Abstract
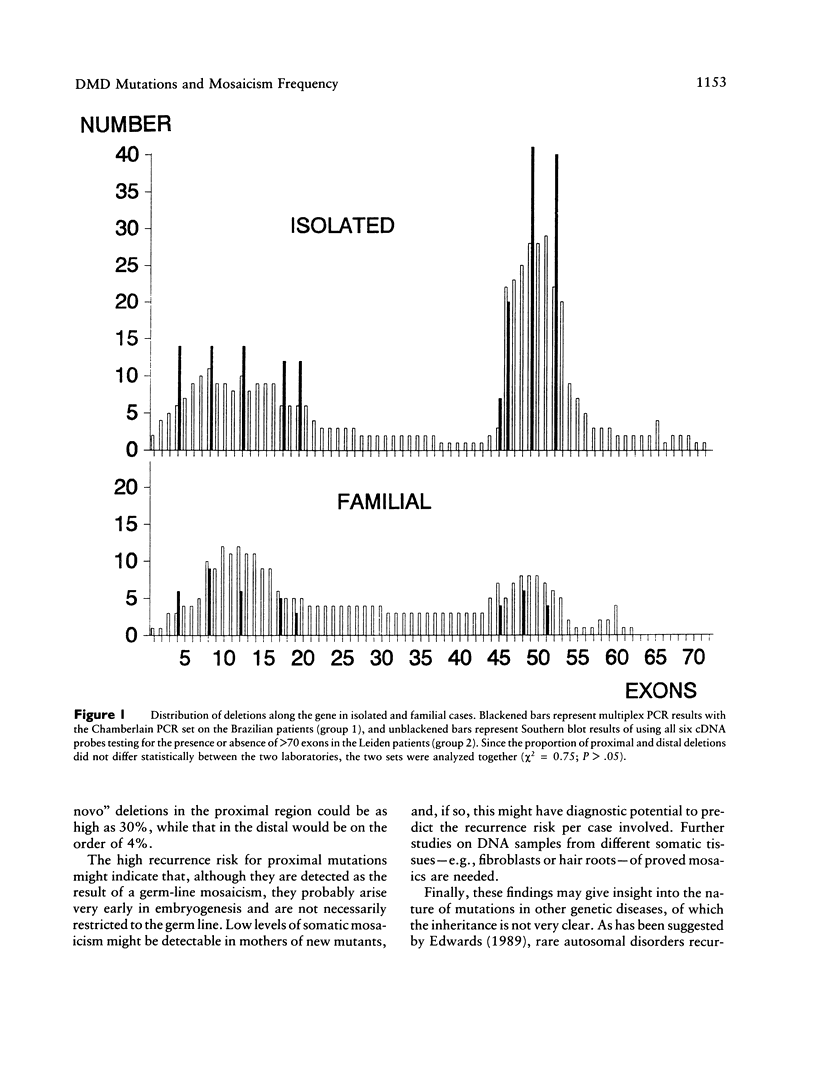
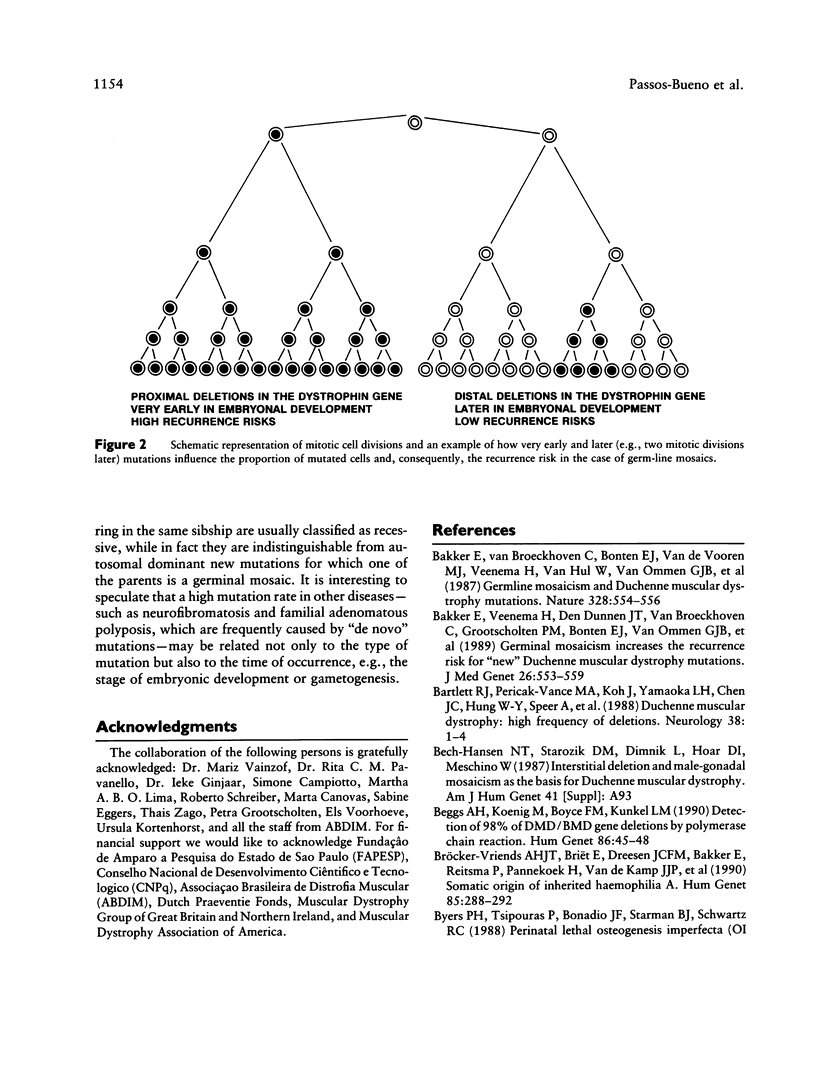
In about 65% of the cases of Duchenne muscular dystrophy (DMD) a partial gene deletion or duplication in the dystrophin gene can be detected. These mutations are clustered at two hot spots: 30% at the hot spot in the proximal part of the gene and about 70% at a more distal hot spot. Unexpectedly we observed a higher frequency of proximal gene rearrangements among proved "germ line" mosaic cases. Of the 24 mosaic cases we are aware of, 19 (79%) have a proximal mutation, while only 5 (21%) have a distal mutation. This finding indicates that the mutations at the two hot spots in the dystrophin gene differ in origin. Independent support for the different mosaicism frequency was found by comparing the mutation spectra observed in isolated cases of DMD and familial cases of DMD. In a large two-center study of 473 patients from Brazil and the Netherlands, we detected a significant difference in the deletion distribution of isolated (proximal:distal ratio 1:3) and familial cases (ratio 1:1). We conclude from these data that proximal deletions most likely occur early in embryonic development, causing them to have a higher chance of becoming familial, while distal deletions occur later and have a higher chance of causing only isolated cases. Finally, our findings have important consequences for the calculation of recurrence-risk estimates according to the site of the deletion: a "proximal" new mutant has an increased recurrence risk of approximately 30%, and a "distal" new mutant has a decreased recurrence risk of approximately 4%.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakker E., Veenema H., Den Dunnen J. T., van Broeckhoven C., Grootscholten P. M., Bonten E. J., van Ommen G. J., Pearson P. L. Germinal mosaicism increases the recurrence risk for 'new' Duchenne muscular dystrophy mutations. J Med Genet. 1989 Sep;26(9):553–559. doi: 10.1136/jmg.26.9.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett R. J., Pericak-Vance M. A., Koh J., Yamaoka L. H., Chen J. C., Hung W. Y., Speer M. C., Wapenaar M. C., Van Ommen G. J., Bakker E. Duchenne muscular dystrophy: high frequency of deletions. Neurology. 1988 Jan;38(1):1–4. doi: 10.1212/wnl.38.1.1. [DOI] [PubMed] [Google Scholar]
- Beggs A. H., Koenig M., Boyce F. M., Kunkel L. M. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet. 1990 Nov;86(1):45–48. doi: 10.1007/BF00205170. [DOI] [PubMed] [Google Scholar]
