Abstract
Here, we represent protein structures as residue interacting networks, which are assumed to involve a permanent flow of information between amino acids. By removal of nodes from the protein network, we identify fold centrally conserved residues, which are crucial for sustaining the shortest pathways and thus play key roles in long-range interactions. Analysis of seven protein families (myoglobins, G-protein-coupled receptors, the trypsin class of serine proteases, hemoglobins, oligosaccharide phosphorylases, nuclear receptor ligand-binding domains and retroviral proteases) confirms that experimentally many of these residues are important for allosteric communication. The agreement between the centrally conserved residues, which are key in preserving short path lengths, and residues experimentally suggested to mediate signaling further illustrates that topology plays an important role in network communication. Protein folds have evolved under constraints imposed by function. To maintain function, protein structures need to be robust to mutational events. On the other hand, robustness is accompanied by an extreme sensitivity at some crucial sites. Thus, here we propose that centrally conserved residues, whose removal increases the characteristic path length in protein networks, may relate to the system fragility.
Keywords: allosteric communications, conserved interconnectivity determinants, long-range interactions, network, robustness
Introduction
Protein topology has been shown to play an important role in the determination of protein function and folding kinetics. The representation of protein structures as networks of interactions between amino acids has proven to be useful in a number of studies, such as protein folding (Vendruscolo et al, 2002), residue contribution to the protein–protein binding free energy in given complexes (del Sol and O'Meara, 2004) and prediction of functionally important residues in enzyme families (Amitai et al, 2004). It has further been shown that protein structures can be represented as graphs corresponding to small-world networks (Greene and Higman, 2003) describing complex systems such as cellular, metabolic and transcriptional regulatory processes (Ravasz et al, 2002), the nervous system of Caenorhabditis elegans (Achacoso and Yamamoto, 1992) and protein domain networks in proteomes of different organisms (Wuchty, 2001). These networks are usually highly clustered with a few links connecting any pair of nodes (Watts and Strogatz, 1998). Consequently, there are relatively few residues located at these short cuts, serving as interconnections between all residues in the structure.
A key feature of many complex systems is their robustness. Robustness is the system's ability to keep functioning despite perturbations. On the other hand, robustness is coupled with fragility toward non-trivial rearrangements of the connections between the system's internal parts (Jeong et al, 2001). Protein structures are no exception. They have evolved toward a robust design, tolerating mutations and environmental changes. At the same time, they are vulnerable to perturbations at key positions or to drastic changes in the environment (Taverna and Goldstein, 2002). Experimental results show that a significant number of single-site mutations have little effect on the protein function (Rennell et al, 1991). Further, these mutations may lead to an appearance of promiscuous functions (Aharoni et al, 2005). This robustness is expected to be reflected in the protein topology. Yet, if we think of protein structures as information processing networks, it would be reasonable to assume that mutations of amino acids crucial for network communications could impair function. The communicated information can be transmitted in a physical (or chemical) form. It is conceivable that residues that are presumed to receive and propagate the information should be central in the interaction network, lying on the shortest pathways between most residue pairs in the protein. The propagation of the information in protein structures is a poorly understood complex process. Yet, a number of theoretical results have suggested the crucial role of the central residues. Vendruscolo et al (2002) showed that a few highly connected amino acids act as a nucleation center for protein folding. Dokholyan et al (2002) supported this finding, showing that a weak participation of residues in the interaction network in pre- and post-transition states is usually associated with a weak impact on protein folding kinetics, and on the native state. More recently, del Sol and O'Meara (2004) observed a correlation between the most interconnected residues at protein–protein interfaces and residues that contribute the most to the binding free energy. Based on a large set of enzymes, Amitai et al (2004) have shown that active site residues tend to be highly central in the structure, suggesting that these positions are crucial for the transmission of information between the residues in the protein. Below, we address system robustness, focusing on identification of residues responsible for maintaining short communiction paths.
Allostery and network robustness
Allosteric communication is an example of propagation of information transmitting signals from one functional site to another. Although the conformational changes in protein structures associated with this process remain unknown, experimental methods, such as double mutant cycle analysis (Schreiber and Fersht, 1995), have provided some insight into this problem. Sequence-based evolutionary methods have been proposed to identify important residues for long-range communications (Kass and Horovitz, 2002). An interesting sequence-based statistical method has been recently introduced by Ranganathan and collaborators for estimating thermodynamic coupling between residues in different protein families (Lockless and Ranganathan, 1999; Süel et al, 2002; Hatley et al, 2003; Shulman et al, 2004). Our network model of protein structures resembles a robust communication system, where the removal of most of the nodes, with their corresponding edges, does not affect significantly the network's interconnectedness as described by the characteristic path length. However, when those residues making the most important contribution to generating the small-world character of the network are computationally removed (including their links), the interconnectedness is remarkably affected by a statistically significant increase in the characteristic path length (below, these residues are termed the network's ‘interconnectivity determinants' or ICDs). Interestingly, our results showed that random rewiring of the edges of the protein networks led to more homogeneous residue centrality distribution, showing that the communications are no longer maintained by just a few key residues. This indicates that these small-world networks have lapsed into randomness.
Allosteric regulation is a dynamic process, which implies equilibrium between the active and inactive conformational states (Volkman et al, 2001; Kern and Zuiderweg, 2003; Gunasekaran et al, 2004). To get an insight into allostery in terms of network communications, we compared the inactive and active conformations of hemoglobin and of the nitrogen regulatory protein C (NtrC). Our analysis showed that structural changes between the active and inactive conformations may lead to a rearrangement of the central residues in the two states. This underscores the fact that network communication is dynamic, with altered preferred routes and key residues in different conformational states. Alternate network communications in different regulatory states are advantageous, probably leading to higher efficiency and better control of the transmission of the information. As these key positions, which are crucial for maintaining the short paths, are centrally conserved in the protein fold (i.e., are a conserved topological characteristic of the fold rather than being conserved in sequence), it further suggests that it is not necessarily specific residue interactions that are important for regulation. Rather, it is the network characteristics, making the system less sensitive to mutations. In particular, this property of the multiplicity of pathways in the ensembles of different regulatory states confers robustness on the system.
The protein families
We carried out a detailed analysis of seven allosteric protein families (myoglobins, G-protein-coupled receptors, the trypsin class of serine proteases, hemoglobins, oligosaccharide phosphorylases, nuclear receptor ligand-binding domains and retroviral proteases). The family structural alignments identified positions corresponding to the ICDs in the structures of most family members (below, these residues are termed ‘conserved interconnectivity determinants' or CICD residues). We examined whether CICD residues are related to residues with experimentally demonstrated roles in signal transmission in the seven families. Our results revealed a general correspondence between many of these positions and key residues in allosteric communication. Interestingly, some of the CICD residues in four of the analyzed examples (G-protein-coupled receptors, the trypsin class of serine proteases, hemoglobins and nuclear receptor ligand-binding domains) were found to be amino acids involved in the networks of statistically coupled residues as predicted by Ranganathan and co-workers (Süel et al, 2002). We note that here it is not our intention to find networks of important residues possibly involved in allosteric communication. Rather, we show that CICD residues, that is, centrally conserved residues crucial for maintaining shorter path lengths in the protein network, mediate the signaling process in protein families. The myoglobin family is a particularly interesting example in our analysis. Recent experiments revealed a level of complexity in myoglobin that was not considered previously, showing that this oxygen-binding protein is an allosteric enzyme that participates in the catalysis of small molecules (Frauenfelder et al, 2001, 2003; Kuriyan, 2004). All the CICD residues predicted in this case were identified as amino acids involved in the myoglobin roles. The HIV-1 protease further constitutes an example where new insights might be gained from an analysis such as the one presented here. Our study detected two CICD residues that are likely to be involved in the communications between non-active site residues and the active site. Mutations of these non-active site residues were reported to confer drug resistance on the HIV-1 protease even though they are away from the active site (Olsen et al, 1999). Further experiments are required to test our predictions.
