Abstract
Our computational model of the circadian clock comprised the feedback loop between LATE ELONGATED HYPOCOTYL (LHY), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and TIMING OF CAB EXPRESSION 1 (TOC1), and a predicted, interlocking feedback loop involving TOC1 and a hypothetical component Y. Experiments based on model predictions suggested GIGANTEA (GI) as a candidate for Y. We now extend the model to include a recently demonstrated feedback loop between the TOC1 homologues PSEUDO-RESPONSE REGULATOR 7 (PRR7), PRR9 and LHY and CCA1. This three-loop network explains the rhythmic phenotype of toc1 mutant alleles. Model predictions fit closely to new data on the gi;lhy;cca1 mutant, which confirm that GI is a major contributor to Y function. Analysis of the three-loop network suggests that the plant clock consists of morning and evening oscillators, coupled intracellularly, which may be analogous to coupled, morning and evening clock cells in Drosophila and the mouse.
Keywords: circadian rhythm, genetic network, photoperiod, mathematical model, systems biology
Introduction
The circadian clock generates 24-h rhythms in most eukaryotes and in cyanobacteria (Dunlap et al, 2003), including the rhythmic expression of 5–15% of genes in eukaryotes (Duffield, 2003). Circadian rhythms are generated by a central network of 6–12 genes that form interlocked feedback loops (Glossop et al, 1999). The relatively small number of components involved in the circadian clock network makes it an ideal candidate for mathematical modelling of complex biological regulation (Ruoff and Rensing, 1996; Leloup and Goldbeter, 1998; Forger and Peskin, 2003).
The clock mechanism in the model plant, Arabidopsis thaliana, was first proposed to comprise a feedback loop in which two partially redundant genes, LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), repress the expression of their activator, TIMING OF CAB EXPRESSION 1 (TOC1) (Alabadi et al, 2001). This circuit cannot fit all experimental data (Locke et al, 2005a), as a short-period rhythm persists for several cycles both in lhy;cca1 (Alabadi et al, 2002; Locke et al, 2005b) and in toc1 mutant plants (Mas et al, 2003a). Previously we used mathematical modelling to propose a new circuit comprising two interlocking feedback loops in order to explain the residual rhythm in the lhy;cca1 plant (Locke et al, 2005b). This model predicted the existence and expression patterns of two hypothetical components X and Y. X is proposed to be activated by TOC1, and X protein then activates LHY transcription, as required by the expression profile of TOC1 protein (Mas et al, 2003b). Y forms a second loop with TOC1, which is responsible for the short-period oscillation in the lhy;cca1 mutant. Based on the similarity of predicted and observed expression patterns, GI was identified as a candidate for Y (Locke et al, 2005b).
Here we have extended our model to include the recently proposed feedback loop between PSEUDO-RESPONSE REGULATOR 7 (PRR7), PRR9 and LHY/CCA1 (Farre et al, 2005; Salome and McClung, 2005), resulting in a three-loop circuit (Figure 1A). We first validate this new model against existing and new experimental data. We then experimentally confirm our prediction that GI functions as a component of Y in a feedback loop with TOC1, and investigate the regulatory properties of the three-loop network.
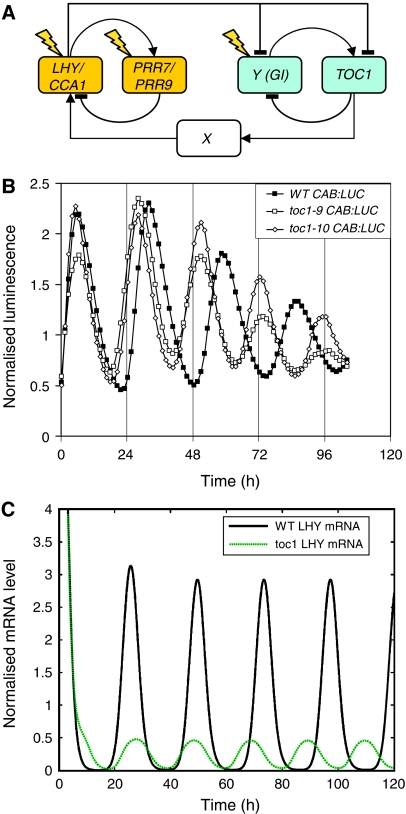
Figure 1.