- Bröcker-Vriends A. H., Briët E., Dreesen J. C., Bakker B., Reitsma P., Pannekoek H., van de Kamp J. J., Pearson P. L. Somatic origin of inherited haemophilia A. Hum Genet. 1990 Aug;85(3):288–292. doi: 10.1007/BF00206748. [DOI] [PubMed] [Google Scholar]
- Byers P. H., Tsipouras P., Bonadio J. F., Starman B. J., Schwartz R. C. Perinatal lethal osteogenesis imperfecta (OI type II): a biochemically heterogeneous disorder usually due to new mutations in the genes for type I collagen. Am J Hum Genet. 1988 Feb;42(2):237–248. [PMC free article] [PubMed] [Google Scholar]
- Claustres M., Kjellberg P., Desgeorges M., Bellet H., Demaille J. Germinal mosaicism from grand-paternal origin in a family with Duchenne muscular dystrophy. Hum Genet. 1990 Dec;86(2):241–243. doi: 10.1007/BF00197714. [DOI] [PubMed] [Google Scholar]
- Darras B. T., Francke U. A partial deletion of the muscular dystrophy gene transmitted twice by an unaffected male. Nature. 1987 Oct 8;329(6139):556–558. doi: 10.1038/329556a0. [DOI] [PubMed] [Google Scholar]
- Den Dunnen J. T., Grootscholten P. M., Bakker E., Blonden L. A., Ginjaar H. B., Wapenaar M. C., van Paassen H. M., van Broeckhoven C., Pearson P. L., van Ommen G. J. Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet. 1989 Dec;45(6):835–847. [PMC free article] [PubMed] [Google Scholar]
- Edwards J. H. Familiarity, recessivity and germline mosaicism. Ann Hum Genet. 1989 Jan;53(Pt 1):33–47. doi: 10.1111/j.1469-1809.1989.tb01120.x. [DOI] [PubMed] [Google Scholar]
- Forrest S. M., Cross G. S., Speer A., Gardner-Medwin D., Burn J., Davies K. E. Preferential deletion of exons in Duchenne and Becker muscular dystrophies. Nature. 1987 Oct 15;329(6140):638–640. doi: 10.1038/329638a0. [DOI] [PubMed] [Google Scholar]
- Hogan M. E., Rooney T. F., Austin R. H. Evidence for kinks in DNA folding in the nucleosome. Nature. 1987 Aug 6;328(6130):554–557. doi: 10.1038/328554a0. [DOI] [PubMed] [Google Scholar]
- Koenig M., Beggs A. H., Moyer M., Scherpf S., Heindrich K., Bettecken T., Meng G., Müller C. R., Lindlöf M., Kaariainen H. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet. 1989 Oct;45(4):498–506. [PMC free article] [PubMed] [Google Scholar]