Results
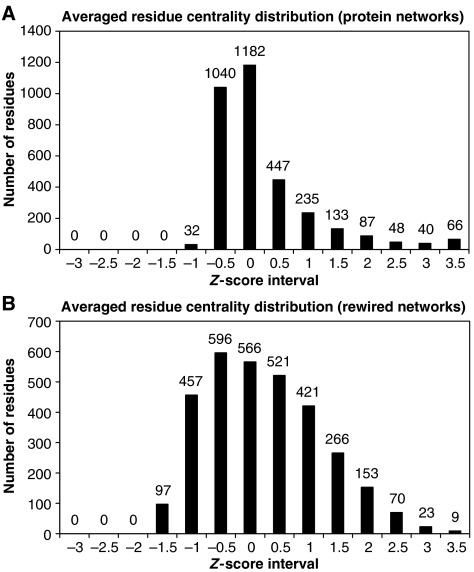
The protein structures of seven structurally and functionally distinct protein families were represented as residue interacting networks. A random rewiring of the residue contacts of the networks of each of the representatives of the protein families decreased the network characteristic path length (averaged shortest distance between all pairs of residues) and the clustering coefficient (averaged value of residue clustering). The residue centrality distribution became more homogeneous, illustrating the transition from small world to random networks (Figure 1 ).
Figure 1.
(A) Averaged distribution of residue centrality of the seven representative protein structures. The average number of residues for each residue centrality z-score interval is indicated at the top of each bar. The average values of the network characteristic path length and clustering coefficient are L=4.21 and C=0.52, respectively. (B) Averaged distribution of residue centrality of the randomly rewired networks of the seven family representative protein structures. The average values of the network characteristic path length and clustering coefficient are L=2.62 and C=0.04, respectively.
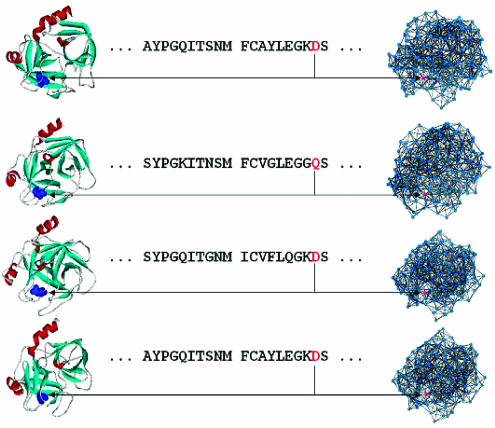
Using the family structural alignments, we carried out an analysis of the transmission of signals initiated at one site in the protein to a distant functional site in those seven structurally and functionally distinct protein families. For each family, we identified the CICD residues (Supplementary Table I) and analyzed their potential role in mediating allosteric regulation and specificity in molecular recognition. To determine the CICD residues, we calculated the changes in the characteristic path length when each node (amino acid) and its links (inter-atomic contacts) are removed from the structure of each family member. Those positions in the family alignments exhibiting a statistically significant change in the characteristic path length ΔL (z-score⩾2.0) in at least 70% of the family members were labeled CICD residues (Figure 2 ). As detailed below, experimental data obtained from databases and from the literature confirmed the direct participation of many of the CICD residues in the propagation of the information in signaling. Interestingly, only about 5% of the sequence conserved residues are CICDs, whereas nearly 70% of the CICD residues of all families are conserved in sequence (Supplementary Table II). Most of the remaining 30% of CICD amino acids are in direct contact with at least one CICD conserved in sequence. Several of these residues have been reported as important for the allosteric communications or protein binding, for example, residues Ile138 and Asp189 of the trypsin family, respectively. Thus, our network analysis captures information about highly cooperative residues important for the protein function, fold or allosteric communications, which cannot be provided solely by a sequence conservation analysis. The network representation of protein structures and the statistical analysis are described in the Materials and methods section. Interestingly, our results for five proteins, which as far as known are non-allosteric, revealed that the CICD residues cluster and largely coincide with experimentally identified key amino acids in folding nuclei (see Supplementary Table III and the table legend for references), whereas the predicted CICDs for the studied allosteric proteins tend to be more distributed over the structure.
Figure 2.
Schematic representation of the analysis for determining the conserved central positions based on an example of a protein family comprising four proteins. The position shown in red in the family structural alignment is central in the network representation of each family member. In the family member structures, this same position is represented in blue.
I. The Myoglobin family (representative structure: 101m, sperm whale myoglobin)
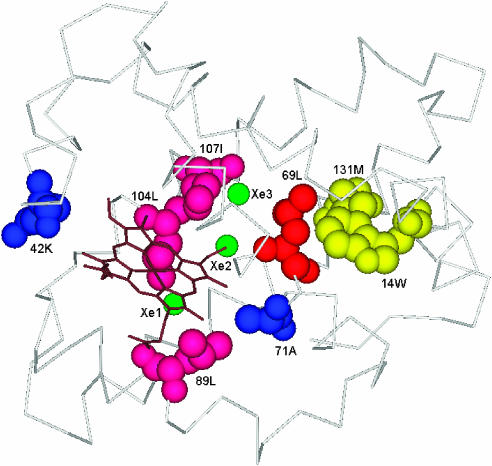
Myoglobin deserves special attention as it has long been thought that this close relative of hemoglobin was a non-allosteric protein capable only of storing dioxygen at the heme iron. However, recent studies point to a more complex picture of myoglobin as an allosteric enzyme that reacts with different small molecules (Frauenfelder et al, 2001). Myoglobin carries out at least two functions: O2 storage and catalysis for the conversion of NO to NO3−. Frauenfelder et al (2003) have identified two properties that characterize myoglobin as an allosteric enzyme: the presence of connected and conserved cavities in the structure and the existence of taxonomic sub-states. X-ray crystallography indicates the existence of five cavities, the heme cavity and four cavities determined by xenon binding Xe1–Xe4 (Tilton et al, 1984). The connected xenon cavities are involved in different chemical reactions, concentrating the reactants, and then modulating their concentration. The residues lining these cavities tend to be conserved and are likely to be functionally important. Structural changes involving these residues modify the connections between the cavities to control the reaction rate (Frauenfelder et al, 2001). On the other hand, there is experimental evidence corroborating the fact that myoglobin can exist in different taxonomic sub-states, with different reactive properties. Two such sub-states (A0 and A1) perform two different functions (Frauenfelder et al, 2003). Myoglobin is able to catalyze different redox reactions, as well as perform its well-known function of O2 storage.