The three-loop Arabidopsis clock model accounts for 20 h rhythms in toc1 mutants. (A) Summary of the three-loop network, showing only genes (boxed), regulatory interactions (arrows) and the locations of light input (flashes). Two-component oscillators are distinguished by shading the gene names in yellow or blue. (B) CAB:LUCIFERASE (CAB:LUC) rhythms in WT (filled squares), toc1-9 (open squares) and toc1-10 (open diamonds) under constant red light (10 μmol m−2 s−1). Luminescence values were normalised to the average over the whole time course. Time zero is the onset of constant light (LL). (C) Simulated expression levels of LHY mRNA in the WT (black solid line) and toc1 backgrounds (green dotted line) in LL. Expression levels were normalised to the average level of expression. Translation rate of TOC1 mRNA in the simulated mutant is 1/1000 WT value.
Results
A three-loop clock network accounts for additional experimental data
A short-period rhythm can exist in mutants with reduced TOC1 function in some conditions (Alabadi et al, 2001; Mas et al, 2003a). In order to test whether such residual rhythmicity was due to residual wild-type (WT) TOC1 mRNA, we tested a TOC1 deletion mutant (Supplementary information) for rhythmic expression of CHLOROPHYLL A/B-BINDING PROTEIN2 (CAB2, also known as LHCB1*1), a morning-expressed clock output gene (Figure 1B). Plants of the Ws accession carrying the toc1-10 deletion had a rhythm of 20 h period and reduced amplitude under constant light (LL) conditions. This was identical in timing to the rhythm of toc1-9 plants, which carry a termination codon within the first domain of the predicted TOC1 protein. Taken together, these data confirm previous suggestions that a TOC1-independent oscillator can persist in toc1 plants (Mas et al, 2003a).
The proposed PRR7/PRR9–LHY/CCA1 feedback loop provided a candidate mechanism to account for this oscillation. We therefore added this loop to the interlocked feedback model (Figure 1A; Supplementary information) to create a three-loop model. As the mutant phenotypes of PRR7 and 9 are weak, apparently less than 1 h different from WT (Nakamichi et al, 2005), we grouped these genes together as one gene, PRR7/9, in our network equations (Supplementary information). LHY and CCA1 were grouped together as LHY (Locke et al, 2005b). The first feedback loop involves LHY activating PRR7/9 transcription (Farre et al, 2005), with PRR7/9 protein going on to repress LHY activation. The remainder of the network follows our previous model (Locke et al, 2005b). LHY represses TOC1 and Y transcription; the dual, repressing and activating role of LHY has experimental support (Harmer and Kay, 2005). TOC1 protein activates X transcription, with X activating LHY transcription to form a second feedback loop. Y activates TOC1 expression and TOC1 represses Y expression, forming the third feedback loop. Light activates expression of LHY, Y, and now also PRR7/9, because PRR9 has been shown to be acutely light-activated (Ito et al, 2003).
We used an extensive parameter search for the new and altered components to test whether the three-loop network could account for the residual oscillations of a toc1 deletion mutant (Supplementary information). Our simulations show that the PRR7/PRR9–LHY/CCA1 loop can generate the short-period rhythm of toc1 plants (Figure 1C), and its absence can result in the very long period of prr7;prr9 double mutants (Supplementary Figure 1; Farre et al, 2005). Neither of these observations could be accounted for with our interlocked feedback loop model (Locke et al, 2005b), which predicted arrhythmia or a long period under all conditions in simulations of a toc1 null or loss-of-function mutants such as toc1-2 (6% of WT RNA levels; Strayer et al, 2000) or the toc1 RNAi lines (10–15%, Mas et al, 2003a). Sensitivity analysis shows that the three-loop model is similarly tolerant of parameter changes as the interlocking-loop model (Supplementary information; Supplementary Figure 2).