- Koenig M., Hoffman E. P., Bertelson C. J., Monaco A. P., Feener C., Kunkel L. M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987 Jul 31;50(3):509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Lanman J. T., Jr, Pericak-Vance M. A., Bartlett R. J., Chen J. C., Yamaoka L., Koh J., Speer M. C., Hung W. Y., Roses A. D. Familial inheritance of a DXS164 deletion mutation from a heterozygous female. Am J Hum Genet. 1987 Aug;41(2):138–144. [PMC free article] [PubMed] [Google Scholar]
- Lebo R. V., Olney R. K., Golbus M. S. Somatic mosaicism at the Duchenne locus. Am J Med Genet. 1990 Oct;37(2):187–190. doi: 10.1002/ajmg.1320370206. [DOI] [PubMed] [Google Scholar]
- Malhotra S. B., Hart K. A., Klamut H. J., Thomas N. S., Bodrug S. E., Burghes A. H., Bobrow M., Harper P. S., Thompson M. W., Ray P. N. Frame-shift deletions in patients with Duchenne and Becker muscular dystrophy. Science. 1988 Nov 4;242(4879):755–759. doi: 10.1126/science.3055295. [DOI] [PubMed] [Google Scholar]
- Monaco A. P., Bertelson C. J., Colletti-Feener C., Kunkel L. M. Localization and cloning of Xp21 deletion breakpoints involved in muscular dystrophy. Hum Genet. 1987 Mar;75(3):221–227. doi: 10.1007/BF00281063. [DOI] [PubMed] [Google Scholar]
- Passos-Bueno M. R., Lima M. A., Zatz M. Estimate of germinal mosaicism in Duchenne muscular dystrophy. J Med Genet. 1990 Nov;27(11):727–728. doi: 10.1136/jmg.27.11.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos-Bueno M. R., Rapaport D., Love D., Flint T., Bortolini E. R., Zatz M., Davies K. E. Screening of deletions in the dystrophin gene with the cDNA probes Cf23a, Cf56a, and Cf115. J Med Genet. 1990 Mar;27(3):145–150. doi: 10.1136/jmg.27.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapenaar M. C., Kievits T., Hart K. A., Abbs S., Blonden L. A., den Dunnen J. T., Grootscholten P. M., Bakker E., Verellen-Dumoulin C., Bobrow M. A deletion hot spot in the Duchenne muscular dystrophy gene. Genomics. 1988 Feb;2(2):101–108. doi: 10.1016/0888-7543(88)90090-0. [DOI] [PubMed] [Google Scholar]
- Wood S., McGillivray B. C. Germinal mosaicism in Duchenne muscular dystrophy. Hum Genet. 1988 Mar;78(3):282–284. doi: 10.1007/BF00291677. [DOI] [PubMed] [Google Scholar]
- van Essen A. J., Abbs S., Baiget M., Bakker E., Boileau C., van Broeckhoven C., Bushby K., Clarke A., Claustres M., Covone A. E. Parental origin and germline mosaicism of deletions and duplications of the dystrophin gene: a European study. Hum Genet. 1992 Jan;88(3):249–257. doi: 10.1007/BF00197255. [DOI] [PubMed] [Google Scholar]