Our network analysis identified eight CICD residues in the myoglobin structure (Trp14, Lys42, Leu69, Ala71, Leu89, Leu104, Ile107, Met131), which are distributed among the heme-binding site, the residues adjacent to the xenon cavities and the experimentally annotated redox-active amino acids (see Table I)(Tilton et al, 1984; Frauenfelder et al, 2001; Pfister et al, 2001). Figure 3 shows the structure of the sperm whale myoglobin (PDB code: 1j52) in the presence of three xenon atoms (green) located at the cavities. Residues lining these cavities are shown in pink and red. The heme group (brown) and the residues in contact with the heme are also represented (pink and blue). The redox-active amino acids are displayed in yellow. Trp14 and Lys42 are structurally conserved residues predicted as important for protein folding kinetics, stability or function according to the CoC database (Donald et al, 2005).
Table 1.
CICD residues of myoglobin distributed among the heme-binding sites, the residues lining the xenon cavities (Xe1, Xe2, Xe4) and the experimentally annotated redox-active amino acids
| Heme-binding sites | Xe1 | Xe2 | Xe4 | Redox-active residues |
|---|---|---|---|---|
| Lys42 | Leu89 | Leu104 | Leu69 | Trp14 |
| Ala71 | Leu104 | Ile107 | Ile107 | Met131 |
| Leu89 | ||||
| Leu104 | ||||
| Ile107 |
Figure 3.
Mapping of CICD residues onto the structure of the sperm whale myoglobin. The heme group is represented in brown and the atoms located at the xenon cavities are shown in green. Residues binding the heme group are shown in pink and blue, those lining the xenon cavities are colored pink and red and the redox-active residues are represented in yellow.
These results clearly show that the crucial amino acids that are involved in network connectivity in the myoglobin structure can be directly involved in one or more catalytic reactions carried out by this allosteric enzyme. These highly cooperative residues are located in regions important for allosteric communications.
II. The G-protein-coupled receptor family (representative structure: 1l9h(A), bovine rhodopsin)
Rhodopsin belongs to the superfamily of G-protein-coupled receptors. It is a good example of a signaling protein with three functional regions: ligand binding, an allosteric linking core and a G-protein-coupling region (Madabushi et al, 2004). Light activation of the rhodopsin receptor induces the disruption of a salt bridge existing between glutamic acid 113 in helix 3 and lysine 296 in helix 7, resulting in the formation of a Schiff base with retinal. As a result, conformational changes transmitted through the linking core reach the coupling region leading to activation of the G protein (Porter et al, 1996). Although this signal transduction mechanism is poorly understood, different residues involved in the allosteric communications have been experimentally verified (Ballesteros et al, 2001; Madabushi et al, 2004).
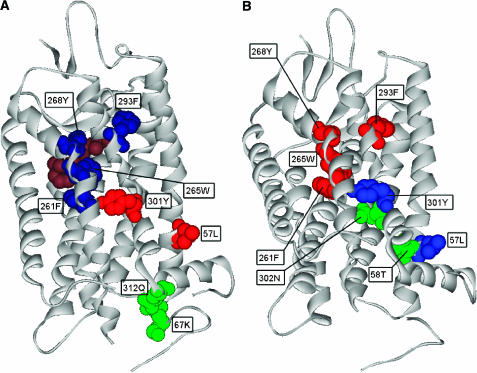
Our network analysis of the rhodopsin structure (PDB code: 1l9h) based on structural alignment identified residues Leu57, Lys67, Phe261, Trp265, Tyr268, Phe293, Tyr301 and Gln312 as the most contributing to the network interconnectedness. Figure 4A shows the mapping of these residues onto the three functional regions of the rhodopsin structure. The group of residues Phe261, Trp265, Tyr268 in helix 6 (blue, Figure 4A) forms a cluster of aromatic residues lining the bottom of the ligand-binding pocket and is protected from water by binding the cyclohexenyl ring of retinal (brown, Figure 4A) (Ballesteros et al, 2001). Residue Phe293 (blue, Figure 4A) located in helix 7 binds retinal and is also in direct contact with Lys296, which is known to be critical for the receptor activation (Ballesteros et al, 2001). Phe261 has been proposed to be functionally coupled to Gly121 in helix 3 (Han et al, 1996), and its mutation has been demonstrated to affect the receptor activity (Garriga et al, 1996; Yano et al, 1997; Andres et al, 2001). On the other hand, mutations of positions 265 and 268 affect ligand binding in different receptor families (Madabushi et al, 2004). Therefore, these four predicted sites belong to the ligand-binding pocket, which is thought to be the initial region involved in signal transduction following ligand binding. Residue Leu57 (red, Figure 4A) is located in a strategic position in helix 1, possibly belonging to the allosteric linking core. Leu57 contacts residues Phe56 and Leu321, which are the binding sites for palmitoyl. At the same time, it is in contact with Thr58 in helix 1 and with Met317 in the carboxy terminus, located in regions that undergo structural changes upon light activation, which possibly contact the G-protein alpha subunit and display some allosteric control (Menon et al, 2001). Tyr301 in helix 7 (red, Figure 4A) represents another position that can be included in the linking core. This residue is part of the binding site of heptane-1,2,3-triol, and is in contact with residue Phe261, which has been previously remarked as functionally important. Tyr301 is also a neighbor of position 302, which has been reported to affect the stability of the inactive conformation and the folding in different receptor families (Han et al, 1998; Madabushi et al, 2004). Finally, positions Lys67 and Gln312 (green, Figure 4A) are located in the coupling region and are in contact with each other. Lys67 belongs to the first intracellular loop and interacts with several residues at the carboxy terminus, and is also in contact with Arg69, located in the binding site of B-nonylglucoside. Gln312 is positioned at the carboxy terminus and is a mercury ion-binding site. Gln312 is also a neighbor of Phe313, and together with Tyr306 is a critical residue for proper light-induced conformational changes in the well-known NPXXY region in GPCRs (Fritze et al, 2003). The CoC database (Donald et al, 2005) annotates the structurally conserved Trp265 and Tyr268 as potentially important for kinetics, stability or function.
Figure 4.
(A) CICD residues located in the three functional regions of the bovine rhodopsin structure. The cyclohexenyl ring of retinal is depicted in brown. Residues in blue are clustered at the bottom of the ligand-binding pocket, whereas those shown in red and green are located in the linking core and the G-protein-coupling region, respectively. (B) CID residues forming part of the network of coupling between positions in the GPCR family, as identified by Ranganathan and co-workers. CICD residues shown in red are part of the network of statistically coupled residues, whereas those represented in blue are neighbors of the residues colored in green belonging to this network.
These results show that the CICD residues in the G-protein-coupled receptor family are distributed among the three most important regions for signal transmission, starting at the ligand-binding pocket, passing through the linking core and finally ending at the G-protein binding region. Experimental data revealed that mutations of some of these residues lead to the loss of allosteric control and constitutive receptor activity (Han et al, 1996; Ballesteros et al, 2001). Other CICD residues are shown to interact directly with key residues for allostery, and are therefore considered as potential candidates for allosteric communication.