We now use the more realistic three-loop model to make further predictions for Y's role in the clock, and test these predictions against the experimental manipulation of GI.
GI is a component of Y
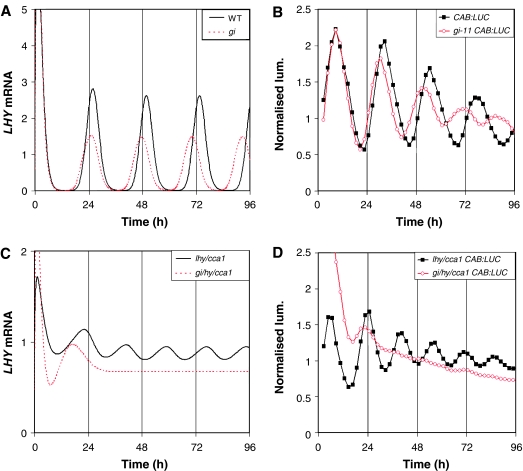
A simulated gi mutation (modelled by reducing Y translation by 70%) gives a 1 h reduction in the period of LHY mRNA oscillations (Figure 2A), which matches well with the observed period of CAB expression rhythms in a gi null mutant background (Figure 2B). According to our models, the Y–TOC1 feedback loop generates the 18 h rhythm seen in an lhy;cca1 mutant (Figure 2C and D). A reduction in Y function in the lhy;cca1 mutant background should therefore reduce the robustness of this residual rhythm. In fact, simulation of the gi;lhy;cca1 triple mutation results in a rapid loss of rhythmicity, reaching a negligible amplitude during the second cycle in LL (Figure 2C). The very strong phenotype encouraged us to test the rhythms of gi;lhy;cca1 triple mutant plants (Figure 2D). An almost exact match is made to the simulation; in the gi;lhy;cca1 triple mutant, the rhythmic amplitude collapses to insignificance during the second cycle.
Figure 2.
GI acts as Y in a feedback loop with TOC1. (A) Simulation of LHY mRNA levels in the WT (black solid line) and gi backgrounds (Y translation rate reduced by 70%, red dotted line) under constant light (LL). (B) Corresponding experimental data assaying circadian control of WT CAB:LUC expression by video imaging. (C) Simulation of LHY mRNA under LL in lhy;cca1 (translation rate of LHY mRNA in simulated mutant is 1/1000 WT value, black line) and gi;lhy;cca1 mutants (red dotted line). (D) Corresponding experimental data assaying CAB:LUC expression. The gi;lhy;cca1 mutant is severely damped (only four out of 23 plants gave a period estimate within the circadian range, and those estimates had an average relative amplitude error of 0.86). All data were normalised to the average level of expression.
An identical, catastrophic damping is also seen experimentally in the rhythmic expression of TOC1 and of COLD AND CIRCADIAN REGULATED 2 (CCR2), an evening-expressed clock output gene, in the triple mutant under LL and constant darkness (DD) (Supplementary Figure 3), whereas the lhy;cca1 double mutant retains short-period rhythms as described (Locke et al, 2005b). The mean level of TOC1 expression is significantly reduced in the gi;lhy;cca1 triple mutant compared with the lhy;cca1 double mutant (Supplementary Figure 4A). This is consistent with GI's functioning in the predicted role of Y, activating TOC1, and matches well to the expression levels in the simulated double and triple mutants (Supplementary Figure 4B). Our predictions also fit with experimental work showing that GI expression is light-responsive (Fowler et al, 1999; Paltiel et al, 2006), and are consistent with GI function in balancing other clock components to generate temperature compensation (Gould et al, 2006). GI is a component of a light-activated feedback loop, separate from LHY and CCA1, which is required for the maintenance of residual rhythms in the lhy;cca1 background.
