Abstract
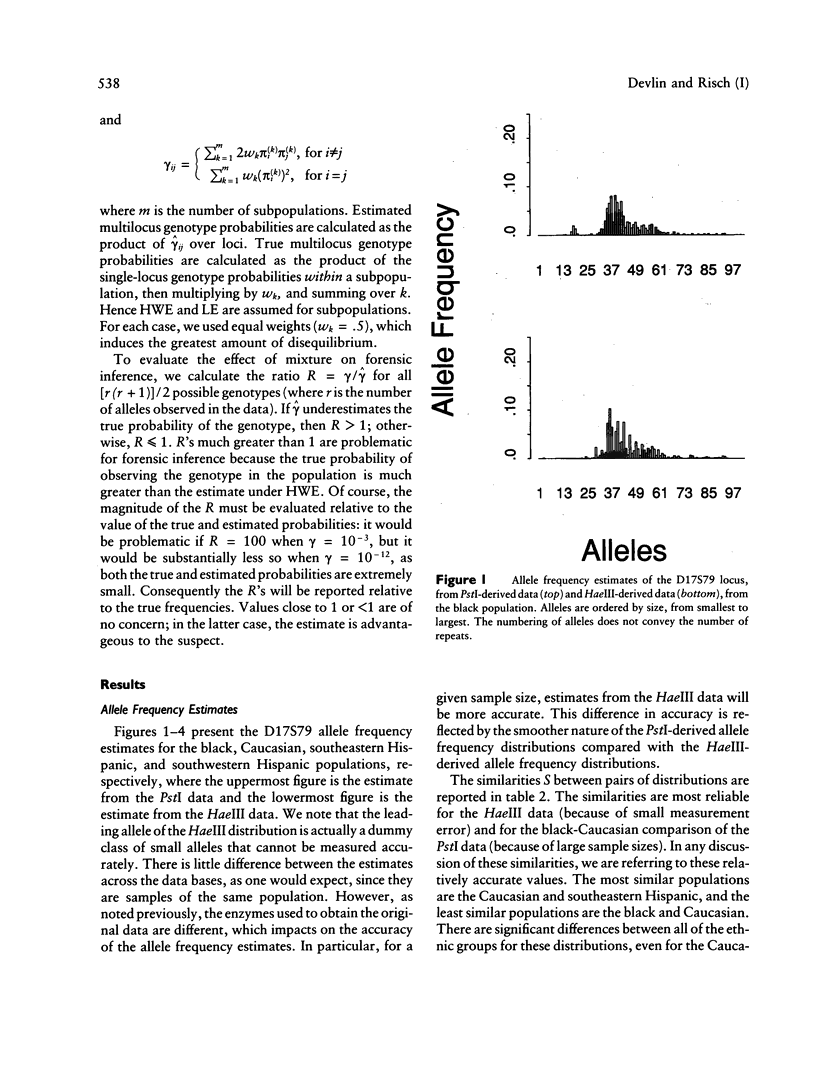
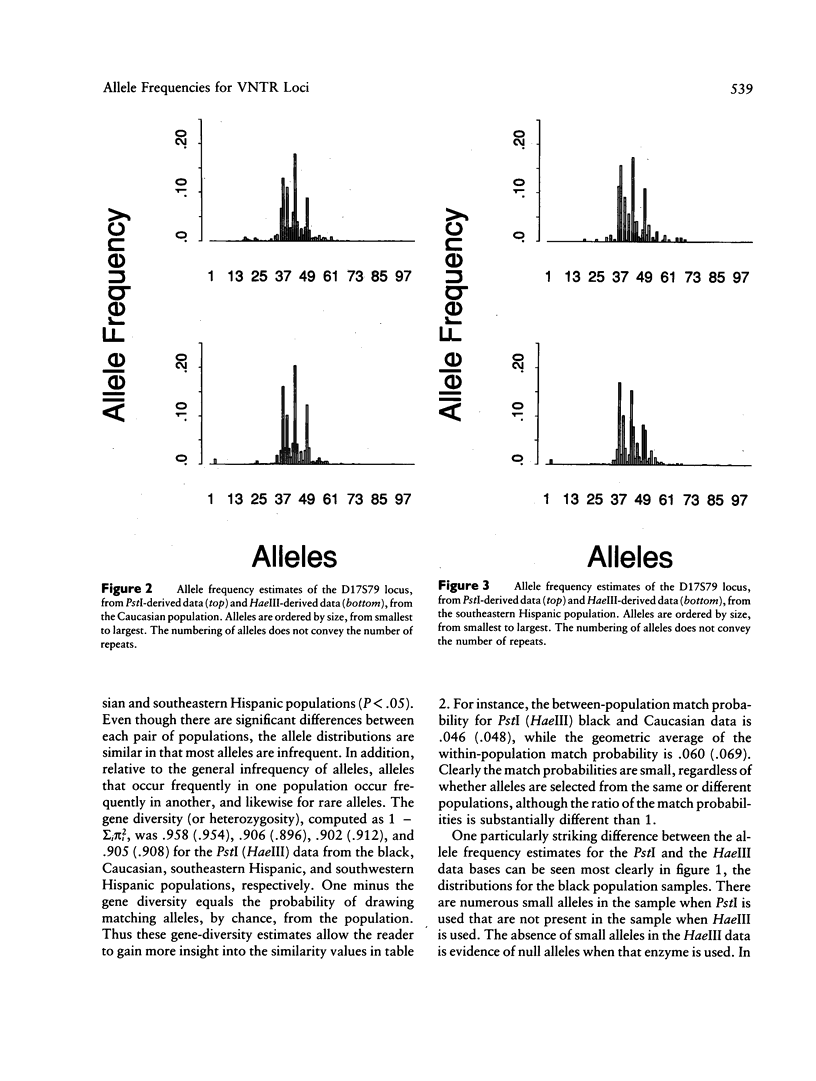
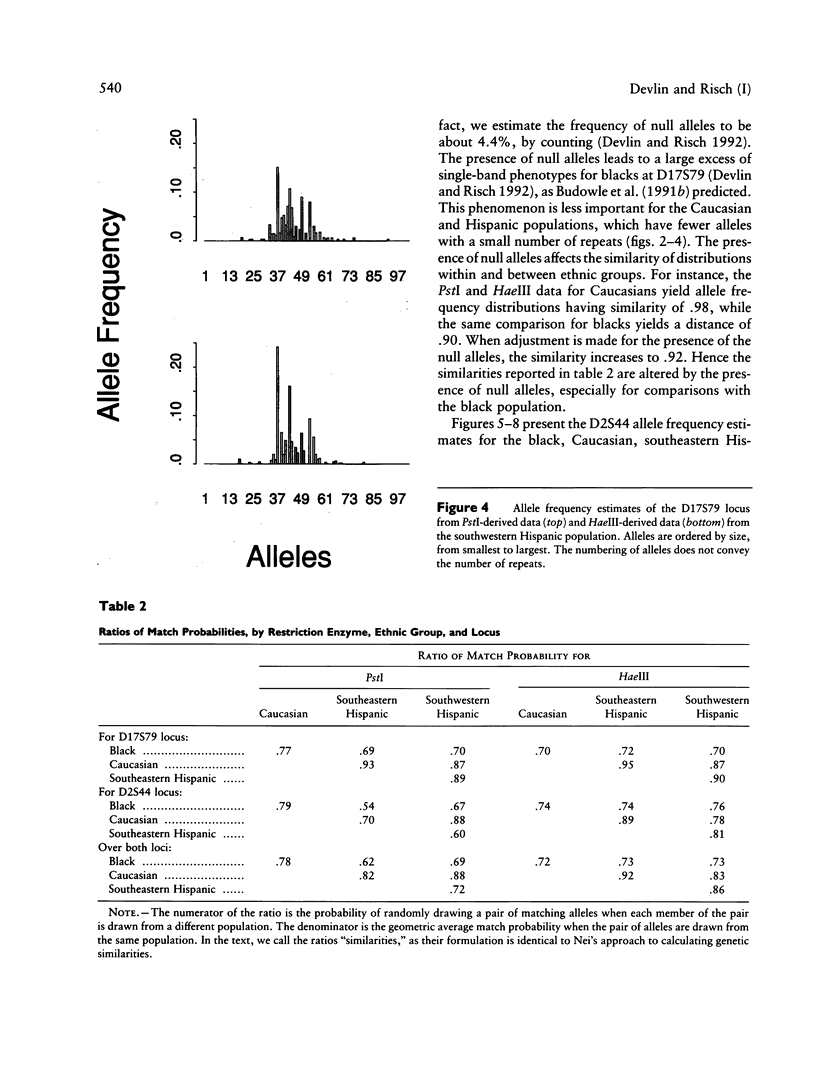
Allele-rich VNTR loci provide valuable information for forensic inference. Interpretation of this information is complicated by measurement error, which renders discrete alleles difficult to distinguish. Two methods have been used to circumvent this difficulty--i.e., binning methods and direct evaluation of allele frequencies, the latter achieved by modeling the data as a mixture distribution. We use this modeling approach to estimate the allele frequency distributions for two loci--D17S79 and D2S44--for black, Caucasian, and Hispanic samples from the Lifecodes and FBI data bases. The data bases are differentiated by the restriction enzyme used: PstI (Lifecodes) and HaeIII (FBI). Our results show that alleles common in one ethnic group are almost always common in all ethnic groups, and likewise for rare alleles; this pattern holds for both loci. Gene diversity, or heterozygosity, measured as one minus the sum of the squared allele frequencies, is greater for D2S44 than for D17S79, in both data bases. The average gene diversity across ethnic groups when PstI (HaeIII) is used is .918 (.918) for D17S79 and is .985 (.983) for D2S44. The variance in gene diversity among ethnic groups is greater for D17S79 than for D2S44. The number of alleles, like the gene diversity, is greater for D2S44 than for D17S79. The mean numbers of alleles across ethnic groups, estimated from the PstI (HaeIII) data, are 40.25 (41.5) for D17S79 and 104 (103) for D2S44. The number of alleles is correlated