In a recent study, using a sequence-based statistical method Ranganathan and co-workers (Süel et al, 2002) were able to identify positions in an alignment of GPCR family members that exhibited some sequence interdependence with the functionally important position Tyr296. The authors showed that the networks of residues statistically coupled to Tyr296 represented structural motifs for signaling communications in the GPCR family. Some of these statistically coupled residues (Phe261, Trp265, Tyr268 and Phe293) correspond to the CICD residues established in our analysis (red, Figure 4B). Residues Leu57 and Tyr301 (blue, Figure 4B) are neighbors of the coupled positions, Thr58 and Asn302, respectively (green, Figure 4B).
III. The trypsin family of serine proteases (representative structure: 2ptc(E), bovine beta-trypsin complex with pancreatic trypsin inhibitor)
Trypsin is an illustrative example of cooperative interactions between residues belonging to different regions. Trypsin hydrolyzes peptides with arginine or lysine residues at the so-called P1 position, whereas chymotrypsin prefers large hydrophobic residues at the same position. It is well known that the negatively charged residue Asp189 in the bottom of the binding pocket of trypsin accounts for the enzyme's specificity, and it has long been thought to be responsible for the specificity difference between trypsin and chymotrypsin (the analogous residue in chymotrypsin is Ser189) (Szabo et al, 1999). However, site-directed mutagenesis analyses have shown that the conversion of trypsin into a chymotrypsin-like protease requires substitutions of different residues from the S1 binding pocket, in addition to mutations of residues belonging to three surface loops (Hedstrom et al, 1994). Surface loops 1 and 2 connect the walls of the S1 pocket, but do not contact the substrate, whereas loop 3 is more distant from the S1 pocket. On the other hand, it has been reported that mutations at selected positions within loops 1, 2 and 3, together with substitutions at the S1 site and residue Ile138, convert trypsin into a protease with elastase-like specificity (Hung and Hedstrom, 1998). These experimental results show that the substrate-binding specificity is regulated by a set of distributed residues in the structure of trypsin, acting in a cooperative manner by interchanging information.
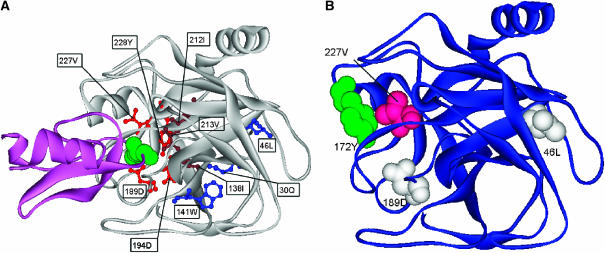
We found a first group of CICD residues located at the S1 site: Asp189, Asp194, Val227 and Tyr228. All these positions interact with the P1 position Lys15 of the pancreatic trypsin inhibitor (chain I). Particularly, Asp189 is known to be crucial in the trypsin binding specificity, contacting Ser195 from the catalytic triad (Figure 5A ) (Szabo et al, 2003). The second group of CICD residues was found to comprise Ile212, Val213 and Ile138. Residue Val213, which is in contact with Ile212 and Ile138, interacts with Lys15 of the pancreatic trypsin inhibitor, and also with His57 and Ser195 belonging to the catalytic triad. Position Ile212, on the other hand, is in contact with Asp102 from the catalytic site. Mutation of residue Ile138, which is not part of the binding site, is one of the known important substitutions for converting the trypsin specificity into the esterase specificity (Figure 5A) (Hung and Hedstrom, 1998). A third group of CICD residues includes positions Gln30, Leu46 and Trp141. Residues Gln30 (E) and Trp141 (E), which are in contact with each other, are located in the core of the protein, and could be important for folding and stability (Figure 5A). These findings illustrate that here many of our predicted CICD residues correspond to residues that act in a cooperative manner for determining the specificity at the S1 site. Asp194 is a structurally conserved residue. It is also annotated by the CoC database (Donald et al, 2005) as having a possible role in function, stability or folding kinetics.
Figure 5.
(A) Structural mapping of CICD residues in the bovine beta-trypsin complex (gray) with pancreatic trypsin inhibitor (magenta). Residues belonging to the trypsin S1 pocket (red) are in contact with Lys15 of the pancreatic trypsin inhibitor (green). CICD residues (brown) located further from the binding site are likely to be important for the binding specificity, whereas those shown in blue reside in the core of the protein. (B) Correspondence between CICD residues and statistically coupled positions for trypsin, as detected by Ranganathan and co-workers. CICD residues (white) belong to the network of statistically coupled residues, whereas Val227 (pink) interacts with the statistically coupled residue Y172 (green).
The trypsin family of serine proteases is another example studied by Ranganathan and co-workers (Süel et al, 2002). Two of our predicted CICD residues, Leu46 and Asp189, correspond to statistically coupled residues in the analysis of different site-specific perturbations carried out by these authors (Figure 5B). The distantly positioned Tyr172 on loop 3, which has been shown to influence specificity, is again one of their detected coupled residues. This residue is in contact with one of our predicted CICD residues Val227, which is part of the binding site (Figure 5B). This interaction could be important for Tyr172 in determining specificity at the S1 site.
IV. The hemoglobin family (representative structure: 1bz0(ABCD), human hemoglobin)
Hemoglobin is a tetramer with two α and two β subunits symmetrically positioned around a central water-filled cavity. According to the Monod, Wyman and Changeux model (Paoli et al, 1998), hemoglobin can exist in two conformations in rapid equilibrium: the T state with low-affinity oxygen binding and the R state with high-affinity oxygen binding. Crystallographic studies have shown structural differences between these two states, characterized by a rotation and translation of one αβ dimer with respect to the other. Cooperativity results from the information transmitted between subunits through the tetramerization interface α1β2 (α2β1) as a consequence of conformational changes in the heme groups. The oxygen ligation to one subunit in the T state induces structural changes in the heme-binding site, which are propagated to the neighboring subunits via the α1β2 (α2β1) interface, allowing the transition to the R state (Perutz et al, 1998).