Morning and evening oscillators allow tracking of dawn and dusk
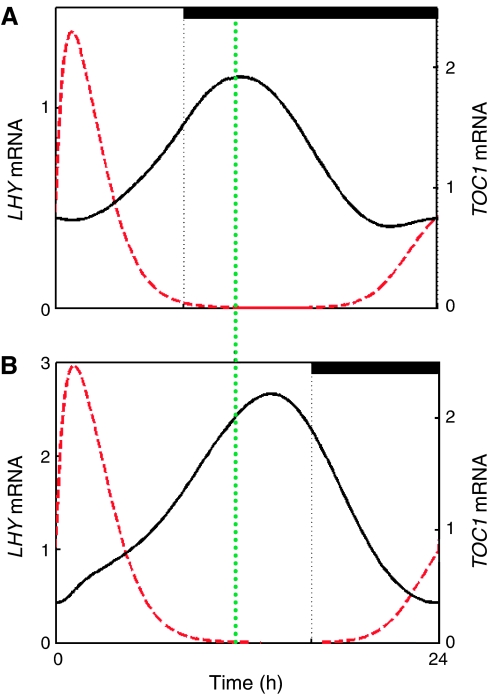
Our three-loop model suggests a symmetrical structure for the Arabidopsis clock circuit. The model predicts that two short-period oscillators, the morning-expressed PRR7/9–LHY/CCA1 loop and the evening-expressed TOC1–Y/GI loop, are coupled together by the LHY/CCA1–TOC1–X loop (Figure 1A). We investigated the effect of a change of photoperiod on the phase of the clock components of our three-loop network (Figure 3). The clock-regulated expression of LHY mRNA before dawn (20–24 h) remains at a fixed phase relative to dawn. In contrast, the peak of TOC1 mRNA is delayed under long photoperiod conditions, showing that its phase also responds to the time of dusk. This flexibility is not seen in our one-loop or interlocked-loop models (Supplementary Figures 5 and 6), in which clock-regulated LHY and TOC1 expressions are fixed relative to dawn, or both move with the time of dusk. Note that LHY is light-induced in all the models, so its peak phase is forced by dawn. The three-loop structure of the clock provides the flexibility to track multiple phases (Rand et al, 2004).
Figure 3.
Three-loop network can track dawn and dusk. Simulations of TOC1 mRNA (black solid line) and LHY mRNA (red dotted line) using the three-loop network under photoperiods of (A) LD8:16 and (B) LD16:8. The vertical dotted line highlights the shift in the peak phase of TOC1 mRNA levels from LD8:16 to LD16:8. The peak phase of LHY mRNA is not shifted.
The three-loop model also predicts that, if the coupling between PRR7/9–LHY/CCA1 loop and the evening-expressed TOC1–Y/GI loop were impaired, the two oscillators might run with different periods within one cell. This is predicted by simulation of an x mutant (Supplementary Figure 7), where LHY mRNA levels oscillated with a 20.4 h period under LL conditions and TOC1 levels oscillated with a 17.3 h period.
Discussion
We present evidence that GI acts with TOC1 in a feedback loop of the circadian clock in A. thaliana. This marks an advance in systems biology, because GI was identified as a candidate gene in this loop using experiments based directly on predictions from mathematical modelling. The three-loop model has greater realism, as it can simulate the short-period rhythms of toc1 and gi mutant plants and the long-period rhythms of prr7;prr9 double mutants, while still correctly matching the mutant phenotypes accounted for by the previous model. Understanding the Arabidopsis clock as a system of coupled, morning and evening oscillators provides a new intellectual framework that may persist over multiple incremental advances in biochemical and genetic realism.
The three-loop model is not yet complete, as it does not incorporate known clock-affecting genes such as PRR3, PRR5, TIME FOR COFFEE (TIC), EARLY FLOWERING 4 (ELF4) and LUX ARRHYTHMO (LUX) (reviewed by McClung, 2006). Rather than a weakness, this indicates three important uses of even incomplete mathematical models, in providing a framework to understand the existing experimental results, in focusing future experimental work on key regulatory interactions that reveal the location of the additional genes within the network and in informing the detailed design of these experiments, specifically to test any unusual aspect of regulation that has been predicted by simulation (Locke et al, 2005b).