with sample size. We use the estimated allele frequency distributions for each ethnic group to explore the effects of unwittingly mixing populations and thereby violating independence assumptions. We show that, even in extreme cases of mixture, the estimated genotype probabilities are good estimates of the true probabilities, contradicting recent claims. Because the binning methods currently used for forensic inference show even less differentiation among ethnic groups, we conclude that mixture has little or no impact on the use of VNTR loci for forensics.
Full text
PDF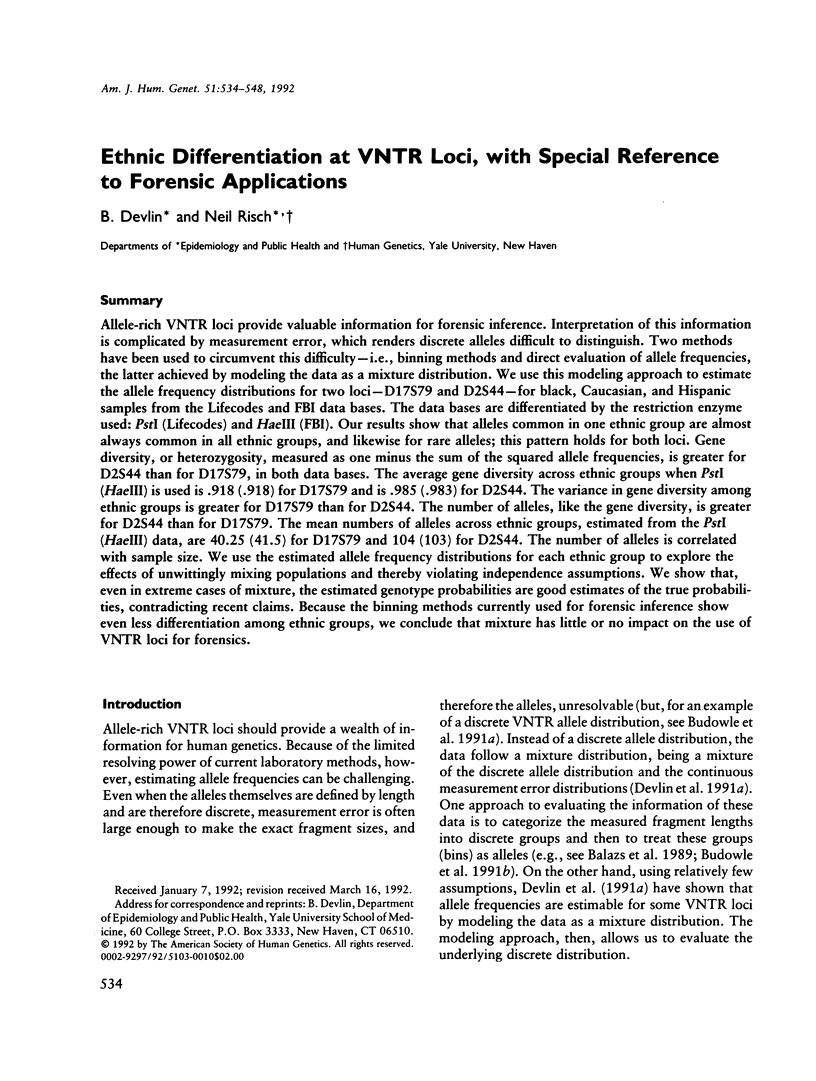








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balazs I., Baird M., Clyne M., Meade E. Human population genetic studies of five hypervariable DNA loci. Am J Hum Genet. 1989 Feb;44(2):182–190. [PMC free article] [PubMed] [Google Scholar]
- Budowle B., Chakraborty R., Giusti A. M., Eisenberg A. J., Allen R. C. Analysis of the VNTR locus D1S80 by the PCR followed by high-resolution PAGE. Am J Hum Genet. 1991 Jan;48(1):137–144. [PMC free article] [PubMed] [Google Scholar]
- Budowle B., Giusti A. M., Waye J. S., Baechtel F. S., Fourney R. M., Adams D. E., Presley L. A., Deadman H. A., Monson K. L. Fixed-bin analysis for statistical evaluation of continuous distributions of allelic data from VNTR loci, for use in forensic comparisons. Am J Hum Genet. 1991 May;48(5):841–855. [PMC free article] [PubMed] [Google Scholar]
- Cerda-Flores R. M., Kshatriya G. K., Bertin T. K., Hewett-Emmett D., Hanis C. L., Chakraborty R. Gene diversity and estimation of genetic admixture among Mexican-Americans of Starr County, Texas. Ann Hum Biol. 1992 Jul-Aug;19(4):347–360. doi: 10.1080/03014469200002222. [DOI] [PubMed] [Google Scholar]