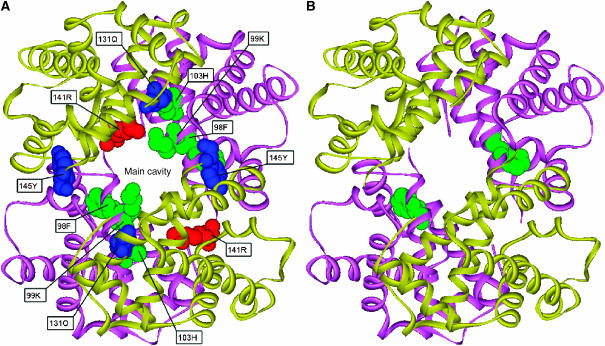
Our network analysis detected CICD residues, which were found to be located at regions important for allosteric communication. Specifically, positions Phe98, Lys99 and His103 belonging to the α subunits are located at the α1β1 (α2β2) interfaces. Phe98 is part of the heme-binding site, whereas Lys99 and His103 are neighbors of heme-binding residues. These residues are situated inside the central cavity of hemoglobin, which involves an excess of positively charged ionizable groups (Figure 6A ). It has been suggested (Bonaventura and Bonaventura, 1978) and experimentally confirmed (Perutz et al, 1998) that the mutual repulsion of these ionizable groups increases the oxygen affinity by raising the free energy of the T structure. Positions Arg141 from both α subunits are situated at the tetramerization interfaces α1β2 (α2β1). These interfaces, and specifically these residues, have been reported to be involved in the structural changes taking place in the switch from the T to the R states (Paoli et al, 1996). Two other relevant positions determined from our analysis are Gln131 and Tyr145 from the two β subunits. Gln131 belongs to the α1β1 (α2β2) interface, and is in contact with the previously analyzed His103 from the α subunits. Finally, residue Tyr145 is located in regions at the α1β2 (α2β1) interface and undergoes drastic structural changes in the switch from T to R states. Phe98, Lys99, His103 and Arg141 are structurally conserved residues, again predicted as important according to the CoC database (Donald et al, 2005). Finally, it is interesting to notice that Süel et al (2002) studied the hemoglobin family and identified Phe98 of the α subunits as statistically coupled residues resulting from a statistical perturbation scan (Figure 6B).
Figure 6.
(A) Representation of CICD residues in the structure of human hemoglobin. The two α and two β subunits are colored in magenta and yellow, respectively. CICD residues belonging to α subunits are located at the α1β1 (α2β2) interfaces (inside the hemoglobin central cavity, green) and at the interfaces α1β2 (α2β1) (red), whereas those from β subunits are part of the α1β1 (α2β2) and α1β2 (α2β1) interfaces (blue). (B) CICD residues forming part of the network of statistically coupled residues, as identified by Ranganathan and collaborators. The two α and two β subunits are colored in magenta and yellow, respectively. CICD residue Phe98 belonging to both α subunits is shown in green, and forms part of the network of coupled residues.
V. The oligosaccharide phosphorylase family (representative structure: 1gpa(AB), rabbit muscle glycogen phosphorylase)
Glycogen phosphorylase is one of the phosphorylase enzymes, which break up glycogen into glucose subunits (Johnson, 1992). This protein is a dimer composed of two identical subunits regulated by phosphorylation and by allosteric effectors such as AMP. According to the Monod–Wyman–Changeux model, it can exist in two states in equilibrium: the inactive (T state) and the active state (R state). The covalently attached phosphate group and other non-covalently bound allosteric effectors lead to conformational changes, which are transmitted from the phosphorylation and allosteric sites to the catalytic site (Johnson, 1992; Buchbinder and Fletterick, 1996). The communication from these sites and the catalytic site results in the activation of the enzyme. Activation occurs by unblocking the access from the solvent to the catalytic site and by creating the substrate phosphate recognition site through an interchange of an acidic group with a basic group (Johnson, 1992).
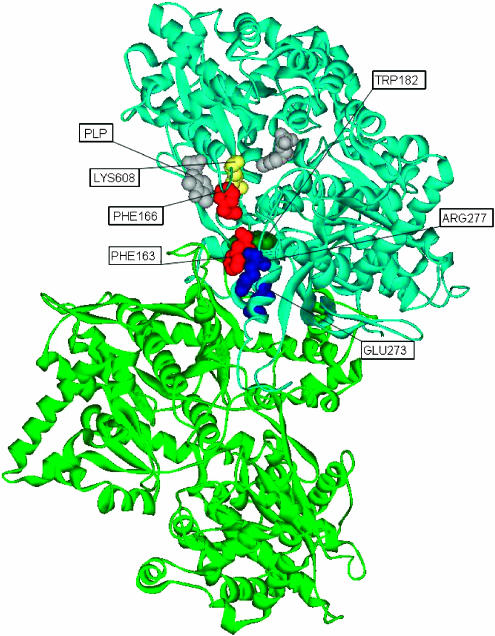
We identified six CICD residues in the glycogen phosphorylase monomeric structure (Phe163, Phe166, Trp182, Glu273, Arg277, Lys608) (Figure 7 ). Amino acids Phe163 and Phe166 belong to the β turn (residues 162–166), which exhibits a structural change in the transition from the T state to the R state. In the transition, the packing of Ile165 with residues belonging to the 280s loop is disrupted, modifying the catalytic site (Johnson, 1992; Buchbinder and Fletterick, 1996). Trp182 contacts directly Phe163 and is possibly involved in the transmission of the conformational changes from the tower/tower interface to the catalytic site. Residue Arg277 is located at the end of the tower helix, which is packed against the tower helix of the symmetry-related unit. On the T to R transition, the tower helices change their angle, and this amino acid shifts to allow structural changes in the catalytic site (Johnson, 1992). Residue Glu273, located at the tower helices, is part of the new allosteric binding site for the CP320626 inhibitor (Oikonomakos et al, 2000). Thus, events in the catalytic site are linked to events in the tower/tower interface. On the other hand, the T to R transition involves the replacement of the hydrogen bond established between Lys608 and the catalytic site residue Arg569 by a new hydrogen bond between Lys608 and the 280s loop residue Asp283, illustrating the important role of Lys608 in the T to R conversion (Johnson, 1992; Mitchell et al, 1996).
Figure 7.
Mapping of CICD residues onto the homodimer structure of the rabbit muscle glycogen phosphorylase. The two subunits of the homodimer are represented in blue (chain A) and green (chain B). The PLP cofactor is represented in gray. Predicted residues binding the cofactor are colored in yellow. Residues located in one of the tower helices are shown in blue, those belonging to the beta turn are colored in red and the more hidden residue Trp182 is represented in dark green.
VI. The nuclear receptor ligand-binding domain family (representative structure: 1g5y(AB), human retinoic acid receptor RXR-alpha)
The retinoic acid receptor RXR-alpha serves as a common dimerization partner for several nuclear receptors. These receptors are modular transcription factors, which are activated through the ligand-binding domain composed of four functionally linked surfaces: the ligand-binding pocket, an activation function 2 (AF2) helix, a cofactor binding surface and a dimerization surface (Shulman et al, 2004). An allosteric interaction between all these surfaces is needed for the nuclear receptor function. Ligand binding influences the transmission of signals across the dimerization interface, illustrating that the ligand-binding pocket and the dimerization interface are allosterically coupled. In such a way, ligands of one member of an RXR dimer can regulate the activity of its partner (‘phantom ligand effect') (Shulman et al, 2004).