The three-loop circuit contributes to the apparent robustness of the Arabidopsis clock, along with the partial redundancy of some genes: few single mutations alter the clock period by more than 3–4 h and arrhythmic mutations are rare (McClung, 2006). GI, one of the first characterised clock-affecting genes (Fowler et al, 1999; Park et al, 1999) with complex functions in both flowering and circadian regulation (Mizoguchi et al, 2005; Gould et al, 2006), illustrates the difficulty of understanding the effect of one component upon a complex network. The gi single mutant had a relatively weak phenotype, whereas our assays of the triple gi;lhy;cca1 mutant demonstrate GI's importance (Figure 2 and Supplementary Figure 3) as one component of Y in the three-loop network. It is likely that other components participate in the evening feedback loop with TOC1, because our current model indicates that the circadian phenotypes of the gi single mutant and the gi;lhy;cca1 triple mutant are accurately simulated by a 70% reduction in Y translation, rather than a complete absence of Y (Figure 2 and Supplementary Figure 3). PRR5 is a candidate component of Y that should now be tested, perhaps in combination with the gi mutation. If PRR5 is indeed part of Y, then our model could explain the arrhythmicity of the prr7;prr9;prr5 triple mutant (Nakamichi et al, 2005): the triple mutation not only removes the PRR7/9 feedback loop, but also impairs the TOC1–Y feedback loop. Constructing such multiple mutants, in combination with reporter genes, is and will remain laborious. Insertional mutants in most Arabidopsis genes are publicly available, but there is no prospect of a comprehensive bank of double mutants. Modelling offers a crucial tool for targeting future mutant construction as well as for extracting the maximum value from time-series studies using existing genetic resources.
Analysis of the three-loop network suggests new avenues for experiments. For example, the prediction that an x mutation could lead to desynchronisation of two short-period clocks (Supplementary Figure 7) suggests that future research could target mutations or chemical manipulations that cause desynchronisation of LHY and TOC1 mRNA rhythms. Period differences among rhythms in the same plant have been observed repeatedly and in some cases can be interpreted as evidence for desynchronisation of two intracellular oscillators, although cell-type-specific effects cannot be excluded (Hall et al, 2002; Michael et al, 2003). The three-loop model provides a mechanism for such intracellular desynchronisation, if the various rhythmic processes are controlled by different loops and coupling between loops is weakened in some conditions. This flexibility of circadian regulation is expected to offer a selective advantage, particularly where seasonal changes in photoperiod vary the relative timing of dawn and dusk (Pittendrigh and Daan, 1976). There is strong evidence in Drosophila (Stoleru et al, 2004) and mammals (Jagota et al, 2000) for separate control of morning and evening processes by oscillators in different cells, which are coupled together by cell–cell signalling. Plant clocks are coupled only weakly between cells, if at all (Thain et al, 2000), but the three-loop circuit suggests that an analogous architecture can be constructed within a single cell, by coupling the loop of morning-expressed genes LHY/CCA1 and APPR7/9 to the evening-expressed TOC1–GI loop. It will now be important to understand the role and balance of the light inputs into each of the feedback loops of the clock, firstly to determine what flexibility the three-loop circuit could provide and then to understand how the plant has evolved to exploit this flexibility in controlling rhythmic functions at different times of day.
Note added in proof
Zeilinger et al, in a study published simultaneously in Molecular Systems Biology, add PRR7 and PRR9 in parallel feedback loops to the interlocked loop network, with an alternative parameter set and light input mechanisms to PRR9 and Y (Zeilinger et al, 2006).