- Chakraborty R., Ferrell R. E., Stern M. P., Haffner S. M., Hazuda H. P., Rosenthal M. Relationship of prevalence of non-insulin-dependent diabetes mellitus to Amerindian admixture in the Mexican Americans of San Antonio, Texas. Genet Epidemiol. 1986;3(6):435–454. doi: 10.1002/gepi.1370030608. [DOI] [PubMed] [Google Scholar]
- Chakraborty R., Jin L. Heterozygote deficiency, population substructure and their implications in DNA fingerprinting. Hum Genet. 1992 Jan;88(3):267–272. doi: 10.1007/BF00197257. [DOI] [PubMed] [Google Scholar]
- Cockerham C. C. Drift and mutation with a finite number of allelic states. Proc Natl Acad Sci U S A. 1984 Jan;81(2):530–534. doi: 10.1073/pnas.81.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. E. DNA fingerprinting for forensic identification: potential effects on data interpretation of subpopulation heterogeneity and band number variability. Am J Hum Genet. 1990 Feb;46(2):358–368. [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Risch N. A note on Hardy-Weinberg equilibrium of VNTR data by using the Federal Bureau of Investigation's fixed-bin method. Am J Hum Genet. 1992 Sep;51(3):549–553. [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Risch N., Roeder K. No excess of homozygosity at loci used for DNA fingerprinting. Science. 1990 Sep 21;249(4975):1416–1420. doi: 10.1126/science.2205919. [DOI] [PubMed] [Google Scholar]
- Efron B., Tibshirani R. Statistical data analysis in the computer age. Science. 1991 Jul 26;253(5018):390–395. doi: 10.1126/science.253.5018.390. [DOI] [PubMed] [Google Scholar]