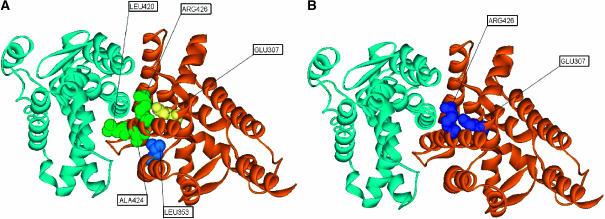
Our network analysis identified five CICD residues in the ligand-binding domain of the retinoic acid receptor RXR-alpha structure: Glu307, Leu353, Leu420, Ala424 and Arg426 (Figure 8A ). Residues Leu420, Ala424 and Arg426 are part of the dimerization interface, which is a key region for the allosteric communications (Figure 8A) (Gampe et al, 2000a, 2000b; Shulman et al, 2004). Specifically, Arg426 has been experimentally reported to be important in nuclear receptor ligand activation (Shulman et al, 2004). Although position Glu307 does not participate directly in ligand recognition, cofactor binding or dimerization, mutation of its corresponding position Glu296 in the liver X receptor (LXR) leads to a loss of the heterodimer's (RXR/LXR) ability to respond to the synthetic RXR agonist LG268 (Shulman et al, 2004). This finding implies that this mutation affects the signaling transmission in the heterodimer. Residue Leu353 has not been reported as important for the allosteric communications; however, it is strategically located between residue Ile310 from the ligand-binding site and residues Ala424 and Glu352 from the dimerization interface (Gampe et al, 2000a, 2000b; Shulman et al, 2004). This residue might be involved in the signaling transmission between these two functional regions.
Figure 8.
(A) CICD residues located in the structure of the homodimer human retinoic acid receptor RXR-alpha. The two subunits are represented in orange and blue, respectively. Residues forming part of the dimerization interface are depicted in green. Residue Glu307, important for the dimerization of LXR receptor, is colored in yellow. Residue Leu353, located between the ligand-binding site and the dimerization interface, is represented in blue. (B) The CICD amino acids corresponding to statistically coupled residues predicted by Ranganathan and co-workers are colored in blue.
Interestingly, Ranganathan and co-workers (Shulman et al, 2004) carried out a sequence-based statistical method for this protein family and found a statistical coupling between two of our CICD residues, Glu307 and Arg426 (Figure 8B).
VII. The retroviral protease family (representative structure: 1kzk(AB), HIV-1 protease complex)
The HIV-1 protease, an enzyme essential for viral replication, has been one of the main drug targets against which several inhibitors have been developed. The appearance of drug-resistant strains of HIV has become one of the major factors in achieving long-term viral suppression (Olsen et al, 1999; Perryman et al, 2004; Bowman et al, 2005). Active site mutations in HIV-1 protease, decreasing binding of different inhibitors, have been well studied, whereas the effect of non-active site mutations on the inhibitor binding affinity is less understood. Several non-active site mutations that compensate active site changes affecting the enzyme catalysis have been reported. However, the role of non-active site residues in the inhibitor binding requires further studies (Perryman et al, 2004).
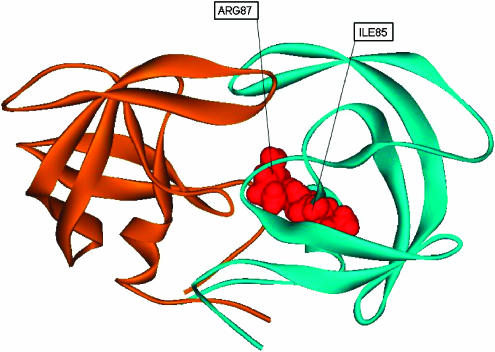
Our network analysis identified two CICD residues in contact with each other (Ile85, Arg87) (Figure 9 ), which to our knowledge have not been reported as important mutations affecting the inhibitor binding affinity. The location of these amino acids in the protease structure suggests that they might play an important role in the transmission of the information between certain non-active site mutations, known to affect the protease enzymatic activity and to contribute to the destabilization of inhibitor binding, and some active site residues, whose mutations were reported to affect the catalytic activity as well as the binding affinity. Residue Ile85 is in contact with two important active site residues: Asp25 and Ile84. Asp25 is known to be a key residue in ligand recognition (Perryman et al, 2004), whereas Ile84 is one of the most studied active site mutations affecting the catalytic efficiency (Olsen et al, 1999; Perryman et al, 2004). On the other hand, Ile85 interacts with the non-active site residues Leu24, Val64, Leu90 and Ile93, whose substitutions were reported to confer drug resistance on the HIV-1 protease (Olsen et al, 1999). Arg87 also interacts with Asp25 and Leu90 (Figure 9). Thus, Ile85 and Arg87 act as connections between non-active site and key active site residues. Mutations of these CICD amino acids could impair the compensating role of the non-active site mutations.
Figure 9.
Representation of CICD residues in the structure of HIV-1 protease complex (red color). Monomers are colored in blue and orange, respectively.
Discussion
Evolution has led to a robust architecture of proteins, with an extraordinary tolerance to mutations at many sites, and an extreme sensitivity to some substitutions at others. This robustness to environmental perturbations is crucial for protein function. Here, we describe protein structures as interacting networks. Such a description facilitates the investigation of their topological characteristics, and represents a simplified model of a robust yet fragile communication system. As expected, we find that removal of the majority of nodes (residues) does not affect the network interconnectedness substantially, yet the absence of a few key vertices drastically changes the system's connectivity. When residue contacts are randomly rewired, these small-world networks become random, exhibiting a more homogeneous distribution of the residue centrality. Interestingly, when comparing the inactive and active conformations in the hemoglobin and NtrC cases, we observed a redistribution of central residues (Supplementary Figures 1 and 2). The fact that there may be different sets of central residues in the two states emphasizes the importance of protein network dynamics. Activation/inactivation transition does not involve a change in the information flow in one specific static network. Rather, it underscores the involvement of multiplicity of networks, contributing to robustness and efficiency in the regulation.
The most important result of our study relates to measuring the contribution of a node to the network's connectivity by considering the change in the characteristic path length following removal of each vertex. We carried out a study of seven experimentally well-characterized protein families (myoglobins, G-protein-coupled receptors, trypsin class of serine proteases, hemoglobins, oligosaccharide phosphorylases, nuclear receptor ligand-binding domains and retroviral proteases). Through an analysis of structural alignments, we identified the key positions for the network's connectivity. We show that many of these centrally conserved residues (the CICD residues) crucial for maintaining the shortest path lengths mediate the efficiency of the signaling process in protein families. Available experimental data in all seven families support our proposition.
Our predictions for the families of G-protein-coupled receptors, trypsin class of serine proteases, hemoglobins and nuclear receptor ligand-binding domains were compared with the results of the statistical method recently introduced by Ranganathan and collaborators (Süel et al, 2002). Despite the fact that our goal differs from the main purpose of these authors, some of the key CICD residues in these examples form part of the networks of statistically coupled residues identified by their method.
Recent findings on the allosteric nature of myoglobin make the myoglobin family an additional, particularly interesting example for an analysis. Frauenfelder et al (2003) have aptly called this protein the hydrogen atom of biology. Myoglobin illustrates that certain characteristics of a protein design may be involved in new functions. Interestingly, all the key residues whose removal significantly elongates the path length in the network correspond to either residues binding the heme group, amino acids lining three of the main xenon cavities and thus likely to be important for the myoglobin allostery, or to redox-active residues, which act in a cooperative way for optimal protein function. Experimental evidence, together with the strategic positioning of these residues, suggests their participation in one or more functions of myoglobin.
