Shuman S, Bentley D L. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho E J, Rodriguez C R, Takagi T, Buratowski S. Genes Dev. 1998;12:3482–3487. doi: 10.1101/gad.12.22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho C K, Shuman S. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 19.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. Nature (London) 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 20.Morris D P, Phatnani H, Greenleaf A L. J Biol Chem. 1999;274:31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- 21.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinmetz E J, Brow D A. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steinmetz E J, Brow D A. Proc Natl Acad Sci USA. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourquin J P, Stagljar I, Meier P, Moosmann P, Silke J, Baechi T, Georgiev O, Schaffner W. Nucleic Acids Res. 1997;25:2055–2061. doi: 10.1093/nar/25.11.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanner S, Stagljar I, Georgiev O, Schaffner W, Bourquin J P. Biol Chem. 1997;378:565–571. doi: 10.1515/bchm.1997.378.6.565. [DOI] [PubMed] [Google Scholar]
- 26.Sune C, Hayashi T, Liu Y, Lane W S, Young R A, Garcia-Blanco M A. Mol Cell Biol. 1997;17:6029–6039. doi: 10.1128/mcb.17.10.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sune C, Garcia-Blanco M A. Mol Cell Biol. 1999;19:4719–4728. doi: 10.1128/mcb.19.7.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bedford M T, Leder P. Trends Biochem Sci. 1999;24:264–265. doi: 10.1016/s0968-0004(99)01417-6. [DOI] [PubMed] [Google Scholar]
- 29.Longley M J, Pierce A J, Modrich P. J Biol Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 30.Morris D P, Lee J M, Sterner D E, Brickey W J, Greenleaf A L. Methods Companion Methods Enzymol. 1997;12:264–275. doi: 10.1006/meth.1997.0478. [DOI] [PubMed] [Google Scholar]
- 31.Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. [Google Scholar]
- 32.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J M, Greenleaf A L. Proc Natl Acad Sci USA. 1989;86:3624–3628. doi: 10.1073/pnas.86.10.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wuarin J, Schibler U. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huibregtse J M, Yang J C, Beaudenon S L. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang A, Cheang S, Espanel X, Sudol M. J Biol Chem. 2000;275:20562–20571. doi: 10.1074/jbc.M002479200. [DOI] [PubMed] [Google Scholar]
- 37.Abovich N, Rosbash M. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 38.Bedford M T, Reed R, Leder P. Proc Natl Acad Sci USA. 1998;95:10602–10607. doi: 10.1073/pnas.95.18.10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kao H Y, Siliciano P G. Mol Cell Biol. 1996;16:960–967. doi: 10.1128/mcb.16.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neubauer G, Gottschalk A, Fabrizio P, Seraphin B, Luhrmann R, Mann M. Proc Natl Acad Sci USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bedford M T, Chan D C, Leder P. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]