Supplementary Material
Supplemental Methods
Supplemental Figures
Supplemental Table 1
Supplemental Figure and Table Legends
SBML file for 3loop model
Acknowledgments
We thank V Hibberd and A Thomson for technical assistance, and P Brown for constructing the SBML file of the three-loop model. JCWL was supported by a postgraduate studentship from the Gatsby Charitable Foundation. LKB was supported by a Marie Curie Fellowship. Computer facilities were provided by the Centre for Scientific Computing at the University of Warwick; low-light imaging facilities were supported in part by the University of Edinburgh. Research in Warwick and Edinburgh was funded by BBSRC grants G13967 and G19886 to AJM. Research at Liverpool was funded by BBSRC grant BBS/B/111125 and Royal Society grant R4917/1 award to AH. Research in Szeged was supported by a Howard Hughes Medical Institute International Scholarship to FN (grant no. HHMI 55005620). The authors declare that they have no competing financial interests.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Alabadi D, Yanovsky MJ, Mas P, Harmer SL, Kay SA (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in arabidopsis. Curr Biol 12: 757–761 [DOI] [PubMed] [Google Scholar]
- Duffield GE (2003) DNA microarray analyses of circadian timing: the genomic basis of biological time. J Neuroendocrinol 15: 991–1002 [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, De Coursey PJ (2003) Chronobiology: biological timekeeping. Sunderland, MA, USA: Sinauer Associates
- Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Forger DB, Peskin CS (2003) A detailed predictive model of the mammalian circadian clock. Proc Natl Acad Sci USA 100: 14806–14811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glossop NRJ, Lyons LC, Hardin PE (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science 286: 766–768 [DOI] [PubMed] [Google Scholar]
- Gould PD, Locke JCW, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, Hall A (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognar L, Bastow RM, Nagy F, Millar AJ (2002) Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J 32: 529–537 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA (2005) Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Matsushika A, Yamada H, Sato S, Kato T, Tabata S, Yamashino T, Mizuno T (2003) Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 44: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Jagota A, de la Iglesia HO, Schwartz WJ (2000) Morning and evening circadian oscillations in the suprachiasmatic nucleus in vitro. Nat Neurosci 3: 372–376 [DOI] [PubMed] [Google Scholar]
- Leloup JC, Goldbeter A (1998) A model for circadian rhythms in Drosophila incorporating the formation of a complex between the PER and TIM proteins. J Biol Rhythms 13: 70–87 [DOI] [PubMed] [Google Scholar]
- Locke JCW, Millar AJ, Turner MS (2005a) Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J Theor Biol 234: 383–393 [DOI] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1: 2005.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA (2003a) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA (2003b) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- McClung CR (2006) Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salome PA, McClung CR (2003) Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci USA 100: 6878–6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, Coupland G (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17: 2255–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T (2005) PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol 46: 686–698 [DOI] [PubMed] [Google Scholar]
- Paltiel J, Amin R, Gover A, Ori N, Samach A (2006) Novel roles for GIGANTEA revealed under environmental conditions that modify its expression in Arabidopsis and Medicago truncatula. Planta advance online publication, 15 June 2006 [DOI] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan S (1976) A functional analysis of circadian pacemakers in nocturnal rodents. V. A clock for all seasons. J Comp Physiol A 106: 333–355 [Google Scholar]
- Rand DA, Shulgin BV, Salazar D, Millar AJ (2004) Design principles underlying circadian clocks. J Roy Soc Interface 1: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoff P, Rensing L (1996) The temperature-compensated Goodwin model simulates many circadian clock properties. J Theor Biol 179: 275–285 [Google Scholar]
- Salome PA, McClung CR (2005) PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M (2004) Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431: 862–868 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Thain SC, Hall A, Millar AJ (2000) Functional independence of circadian clocks that regulate plant gene expression. Curr Biol 10: 951–956 [DOI] [PubMed] [Google Scholar]
- Zeilinger MN, Farr é EM, Taylor SR, Kay SA, Doyle FJ III (2006) A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol 2: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Methods
Supplemental Figures
Supplemental Table 1
Supplemental Figure and Table Legends
SBML file for 3loop model