As in the HIV-1 protease example, our predictions may shed light on the identification of residues important for maintaining long-range communications between non-active site residues conferring drug resistance and the active site. In summary, the analysis of the change in the characteristic path length through node removal provides an insight into residues important for the long-range communications in protein families.
Materials and methods
Protein structure and sequence analysis
We compiled seven protein families, with all their members having a known structure in the PDB database. The family alignments (Supplementary Figure 3) were generated using 3Dcoffee, which is a method that combines protein sequences and structures (Poirot et al, 2004). Protein structures are shown with the DS ViewerPro 6.0 (http://www.accelrys.com/dstudio/ds_viewer/index.html). Sequence conservation of multiple alignments was calculated using the ConSurf server (Glaser et al, 2003). Sequence conserved residues were considered as those with a color-coded score equal to nine.
Network representation of protein structures
Each protein structure was modeled as an undirected graph, where amino-acid residues corresponded to vertices, and contacts between them were represented as edges. Residues i and j were considered to be in contact if at least one atom corresponding to residue i was at a distance of less than or equal to 5.0 Å to an atom from residue j. This value approximates the upper limit for attractive London–van der Waals forces (Greene and Higman, 2003), and reveals the highest percentage of overlapping of detected CICD residues with other cutoffs (Supplementary Figure 4).
The residue centrality was calculated using the change of the characteristic path length under removal of node k (with its links). Namely, 
where L is the characteristic path length defined as 
with Np being the number of residue pairs and d(i,j) being the shortest path distance between residues i and j. Lrem.k represents the characteristic path length after the removal of node k and its corresponding links from the network.
Rewiring of protein structures
We randomly rewired 100 times each family representative protein structure, keeping the residue number of contacts unchanged. We then calculated the averaged residue centrality distribution for each family representative protein structure (Supplementary Figure 5). The mean of the averaged distributions is shown in Figure 1B.
Statistical analysis
The statistically significant central residues were evaluated using the z-score values of the residue centrality, defined as

where ΔLk is the change of the characteristic path length under removal of node k,
is the change of the characteristic path length under node removal averaged over all protein residues and σ is the corresponding standard deviation. The z-score distribution of residue centrality for all members of the studied families is shown in Supplementary Figure 6.
Supplementary Material
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. One of the authors (AS) thanks Tara C Marshall for her help in editing of this manuscript.
References
- Achacoso TB, Yamamoto WS (1992) AY's Neuroanatomy of C. elegans for Computation. Boca Raton, FL: CRC Press [Google Scholar]
- Aharoni A, Gaidukov L, Khersonsky O, Goulds MMcQ, Roodveldt C, Tawfik DS (2005) The evolvability of promiscuous protein functions. Nat Genet 37: 73–76 [DOI] [PubMed] [Google Scholar]
- Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S (2004) Network analysis of protein structures identifies functional residues. J Mol Biol 344: 1135–1146 [DOI] [PubMed] [Google Scholar]
- Andres A, Kosoy A, Garriga P, Manyosa J (2001) Mutations at position 125 in transmembrane helix III of rhodopsin affect the structure and signalling of the receptor. Eur J Biochem 268: 5696–5704 [DOI] [PubMed] [Google Scholar]
- Ballesteros JA, Shi L, Javitch JA (2001) Structural mimicry in G protein-coupled receptors: implications of the high-resolution structure of rhodopsin for structure–function analysis of rhodopsin-like receptors. Mol Pharmacol 60: 1–19 [PubMed] [Google Scholar]
- Bonaventura C, Bonaventura J (1978) Anionic control of hemoglobin function. In Biochemical and Clinical Aspects of Hemoglobin Abnormalities, Caughey WS (ed) pp 647–663. New York: Academic [Google Scholar]
- Bowman MJ, Byrne S, Chmielewski J (2005) Switching between allosteric and dimerization inhibition of HIV-1 protease. Chem Biol 12: 439–444 [DOI] [PubMed] [Google Scholar]
- Buchbinder JL, Fletterick RJ (1996) Role of the active site gate of glycogen phosphorylase in allosteric inhibition and substrate binding. J Biol Chem 271: 22305–22309 [DOI] [PubMed] [Google Scholar]
- del Sol A, O'Meara P (2004) Small-world network approach to identify key residues in protein–protein interaction. Proteins 58: 672–682 [DOI] [PubMed] [Google Scholar]
- Dokholyan NV, Li L, Ding F, Shakhnovich EI (2002) Topological determinants of protein folding. Proc Natl Acad Sci USA 99: 8637–8641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald JE, Hubner IA, Rotemberg VM, Shakhnovich EI, Mirny LA (2005) CoC: a database of universally conserved residues in protein folds. Bioinformatics 21: 2539–2540 [DOI] [PubMed] [Google Scholar]
- Frauenfelder H, McMahon BH, Austin RH, Chu K, Groves JT (2001) The role of structure, energy landscape, dynamics, and allostery in enzymatic function of myoglobin. Proc Natl Acad Sci USA 98: 2370–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauenfelder H, McMahon, Fenimore PW (2003) Myoglobin: the hydrogen atom of biology and a paradigm of complexity. Proc Natl Acad Sci USA 100: 8615–8617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, Ernst OP (2003) Role of the conserved NPxxY(x)5, 6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci USA 100: 2290–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampe RT, Montana VG, Lambert MH, Miller AB, Bledsoe RK, Milburn MV, Kliewer SA, Willson TM, Xu E (2000a) Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell 5: 545–555 [DOI] [PubMed] [Google Scholar]
- Gampe RT, Montana VG, Lambert MH, Wisely GB, Milburn MV, Xu E (2000b) Structural basis for auto-repression of retinoid X receptor by tetramer formation and the AF-2 helix. Genes Dev 14: 2229–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga P, Liu X, Khorana HG (1996) Structure and function in rhodopsin: correct folding and misfolding in point mutants at and in proximity to the site of the retinitis pigmentosa mutation Leu-125 → Arg in the transmembrane helix C. Proc Natl Acad Sci USA 93: 4560–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N (2003) ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19: 163–164 [DOI] [PubMed] [Google Scholar]
- Greene L, Higman V (2003) Uncovering network systems within protein structures. J Mol Biol 334: 781–791 [DOI] [PubMed] [Google Scholar]
- Gunasekaran K, Ma B, Nussinov R (2004) Is allostery an intrinsic property of all dynamic proteins? Proteins 57: 433–443 [DOI] [PubMed] [Google Scholar]