- Green P., Lander E. S. Forensic DNA tests and hardy-weinberg equilibrium. Science. 1991 Aug 30;253(5023):1038–1039. doi: 10.1126/science.253.5023.1038. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Royle N. J., Wilson V., Wong Z. Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature. 1988 Mar 17;332(6161):278–281. doi: 10.1038/332278a0. [DOI] [PubMed] [Google Scholar]
- Lander E. S. DNA fingerprinting on trial. Nature. 1989 Jun 15;339(6225):501–505. doi: 10.1038/339501a0. [DOI] [PubMed] [Google Scholar]
- Lewontin R. C., Hartl D. L. Population genetics in forensic DNA typing. Science. 1991 Dec 20;254(5039):1745–1750. doi: 10.1126/science.1845040. [DOI] [PubMed] [Google Scholar]
- Li C. C. Population subdivision with respect to multiple alleles. Ann Hum Genet. 1969 Jul;33(1):23–29. doi: 10.1111/j.1469-1809.1969.tb01625.x. [DOI] [PubMed] [Google Scholar]
- Reed T. E. Ethnic classification of Mexican-Americans. Science. 1974 Jul 19;185(4147):283–283. doi: 10.1126/science.185.4147.283. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and selection in a cline. Genetics. 1973 Dec;75(4):733–756. doi: 10.1093/genetics/75.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smouse P. E., Chakraborty R. The use of restriction fragment length polymorphisms in paternity analysis. Am J Hum Genet. 1986 Jun;38(6):918–939. [PMC free article] [PubMed] [Google Scholar]
- Weir B. S. Independence of VNTR alleles defined as fixed bins. Genetics. 1992 Apr;130(4):873–887. doi: 10.1093/genetics/130.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]