- Han M, Lin SW, Minkova M, Smith SO, Sakmar TP (1996) Functional interaction of transmembrane helices 3 and 6 in rhodopsin. Replacement of phenylalanine 261 by alanine causes reversion of phenotype of a glycine 121 replacement mutant. J Biol Chem 271: 32337–32342 [DOI] [PubMed] [Google Scholar]
- Han M, Smith SO, Sakmar TP (1998) Constitutive activation of opsin by mutation of methionine 257 on transmembrane helix 6. Biochemistry 37: 8253–8261 [DOI] [PubMed] [Google Scholar]
- Hatley ME, Lockless SW, Gibson SK, Gilman AG, Ranganathan R (2003) Allosteric determinants in guanine nucleotide-binding proteins. Proc Natl Acad Sci USA 100: 14445–14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom L, Perona JJ, Rutter WJ (1994) Converting trypsin to chymotrypsin: residue 172 is a substrate specificity determinant. Biochemistry 33: 8757–8763 [DOI] [PubMed] [Google Scholar]
- Hung SH, Hedstrom L (1998) Converting trypsin to elastase: substitution of the S1 site and adjacent loops reconstitutes esterase specificity but not amidase activity. Protein Eng 11: 669–673 [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411: 41–42 [DOI] [PubMed] [Google Scholar]
- Johnson LN (1992) Glycogen phosphorylase: control by phosphorylation and allosteric effectors. FASEB J 6: 2274–2282 [DOI] [PubMed] [Google Scholar]
- Kass I, Horovitz A (2002) Mapping pathways of allosteric communication in GroEL by analysis of correlated mutations. Proteins 48: 611–617 [DOI] [PubMed] [Google Scholar]
- Kern D, Zuiderweg ERP (2003) The role of dynamics in allosteric regulation. Curr Opin Struct Biol 13: 748–757 [DOI] [PubMed] [Google Scholar]
- Kuriyan J (2004) Allostery and coupled sequence variation in nuclear hormone receptors. Cell 116: 354–356 [DOI] [PubMed] [Google Scholar]
- Lockless SW, Ranganathan R (1999) Evolutionary conserved pathways of energetic connectivity in protein families. Science 286: 295–299 [DOI] [PubMed] [Google Scholar]
- Madabushi S, Gross AK, Philippi A, Meng EC, Wensel TG, Lichtarge O (2004) Evolutionary trace of G protein-coupled receptors reveals clusters of residues that determine global and class-specific functions. J Biol Chem 279: 8126–8132 [DOI] [PubMed] [Google Scholar]
- Menon ST, Han M, Sakmar TP (2001) Rhodopsin: structural basis of molecular physiology. Physiol Rev 81: 1659–1688 [DOI] [PubMed] [Google Scholar]
- Mitchell EP, Withers SG, Ermert P, Vasella AT, Garman EF, Oikonomakos NG, Johnson LN (1996) Ternary complex crystal structures of glycogen phosphorylase with the transition state analogue Nojirimycin Tetrazole and phosphate in the T and R states. Biochemistry 35: 7341–7355 [DOI] [PubMed] [Google Scholar]
- Oikonomakos NG, Skamnaki VT, Tsitsanou KE, Gavalas NG, Johnson LN (2000) A new allosteric site in glycogen phosphorylase b as a target for drug interactions. Structure 8: 575–584 [DOI] [PubMed] [Google Scholar]
- Olsen DB, Stahlhut MW, Rutkowski CA, Schock HB, vanOlden AL, Kuo LC (1999) Non-active site changes elicit broad-based cross-resistance of the HIV-1 protease to inhibitors. J Biol Chem 274: 23699–23701 [DOI] [PubMed] [Google Scholar]
- Paoli M, Liddington R, Tame J, Wilkinson A, Dodson G (1996) Crystal structure of T state haemoglobin with oxygen bound at all four haems. J Mol Biol 256: 775–792 [DOI] [PubMed] [Google Scholar]
- Perryman AL, Lin J-H, Mccammon JA (2004) HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contribution to drug resistance and a potential new target site for drugs. Protein Sci 13: 1108–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz MF, Wilkinson AJ, Paoli M, Dodson GG (1998) The stereochemical mechanism of the cooperative effects in hemoglobin revisited. Annu Rev Biophys Biomol Struct 27: 1–34 [DOI] [PubMed] [Google Scholar]
- Pfister TD, Gengenbach AJ, Syn S, Lu Y (2001) The role of redox-active amino acids on compound I stability, substract oxidation, and protein cross-linking in yeast cytochrome C peroxidase. Biochemistry 40: 14942–14951 [DOI] [PubMed] [Google Scholar]
- Poirot O, Suhre K, Abergel C, O'Toole E, Notredame C (2004) Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res 32: 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JE, Hwa J, Perez DM (1996) Activation of the α1b-adrenergic receptor is initiated by disruption of an interhelical salt bridge constraint. J Biol Chem 271: 28318–28323 [DOI] [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi A-L (2002) Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555 [DOI] [PubMed] [Google Scholar]
- Rennell D, Bouvier SE, Hardy LW, Poteete AR (1991) Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol 222: 67–88 [DOI] [PubMed] [Google Scholar]
- Schreiber G, Fersht AR (1995) Energetics of protein–protein interactions: analysis of the barnase–barstar interface by single mutations and double mutant cycles. J Mol Biol 248: 478–486 [DOI] [PubMed] [Google Scholar]
- Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R (2004) Structural determinants of allosteric ligands activation in RXR heterodimers. Cell 116: 417–429 [DOI] [PubMed] [Google Scholar]
- Süel GM, Lockless SW, Wall MA, Ranganathan R (2002) Evolutionary conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol 10: 59–69 [DOI] [PubMed] [Google Scholar]
- Szabo E, Bocskei Z, Naray-Szabo G, Graf L (1999) The three-dimensional structure of Asp189Ser trypsin provides evidence for an inherent structural plasticity of the protease. Eur J Biochem 263: 20–26 [DOI] [PubMed] [Google Scholar]
- Szabo E, Venekei I, Bocskei Z, Naray-Szabo G, Graf L (2003) Three dimensional structures of S189D chymotrypsin and D189S trypsin mutants: the effect of polarity at site 189 on a protease-specific stabilization of the substrate-binding site. J Mol Biol 331: 1121–1130 [DOI] [PubMed] [Google Scholar]
- Taverna DM, Goldstein RA (2002) Why are proteins so robust to site mutations? J Mol Biol 315: 479–484 [DOI] [PubMed] [Google Scholar]
- Tilton RF, Kuntz ID, Petsko GA (1984) Cavities in proteins: structure of metmyoglobin–xenon complex solved to 1.9 Å. Biochemistry 23: 2849–2857 [DOI] [PubMed] [Google Scholar]
- Vendruscolo M, Dokholyan NV, Paci E, Karplus M (2002) Small-world view of the amino acids that play a key role in protein fold. Phys Rev E 65: 0619101–0619104 [DOI] [PubMed] [Google Scholar]
- Volkman BF, Lipson D, Wemmer DE, Kern D (2001) Two-state allosteric behavior in a single-domain signaling protein. Science 291: 2429–2433 [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH (1998) Collective dynamics of small-world networks. Nature 393: 440–442 [DOI] [PubMed] [Google Scholar]
- Wuchty S (2001) Scale-free behavior in protein domain networks. Mol Biol Evol 18: 1694–1702 [DOI] [PubMed] [Google Scholar]
- Yano K, Kohn LD, Saji M, Okuno A, Cutler GB (1997) Phe576 plays an important role in the secondary structure and intracellular signaling of the human luteinizing hormone/chorionic gonadotropin receptor. J Clin Endocrinol Metab 82: 2586–2591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material